Unveiling the Full Protein Effectorome of the Black Sigatoka Pathogen Pseudocercospora fijiensis—An In Silico Approach
Abstract
1. Introduction
2. Materials and Methods
3. Results
3.1. Effectorome Generalities
3.2. Effector Functions of P. fijiensis Effectors Are Associated with Stages of Biotrophy and Necrotrophy
3.3. Predicted P. fijiensis Effectors Are Expressed in Mycelia and Conidia, and in Interaction with the Plant Host
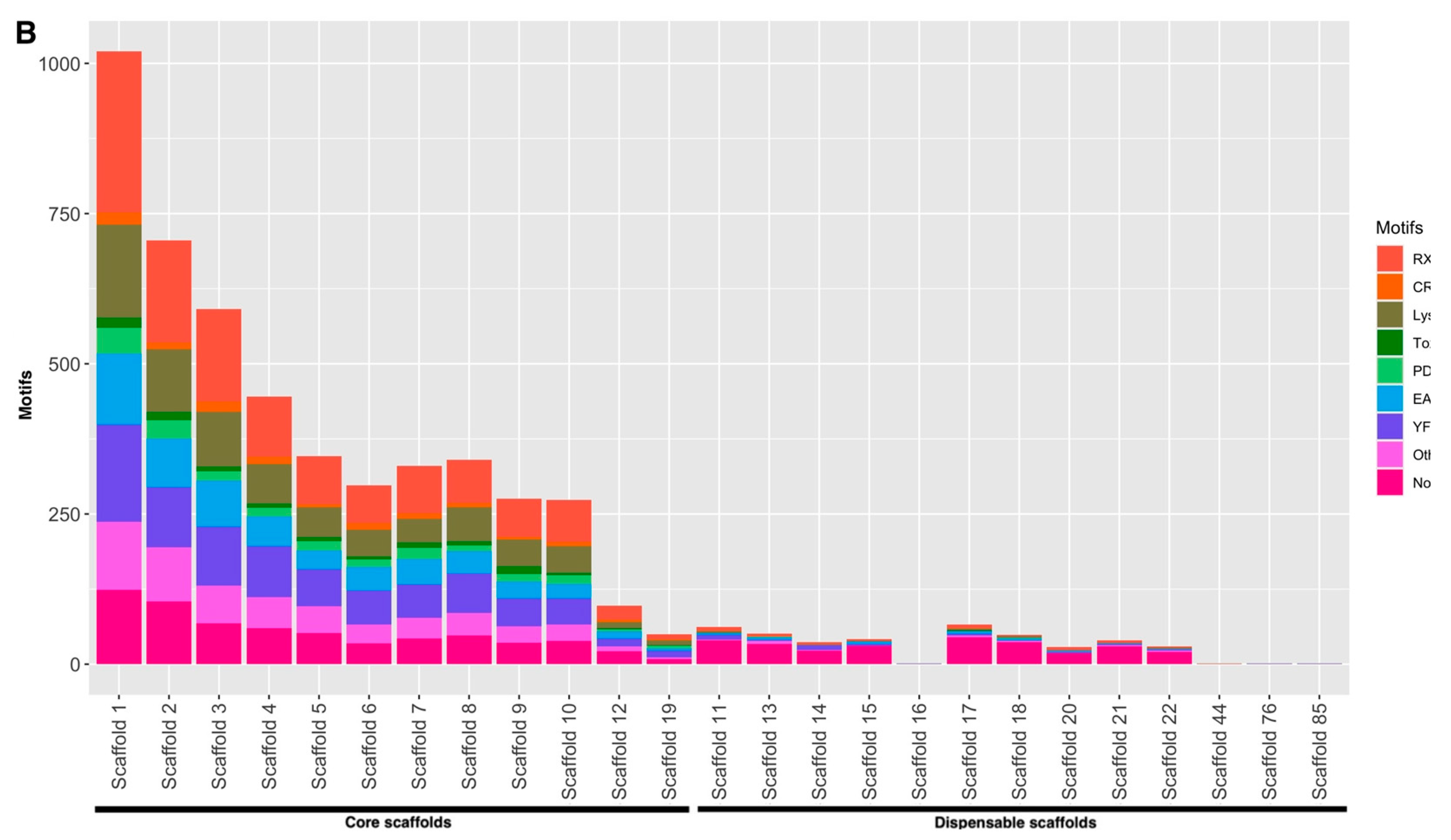
3.4. Effectors Are Distributed Throughout the P. fijiensis Genome
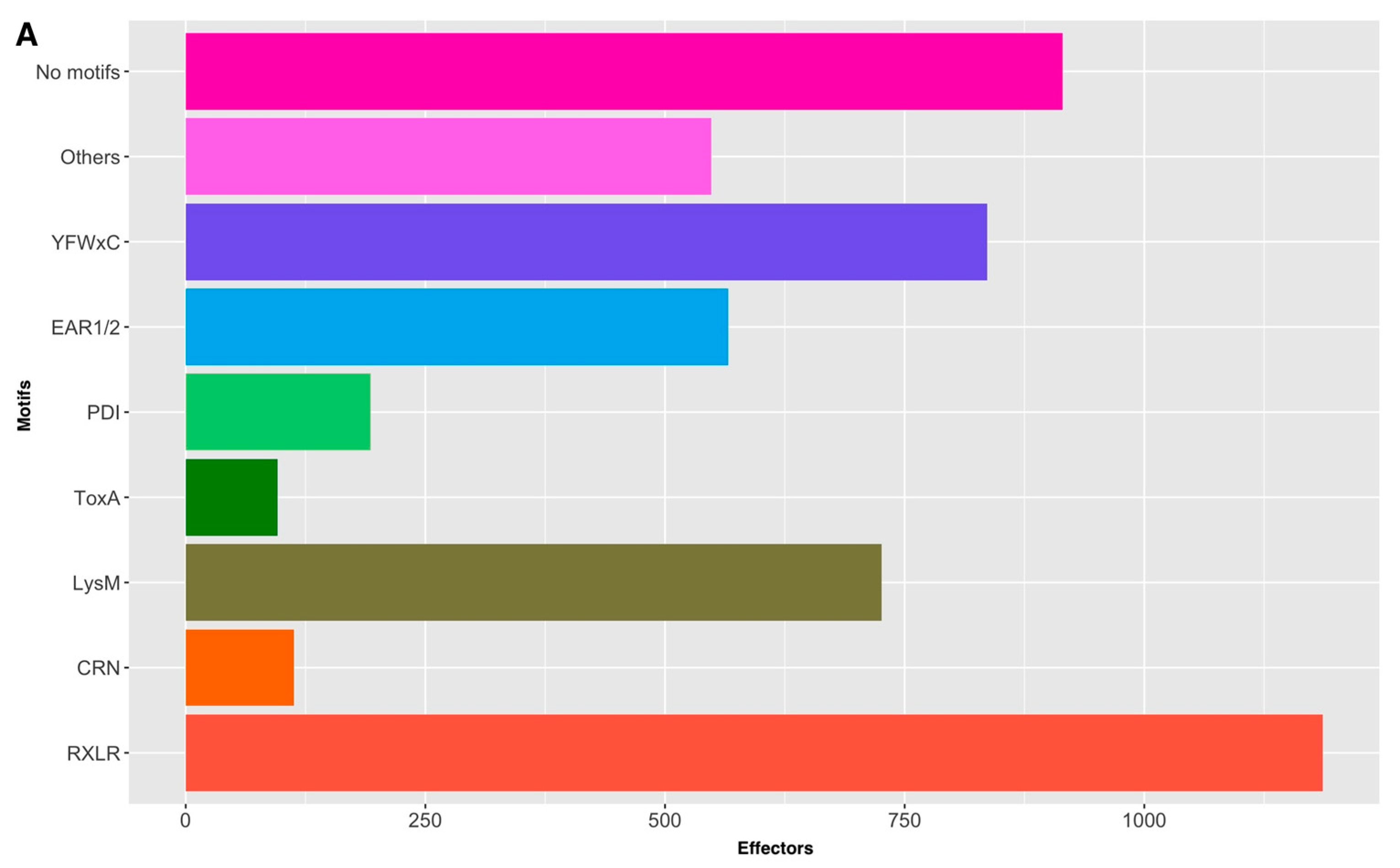
3.5. Motif Screening in P. fijiensis Effectors
3.6. Presence of Motifs in Other Fungal Effectors
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Churchill, A.C.L. Mycosphaerella Fijiensis, the Black Leaf Streak Pathogen of Banana: Progress towards Understanding Pathogen Biology and Detection, Disease Development, and the Challenges of Control. Mol. Plant Pathol. 2011, 12, 307–328. [Google Scholar] [CrossRef]
- Arango Isaza, R.E.; Diaz-Trujillo, C.; Dhillon, B.; Aerts, A.; Carlier, J.; Crane, C.F.; V. De Jong, T.; De Vries, I.; Dietrich, R.; Farmer, A.D.; et al. Combating a Global Threat to a Clonal Crop: Banana Black Sigatoka Pathogen Pseudocercospora Fijiensis (Synonym Mycosphaerella Fijiensis) Genomes Reveal Clues for Disease Control. PLoS Genet. 2016, 12, e1005876. [Google Scholar] [CrossRef]
- Chang, T.-C.; Salvucci, A.; Crous, P.W.; Stergiopoulos, I. Comparative Genomics of the Sigatoka Disease Complex on Banana Suggests a Link between Parallel Evolutionary Changes in Pseudocercospora fijiensis and Pseudocercospora eumusae and Increased Virulence on the Banana Host. PLoS Genet. 2016, 12, e1005904. [Google Scholar] [CrossRef]
- Soares, J.M.D.S.; Rocha, A.D.J.; Nascimento, F.D.S.; Amorim, V.B.O.D.; Ramos, A.P.D.S.; Ferreira, C.F.; Haddad, F.; Amorim, E.P. Gene Expression, Histology and Histochemistry in the Interaction between Musa Sp. and Pseudocercospora fijiensis. Plants 2022, 11, 1953. [Google Scholar] [CrossRef]
- Todd, J.N.A.; Carreón-Anguiano, K.G.; Couoh-Dzul, O.J.; De Los Santos-Briones, C.; Canto-Canché, B. Effectors: Key Actors in Phytopathology. Rev. Mex. Fitopatol. 2023, 41, 203–228. [Google Scholar] [CrossRef]
- Derbyshire, M.C.; Raffaele, S. Surface Frustration Re-Patterning Underlies the Structural Landscape and Evolvability of Fungal Orphan Candidate Effectors. Nat. Commun. 2023, 14, 5244. [Google Scholar] [CrossRef]
- De Wit, P.J.G.M.; Mehrabi, R.; Van Den Burg, H.A.; Stergiopoulos, I. Fungal Effector Proteins: Past, Present and Future. Mol. Plant Pathol. 2009, 10, 735–747. [Google Scholar] [CrossRef]
- Stergiopoulos, I.; Van Den Burg, H.A.; Ökmen, B.; Beenen, H.G.; Van Liere, S.; Kema, G.H.J.; De Wit, P.J.G.M. Tomato Cf Resistance Proteins Mediate Recognition of Cognate Homologous Effectors from Fungi Pathogenic on Dicots and Monocots. Proc. Natl. Acad. Sci. USA 2010, 107, 7610–7615. [Google Scholar] [CrossRef]
- Van Den Burg, H.A.; Harrison, S.J.; Joosten, M.H.A.J.; Vervoort, J.; De Wit, P.J.G.M. Cladosporium Fulvum Avr4 Protects Fungal Cell Walls Against Hydrolysis by Plant Chitinases Accumulating during Infection. MPMI 2006, 19, 1420–1430. [Google Scholar] [CrossRef]
- Kohler, A.C.; Chen, L.-H.; Hurlburt, N.; Salvucci, A.; Schwessinger, B.; Fisher, A.J.; Stergiopoulos, I. Structural Analysis of an Avr4 Effector Ortholog Offers Insight into Chitin Binding and Recognition by the Cf-4 Receptor. Plant Cell 2016, 28, 1945–1965. [Google Scholar] [CrossRef]
