Plant Growth-Promoting and Tequila Vinasse-Resistant Bacterial Strains
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation of Bacterial Strains
2.2. Identification of Bacterial Strains
2.3. Plant Growth Promotion and Biocontrol Traits of the Bacterial Strains
2.4. Tequila Vinasses’ Minimal Inhibitory Concentration (MIC) on Bacterial Strains
2.5. Plant-Growth Promotion by Bacterial Strains
2.6. Effect of Soils Irrigated with Vinasse on Plant Growth
2.7. Plant Growth Promotion by Bacterial Strains in the Presence of Tequila Vinasse
3. Results
3.1. Selection and Identification of Bacterial Strains
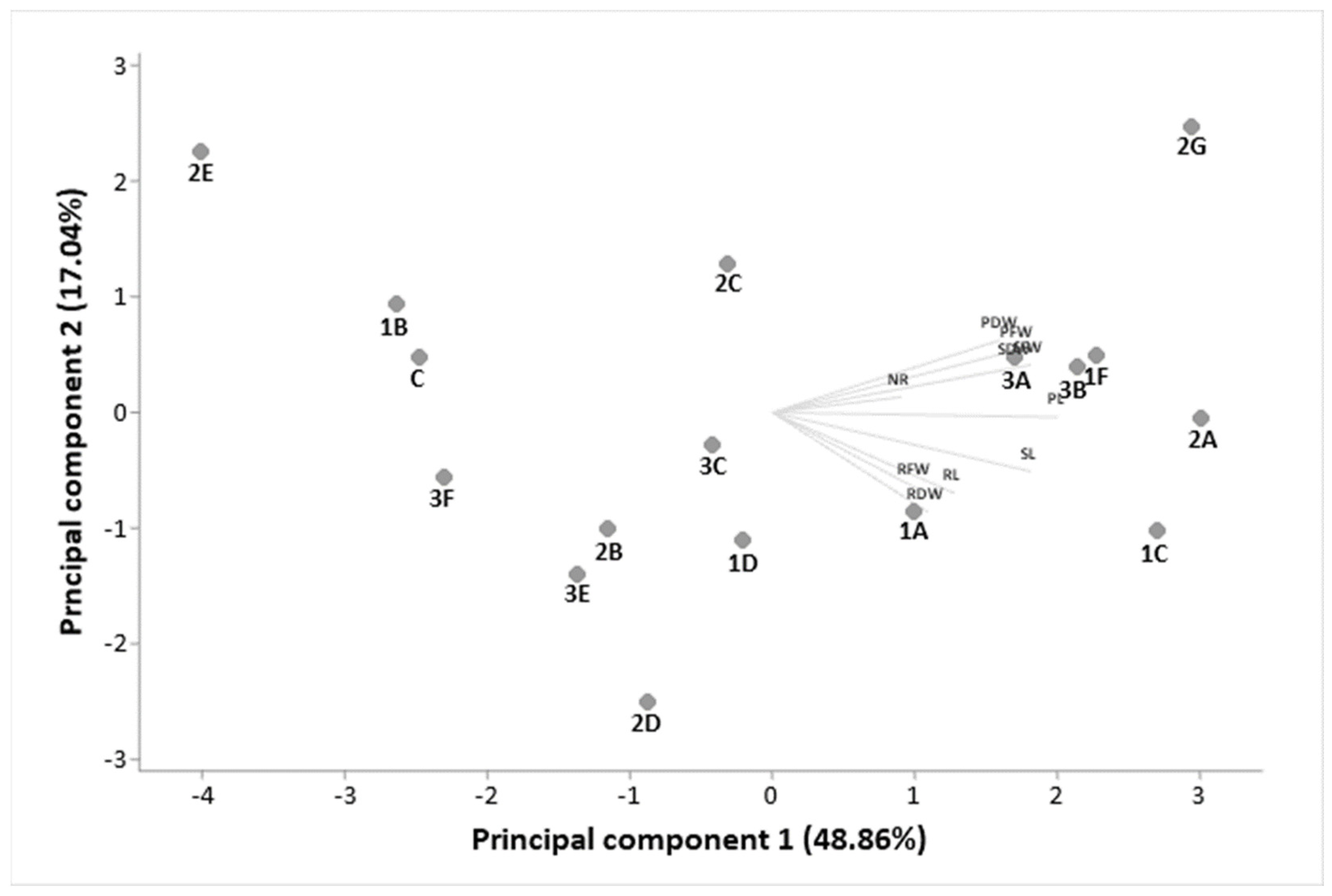
3.2. Plant Growth Promotion and Biocontrol Capacity of Bacterial Strains
3.3. Tequila Vinasses’ Minimal Inhibitory Concentrations (MICs)
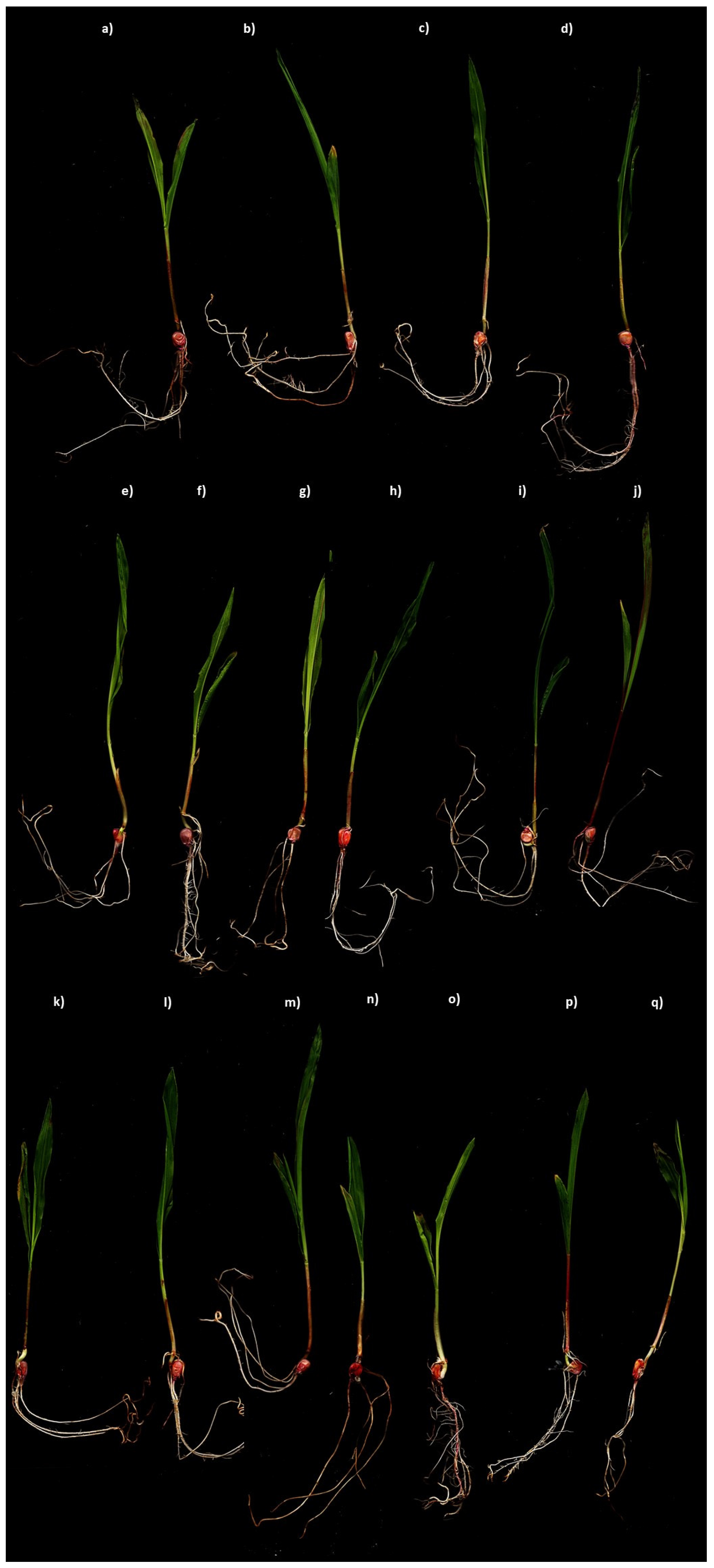
3.4. Plant Growth-Promoting Bioassay
3.5. Maize Germination on Soils Irrigated with Vinasse
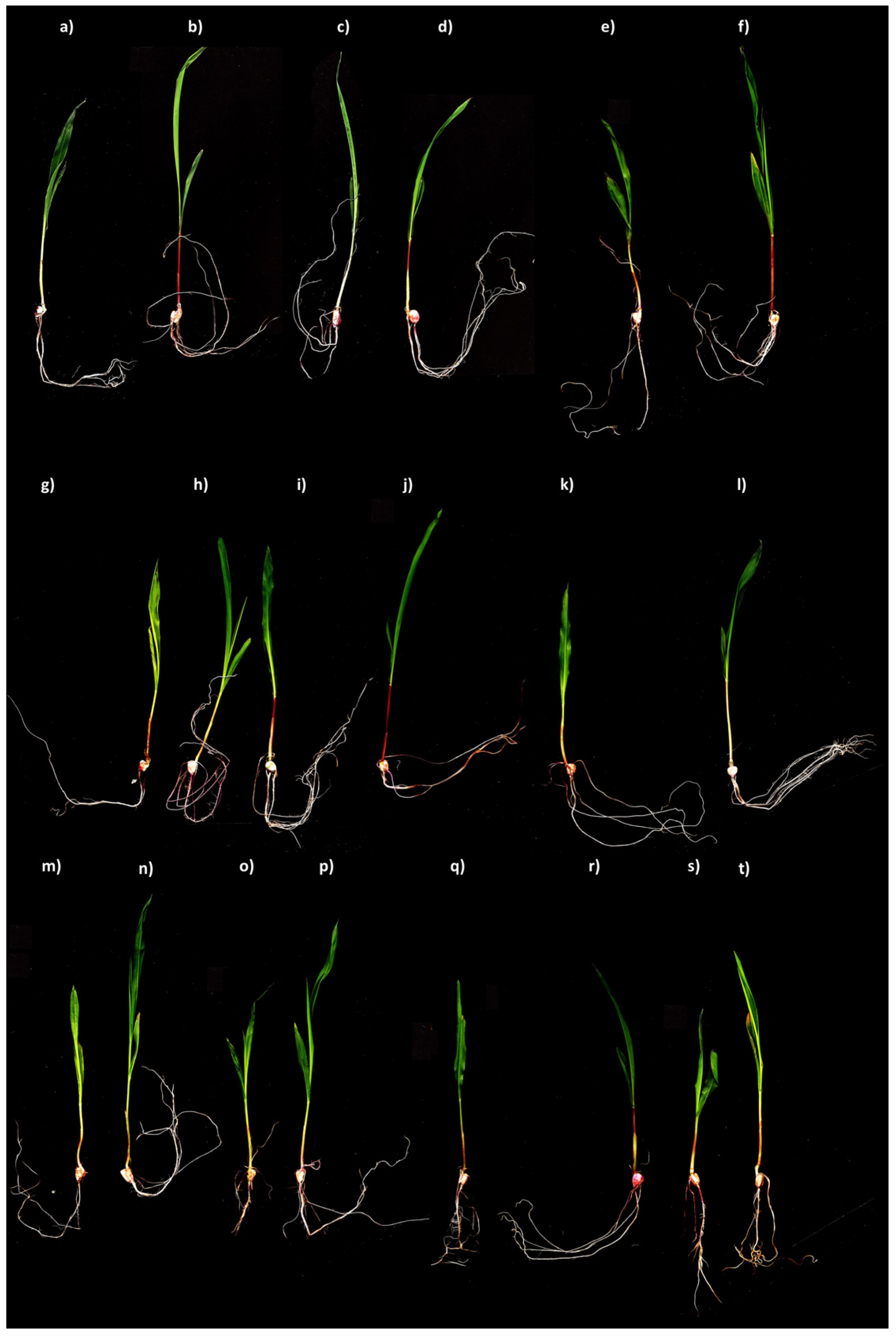
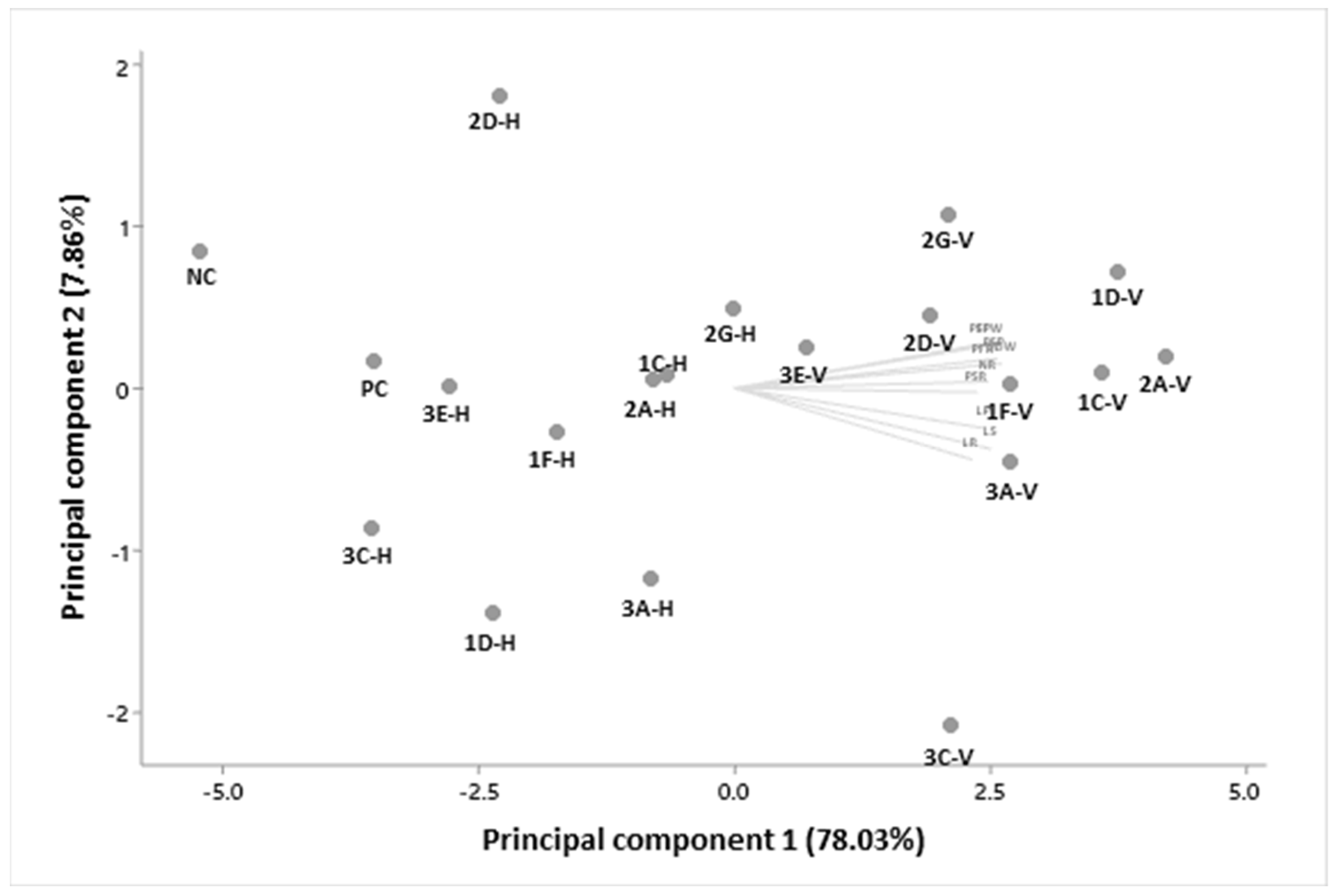
3.6. Plant Growth-Promoting Bioassay under Tequila Vinasse and Bacterial Strain Influence
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cea-Barcia, G.E.; Imperial-Cervantes, R.A.; Torres-Zuniga, I.; Van Den-Hende, S. Converting tequila vinasse diluted with tequila process water into microalgae-yeast flocs and dischargeable effluent. Bioresour. Technol. 2020, 300, 122644. [Google Scholar] [CrossRef] [PubMed]
- Zurita, F.; Tejeda, A.; Montoya, A.; Carrillo, I.; Sulbarán-Rangel, B.; Carreón-Álvarez, A. Generation of tequila vinasses, characterization, current disposal practices and study cases of disposal methods. Water 2022, 14, 1395. [Google Scholar] [CrossRef]
- Moran-Salazar, R.G.; Sanchez-Lizarraga, A.L.; Rodriguez-Campos, J.; Davila-Vazquez, G.; Marino-Marmolejo, E.N.; Dendooven, L.; Contreras-Ramos, S.M. Utilization of vinasses as soil amendment: Consequences and perspectives. Springerplus 2016, 5, 1007. [Google Scholar] [CrossRef] [PubMed]
- Robertiello, A. Upgrading of agricultural and agroindustrial wastes: The treatment of distillery effluents (vinasses) in Italy. Agric. Wastes 1983, 4, 387–395. [Google Scholar] [CrossRef]
- Rodrigues-Reis, C.E.; Hu, B. Vinasse from sugarcane ethanol production: Better treatment or better utilization? Front. Energy Res. 2017, 5, 7. [Google Scholar] [CrossRef]
