Altered Cytostructure and Lignolytic Enzymes of Ganoderma boninense in Response to Phenolic Compounds
Abstract
1. Introduction
2. Materials and Methods
2.1. Fungi, Culture Maintenance, and Treatments
2.2. Screening of Phenolic Compounds for the Production of Hydrolytic and Lignolytic Enzymes by G. boninense per 71
2.2.1. Hydrolytic Enzymes
2.2.2. Ligninolytic Enzymes
2.3. Quantification of Hydrolytic and Ligninolytic Enzymes Produced by G. boninense per 71
2.3.1. Cellulase Activity
2.3.2. Amylase Activity
2.3.3. Xylanase Activity
2.3.4. Laccase Activity
2.3.5. Lignin Peroxidase Activity
2.3.6. Manganese Peroxidase Activity
2.4. Scanning Electron Microscopy
2.5. High-Resolution Transmission Electron Microscopy (HR-TEM)
2.6. Statistical Analysis
3. Results
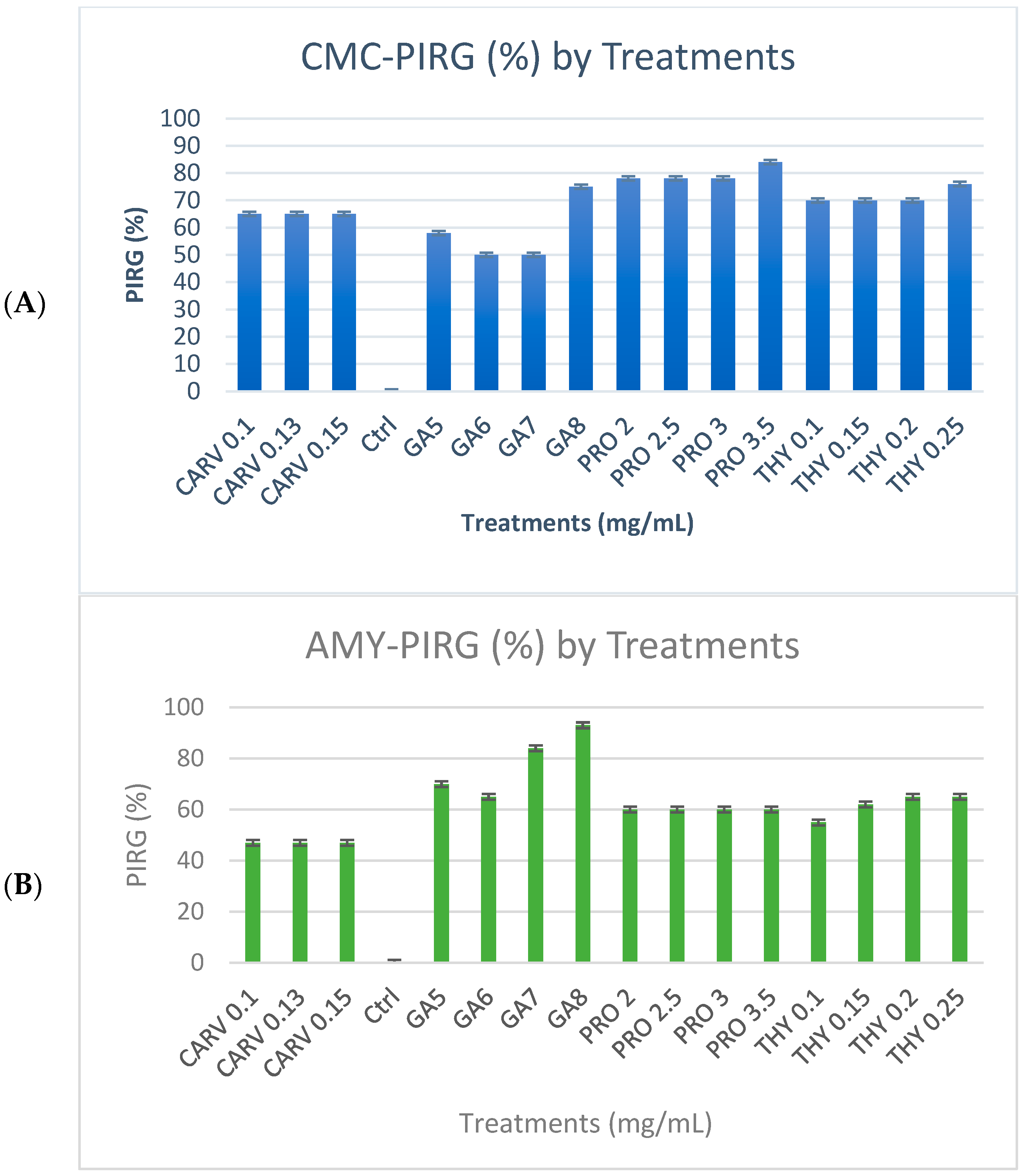
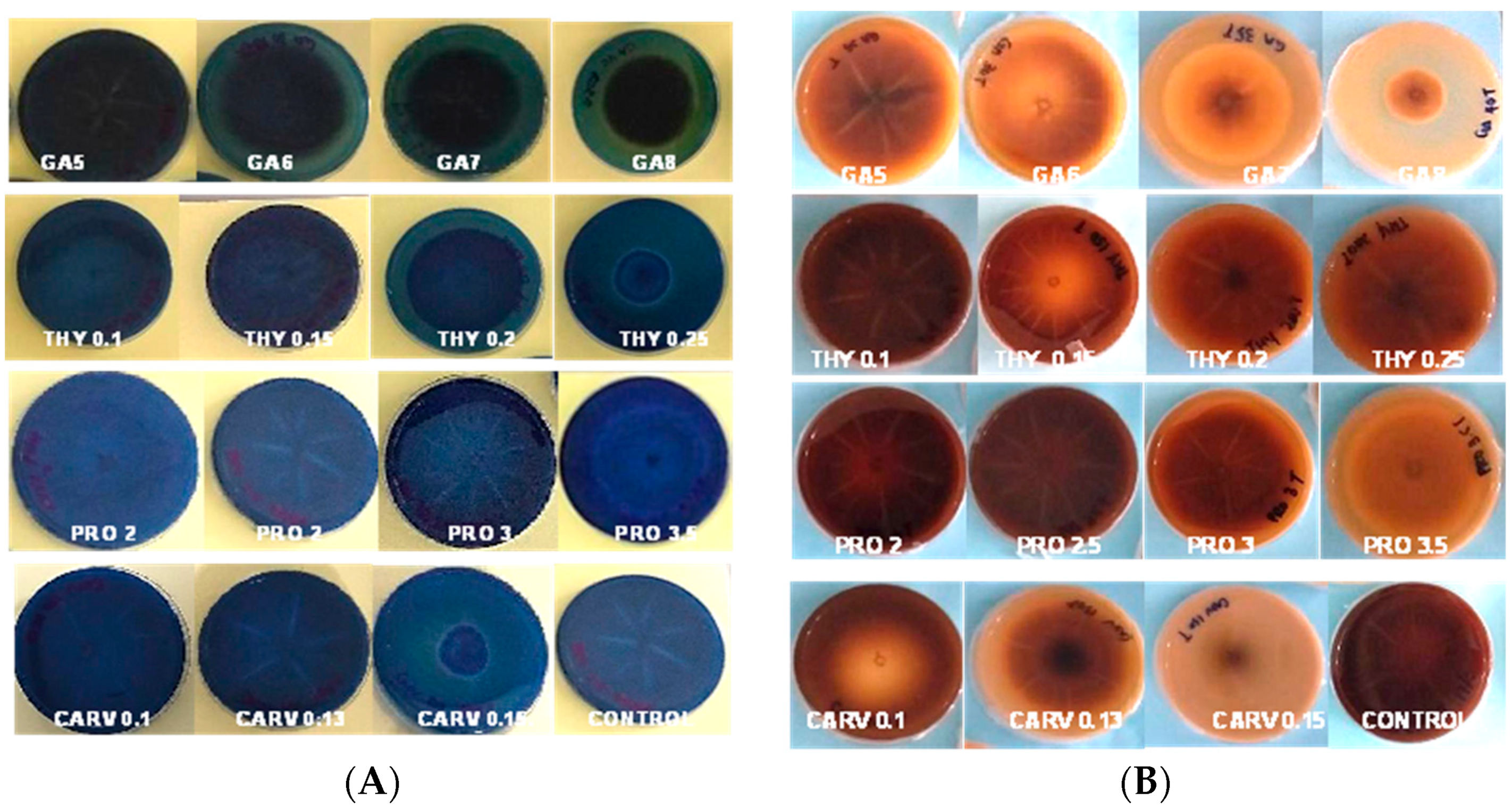
3.1. Qualitative and Quantitative Analyses of Hydrolytic Enzymes
Cellulase, Amylase, and Xylanase Activity
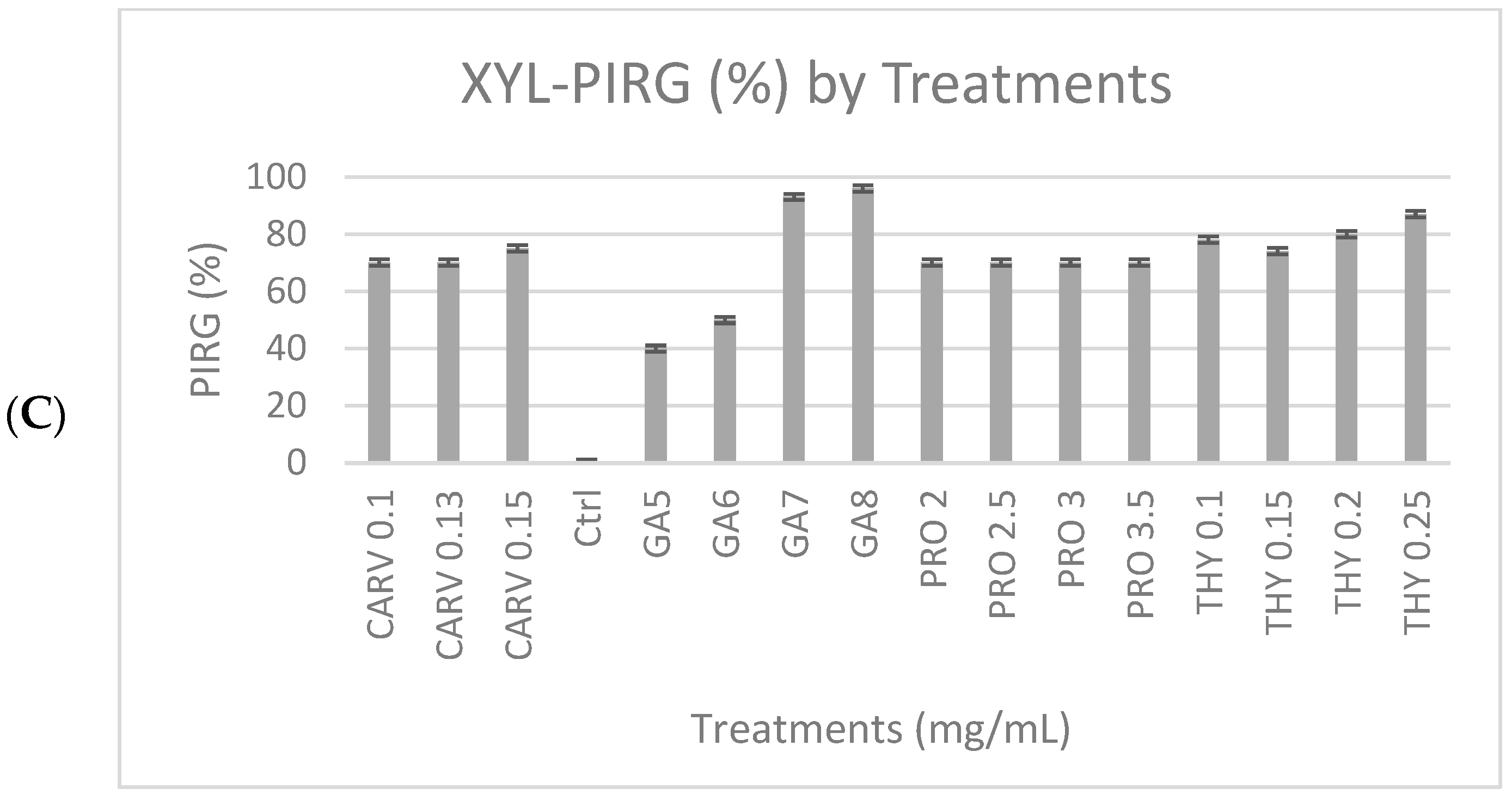
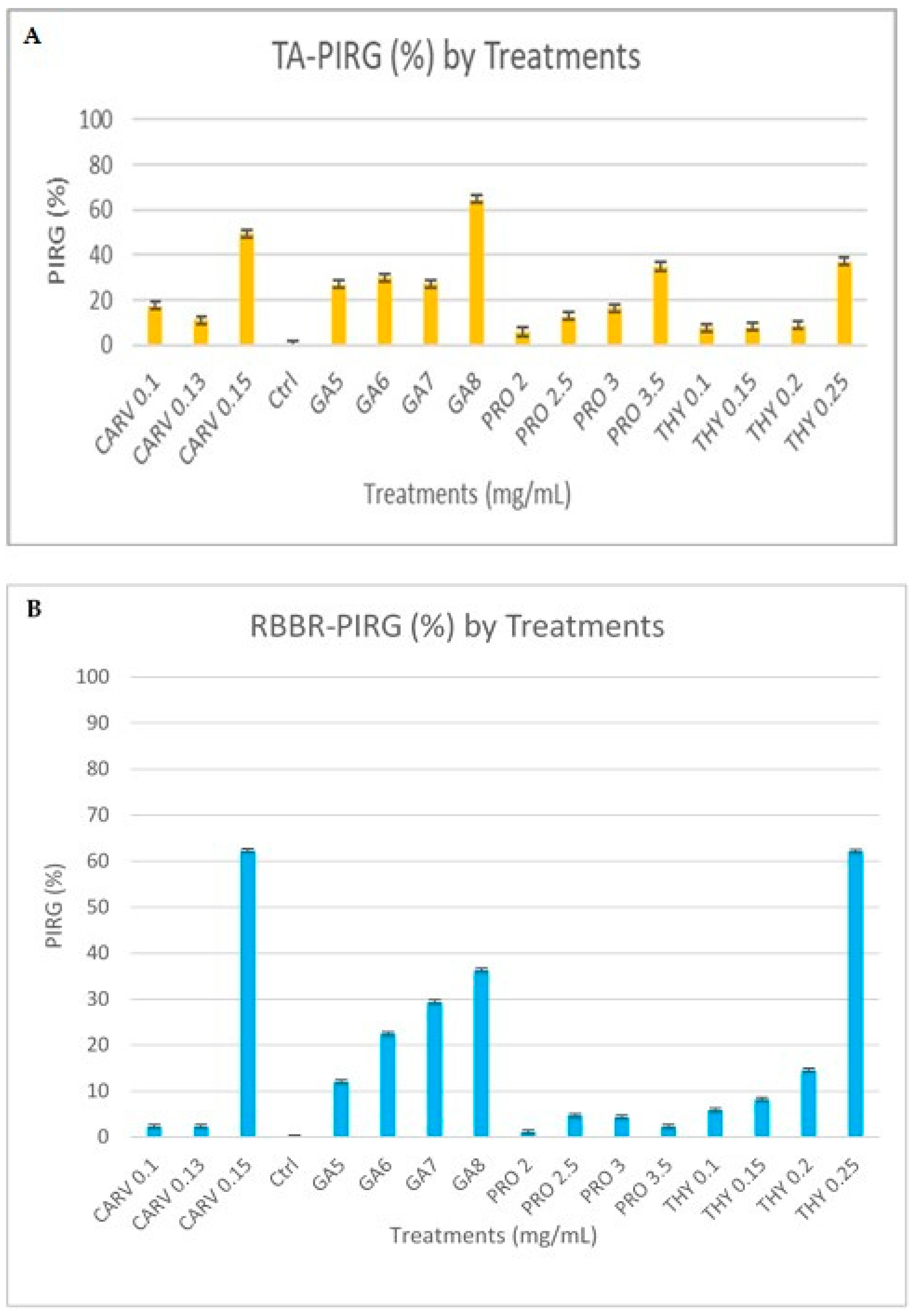
3.2. Qualitative and Quantitative Analyses of Ligninolytic Enzymes
Laccase, Lignin Peroxidase, and Manganese Peroxidase Activity
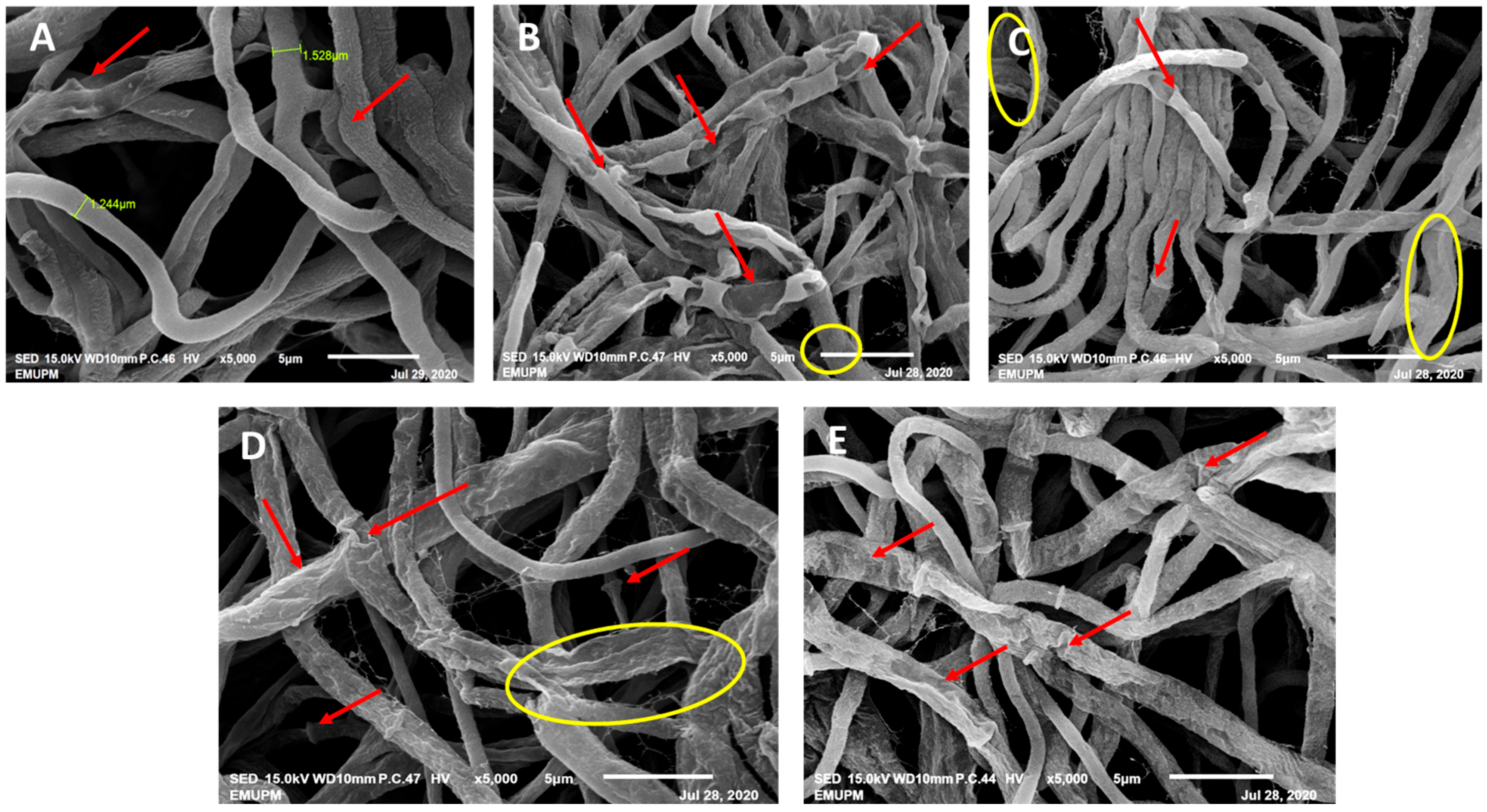
3.3. Scanning Electron Microscopy (SEM)
3.4. High-Resolution Transmission Electron Microscopy (HR-TEM)
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| GA | Gallic acid |
| THY | Thymol |
| PRO | Propolis |
| CARV | Carvacrol |
| BSR | Basal stem rot |
| Lac | Laccase |
| MnP | Manganese peroxidase |
| LiP | Lignin peroxidase |
| SEM | Scanning Electron Microscopy |
| HR-TEM | High-resolution transmission electron microscopy |
| LME | Lignin-modifying enzymes |
| OP | Oil palm |
References