- Santos Rezende, J.; Zivanovic, M.; Costa De Novaes, M.I.; Chen, Z. The AVR4 Effector Is Involved in Cercosporin Biosynthesis and Likely Affects the Virulence of Cercospora Cf. flagellaris on Soybean. Mol. Plant Pathol. 2020, 21, 53–65. [Google Scholar] [CrossRef]
- Van Den Burg, H.A.; Westerink, N.; Francoijs, K.-J.; Roth, R.; Woestenenk, E.; Boeren, S.; De Wit, P.J.G.M.; Joosten, M.H.A.J.; Vervoort, J. Natural Disulfide Bond-Disrupted Mutants of AVR4 of the Tomato Pathogen Cladosporium Fulvum Are Sensitive to Proteolysis, Circumvent Cf-4-Mediated Resistance, but Retain Their Chitin Binding Ability. J. Biol. Chem. 2003, 278, 27340–27346. [Google Scholar] [CrossRef]
- Mesarich, C.H.; Stergiopoulos, I.; Beenen, H.G.; Cordovez, V.; Guo, Y.; Karimi Jashni, M.; Bradshaw, R.E.; De Wit, P.J.G.M. A Conserved Proline Residue in Dothideomycete Avr4 Effector Proteins Is Required to Trigger a Cf-4-dependent Hypersensitive Response. Mol. Plant Pathol. 2016, 17, 84–95. [Google Scholar] [CrossRef] [PubMed]
- Laugé, R.; Joosten, M.H.A.J.; Van Den Ackerveken, G.F.J.M.; Van Den Broek, H.W.J.; De Wit, P.J.G.M. The In Planta-Produced Extracellular Proteins ECP1 and ECP2 of Cladosporium fulvum Are Virulence Factors. MPMI 1997, 10, 725–734. [Google Scholar] [CrossRef]
- Zhang, M.; Xie, S.; Zhao, Y.; Meng, X.; Song, L.; Feng, H.; Huang, L. Hce2 Domain-containing Effectors Contribute to the Full Virulence of Valsa Mali in a Redundant Manner. Mol. Plant Pathol. 2019, 20, 843–856. [Google Scholar] [CrossRef] [PubMed]
- Stergiopoulos, I.; Kourmpetis, Y.A.I.; Slot, J.C.; Bakker, F.T.; De Wit, P.J.G.M.; Rokas, A. In Silico Characterization and Molecular Evolutionary Analysis of a Novel Superfamily of Fungal Effector Proteins. Mol. Biol. Evol. 2012, 29, 3371–3384. [Google Scholar] [CrossRef]
- Guo, Y.; Hunziker, L.; Mesarich, C.H.; Chettri, P.; Dupont, P.-Y.; Ganley, R.J.; McDougal, R.L.; Barnes, I.; Bradshaw, R.E. DsEcp2-1 Is a Polymorphic Effector That Restricts Growth of Dothistroma septosporum in Pine. Fungal Genet. Biol. 2020, 135, 103300. [Google Scholar] [CrossRef]
- De Jonge, R.; Peter Van Esse, H.; Kombrink, A.; Shinya, T.; Desaki, Y.; Bours, R.; Van Der Krol, S.; Shibuya, N.; Joosten, M.H.A.J.; Thomma, B.P.H.J. Conserved Fungal LysM Effector Ecp6 Prevents Chitin-Triggered Immunity in Plants. Science 2010, 329, 953–955. [Google Scholar] [CrossRef]
- Bolton, M.D.; Van Esse, H.P.; Vossen, J.H.; De Jonge, R.; Stergiopoulos, I.; Stulemeijer, I.J.E.; Van Den Berg, G.C.M.; Borrás-Hidalgo, O.; Dekker, H.L.; De Koster, C.G.; et al. The Novel Cladosporium fulvum Lysin Motif Effector Ecp6 Is a Virulence Factor with Orthologues in Other Fungal Species. Mol. Microbiol. 2008, 69, 119–136. [Google Scholar] [CrossRef]
- Ohm, R.A.; Feau, N.; Henrissat, B.; Schoch, C.L.; Horwitz, B.A.; Barry, K.W.; Condon, B.J.; Copeland, A.C.; Dhillon, B.; Glaser, F.; et al. Diverse Lifestyles and Strategies of Plant Pathogenesis Encoded in the Genomes of Eighteen Dothideomycetes Fungi. PLoS Pathog. 2012, 8, e1003037. [Google Scholar] [CrossRef]
- Noar, R.D.; Daub, M.E. Transcriptome Sequencing of Mycosphaerella fijiensis during Association with Musa acuminata Reveals Candidate Pathogenicity Genes. BMC Genom. 2016, 17, 690. [Google Scholar] [CrossRef] [PubMed]
- Rep, M. Small Proteins of Plant-Pathogenic Fungi Secreted during Host Colonization. FEMS Microbiol. Lett. 2005, 253, 19–27. [Google Scholar] [CrossRef]
- Giraldo, M.C.; Valent, B. Filamentous Plant Pathogen Effectors in Action. Nat. Rev. Microbiol. 2013, 11, 800–814. [Google Scholar] [CrossRef]
- Carreón-Anguiano, K.G.; Islas-Flores, I.; Vega-Arreguín, J.; Sáenz-Carbonell, L.; Canto-Canché, B. EffHunter: A Tool for Prediction of Effector Protein Candidates in Fungal Proteomic Databases. Biomolecules 2020, 10, 712. [Google Scholar] [CrossRef]