- Díaz-Vázquez, D.; Garibay, M.V.; Fernández-del Castillo, A.; Orozco-Nunnelly, D.A.; Senés-Guerrero, C.; Gradilla-Hernández, M.S. Yeast community composition impacts on tequila industry waste treatment for pollution control and waste-to-product synthesis. Front. Chem. Eng. 2022, 4, 1013873. [Google Scholar] [CrossRef]
- Prasad, K.R.; Kumar, R.R.; Srivastava, S.N. Design of optimum response surface experiments for electro-coagulation of distillery spent wash. Wat. Air Soil Pollut. 2008, 191, 5–13. [Google Scholar] [CrossRef]
- de Resende, A.S.; Xavier, R.P.; de Oliveira, O.C.; Urquiaga, S.; Alves, B.J.R.; Boddey, R.M. Long-term effects of pre-harvest burning and nitrogen and vinasse applications on yield of sugar cane and soil carbon and nitrogen stocks on plantation in Pernambuco, N.E. Brazil. Plant Soil 2006, 281, 339–351. [Google Scholar] [CrossRef]
- Leal, J.R.; Amaral-Sobrinho, N.M.B.; Velloso, A.C.X.; Rossielo, R.O.P. Potencial redox e pH: Variação em um solo tratado com vinhaça. Rev. Bras. Ciênc. Solo 1983, 7, 257–261. [Google Scholar]
- Silva, M.A.S.; Griebeler, N.P.; Borges, L.C. Uso de vinhaça e impactos nas propriedades do solo e lençol freático. Ver. Bras. Eng. Agríc. Ambient. 2007, 11, 108–114. [Google Scholar] [CrossRef]
- Oliveira, B.G.; Carvalho, J.L.N.; Cerri, C.E.P.; Cerri, C.C.; Feigl, B.J. Soil greenhouse gas fluxes from vinasse application in Brazilian sugarcane areas. Geoderma 2013, 200–201, 77–84. [Google Scholar] [CrossRef]
- Santos, T.M.C.; Santos, M.A.L.; Santos, C.G.; Santos, V.R. Efeito da fertirrigação com vinhaça nos microrganismos do solo. Rev. Caatinga 2009, 22, 155–160. [Google Scholar]
- Camargo, R. O desenvolvimento da Flora Microbiana nos solos Tratados com Vinhaça. Ph.D. Dissertation, Universidade de São Paulo, São Paulo, Brazil, 1953; pp. 30–35. [Google Scholar]
- Tejada, M.; Moreno, J.L.; Hernandez, M.T.; Garcia, C. Application of two beet vinasse forms in soil restoration: Effects on soil properties in an arid environment in southern Spain. Agric. Ecosys. Environ. 2007, 119, 289–298. [Google Scholar] [CrossRef]
- Atlas, R.M. Handbook of Microbiological Media, 4th ed.; CRC Press: Boca Raton, FL, USA, 2010; pp. 50–200. [Google Scholar]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program Windows 95/98/NT. Nucl. Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Ashfaq, M.; Hassan, H.M.; Ghazali, A.H.A.; Ahmad, M. Halotolerant potassium solubilizing plant growth promoting rhizobacteria may improve potassium availability under saline conditions. Environ. Monit. Assess. 2020, 192, 697. [Google Scholar] [CrossRef]