- Surendran, A.; Siddiqui, Y.; Saud, H.M.; Ali, N.S.; Manickam, S. The antagonistic effect of phenolic compounds on ligninolytic and cellulolytic enzymes of Ganoderma boninense, causing basal stem rot in oil palm. Int. J. Agric. Biol. 2017, 19, 1437–1446. [Google Scholar]
- Surendran, A.; Siddiqui, Y.; Manickam, S.; Ali, A. Role of benzoic and salicylic acids in the immunization of oil palm seedlings challenged by Ganoderma boninense. Ind. Crops Prod. 2017, 122, 358–365. [Google Scholar] [CrossRef]
- Surendran, A.; Siddiqui, Y.; Ahmad, K.; Fernanda, R. Deciphering the Physicochemical and Microscopical Changes in Ganoderma boninense-Infected Oil PalmWoodblocks under the Influence of Phenolic Compounds. Plants 2021, 10, 1797. [Google Scholar] [CrossRef] [PubMed]
- Lundell, T.K.; Mäkelä, M.R.; de Vries, R.P.; Hildén, K.S. Chapter Eleven. Genomics, Lifestyles and Future Prospects of Wood-Decay and Litter-Decomposing Basidiomycota. In Advances in Botanical Research; Martin, F.M., Ed.; Academic Press: Cambridge, MA, USA, 2014; Volume 70, pp. 329–370. [Google Scholar]
- Koyani, R.D.; Bhatt, I.M.; Patel, H.R.; Vasava, A.M.; Rajput, K.S. Evaluation of Schizophyllum commune Fr. potential for biodegradation of lignin: A light microscopic analysis. Wood Mater. Sci. Eng. 2016, 11, 46–56. [Google Scholar] [CrossRef]
- Erwin Takemoto, S.; Hwang, W.J.; Takeuchi, M.; Itoh, T.; Imamura, Y. Anatomical characterization of decayed wood in standing light red meranti and identification of the fungi isolated from the decayed area. J. Wood Sci. 2008, 54, 233–241. [Google Scholar] [CrossRef]
- Rees, R.W.; Flood, J.; Hasan, Y.; Potter, U.; Cooper, R.M. Basal stem rot of oil palm (Elaeis guineensis); Mode of root infection and lower stem invasion by Ganoderma boninense. Plant Pathol. 2009, 58, 982–989. [Google Scholar] [CrossRef]