- Ghareeb, H.; Drechsler, F.; Löfke, C.; Teichmann, T.; Schirawski, J. SUPPRESSOR OF APICAL DOMINANCE 1 of Sporisorium Reilianum Modulates Inflorescence Branching Architecture in Maize and Arabidopsis. Plant Physiol. 2015, 169, 2789–2804. [Google Scholar] [CrossRef]
- Liu, T.; Song, T.; Zhang, X.; Yuan, H.; Su, L.; Li, W.; Xu, J.; Liu, S.; Chen, L.; Chen, T.; et al. Unconventionally Secreted Effectors of Two Filamentous Pathogens Target Plant Salicylate Biosynthesis. Nat. Commun. 2014, 5, 4686. [Google Scholar] [CrossRef]
- Pennington, H.G.; Jones, R.; Kwon, S.; Bonciani, G.; Thieron, H.; Chandler, T.; Luong, P.; Morgan, S.N.; Przydacz, M.; Bozkurt, T.; et al. The Fungal Ribonuclease-like Effector Protein CSEP0064/BEC1054 Represses Plant Immunity and Interferes with Degradation of Host Ribosomal RNA. PLoS Pathog. 2019, 15, e1007620. [Google Scholar] [CrossRef]
- Carreón-Anguiano, K.G.; Todd, J.N.A.; Chi-Manzanero, B.H.; Couoh-Dzul, O.J.; Islas-Flores, I.; Canto-Canché, B. WideEffHunter: An Algorithm to Predict Canonical and Non-Canonical Effectors in Fungi and Oomycetes. IJMS 2022, 23, 13567. [Google Scholar] [CrossRef] [PubMed]
- Bertazzoni, S.; Williams, A.H.; Jones, D.A.; Syme, R.A.; Tan, K.-C.; Hane, J.K. Accessories Make the Outfit: Accessory Chromosomes and Other Dispensable DNA Regions in Plant-Pathogenic Fungi. MPMI 2018, 31, 779–788. [Google Scholar] [CrossRef]
- Chen, H.; King, R.; Smith, D.; Bayon, C.; Ashfield, T.; Torriani, S.; Kanyuka, K.; Hammond-Kosack, K.; Bieri, S.; Rudd, J. Combined Pangenomics and Transcriptomics Reveals Core and Redundant Virulence Processes in a Rapidly Evolving Fungal Plant Pathogen. BMC Biol. 2023, 21, 24. [Google Scholar] [CrossRef]
- Dutheil, J.Y.; Mannhaupt, G.; Schweizer, G.; Sieber, C.M.K.; Münsterkötter, M.; Güldener, U.; Schirawski, J.; Kahmann, R. A Tale of Genome Compartmentalization: The Evolution of Virulence Clusters in Smut Fungi. Genome Biol. Evol. 2016, 8, 681–704. [Google Scholar] [CrossRef] [PubMed]
- Gout, L.; Fudal, I.; Kuhn, M.; Blaise, F.; Eckert, M.; Cattolico, L.; Balesdent, M.; Rouxel, T. Lost in the Middle of Nowhere: The AvrLm1 Avirulence Gene of the Dothideomycete Leptosphaeria maculans. Mol. Microbiol. 2006, 60, 67–80. [Google Scholar] [CrossRef] [PubMed]
- Plissonneau, C.; Stürchler, A.; Croll, D. The Evolution of Orphan Regions in Genomes of a Fungal Pathogen of Wheat. mBio 2016, 7, e01231-16. [Google Scholar] [CrossRef] [PubMed]
- Torres, D.E.; Oggenfuss, U.; Croll, D.; Seidl, M.F. Genome Evolution in Fungal Plant Pathogens: Looking beyond the Two-Speed Genome Model. Fungal Biol. Rev. 2020, 34, 136–143. [Google Scholar] [CrossRef]
- Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Gonzales, N.R.; Gwadz, M.; Lu, S.; Marchler, G.H.; Song, J.S.; Thanki, N.; Yamashita, R.A.; et al. The Conserved Domain Database in 2023. Nucleic Acids Res. 2023, 51, D384–D388. [Google Scholar] [CrossRef] [PubMed]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The Protein Families Database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. 20 Years of the SMART Protein Domain Annotation Resource. Nucleic Acids Res. 2018, 46, D493–D496. [Google Scholar] [CrossRef]
- Tatusov, R.L. The COG Database: New Developments in Phylogenetic Classification of Proteins from Complete Genomes. Nucleic Acids Res. 2001, 29, 22–28. [Google Scholar] [CrossRef]
- Haft, D.H.; Selengut, J.D.; Richter, R.A.; Harkins, D.; Basu, M.K.; Beck, E. TIGRFAMs and Genome Properties in 2013. Nucleic Acids Res. 2012, 41, D387–D395. [Google Scholar] [CrossRef]