- Mehta, S.; Nautiyal, C.S. An efficient method for qualitative screening of phosphate solubilizing bacteria. Curr. Microbiol. 2001, 43, 51–56. [Google Scholar] [CrossRef]
- Fasim, F.; Ahmed, N.; Parsons, R.; Gadd, G.M. Solubilization of zinc salts by a bacterium isolated from the air environment of a tannery. FEMS Microbiol. Lett. 2002, 213, 1–6. [Google Scholar] [CrossRef]
- Schwyn, B.; Neilands, J.B. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 1987, 160, 47–56. [Google Scholar] [CrossRef]
- Hankin, L.; Anagnostakis, S. Solid media containing carboxy methyl cellulose to detect CM cellulase activity of microorganisms. Microbiology 1977, 98, 109–115. [Google Scholar]
- Al Mohaini, M.; Farid, A.; Muzammal, M.; Ghazanfar, S.; Dadrasnia, A.; Alsalman, A.J.; Al Hawaj, M.A.; Alhashem, Y.N.; Ismail, S. Enhancing lipase production of Bacillus salmalaya strain 139SI using different carbon sources and surfactants. Appl. Microbiol. 2022, 2, 237–247. [Google Scholar] [CrossRef]
- Ramos-Garza, J.; Bustamante-Brito, R.; Angeles-de Paz, G.; Medina-Canales, M.G.; Vásquez-Murrieta, M.S.; Tao-Wang, E.; Rodríguez-Tovar, V.A. Isolation and characterization of yeasts associated with plants growing in heavy-metal and arsenic-contaminated soils. Can. J. Microbiol. 2016, 62, 307–319. [Google Scholar] [CrossRef] [PubMed]
- Calvillo-Aguilar, F.F.; Cruz-Cárdenas, C.I.; Chávez-Díaz, I.F.; Sandoval-Cancino, G.; Ruiz-Ramírez, S.; Bautista-Ramírez, E.; Ramos-Garza, J.; Hernández-Rodríguez, C.H.; Zelaya-Molina, L.X. Germination test for the evaluation of plant-growth promoting microorganisms. J. Microbiol. Methods 2023, 207, 106708. [Google Scholar] [CrossRef] [PubMed]
- Karmakar, S.; Billah, M.; Hasan, M.; Sohan, S.R.; Hossain, M.F.; Hoque, K.M.F.; Kabir, A.H.; Rashid, M.; Talukder, M.R.; Reza, M.A. Impact of LFGD (Ar+ O2) plasma on seed surface, germination, plant growth, productivity and nutritional composition of maize (Zea mays L.). Heliyon 2021, 7, e06458. [Google Scholar] [CrossRef] [PubMed]
- Zizumbo-Villarreal, D.; Colunga-GarcíaMarín, P. Early coconut distillation and the origins of mezcal and tequila spirits in west-central Mexico. Genet. Resour. Crop Evol. 2008, 55, 493–510. [Google Scholar] [CrossRef]
- CRT (Consejo Regulador de Tequila). Información Estadística. Available online: http://www.crt.org.mx/EstadisticasCRTweb/ (accessed on 20 May 2024).