- Surendran, A.; Siddiqui, Y.; Saud, H.M.; Ali, N.S.; Manickam, S. Inhibition and kinetic studies of lignin-degrading enzymes of Ganoderma boninense by naturally occurring phenolic compounds. J. Appl. Microbiol. 2018, 125, 876–887. [Google Scholar] [CrossRef]
- Bucher, V.V.C.; Hyde, K.D.; Pointing, S.B.; Reddy, C.A. Production of wood decay enzymes, mass loss and lignin solubilization in wood by marine ascomycetes and their anamorphs. Fungal Divers. 2004, 15, 14. [Google Scholar]
- Ganapathy, D.; Siddiqui, Y.; Ahmad, K.; Adzmi, F.; Ling, K.L. Alterations in Mycelial Morphology and Flow Cytometry Assessment of Membrane Integrity of Ganoderma boninense Stressed by Phenolic Compounds. Biology 2021, 10, 930. [Google Scholar] [CrossRef]
- Witzell, J.; Martín, J.A. Phenolic metabolites in the resistance of northern forest trees to pathogens—Past experiences and future prospects. Can. J. For. Res. 2008, 38, 2711–2727. [Google Scholar] [CrossRef]
- Paterson, R.R.; Meon, S.; Abidin, M.Z.; Lima, N. Prospects for inhibition of lignin degrading enzymes to control Ganoderma white rot of oil palm. Curr. Enzym. Inhib. 2008, 4, 172–179. [Google Scholar] [CrossRef]
- Machado, K.M.; Matheus, D.R.; Bononi, V.L. Ligninolytic enzymes production and Remazol Brilliant Blue R decolorization by tropical Brazilian basidiomycetes fungi. Braz. J. Microbiol. 2005, 36, 246–252. [Google Scholar] [CrossRef]
- Srivastava, N.; Rawat, R.; Singh Oberoi, H.; Ramteke, P.W. A review on fuel ethanol production from lignocellulosic biomass. Int. J. Green Energy 2015, 2, 949–960. [Google Scholar] [CrossRef]
- Miller, G.L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Naidu, Y.; Siddiqui, Y.; Rafii, M.Y.; Saud, H.M.; Idris, A.S. Investigating the effect of white-rot hymenomycetes biodegradation on basal stem rot-infected oil palm wood blocks: Biochemical and anatomical characterisation. Ind. Crops Prod. 2017, 108, 872–882. [Google Scholar] [CrossRef]