- Klimke, W.; Agarwala, R.; Badretdin, A.; Chetvernin, S.; Ciufo, S.; Fedorov, B.; Kiryutin, B.; O’Neill, K.; Resch, W.; Resenchuk, S.; et al. The National Center for Biotechnology Information’s Protein Clusters Database. Nucleic Acids Res. 2009, 37, D216–D223. [Google Scholar] [CrossRef]
- Li, W.; O’Neill, K.R.; Haft, D.H.; DiCuccio, M.; Chetvernin, V.; Badretdin, A.; Coulouris, G.; Chitsaz, F.; Derbyshire, M.K.; Durkin, A.S.; et al. RefSeq: Expanding the Prokaryotic Genome Annotation Pipeline Reach with Protein Family Model Curation. Nucleic Acids Res. 2021, 49, D1020–D1028. [Google Scholar] [CrossRef]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The Conserved Domain Database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef]
- Madeira, F.; Madhusoodanan, N.; Lee, J.; Eusebi, A.; Niewielska, A.; Tivey, A.R.N.; Lopez, R.; Butcher, S. The EMBL-EBI Job Dispatcher Sequence Analysis Tools Framework in 2024. Nucleic Acids Res. 2024, 2024, gkae241. [Google Scholar] [CrossRef]
- Noar, R.D.; Daub, M.E. Bioinformatics Prediction of Polyketide Synthase Gene Clusters from Mycosphaerella fijiensis. PLoS ONE 2016, 11, e0158471. [Google Scholar] [CrossRef]
- Carreón-Anguiano, K.G.; Gómez-Tah, R.; Pech-Balan, E.; Ek-Hernández, G.E.; De Los Santos-Briones, C.; Islas-Flores, I.; Canto-Canché, B. Pseudocercospora fijiensis Conidial Germination Is Dominated by Pathogenicity Factors and Effectors. JoF 2023, 9, 970. [Google Scholar] [CrossRef]
- Carreón-Anguiano, K.G.; Vila-Luna, S.E.; Sáenz-Carbonell, L.; Canto-Canche, B. PhyEffector, the First Algorithm That Identifies Classical and Non-Classical Effectors in Phytoplasmas. Biomimetics 2023, 8, 550. [Google Scholar] [CrossRef]
- Sonah, H.; Deshmukh, R.K.; Bélanger, R.R. Computational Prediction of Effector Proteins in Fungi: Opportunities and Challenges. Front. Plant Sci. 2016, 7, 126. [Google Scholar] [CrossRef]
- Sperschneider, J.; Dodds, P.N. EffectorP 3.0: Prediction of Apoplastic and Cytoplasmic Effectors in Fungi and Oomycetes. Mol. Plant-Microbe Interact. 2021, 35, 146–156. [Google Scholar] [CrossRef]
- Huang, Z.; Li, H.; Zhou, Y.; Bao, Y.; Duan, Z.; Wang, C.; Powell, C.A.; Chen, B.; Zhang, M.; Yao, W. Predication of the Effector Proteins Secreted by Fusarium sacchari Using Genomic Analysis and Heterogenous Expression. JoF 2022, 8, 59. [Google Scholar] [CrossRef]
- Zhao, S.; Ye, Z.; Stanton, R. Misuse of RPKM or TPM Normalization When Comparing across Samples and Sequencing Protocols. RNA 2020, 26, 903–909. [Google Scholar] [CrossRef]
- Duplessis, S.; Cuomo, C.A.; Lin, Y.-C.; Aerts, A.; Tisserant, E.; Veneault-Fourrey, C.; Joly, D.L.; Hacquard, S.; Amselem, J.; Cantarel, B.L.; et al. Obligate Biotrophy Features Unraveled by the Genomic Analysis of Rust Fungi. Proc. Natl. Acad. Sci. USA 2011, 108, 9166–9171. [Google Scholar] [CrossRef]
- Chen, J.; Liu, C.; Gui, Y.; Si, K.; Zhang, D.; Wang, J.; Short, D.P.G.; Huang, J.; Li, N.; Liang, Y.; et al. Comparative Genomics Reveals Cotton-specific Virulence Factors in Flexible Genomic Regions in Verticillium dahliae and Evidence of Horizontal Gene Transfer from Fusarium. New Phytol. 2018, 217, 756–770. [Google Scholar] [CrossRef]
- Marton, K.; Flajšman, M.; Radišek, S.; Košmelj, K.; Jakše, J.; Javornik, B.; Berne, S. Comprehensive Analysis of Verticillium nonalfalfae in Silico Secretome Uncovers Putative Effector Proteins Expressed during Hop Invasion. PLoS ONE 2018, 13, e0198971. [Google Scholar] [CrossRef]
- Wang, D.; Tian, L.; Zhang, D.; Song, J.; Song, S.; Yin, C.; Zhou, L.; Liu, Y.; Wang, B.; Kong, Z.; et al. Functional Analyses of Small Secreted Cysteine-rich Proteins Identified Candidate Effectors in Verticillium dahliae. Mol. Plant Pathol. 2020, 21, 667–685. [Google Scholar] [CrossRef]