- Navarrete-Torres, M.C. Female leadership in the tequila industry in Mexico. Soc. Econ. Rev. 2023, 21, 49–56. [Google Scholar]
- Christofoletti, C.A.; Escher, J.P.; Correia, J.E.; Urbano-Marinho, J.F.; Fontanetti, C.S. Sugarcane vinasse: Environmental implications of its use. Waste Manag. 2013, 33, 2752–2761. [Google Scholar] [CrossRef] [PubMed]
- Navarro, A.R.; Sepúlveda, M.C.; Rubio, M.C. Bio-concentration of vinasse from the alcoholic fermentation of sugar cane molasses. Waste Manag. 2000, 20, 581–585. [Google Scholar] [CrossRef]
- Santana, V.S.; Fernandes-Machado, N.R.C. Photocatalytic degradation of the vinasse under solar radiation. Catal. Today 2008, 133–135, 606–610. [Google Scholar] [CrossRef]
- Ramanathan, T.; Ting, Y.P. Selective copper bioleaching by pure and mixed cultures of alkaliphilic bacteria isolated from a fly ash landfill site. Wat. Air Soil Pollut. 2015, 226, 374. [Google Scholar] [CrossRef]
- De Maria, S.; Rivelli, A.R.; Kuffner, M.; Sessitsch, A.; Wenzel, W.W.; Gorfer, M.; Strauss, J.; Puschenreiter, M. Interactions between accumulation of trace elements and macronutrients in Salix caprea after inoculation with rhizosphere microorganisms. Chemosphere 2011, 84, 1256–1261. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.S.; Wang, K.H.; Cai, M.; Yang, M.L.; Wang, X.K.; Ma, H.L.; Yuan, Y.H.; Wu, L.H.; Li, D.F.; Liu, S.J. Agromyces chromiiresistens sp. nov., Novosphingobium album sp. nov., Sphingobium arseniciresistens sp. nov., Sphingomonas pollutisoli sp. nov., and Salinibacterium metalliresistens sp. nov.: Five new members of Microbacteriaceae and Sphingomonadaceae from polluted soil. Front. Microbiol. 2023, 14, 1289110. [Google Scholar]
- Breton-Deval, L.; Salinas-Peralta, I.; Alarcón-Aguirre, J.S.; Sulbarán-Rangel, B.; Gurubel-Tun, K.J. Taxonomic binning approaches and functional characteristics of the microbial community during the anaerobic digestion of hydrolyzed corncob. Energies 2020, 14, 66. [Google Scholar] [CrossRef]
- Lee, J.C.; Whang, K.S. Agromyces humi sp. nov., actinobacterium isolated from farm soil. Int. J. Syst. Evol. Microbiol. 2020, 70, 5032–5039. [Google Scholar] [CrossRef] [PubMed]
- O’Loughlin, S.N.; Graham, R.L.; McMullan, G.; Ternan, N.G. A role for carbon catabolite repression in the metabolism of phosphonoacetate by Agromyces fucosus Vs2. FEMS Microbiol. Lett. 2006, 261, 133–140. [Google Scholar] [PubMed]
- Al-Awadhi, H.; Al-Mailem, D.; Dashti, N.; Hakam, L.; Eliyas, M.; Radwan, S. The abundant occurrence of hydrocarbon-utilizing bacteria in the phyllospheres of cultivated and wild plants in Kuwait. Int. Biodeterior. Biodegradation 2012, 73, 73–79. [Google Scholar] [CrossRef]
- González-López, J.; Rodelas, B.; Pozo, C.; Salmerón-López, V.; Martínez-Toledo, M.V.; Salmerón, V. Liberation of amino acids by heterotrophic nitrogen fixing bacteria. Amino Acids 2005, 28, 363–367. [Google Scholar] [CrossRef] [PubMed]
- Aquilanti, L.; Favilli, F.; Clementi, F. Comparison of different strategies for isolation and preliminary identification of Azotobacter from soil samples. Soil Biol. 2004, 36, 1475–1483. [Google Scholar] [CrossRef]
- Onwurah, I.N.; Nwuke, C. Enhanced bioremediation of crude oil-contaminated soil by a Pseudomonas species and mutually associated adapted Azotobacter vinelandii. J. Chem. Technol. Biotechnol. Int. Res. Process Environ. Clean Technol. 2004, 79, 491–498. [Google Scholar] [CrossRef]
- Wu, G.; Xu, H.; Jiang, M. Biodegradation of chlorophenols: A review. Chem. J. Internet. 2004, 6, 60–67. [Google Scholar]
- Kumar, A.; Trefault, N.; Olaniran, A.O. Microbial degradation of 2, 4-dichlorophenoxyacetic acid: Insight into the enzymes and catabolic genes involved, their regulation and biotechnological implications. Crit. Rev. Microbiol. 2016, 42, 194–208. [Google Scholar] [CrossRef] [PubMed]
- Anupama, K.S.; Paul, S. Ex situ and in situ biodegradation of lindane by Azotobacter chroococcum. J. Environ. Sci. Health B 2009, 45, 58–66. [Google Scholar] [CrossRef] [PubMed]
- Abo-Amer, A.E.; Abu-Gharbia, M.A.; Soltan, E.S.M.; Abd El-Raheem, W.M. Isolation and molecular characterization of heavy metal-resistant Azotobacter chroococcum from agricultural soil and their potential application in bioremediation. Geomicrobiol. J. 2014, 31, 551–561. [Google Scholar] [CrossRef]
- Hindersah, R.; Handyman, Z.; Indriani, F.N.; Suryatmana, P.; Nurlaeny, N. Azotobacter population, soil nitrogen and groundnut growth in mercury-contaminated tailing inoculated with Azotobacter. J. Degraded Min. Lands Manag. 2018, 5, 1269. [Google Scholar] [CrossRef]
- Piperidou, C.I.; Chaidou, C.I.; Stalikas, C.D.; Soulti, K.; Pilidis, G.A.; Balis, C. Bioremediation of olive oil mill wastewater: Chemical alterations induced by Azotobacter vinelandii. J. Agric. Food Chem. 2000, 48, 1941–1948. [Google Scholar] [CrossRef] [PubMed]