- Calderon-Garciduenas, L.; González-Maciel, A.; Reynoso-Robles, R.; Rodríguez-López, J.L.; Silva-Pereyra, H.G.; Labrada-Delgado, G.J.; Pérez-Guillé, B.; Soriano-Rosales, R.E.; Jimenez-Bravo Luna, M.A.; Brito-Aguilar, R.; et al. Environmental Fe, Ti, Al, Cu, Hg, Bi, and Si nanoparticles in the atrioventricular conduction axis and the associated ultrastructural damage in young urbanites: Cardiac arrhythmias caused by anthropogenic, industrial, E-waste, and indoor nanoparticles. Environ. Sci. Technol. 2021, 55, 8203–8214. [Google Scholar] [CrossRef] [PubMed]
- Sansone, G.; Rezza, I.; Fernández, G.; Calvente, V.; Benuzzi, D.; Sanz, M.I. Inhibitors of polygalacturonase and laccase of Botrytis cinerea and their application to the control of this fungus. Int. Biodeterior. Biodegrad. 2011, 65, 243–247. [Google Scholar] [CrossRef]
- Ahn, J.; Grün, I.U.; Mustapha, A. Effects of plant extracts on microbial growth, color change, and lipid oxidation in cooked beef. Food Microbiol. 2007, 24, 7–14. [Google Scholar] [CrossRef]
- Muniroh, M.; Nusaibah, S.; Vadamalai, G.; Siddique, Y. Proficiency of biocontrol agents as plant growth promoters and hydrolytic enzyme producers in Ganoderma boninense infected oil palm seedlings. Curr. Plant Biol. 2019, 20, 100116. [Google Scholar] [CrossRef]
- Assis, K.; Chong, K.P.; Idris, A.S.; Ho, C.M. Economic loss due to Ganoderma disease in oil palm. In Proceedings of the Indonesia Conference on Mathematics, Statistics and Scientific Computing, Kuala Lumpur, Malaysia, 21–22 December 2016; Volume 11, p. 12. [Google Scholar]
- H’ng, P.S.; Wong, L.J.; Chin, K.L.; Tor, E.S.; Tan, S.E.; Tey, B.T.; Maminski, M. Oil palm (Elaeis guineensis) trunk as a resource of starch and other sugars. J. Appl. Sci. 2011, 11, 3053–3057. [Google Scholar] [CrossRef]