- Morais Do Amaral, A.; Antoniw, J.; Rudd, J.J.; Hammond-Kosack, K.E. Defining the Predicted Protein Secretome of the Fungal Wheat Leaf Pathogen Mycosphaerella graminicola. PLoS ONE 2012, 7, e49904. [Google Scholar] [CrossRef]
- Arroyo-Velez, N.; González-Fuente, M.; Peeters, N.; Lauber, E.; Noël, L.D. From Effectors to Effectomes: Are Functional Studies of Individual Effectors Enough to Decipher Plant Pathogen Infectious Strategies? PLoS Pathog. 2020, 16, e1009059. [Google Scholar] [CrossRef]
- Jones, D.A.B.; Rozano, L.; Debler, J.W.; Mancera, R.L.; Moolhuijzen, P.M.; Hane, J.K. An Automated and Combinative Method for the Predictive Ranking of Candidate Effector Proteins of Fungal Plant Pathogens. Sci. Rep. 2021, 11, 19731. [Google Scholar] [CrossRef]
- De Wit, P.J.G.M.; Van Der Burgt, A.; Ökmen, B.; Stergiopoulos, I.; Abd-Elsalam, K.A.; Aerts, A.L.; Bahkali, A.H.; Beenen, H.G.; Chettri, P.; Cox, M.P.; et al. The Genomes of the Fungal Plant Pathogens Cladosporium fulvum and Dothistroma septosporum Reveal Adaptation to Different Hosts and Lifestyles But Also Signatures of Common Ancestry. PLoS Genet. 2012, 8, e1003088. [Google Scholar] [CrossRef]
- Manning, V.A.; Pandelova, I.; Dhillon, B.; Wilhelm, L.J.; Goodwin, S.B.; Berlin, A.M.; Figueroa, M.; Freitag, M.; Hane, J.K.; Henrissat, B.; et al. Comparative Genomics of a Plant-Pathogenic Fungus, Pyrenophora Tritici-Repentis, Reveals Transduplication and the Impact of Repeat Elements on Pathogenicity and Population Divergence. G3 2013, 3, 41–63. [Google Scholar] [CrossRef]
- Armitage, A.D.; Taylor, A.; Sobczyk, M.K.; Baxter, L.; Greenfield, B.P.J.; Bates, H.J.; Wilson, F.; Jackson, A.C.; Ott, S.; Harrison, R.J.; et al. Characterisation of Pathogen-Specific Regions and Novel Effector Candidates in Fusarium oxysporum f. Sp. Cepae. Sci. Rep. 2018, 8, 13530. [Google Scholar] [CrossRef]
- Nur, M.; Wood, K.; Michelmore, R. EffectorO: Motif-Independent Prediction of Effectors in Oomycete Genomes Using Machine Learning and Lineage Specificity. MPMI 2023, 36, 397–410. [Google Scholar] [CrossRef]
- Liang, P.; Liu, S.; Xu, F.; Jiang, S.; Yan, J.; He, Q.; Liu, W.; Lin, C.; Zheng, F.; Wang, X.; et al. Powdery Mildews Are Characterized by Contracted Carbohydrate Metabolism and Diverse Effectors to Adapt to Obligate Biotrophic Lifestyle. Front. Microbiol. 2018, 9, 3160. [Google Scholar] [CrossRef]
- Li, Q.; Feng, Y.; Li, J.; Hai, Y.; Si, L.; Tan, C.; Peng, J.; Hu, Z.; Li, Z.; Li, C.; et al. Multi-Omics Approaches to Understand Pathogenicity during Potato Early Blight Disease Caused by Alternaria solani. Front. Microbiol. 2024, 15, 1357579. [Google Scholar] [CrossRef]
- Donzelli, B.G.G.; Churchill, A.C.L. A dose-response approach differentiating virulence of Mycosphaerella fijiensis strains on banana leaves uses either spores or mycelia as inocula. Acta Hortic. 2009, 828, 153–160. [Google Scholar] [CrossRef]
- Hubrich, F.; Müller, M.; Andexer, J.N. Chorismate- and Isochorismate Converting Enzymes: Versatile Catalysts Acting on an Important Metabolic Node. Chem. Commun. 2021, 57, 2441–2463. [Google Scholar] [CrossRef]
- Kuhn, H.; Kwaaitaal, M.; Kusch, S.; Acevedo-Garcia, J.; Wu, H.; Panstruga, R. Biotrophy at Its Best: Novel Findings and Unsolved Mysteries of the Arabidopsis-Powdery Mildew Pathosystem. Arab. Book. 2016, 14, e0184. [Google Scholar] [CrossRef]
- Nick, P. Taming the Fire—Transcription Factors for Redox Control in Animals and Plants. Protoplasma 2024, 261, 395–396. [Google Scholar] [CrossRef]