- Omar, M.N.A.; Mostafa, A.T.; Ahmed, A.S. Concentrated vinasse as a novel diazotrophs growth medium (biovinasse inoculant) and soil conditioner to improve faba bean yield under dripping irrigation system. In Proceedings of the 17th World Congress of Soil Science, Bangkok, Thailand, 14–21 August 2002. [Google Scholar]
- Ventorino, V.; Nicolaus, B.; Di Donato, P.; Pagliano, G.; Poli, A.; Robertiello, A.; Iavarone, V.; Pepe, O. Bioprospecting of exopolysaccharide-producing bacteria from different natural ecosystems for biopolymer synthesis from vinasse. Chem. Biol. Technol. Agric. 2019, 6, 18. [Google Scholar] [CrossRef]
- Liu, L.; Yuan, T.; An, Q.; Yang, M.; Mao, X.; Mo, C.; Tang, Z.; Peng, G. Azotobacter bryophylli sp. nov., isolated from the succulent plant Bryophyllum pinnatum. Int. J. Syst. Evol. Microbiol. 2019, 69, 1986–1992. [Google Scholar] [CrossRef] [PubMed]
- Aislabie, J.; Davison, A.D.; Boul, H.L.; Franzmann, P.D.; Jardine, D.R.; Karuso, P. Isolation of Terrabacter sp. strain DDE-1, which metabolizes 1,1-dichloro-2,2-bis(4-chlorophenyl)ethylene when induced with biphenyl. Appl. Environ. Microbiol. 1999, 65, 5607–5611. [Google Scholar] [CrossRef]
- Xue, G.; Zhang, L.; Fan, X.; Luo, K.; Guo, S.; Chen, H.; Li, X.; Jian, Q. Responses of soil fertility and microbiomes of atrazine contaminated soil to remediation by hydrochar and persulfate. J. Hazard. Mater. 2002, 435, 128944. [Google Scholar] [CrossRef]
- Habe, H.; Ide, K.; Yotsumoto, M.; Tsuji, H.; Yoshida, T.; Nojiri, H.; Omori, T. Degradation characteristics of a dibenzofuran-degrader Terrabacter sp. strain DBF63 toward chlorinated dioxins in soil. Chemosphere 2002, 48, 201–207. [Google Scholar] [CrossRef]
- Jiang, Y.; Xu, L.; Wang, K.; Liu, G.; Ma, J.; Zhou, Y.; Xu, Q.; Hong, Q.; He, J.; Qiu, J. Characterization and genetic determination of a newly isolated cotinine-degrading bacterium Terrabacter sp. strain cot-2 from synergistic consortium. J. Clean. Prod. 2024, 454, 142278. [Google Scholar] [CrossRef]
- Iida, T.; Mukouzaka, Y.; Nakamura, K.; Kudo, T. Plasmid-borne genes code for an angular dioxygenase involved in dibenzofuran degradation by Terrabacter sp. strain YK3. Appl. Environ. Microbiol. 2002, 68, 3716–3723. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.F.; Liu, M.Y.; Tian, Z.X.; Xiao, Y.; Zeng, P.; Han, Z.Y.; Zhou, H.; Liao, B.H. Physiological tolerance of black locust (Robinia pseudoacacia L.) and changes of rhizospheric bacterial communities in response to Cd and Pb in the contaminated soil. Environ. Sci. Pollut. R. 2024, 31, 2987–3003. [Google Scholar] [CrossRef] [PubMed]
- Pereira, L.B.; Andrade, G.S.; Meneghin, S.P.; Vicentini, R.; Ottoboni, L.M. Prospecting plant growth-promoting bacteria isolated from the rhizosphere of sugarcane under drought stress. Curr. Microbiol. 2019, 76, 1345–1354. [Google Scholar] [CrossRef] [PubMed]
- Montero-Barrientos, M.; Rivas, R.; Velazquez, E.; Monte, E.; Roig, M.G. Terrabacter terrae sp. nov., a novel actinomycete isolated from soil in Spain. Int. J. Syst. Evol. Microbiol. 2005, 55, 2491–2495. [Google Scholar] [CrossRef]
- Almaki, J.H.; Nasiri, R.; Soon, W.T.; Huyop, F.Z. Identification of novel bacterial species capable of degrading dalapon using 16s rRNA sequencing. J. Teknol. 2016, 78, 77–82. [Google Scholar]
- Li, M.; Cheng, X.; Guo, H.; Yang, Z. Biomineralization of carbonate by Terrabacter tumescens for heavy metal removal and biogrouting applications. J. Environ. Eng. 2016, 142, C4015005. [Google Scholar] [CrossRef]
- Nordstedt, N.P.; Roman-Reyna, V.; Jacobs, J.M.; Jones, M.L. Comparative genomic understanding of gram-positive plant growth-promoting Leifsonia. Phytobiomes J. 2021, 5, 263–274. [Google Scholar] [CrossRef]
- Tchuisseu-Tchakounté, G.V.; Berger, B.; Patz, S.; Fankem, H.; Ruppel, S. Community structure and plant growth-promoting potential of cultivable bacteria isolated from Cameroon soil. Microbiol. Res. 2018, 214, 47–59. [Google Scholar] [CrossRef]
- Cai, Y.; Tao, W.Z.; Ma, Y.J.; Cheng, J.; Zhang, M.Y.; Zhang, Y.X. Leifsonia flava sp. nov., a novel actinobacterium isolated from the rhizosphere of Aquilegia viridiflora. J. Microbiol. 2018, 56, 549–555. [Google Scholar] [CrossRef] [PubMed]
- Barzanti, R.; Ozino, F.; Bazzicalupo, M.; Gabbrielli, R.; Galardi, F.; Gonnelli, C.; Mengoni, A. Isolation and characterization of endophytic bacteria from the nickel hyperaccumulator plant Alyssum bertolonii. Microb. Ecol. 2007, 53, 306–316. [Google Scholar] [CrossRef]
- Shyam, S.; Nava, A.; Sarma, H. Potential microbes for environment and agriculture: Bioengineering strategies for a sustainable future. In Biotechnology of Emerging Microbes; Sarma, H., Joshi, S.J., Eds.; Academic Press: London, UK, 2024; pp. 1–29. [Google Scholar]