- Surendran, A. Effect of Naturally Occuring Phenolic Compound on Cell Wall Degrading Enzymes and Suppression of Ganoderma Boninense Infection in Oil Palm Seedlings. Ph.D. Thesis, University of Putra Malaysia, Kembangan, Malaysia, 2018. [Google Scholar]
- Wang, L.; Fan, D.; Chen, W.; Terentjev, E. Bacterial growth, detachment and cell size control on polyethylene terephthalate surfaces. Sci. Rep. 2015, 5, 15159. [Google Scholar] [CrossRef] [PubMed]
- Camarero, S.; Sarkar, S.; Ruiz-Dueñas, F.J.; Martínez, M.J.; Martínez, A.T. Description of a versatile peroxidase involved in the natural degradation of lignin that has both manganese peroxidase and lignin peroxidase substrate interaction sites. J. Biol. Chem. 1999, 274, 10324–10330. [Google Scholar] [CrossRef] [PubMed]
- Kresnawaty, I.; Eris, D.; Mulyatni, A.; Prakoso, H. Inhibitory effect of phenolic acid on Ganoderma boninense enzyme as an approach on Ganoderma infection. IOP Conf. Ser. Earth Environ. Sci. 2018, 183, 012023. [Google Scholar] [CrossRef]
- Zhang, J.; Ma, S.; Du, S.; Chen, S.; Sun, H. Antifungal activity of thymol and carvacrol against postharvest pathogens Botrytis cinerea. J. Food Sci. Technol. 2019, 56, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, L.; Lv, Q.; Yan, L.; Wang, Y.; Jiang, Y. The fungal CYP51s: Their functions, structures, related drug resistance, and inhibitors. Front. Microbiol. 2019, 10, 691. [Google Scholar] [CrossRef] [PubMed]
- Sahebi, M.; Hanafi, M.M.; Mohidin, H.; Rafii, M.Y.; Azizi, P.; Idris, A.S.; Moradpoor, M. Antioxidant enzyme activities and secondary metabolite profiling of oil palm seedlings treated with combination of NPK fertilizers infected with Ganoderma boninense. BioMed Res. Int. 2018, 2018, 1494157. [Google Scholar] [CrossRef]