- Nagano, N.; Umemura, M.; Izumikawa, M.; Kawano, J.; Ishii, T.; Kikuchi, M.; Tomii, K.; Kumagai, T.; Yoshimi, A.; Machida, M.; et al. Class of Cyclic Ribosomal Peptide Synthetic Genes in Filamentous Fungi. Fungal Genet. Biol. 2016, 86, 58–70. [Google Scholar] [CrossRef]
- Burgess, A.; Mornon, J.-P.; De Saint-Basile, G.; Callebaut, I. A Concanavalin A-like Lectin Domain in the CHS1/LYST Protein, Shared by Members of the BEACH Family. Bioinformatics 2009, 25, 1219–1222. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ohtaki, S.; Maeda, H.; Takahashi, T.; Yamagata, Y.; Hasegawa, F.; Gomi, K.; Nakajima, T.; Abe, K. Novel Hydrophobic Surface Binding Protein, HsbA, Produced by Aspergillus oryzae. Appl. Environ. Microbiol. 2006, 72, 2407–2413. [Google Scholar] [CrossRef]
- Fernandes, T.R.; Segorbe, D.; Prusky, D.; Di Pietro, A. How Alkalinization Drives Fungal Pathogenicity. PLoS Pathog. 2017, 13, e1006621. [Google Scholar] [CrossRef] [PubMed]
- Thynne, E.; Saur, I.M.L.; Simbaqueba, J.; Ogilvie, H.A.; Gonzalez-Cendales, Y.; Mead, O.; Taranto, A.; Catanzariti, A.; McDonald, M.C.; Schwessinger, B.; et al. Fungal Phytopathogens Encode Functional Homologues of Plant Rapid Alkalinization Factor (RALF) Peptides. Mol. Plant Pathol. 2017, 18, 811–824. [Google Scholar] [CrossRef] [PubMed]
- Seidl, M.F.; Faino, L.; Shi-Kunne, X.; Van Den Berg, G.C.M.; Bolton, M.D.; Thomma, B.P.H.J. The Genome of the Saprophytic Fungus Verticillium tricorpus Reveals a Complex Effector Repertoire Resembling That of Its Pathogenic Relatives. MPMI 2015, 28, 362–373. [Google Scholar] [CrossRef] [PubMed]
- Simbaqueba, J.; Rodríguez, E.A.; Burbano-David, D.; González, C.; Caro-Quintero, A. Putative Novel Effector Genes Revealed by the Genomic Analysis of the Phytopathogenic Fungus Fusarium oxysporum f. Sp. Physali (Foph) That Infects Cape Gooseberry Plants. Front. Microbiol. 2021, 11, 593915. [Google Scholar] [CrossRef]
- Covert, S.F. Supernumerary Chromosomes in Filamentous Fungi. Curr. Genet. 1998, 33, 311–319. [Google Scholar] [CrossRef]
- Goodwin, S.B.; Ben M’Barek, S.; Dhillon, B.; Wittenberg, A.H.J.; Crane, C.F.; Hane, J.K.; Foster, A.J.; Van Der Lee, T.A.J.; Grimwood, J.; Aerts, A.; et al. Finished Genome of the Fungal Wheat Pathogen Mycosphaerella graminicola Reveals Dispensome Structure, Chromosome Plasticity, and Stealth Pathogenesis. PLoS Genet. 2011, 7, e1002070. [Google Scholar] [CrossRef]
- Hatta, R.; Ito, K.; Hosaki, Y.; Tanaka, T.; Tanaka, A.; Yamamoto, M.; Akimitsu, K.; Tsuge, T. A Conditionally Dispensable Chromosome Controls Host-Specific Pathogenicity in the Fungal Plant Pathogen Alternaria alternata. Genet. 2002, 161, 59–70. [Google Scholar] [CrossRef]
- Peng, Z.; Oliveira-Garcia, E.; Lin, G.; Hu, Y.; Dalby, M.; Migeon, P.; Tang, H.; Farman, M.; Cook, D.; White, F.F.; et al. Effector Gene Reshuffling Involves Dispensable Mini-Chromosomes in the Wheat Blast Fungus. PLoS Genet. 2019, 15, e1008272. [Google Scholar] [CrossRef]
- Rocafort, M.; Bowen, J.K.; Hassing, B.; Cox, M.P.; McGreal, B.; De La Rosa, S.; Plummer, K.M.; Bradshaw, R.E.; Mesarich, C.H. The Venturia inaequalis Effector Repertoire Is Dominated by Expanded Families with Predicted Structural Similarity, but Unrelated Sequence, to Avirulence Proteins from Other Plant-Pathogenic Fungi. BMC Biol. 2022, 20, 246. [Google Scholar] [CrossRef]
- Queiroz, C.B.D.; Santana, M.F. Prediction of the Secretomes of Endophytic and Nonendophytic Fungi Reveals Similarities in Host Plant Infection and Colonization Strategies. Mycologia 2020, 112, 491–503. [Google Scholar] [CrossRef]