- Davis, M.J.; Gillaspie Jr, A.G.; Vidaver, A.K.; Harris, R.W. Clavibacter: A new genus containing some phytopathogenic coryneform bacteria, including Clavibacter xyli subsp. xyli sp. nov., subsp. nov. and Clavibacter xyli subsp. cynodontis subsp. nov., pathogens that cause ratoon stunting disease of sugarcane and bermudagrass stunting disease. Int. J. Syst. Evol. Microbiol. 1984, 34, 107–117. [Google Scholar]
- Kang, S.M.; Waqas, M.; Hamayun, M.; Asaf, S.; Khan, A.L.; Kim, A.Y.; Park, Y.G.; Lee, I.J. Gibberellins and indole-3-acetic acid producing rhizospheric bacterium Leifsonia xyli SE134 mitigates the adverse effects of copper-mediated stress on tomato. J. Plant Interact. 2017, 12, 373–380. [Google Scholar] [CrossRef]
- Tay, C.C.; Mohamad-Nasir, N.; Hashim, S.N.; Lokman, N.F.; Wong, K.K. Different enzymatic strategy to degrade carbamazepine by Rhodococcus zopfii and Leifsonia shinshuensis. Proc. Natl. Acad. Sci. India Sect. B Biol. Sci. 2024, 94, 383–388. [Google Scholar] [CrossRef]
- Luan, H.; Hu, Y.; Liu, X.; Hao, D.; Yang, L. Purification and characterization of a beta-D-xylosidase from Leifsonia shinshuensis DICP 16. Chin. J. Biotechnol. 2008, 24, 867–873. [Google Scholar]
- Anteneh, Y.S.; Franco, C.M.M. Whole cell actinobacteria as biocatalysts. Front. Microbiol. 2019, 10, 77. [Google Scholar] [CrossRef] [PubMed]
- Busch, H.; Hagedoorn, P.L.; Hanefeld, U. Rhodococcus as a versatile biocatalyst in organic synthesis. Int. J. Mol. Sci. 2019, 20, 4787. [Google Scholar] [CrossRef] [PubMed]
- Benning, S.; Pritsch, K.; Radl, V.; Siani, R.; Wang, Z.; Schloter, M. (Pan)genomic analysis of two Rhodococcus isolates and their role in phenolic compound degradation. Microbiol. Spectr. 2024, 12, e03783-23. [Google Scholar] [CrossRef] [PubMed]
- Ivshina, I.B.; Kuyukina, M.S.; Krivoruchko, A.V.; Tyumina, E.A. Responses to ecopollutants and pathogenization risks of saprotrophic Rhodococcus species. Pathogens 2021, 10, 974. [Google Scholar] [CrossRef]
- Ivshina, I.; Bazhutin, G.; Tyumina, E. Rhodococcus strains as a good biotool for neutralizing pharmaceutical pollutants and obtaining therapeutically valuable products: Through the past into the future. Front. Microbiol. 2022, 13, 967127. [Google Scholar] [CrossRef]
- Kuyukina, M.S.; Ivshina, I.B. Application of Rhodococcus in bioremediation of contaminated environments. In Biology of Rhodococcus; Alvarez, H.M., Ed.; Springer Nature: Cham, Switzerland, 2010; pp. 231–262. [Google Scholar]
- Zampolli, J.; Orro, A.; Vezzini, D.; Di Gennaro, P. Genome-based exploration of Rhodococcus species for plastic-degrading genetic determinants using bioinformatic analysis. Microorganisms 2022, 10, 1846. [Google Scholar] [CrossRef] [PubMed]
- Sangal, V.; Goodfellow, M.; Jones, A.L.; Sutcliffe, I.C. A stable home for an equine pathogen: Valid publication of the binomial Prescottella equi gen nov., comb. nov., and reclassification of four rhodococcal species into the genus Prescottella. Int. J. Syst. Evol. Microbiol. 2022, 72, 005551. [Google Scholar] [CrossRef]
- Nguyen, N.T.; Vo, V.T.; Nguyen, T.H.P.; Kiefer, R. Isolation and optimization of a glyphosate-degrading Rhodococcus soli G41 for bioremediation. Arch. Microbiol. 2022, 204, 252. [Google Scholar] [CrossRef]
- Kim, D.; Choi, K.Y.; Yoo, M.; Zylstra, G.J.; Kim, E. Biotechnological potential of Rhodococcus biodegradative pathways. J. Microbiol. Biotechnol. 2018, 28, 1037–1051. [Google Scholar] [CrossRef] [PubMed]
- Mawang, C.; Azman, A.; Mohd, F.A.; Ahamad, M. Actinobacteria: An eco-friendly and promising technology for the bioaugmentation of contaminants. Biotechnol. Rep. 2021, 32, e00679. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Hernández, M.A.; Reyes-Peralta, J.; Mendoza-Herrera, A.; Rivera, G.; Bocanegra-García, V. Characterization of a Microbacterium sp. strain isolated from soils contaminated with hydrocarbons in the Burgos basin, Mexico. Rev. Int. Contam. Ambie. 2021, 37, 227–235. [Google Scholar] [CrossRef]
- Lee, H.; Han, Y.; Kim, D. Sinomonas terrae sp. nov., isolated from an agricultural soil. J. Microbiol. Biotechnol. 2023, 33, 909–914. [Google Scholar] [CrossRef]
- Pine, O.W.; Ferreira-de Carmago, A.; Stropa, G.K.C.; de Macedo, L.E.G.; Marcondes-de Souza, J.A. Influence of vinasse application in the structure and composition of the bacterial community of the soil under sugarcane cultivation. Int. J. Microbiol. 2016, 2016, 2349514. [Google Scholar]


| Isolation Source | Strain | UFC g-1 TVCS | GenBank Accession Number | TV-MICs (%) | |
|---|---|---|---|---|---|
| WD | W | ||||
| Soil 1 | WCNS1A | 5.0 × 104 | OM060440 | 15.0 | 7.5 |
| (S1) | WCNS1B | 5.0 × 104 | OM060441 | - | 7.5 |
