The Triterpenoid High-Performance Liquid Chromatography Analytical Profiles of the Mycelia of Ganoderma lucidum (lingzhi)
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Apparatus
2.2. Cultivation of Ganoderma Mycelia
2.3. HPLC Analysis of the Triterpenes 1–19
2.4. Isolation of Compounds 1–5 from YK-01 Mycelium
2.5. Determining Ganoderic Acid T (10) and S (12)
2.5.1. Sample Pretreatment
2.5.2. Calibration Curve
2.5.3. Recovery Tests
2.5.4. Limit of Detection and Limit of Quantitation
2.5.5. Accuracy and Precision
3. Results
3.1. Structural Identification of the New Triterpenes 1~5
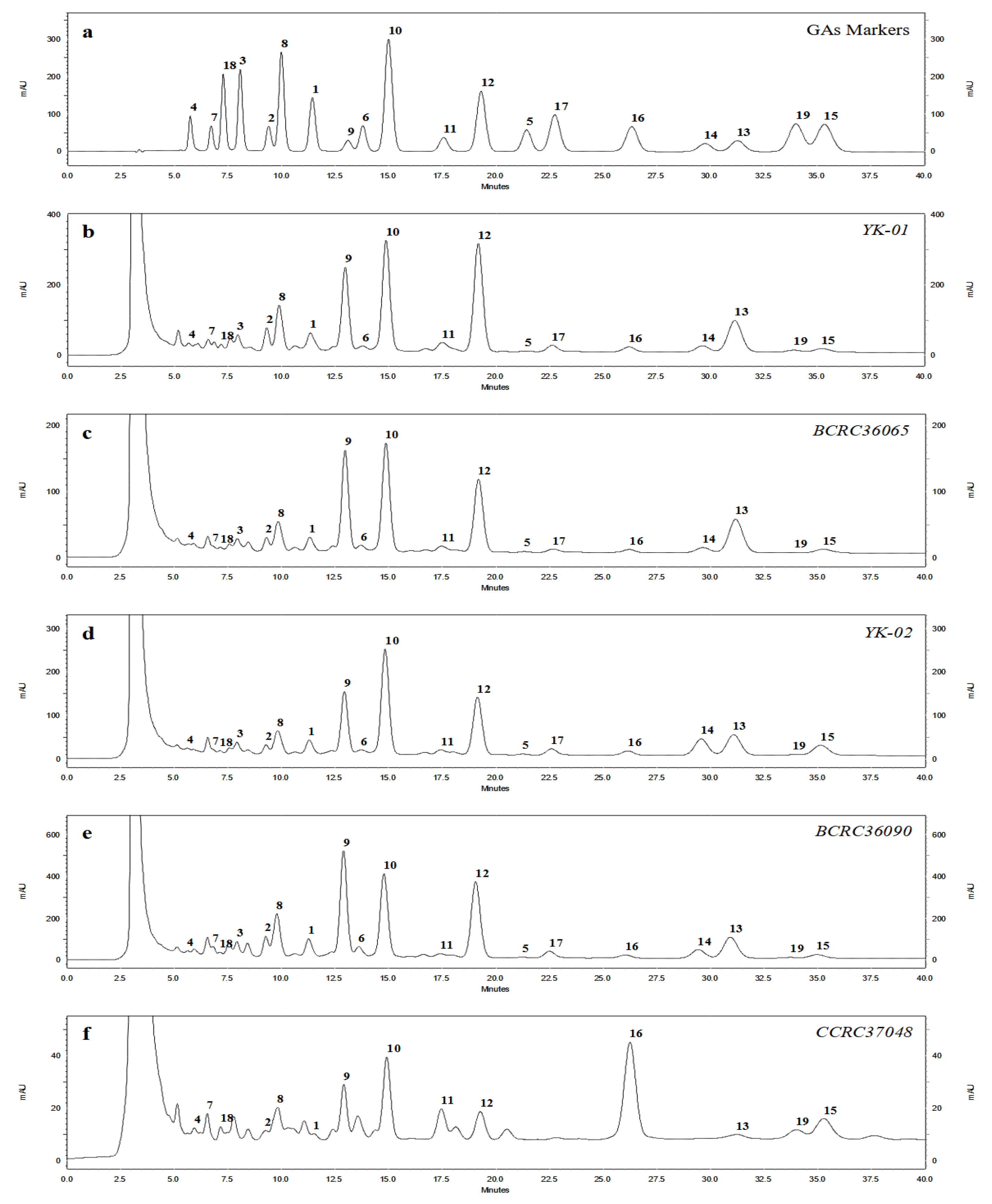
3.2. HPLC Fingerprint Profiles of Triterpenes from Ganoderma Mycelia
3.3. Methods for the Validation and Quantitation of Ganoderic Acid T (10) and S (12)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cör, D.; Knez, Ž.; Hrnčič, M.K. Antitumour, antimicrobial, antioxidant and antiacetylcholinesterase effect of Ganoderma lucidum terpenoids and polysaccharides: A review. Molecules 2018, 23, 649–669. [Google Scholar] [CrossRef]
- Chen, D.H.; Ju, H.Y.; Sheu, K.C. Simple Fourier transform (FT)-IR and reverse-phase HPLC identification methods of commercial Ganoderma products. J. Chin. Chem. Soc. 2001, 48, 1207–1210. [Google Scholar] [CrossRef]
- Xia, Q.; Zhang, H.Z.; Sun, X.F.; Zhao, H.J.; Wu, L.F.; Zhu, D.; Yang, G.H.; Xin, Y.N.; Mao, L.Z.; Zhang, G.M. A comprehensive review of the structure elucidation and biological activity of triterpenoids from Ganoderma spp. Molecules 2014, 19, 17478–17535. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.H.; Chen, K.D. Determination of ganoderic acids in triterpenoid constituents of Ganoderma tsugae. J. Food Drug Anal. 2003, 11, 195–200. [Google Scholar] [CrossRef]