- Gay, E.J.; Soyer, J.L.; Lapalu, N.; Linglin, J.; Fudal, I.; Da Silva, C.; Wincker, P.; Aury, J.-M.; Cruaud, C.; Levrel, A.; et al. Large-Scale Transcriptomics to Dissect 2 Years of the Life of a Fungal Phytopathogen Interacting with Its Host Plant. BMC Biol. 2021, 19, 55. [Google Scholar] [CrossRef] [PubMed]
- Pathak, G.M.; Gurjar, G.S.; Kadoo, N.Y. Insights of Bipolaris sorokiniana Secretome—An in silico Approach. Biologia 2020, 75, 2367–2381. [Google Scholar] [CrossRef]
- Syme, R.A.; Tan, K.-C.; Rybak, K.; Friesen, T.L.; McDonald, B.A.; Oliver, R.P.; Hane, J.K. Pan-Parastagonospora Comparative Genome Analysis—Effector Prediction and Genome Evolution. Genome Biol. Evol. 2018, 10, 2443–2457. [Google Scholar] [CrossRef] [PubMed]
| Homology | Fungi with Hits | Canonical | No Canonical |
|---|---|---|---|
| Wide phylogenetic distribution | Some examples: Aspergillus species, Fusarium species, Verticillium species, Ramularia species, Bipolaris species, Alternaria species, Friedmanniomyces species, Hortae species, etc. | 8 | 1992 |
| Closely related fungal genera | Cercospora species, Dothistroma species, Zymoseptoria species, and other Mycosphaerellaceae fungi | 175 | 2025 |
| Sigatoka complex | Only P. eumusae and P. musae | 14 | 53 |
| Closely related fungal species | Pseudocercospora eumusae or Pseudocercospora musae | 3 | 56 |
| 6 | 27 |
| Cysteine Percentage (%) | Number of Cysteines (Rank) | Length of the Candidates (Rank) | Number of Proteins |
|---|---|---|---|
| Canonical | |||
| 1.00–2.99 | 4–11 | 136–397 | 134 |
| 3.00–4.99 | 4–17 | 87–390 | 66 |
| 5.00–6.99 | 4–12 | 70–212 | 23 |
| 7.00–8.99 | 6–14 | 82–185 | 14 |
| 9.00–11.11 | 11–24 | 108–254 | 3 |
| Non-Canonical | |||
| 0–1.00 | 0–37 | 9–4644 | 1710 |
| 1.00–2.99 | 1–71 | 51–3161 | 2545 |
| 3.00–4.99 | 2–98 | 52–3193 | 528 |
| 5.00–6.99 | 3–46 | 49–819 | 101 |
| 7.00–8.99 | 4–33 | 55–457 | 35 |
| 9.00–11.58 | 5–30 | 54–297 | 17 |
| 12.00–15.69 | 7–8 | 51–55 | 3 |
| Domains | Effectors |
|---|---|
| Cutinase | 5 |
| CFEM domain | 3 |
| Common central domain of tyrosinase | 3 |
| Glycosyl hydrolases family 61 | 3 |
| CAP | 2 |
| GDSL-like lipase/acylhydrolase | 2 |
| Glycosyl hydrolases family 43 | 2 |
| PAN domain | 2 |
| Peptidase_M43 | 2 |
| Rapid Alkalinization Factor (RALF) | 2 |
| S1/P1 nuclease | 2 |
| Ser-Thr-rich glycosyl-phosphatidyl-inositol-anchored membrane | 2 |
| Concanavalin A-like lectin | 2 |
| Domains | Effectors * |
|---|---|
| Fungal_TF_MHR | 42 |
| NADB_Rossmann superfamily | 42 |
| Mycotoxin biosynthesis protein UstYa-like | 32 |
| PKc | 30 |
| Concanavalin A-like lectin/glucanase | 29 |
| Mito_carr | 26 |
| WD40 | 25 |
| GAL4 | 21 |
| AA_permease_2 superfamily | 19 |
| FabG | 19 |
| Abhydrolase superfamily | 16 |
| Hydrophobic surface binding protein A | 15 |
| Sugar_tr | 15 |
| CzcO | 14 |
| HET | 14 |
| RING_Ubox superfamily | 14 |
| UbiH | 13 |
| BetA | 12 |
| NOX_Duox_like_FAD_NADP | 11 |
| RRM_SF superfamily | 11 |
| ANKYR | 10 |
| FAD binding domain | 10 |
| SLC5-6-like_sbd superfamily | 10 |
| Domain | Canonical | Non-Canonical |
|---|---|---|
| CAP/Cysteine-rich secretory protein family | 2 | 2 |
| CFEM | 3 | 8 |
| Protein of unknown function (DUF3176) | 1 | 7 |
| Protein of unknown function (DUF3455) | 1 | 1 |
| LysM | 1 | 6 |
| Hce2 | 1 | 2 |
| Necrosis-inducing protein (NPP1) | 1 | 1 |
| Chitin-binding | 1 | 2 |
| Isochorismatase family | 1 | 7 |
| Cutinases | 5 | 2 |
| FAD binding domain | 1 | 10 |
| Peptidase_S10 | 1 | 4 |
| Concanavalin A-like lectin | 2 | 29 |
| Cupin | 1 | 14 |
| Scaffold | Canonical * | Non-Canonical ** |
|---|---|---|
| Core scaffolds | ||
| 1 | 43 (17.92%) | 978 (21.59%) |