- Choudhary, D.K.; Prakash, A.; Johri, B.N. Induced systemic resistance (ISR) in plants: Mechanism of action. Indian J. Microbiol. 2007, 47, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Bivi, M.S.; Paiko, A.S.; Khairulmazmi, A.; Akhtar, M.S.; Idris, A.S. Control of Basal Stem Rot Disease in Oil Palm by Supplementation of Calcium, Copper, and Salicylic Acid. J. Plant Pathol. 2016, 32, 396. [Google Scholar] [CrossRef]
- Paterson, R.R.M. Ganoderma disease of oil palm—A white rot perspective necessary for integrated control. Crop Prot. 2007, 26, 1369–1376. [Google Scholar] [CrossRef]
- An, P.; Yang, X.; Yu, J.; Qi, J.; Ren, X.; Kong, Q. A-terpineol and terpene-4-ol, the critical components of tea tree, exert antifungal activites in vivo against Aspergillus niger in grapes by inducing morphous damage and metabolic changes of fungus. Food Control 2018, 98, 42–53. [Google Scholar] [CrossRef]
- Fernanda, R.; Siddiqui, Y.; Ganapathy, D.; Ahmad, K.; Surendran, A. Suppression of Ganoderma boninense Using Benzoic Acid: Impact on Cellular Ultrastructure and Anatomical Changes in Oil PalmWood. Forest 2021, 12, 1231. [Google Scholar] [CrossRef]
- Xu, Y.; Luo, L.; Chen, B.; Fu, Y. Recent development of chemical components in propolis. Front. Biol. China 2009, 4, 385–391. [Google Scholar] [CrossRef]
- Liu, J.; Zong, Y.; Qin, G.; Li, B.; Tian, S. Plasma membrane damage contributes to antifungal activity of silicon against penicillium digitatum. Curr. Microbiol. 2010, 61, 274–279. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.Q.; Rao, R. Beyond ergosterol: Linking pH to antifungal mechanisms. Virulence 2010, 1, 551–554. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Liu, M.; Dawuti, G.; Dou, Q.; Ma, Y.; Liu, H.; Aibai, S. Antifungal Activity of Gallic Acid In Vitro and In Vivo. Phytother. Res. 2007, 31, 1039–1045. [Google Scholar] [CrossRef]