| WCNS1C | 1.6 × 105 | OM060442 | 12.5 | >30.0 | |
| WCNS1D | 2.0 × 104 | OM060443 | 17.5 | >30.0 | |
| WCNS1E | 5.1 × 105 | OM060444 | 25.0 | 7.5 | |
| WCNS1F | 9.5 × 105 | OM060445 | 12.5 | 7.5 | |
| Soil 2 | WCNS2A | 5.0 × 104 | OM060446 | >30.0 | >30.0 |
| (S2) | WCNS2B | 2.9 × 105 | OM060447 | 20.0 | 7.5 |
| WCNS2C | 1.6 × 105 | OM060448 | 25.0 | 7.5 | |
| WCNS2D | 1.3 × 105 | OM060449 | 25.0 | >30.0 | |
| WCNS2E | 4.2 × 105 | OM060450 | 22.5 | 7.5 | |
| WCNS2F | 1.7 × 105 | OM060451 | 20.0 | 7.5 | |
| WCNS2G | 1.0 × 105 | OM060452 | 22.5 | 7.5 | |
| Soil 3 | WCNS3A | 4.0 × 104 | OM060453 | 22.5 | >30.0 |
| (S3) | WCNS3B | 1.1 × 105 | OM060454 | >30.0 | 15.0 |
| WCNS3C | 4.0 × 104 | OM060455 | 20.0 | 30.0 | |
| WCNS3D | 1.0 × 104 | OM060456 | 12.5 | 7.5 | |
| WCNS3E | 5.0 × 104 | OM060457 | >30.0 | 15.0 | |
| WCNS3F | 1.0 × 104 | OM060458 | 15.0 | 7.5 | |
| Strain | Species Identified | Identity (%) | Related Type Strain |
|---|---|---|---|
| WCNS1A | Agromyces sp. | 99.9 | Agromyces fucosus NR_104982 |
| WCNS1B | Microbacterium sp. | 99.7 | Microbacterium liquefaciens NR_026162 |
| WCNS1C | Terrabacter sp. | 99.1 | Terrabacter terrae NR_043286 |
| WCNS1D | Azotobacter sp. | 97.6 | Azotobacter bryophylli MF078077 |
| WCNS1E | Pseudomonas sp. | 99.6 | Pseudomonas harudinis NR_181730 |
| WCNS1F | Agromyces sp. | 99.9 | Agromyces fucosus NR_104982 |
| WCNS2A | Azotobacter sp. | 97.5 | Azotobacter bryophylli MF078077 |
| WCNS2B | Azotobacter sp. | 97.4 | Azotobacter bryophylli MF078077 |
| WCNS2C | Sinomonas sp. | 99.5 | Sinomonas atrocyanea CP014518 |
| WCNS2D | Prescottella sp. | 100 | Prescottella equi X80614 |
| WCNS2E | Arthrobacter sp. | 98.4 | Arthrobacter globiformi NR_112192 |
| WCNS2F | Streptomyces sp. | 100 | Streptomyces bungoensis NR_041191 |
| WCNS2G | Leifsonia sp. | 98.9 | Leifsonia xyli CP006734 |
| WCNS3A | Rhodococcus sp. | 100 | Rhodococcus ruber OQ345823 |
| WCNS3B | Azotobacter sp. | 97.5 | Azotobacter bryophylli MF078077 |
| WCNS3C | Rhodococcus sp. | 100 | Rhodococcus ruber OQ345823 |
| WCNS3D | Azotobacter sp. | 97.5 | Azotobacter bryophylli MF078077 |
| WCNS3E | Azotobacter sp. | 97.5 | Azotobacter bryophylli MF078077 |
| WCNS3F | Azotobacter sp. | 97.4 | Azotobacter bryophylli MF078077 |
| Solubilization Halo (cm) | Siderophore Halo (cm) | Hydrolytic Enzyme Halo (cm) | ||||||
|---|---|---|---|---|---|---|---|---|
| Strain | K | PO4 | Amylase | Protease | Cellulase | Lipase | Esterase | |
| WCNS1A | - | - | - | - | 0.96 e | 1.82 a | - | - |
| WCNS1B | - | - | - | - | - | 0.35 l | - | - |
| WCNS1C | - | - | - | - | 0.07 op | 1.06 d | - | - |
| WCNS1D | - | - | - | - | - | - | - | - |
| WCNS1E | 0.10 o | 0.56 j | 0.87 f | - | - | 1.37 b | - | - |
| WCNS1F | 0.37 l | - | 0.24 m | - | - | - | 1.23 c | 1.05 d |
| WCNS2A | - | - | - | - | - | - | - | - |
| WCNS2B | - | - | - | - | - | - | - | - |
| WCNS2C | - | - | - | - | - | - | 0.84 fg | - |
| WCNS2D | - | - | - | - | - | - | - | - |
| WCNS2E | - | - | - | - | - | 0.64 i | - | - |
| WCNS2F | - | - | - | - | - | - | - | - |
| WCNS2G | - | - | 0.09 o | - | - | - | - | - |
| WCNS3A | - | - | 0.07 op | - | - | - | - | 0.81 g |
| WCNS3B | - | - | - | - | - | - | - | - |
| WCNS3C | - | - | 0.17 n | 0.04 p | - | - | - | 0.46 k |
| WCNS3D | - | - | - | - | - | - | - | - |
| WCNS3E | - | - | 0.73 h | - | - | - | - | - |
| WCNS3F | - | - | - | - | - | - | - | - |
| Strain | Seedling Parameters | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| PL (cm) | RL (cm) | SL (cm) | NR | RFW (g) | RDW (g) | PFW (g) | PDW (g) | SFW (g) | SDW (g) | |
| WCNS1A | 20.8 abc | 19.0 a | 39.8 ab | 4.2 abc | 0.49 abcd | 0.12 ab | 3.14 c | 0.48 abc | 3.6 bc | 0.60 abc |
| WCNS1B | 19.7 abcd | 11.8 c | 31.4 de | 4.0 abc | 0.43 cdef | 0.09 bcde | 2.79 e | 0.45 bc | 3.2 def | 0.54 bc |
| WCNS1C | 22.0 ab | 19.3 a | 41.3 ab | 4.5 abc | 0.56 a | 0.13 a | 3.06 d | 0.54 ab | 3.6 b | 0.67 ab |
| WCNS1D | 20.6 abcd | 20.9 a | 41.5 ab | 5.3 a | 0.41 def | 0.10 bcd | 2.45 fg | 0.48 abc | 2.9 gh | 0.58 abc |
| WCNS1F | 20.7 abcd | 19.3 a | 40.0 ab | 4.3 abc | 0.45 bcd | 0.12 ab | 3.14 c | 0.59 a | 3.6 bc | 0.71 a |
| WCNS2A | 22.3 ab | 20.8 a | 43.1 a | 4.2 abc | 0.44 bcde | 0.12 ab | 3.65 a | 0.52 ab | 4.1 a | 0.64 abc |
| WCNS2B | 20.0 abcd | 18.6 a | 38.6 abc | 3.8 abc | 0.49 abcd | 0.09 cde | 2.76 e | 0.44 bc | 3.3 de | 0.53 c |
| WCNS2C | 19.8 abcd | 17.8 ab | 37.6 abc | 5.0 ab | 0.29 g | 0.09 bcde | 2.90 de | 0.48 abc | 3.2 def | 0.67 ab |
| WCNS2D | 19.6 abcd | 17.5 ab | 37.1 bcd | 4.2 abc | 0.55 a | 0.14 a | 2.74 e | 0.40 c | 3.3 de | 0.54 bc |
| WCNS2E | 16.4 d | 12.7 c | 29.2 e | 3.7 bc | 0.36 efg | 0.06 e | 2.73 e | 0.48 abc | 3.1 efg | 0.55 bc |