| 2 | 39 (16.25%) | 666 (14.70%) |
| 3 | 19 (7.92%) | 572 (12.63%) |
| 4 | 22 (9.17%) | 423 (9.33%) |
| 5 | 28 (11.67%) | 318 (7.02%) |
| 6 | 20 (8.33%) | 278 (6.14%) |
| 7 | 17 (7.08%) | 313 (6.91%) |
| 8 | 25 (10.42%) | 315 (6.95%) |
| 9 | 5 (2.08%) | 270 (5.96%) |
| 10 | 10 (4.17%) | 263 (5.80%) |
| 12 | 8 (3.33%) | 89 (1.96%) |
| 19 | 4 (1.67%) | 46 (1.01%) |
| Dispensable scaffolds | ||
| 11 | 0 | 62 (15.17% |
| 13 | 0 | 51 (12.47%) |
| 14 | 0 | 37 (9.05%) |
| 15 | 0 | 42 (10.27%) |
| 16 | 0 | 1 (0.24%) |
| 17 | 0 | 65 (15.89%) |
| 18 | 0 | 49 (11.98%) |
| 20 | 0 | 29 (7.10%) |
| 21 | 0 | 40 (9.78%) |
| 22 | 0 | 30 (7.33%) |
| 44 | 0 | 1 (0.24%) |
| 76 | 0 | 1 (0.24%) |
| 85 | 0 | 1 (0.24%) |
| Function or Domain | Total Members | Set of Candidates | Members Forming Clusters | Clusters | Scaffolds |
|---|---|---|---|---|---|
| DUF | 320 | Canonical, non-canonical | 64 | 8 | 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 12, 19 |
| Fungal_TF_MHR | 42 | Non-canonical | 37 | 5 | 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, |
| Mycotoxin biosynthesis protein UstYa-like | 32 | Non-canonical | 28 | 6 | 1, 2, 3, 4, 5, 7, 9, 10, 12 |
| Concanavalin A-like | |||||
| HsbA | 29 | Non-canonical | 14 | 2 | 1, 2, 3, 4, 5, 6, 7, 8, 9, 10 |
| CFEM | 15 | Non-canonical | 8 | 2 | 1, 2, 3, 4, 5, 6,7, 10, 12 |
| Salicylate hydroxylase | 11 | Canonical, non-canonical | 8 | 2 | 1, 2, 5, 7, 8, 9, 12, 19 |
| Isochorismatase | 7 | Non-canonical | 7 | 1 | 4, 5, 7, 8, 9, 10, 12 |
| LysM | 8 | Canonical, non-canonical | 4 | 1 | 1, 2, 3, 4 |
| Cutinase | 7 | Canonical, non-canonical | 3 | 1 | 2, 4, 5, 6, 7, 8 |
| CAP | 7 | Canonical, non-canonical | 7 | 1 | 1, 2, 4, 6 |
| Hce2 | 4 | Canonical, non-canonical | 4 | 1 | 2, 7 |
| 3 | Canonical, non-canonical | 3 | 1 | 2, 3, 6 |
| Motif | Canonical | Non-Canonical |
|---|---|---|
| [LI]xAR | 7 | 450 |
| [RK]CxC.{12}H | 0 | 2 |
| [RK]VY[LI]R | 0 | 2 |
| [SG]PC[KR]P | 0 | 1 |
| CFEM_2 | 3 | 8 |
| CHxC | 0 | 10 |
| Crinkler | 4 | 109 |
| EAR_1 | 22 | 537 |
| EAR_2 | 0 | 7 |
| G[IFY][ALST]R | 2 | 62 |
| LysM | 16 | 710 |
| PDI | 1 | 192 |
| RXLR | 4 | 1182 |
| ToxA | 0 | 96 |
| YFWxC | 35 | 801 |
| YxSL[RK] | 0 | 11 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carreón-Anguiano, K.G.; Todd, J.N.A.; Santos-Briones, C.D.l.; Peraza-Echeverría, S.; Islas-Flores, I.; Canto-Canché, B. Unveiling the Full Protein Effectorome of the Black Sigatoka Pathogen Pseudocercospora fijiensis—An In Silico Approach. Microbiol. Res. 2024, 15, 1880-1899. https://doi.org/10.3390/microbiolres15030126
Carreón-Anguiano KG, Todd JNA, Santos-Briones CDl, Peraza-Echeverría S, Islas-Flores I, Canto-Canché B. Unveiling the Full Protein Effectorome of the Black Sigatoka Pathogen Pseudocercospora fijiensis—An In Silico Approach. Microbiology Research. 2024; 15(3):1880-1899. https://doi.org/10.3390/microbiolres15030126
Chicago/Turabian StyleCarreón-Anguiano, Karla Gisel, Jewel Nicole Anna Todd, César De los Santos-Briones, Santy Peraza-Echeverría, Ignacio Islas-Flores, and Blondy Canto-Canché. 2024. "Unveiling the Full Protein Effectorome of the Black Sigatoka Pathogen Pseudocercospora fijiensis—An In Silico Approach" Microbiology Research 15, no. 3: 1880-1899. https://doi.org/10.3390/microbiolres15030126
APA StyleCarreón-Anguiano, K. G., Todd, J. N. A., Santos-Briones, C. D. l., Peraza-Echeverría, S., Islas-Flores, I., & Canto-Canché, B. (2024). Unveiling the Full Protein Effectorome of the Black Sigatoka Pathogen Pseudocercospora fijiensis—An In Silico Approach. Microbiology Research, 15(3), 1880-1899. https://doi.org/10.3390/microbiolres15030126