- Pag, U.; Oedenkoven, M.; Sass, V.P.; Shai, Y.; Shamova, O.; Antcheva, N.; Tossi, A.; Sahl, H.-G. Analysis of in vitro activities and modes of action of synthetic antimicrobial peptides derived from a_- helical ‘sequence template’. J. Antimicrob. Chemother. 2008, 61, 341–352. [Google Scholar] [CrossRef]


| Cellulase | Enzyme Activity (U/mL) Amylase | Xylanase | |
|---|---|---|---|
| Control | 1.4349 ± 0.68 a | 1.371 ± 1.3 a | 1.1147 ± 1.25 ab |
| GA-5 | 1.0287 ± 1.25 b | 0.5036 ± 1.2 b | 1.2503 ± 1.4 a |
| GA-6 | 0.5266 ± 1.25 d | 0.4998 ± 1.2 c | 1.032 ± 1.4 ab |
| GA-7 | 0.1966 ± 1.25 e | 0.3389 ± 1.2 d | 0.746 ± 1.4 b |
| GA-8 | 0.3387 ± 1.25 f | 0.3314 ± 1.3 d | 0.2113 ± 1.4 b |
| THY-0.1 | 0.8963 ± 1.3 c | 0.2980 ± 1.7 d | 0.925 ± 1.8 b |
| THY-0.15 | 0.8542 ± 1.3 c | 0.0047 ± 1.7 e | 0.283 ± 1.8 c |
| THY-0.2 | 1.0079 ± 1.3 b | 0.2872 ± 1.7 c | 0.718 ± 1.8 b |
| THY-0.25 | 0.2675 ± 1.3 e | 0.3710 ± 1.7 c | 0.736 ± 1.8 b |
| PRO-2 | 0.7163 ± 1.4 c | 1.1399 ± 1.8 a | 0.933 ± 1.5 b |
| PRO- 2.5 | 0.7203 ± 1.4 c | 0.8949 ± 1.8 a | 1.075 ± 1.5 a |
| PRO-3 | 0.3063 ± 1.4 e | 0.4197 ± 1.8 c | 1.233 ± 1.5 a |
| PRO-3.5 | 0.3025 ± 1.4 e | 0.7082 ± 1.8 b | 0.038 ± 1.5 c |
| CARV-0.1 | 1.7525 ± 1.45 a | 0.6222 ± 1.8 b | 1.035 ± 1.7 b |
| CARV-0.13 | 1.2034 ± 1.45 a | 0.8711 ± 1.8 a | 0.993 ± 1.7 b |
| CARV-0.15 | 1.2356 ± 1.45 a | 0.9519 ± 1.8 a | 1.057 ± 1.7 a |
| Enzyme Activity (U/mL) | |||
|---|---|---|---|
| Laccase | Lignin Peroxidase | Manganese Peroxidase | |
| Control | 0.9865 ± 1.85 a | 1.883 ± 1.5 a | 0.846 ± 1.7 a |
| GA-5 | 0.0844 ± 1.3 c | 1.613 ± 1.8 a | 0.916 ± 1.8 a |
| GA-6 | 0.7255 ± 1.3 a | 1.844 ± 1.8 a | 0.807 ± 1.8 a |