| WCNS2G | 23.3 a | 14.3 bc | 37.7 abc | 5.3 a | 0.52 ab | 0.09 cde | 3.66 a | 0.58 abc | 4.2 a | 0.67 abc |
| WCNS3A | 20.9 abc | 19.5 a | 40.4 ab | 3.5 bc | 0.42 def | 0.11 abcd | 3.37 b | 0.54 ab | 3.8 b | 0.65 abc |
| WCNS3B | 23.1 ab | 18.2 ab | 41.3 ab | 3.8 abc | 0.51 abc | 0.09 bcde | 3.65 b | 0.50 abc | 4.2 a | 0.59 abc |
| WCNS3C | 20.0 abcd | 18.2 ab | 38.2 abc | 3.5 bc | 0.46 bcd | 0.10 bcd | 2.90 de | 0.49 abc | 3.4 cd | 0.59 abc |
| WCNS3E | 19.8 abcd | 18.5 ab | 38.3 abc | 3.8 abc | 0.49 abcd | 0.10 bcd | 2.28 g | 0.48 abc | 2.8 h | 0.58 abc |
| WCNS3F | 18.9 bcd | 14.3 bc | 33.2 cde | 4.0 abc | 0.45 bcde | 0.12 abc | 2.52 f | 0.45 bc | 3.0 fgh | 0.57 bc |
| C | 17.6 cd | 17.3 ab | 36.5 bcd | 3.2 c | 0.35 fg | 0.08 de | 2.75 e | 0.47 abc | 3.1 efg | 0.56 bc |
| Strain | Seedling Parameters | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| PL (cm) | RL (cm) | SL (cm) | NR | RFW (g) | RDW (g) | PFW (g) | PDW (g) | SFW (g) | SDW (g) | |
| WCNS1C-V | 24.2 abc | 23.6 abc | 47.8 ab | 5.3 a | 0.61 abc | 0.13 abc | 4.7 ab | 0.71 abc | 5.3 abc | 0.85 abc |
| WCNS1C-H | 21.3 cdef | 19.8 abc | 41.2 bcdef | 4.4 abc | 0.57 bcde | 0.13 abc | 3.0 e | 0.58 defgh | 3.6 g | 0.71 efghi |
| WCNS1D-V | 24.7 ab | 21.9 ab | 46.5 abc | 5.7 a | 0.70 a | 0.13 abc | 4.6 b | 0.73 ab | 5.3 bcd | 0.85 ab |
| WCNS1D-H | 21.3 cdef | 21.4 cdef | 42.7 abcde | 4.8 abc | 0.39 gh | 0.09 c | 2.7 e | 0.50 ghi | 3.1 gh | 0.60 j |
| WCNS1F-V | 21.8 abcdef | 24.0 abcdef | 45.8 abc | 5.3 ab | 0.57 bcde | 0.15 a | 4.3 bcd | 0.68 abcd | 4.9 bcdef | 0.83 abcd |
| WCNS1F-H | 21.6 bcdef | 19.3 bcdef | 40.9 bcdef | 4.4 abc | 0.48 defg | 0.12 abc | 3.1 e | 0.51 ghi | 3.6 g | 0.63 hij |
| WCNS2A-V | 23.0 abcde | 24.8 abcde | 47.8 ab | 5.2 ab | 0.72 a | 0.15 a | 4.5 bc | 0.75 a | 5.2 abcd | 0.90 a |
| WCNS2A-H | 20.4 efg | 21.4 efg | 41.8 bcde | 4.2 abc | 0.47 efg | 0.12 abc | 3.8 d | 0.55 efghi | 4.3 f | 0.68 efghij |
| WCNS2D-V | 21.1 abcde | 22.5 abcde | 43.6 abcde | 5.5 a | 0.56 cde | 0.15 a | 4.2 bcd | 0.64 bcde | 4.7 cdef | 0.79 cde |
| WCNS2D-H | 19.9 efg | 16.7 efg | 36.6 ef | 3.9 abc | 0.49 defg | 0.11 abc | 3.8 d | 0.55 efghi | 4.3 f | 0.66 fghij |
| WCNS2G-V | 22.7 abcde | 21.5 abcde | 44.1 abcd | 5.0 ab | 0.55 cdef | 0.13 abc | 5.2 a | 0.63 cdef | 5.8 a | 0.76 bcdef |
| WCNS2G-H | 21.6 bcdef | 20.9 bcdef | 42.5 bcde | 4.5 abc | 0.50 defg | 0.12 abc | 4.5 bc | 0.54 efghi | 5.0 bcde | 0.66 fghij |
| WCNS3A-V | 23.9 abcd | 23.9 abcd | 47.7 ab | 5.1 ab | 0.66 a | 0.14 ab | 4.7 ab | 0.58 defgh | 5.3 ab | 0.72 defghi |
| WCNS3A-H | 21.8 abcdef | 22.1 abcdef | 43.9 abcde | 3.9 abc | 0.50 defg | 0.13 abc | 3.0 e | 0.55 efghi | 3.5 g | 0.68 efghij |
| WCNS3C-V | 25.0 a | 25.0 a | 49.9 a | 4.8 abc | 0.49 defg | 0.14 ab | 3.9 cd | 0.60 cdfg | 4.4 def | 0.74 cdefgh |
| WCNS3C-H | 19.2 fg | 20.9 fg | 40.1 cdef | 3.1 c | 0.41 fgh | 0.11 abc | 2.8 e | 0.47 i | 3.2 gh | 0.58 j |
| WCNS3E-V | 22.1 abcdef | 21.5 abcdef | 43.5 abcde | 4.4 abc | 0.66 abc | 0.12 abc | 3.8 d | 0.62 cdef | 4.4 ef | 0.74 cdefg |
| WCNS3E-H | 20.6 defg | 18.8 defg | 39.4 cdef | 3.8 abc | 0.48 defg | 0.10 bc | 2.8 e | 0.53 fghi | 3.3 gh | 0.63 ghij |
| PC | 19.0 fg | 19.0 fg | 38.0 def | 3.8 abc | 0.38 gh | 0.10 bc | 2.9 e | 0.51 ghi | 3.2 gh | 0.61 ij |
| NC | 18.0 g | 16.3 g | 34.2 f | 3.4 bc | 0.31 h | 0.10 bc | 2.5 e | 0.48 hi | 2.9 h | 0.58 j |
| p value of significance | ||||||||||
| Treat | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 |
| Strain | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 |
| Treat × Strain | 0.001 | 0.327 | 0.248 | 0.735 | <0.001 | 0.094 | <0.001 | <0.001 | <0.001 | <0.001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Valencia-Botín, A.J.; Chávez-Díaz, I.F.; Zurita-Martínez, F.; Tejeda-Ortega, A.; Zelaya-Molina, L.X. Plant Growth-Promoting and Tequila Vinasse-Resistant Bacterial Strains. Microbiol. Res. 2024, 15, 1144-1162. https://doi.org/10.3390/microbiolres15030077
Valencia-Botín AJ, Chávez-Díaz IF, Zurita-Martínez F, Tejeda-Ortega A, Zelaya-Molina LX. Plant Growth-Promoting and Tequila Vinasse-Resistant Bacterial Strains. Microbiology Research. 2024; 15(3):1144-1162. https://doi.org/10.3390/microbiolres15030077
Chicago/Turabian StyleValencia-Botín, Alberto J., Ismael F. Chávez-Díaz, Florentina Zurita-Martínez, Allan Tejeda-Ortega, and Lily X. Zelaya-Molina. 2024. "Plant Growth-Promoting and Tequila Vinasse-Resistant Bacterial Strains" Microbiology Research 15, no. 3: 1144-1162. https://doi.org/10.3390/microbiolres15030077
APA StyleValencia-Botín, A. J., Chávez-Díaz, I. F., Zurita-Martínez, F., Tejeda-Ortega, A., & Zelaya-Molina, L. X. (2024). Plant Growth-Promoting and Tequila Vinasse-Resistant Bacterial Strains. Microbiology Research, 15(3), 1144-1162. https://doi.org/10.3390/microbiolres15030077