| GA-7 | 0.6754 ± 1.3 b | 1.655 ± 1.8 a | 0.564 ± 1.8 b |
| GA-8 | 0.4885 ± 1.3 b | 0.218 ± 1.8 c | 0.386 ± 1.8 b |
| THY-0.1 | 0.1321 ± 1.8 c | 2.080 ± 1.5 a | 0.037 ± 1.3 d |
| THY-0.15 | 0.0878 ± 1.8 c | 1.700 ± 1.5 b | 0.056 ± 1.3 d |
| THY-0.2 | 0.0776 ± 1.8 c | 1.563 ± 1.5 b | 0.101 ± 1.3 c |
| THY-0.25 | 0.1184 ± 1.8 c | 2.023 ± 1.5 a | 0.831 ± 1.3 a |
| PRO-2 | 0.1424 ± 1.25 a | 1.660 ± 1.83 b | 0.132 ± 1.5 c |
| PRO-2.5 | 0.0978 ± 1.25 d | 1.607 ± 1.83 b | 0.163 ± 1.5 c |
| PRO-3 | 0.0665 ± 1.25 d | 2.040 ± 1.83 a | 0.048 ± 1.5 d |
| PRO-3.5 | 0.0644 ± 1.25 d | 2.226 ± 1.83 a | 0.275 ± 1.5 c |
| CARV-0.1 | 0.0543 ± 1.4 d | 1.492 ± 1.9 b | 0.005 ± 1.5 e |
| CARV-0.13 | 0.1834 ± 1.4 c | 1.443 ± 1.9 b | 0.039 ± 1.5 d |
| CARV-0.15 | 0.1246 ± 1.4 c | 1.743 ± 1.9 b | 0.056 ± 1.5 d |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Siddiqui, Y.; Ganapathy, D. Altered Cytostructure and Lignolytic Enzymes of Ganoderma boninense in Response to Phenolic Compounds. Microbiol. Res. 2024, 15, 550-566. https://doi.org/10.3390/microbiolres15020036
Siddiqui Y, Ganapathy D. Altered Cytostructure and Lignolytic Enzymes of Ganoderma boninense in Response to Phenolic Compounds. Microbiology Research. 2024; 15(2):550-566. https://doi.org/10.3390/microbiolres15020036
Chicago/Turabian StyleSiddiqui, Yasmeen, and Daarshini Ganapathy. 2024. "Altered Cytostructure and Lignolytic Enzymes of Ganoderma boninense in Response to Phenolic Compounds" Microbiology Research 15, no. 2: 550-566. https://doi.org/10.3390/microbiolres15020036
APA StyleSiddiqui, Y., & Ganapathy, D. (2024). Altered Cytostructure and Lignolytic Enzymes of Ganoderma boninense in Response to Phenolic Compounds. Microbiology Research, 15(2), 550-566. https://doi.org/10.3390/microbiolres15020036