- Chen, D.H.; Wang, J.Y.; Chen, M.T.; Chen, K.D. HPLC fingerprint profiles of lucidenic acids from Ganoderma lucidum (lingzhi). J. Chin. Chem. Soc. 2022, 69, 950–959. [Google Scholar] [CrossRef]
- You, B.J.; Lee, H.Z.; Chung, K.R.; Lee, M.H.; Huang, M.J.; Tien, N.; Chgen, C.W.; Kuo, Y.H. Enhanced production of ganoderic acids and cytotoxicity of Ganoderma lucidum using solid-medium culture. Biosci. Biotechnol. Biochem. 2012, 76, 1529–1534. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.H.; Liu, J.W.; Zhong, J.J. Ganoderic acid T inhibits tumor invasion in vitro and in vivo through inhibition of MMP expression. Pharmacol. Rep. 2010, 62, 150–163. [Google Scholar] [CrossRef] [PubMed]
- Lai, H.H.; Huang, C.P.; Chen, D.H.; Chen, K.D.; Lin, S.B. Ganoderma triterpenoid ganoderic acid T inhibits growth and metastasis of A549 lung adenocarcinoma in vitro and in vivo. J. Chin. Oncl. Soc. 2009, 25, 413–420. [Google Scholar] [CrossRef]
- Chyr, R.; Shiao, M.S. Liquid chromatographic characterization of the triterpenoid patterns in Ganoderma lucidum and related species. J. Chromatogr. 1991, 542, 327–336. [Google Scholar] [CrossRef]
- Hirotani, M.; Asaka, I.; Ino, C.; Furuya, T.; Shiro, M. Ganoderic acid derivatives and ergosta-4,7,22-triene-3,6-dione from Ganoderma lucidum. Phytochemistry 1987, 26, 2797–2803. [Google Scholar] [CrossRef]
- Lin, L.J.; Shiao, M.S. Seven new triterpenes from Ganoderma lucidum. J. Nat. Prod. 1988, 51, 918–924. [Google Scholar] [CrossRef] [PubMed]
- Shiao, M.S.; Lin, L.J.; Yeh, S.F. Triterpenes in Ganoderma lucidum. Phytochemistry 1988, 27, 873–875. [Google Scholar] [CrossRef]
- Zhang, X.Q.; Gao, X.X.; Yang, Y.C.; Chen, G.; Hou, G.L.; Huo, X.T.; Jia, X.M.; Wang, A.H.; Hu, G.S. Lanostane-type triterpenoids from the mycelial mat of Ganoderma lucidum and their hepatoprotective activities. Phytochemistry 2022, 198, 113131. [Google Scholar] [CrossRef] [PubMed]
- Nishitoba, T.; Sato, H.; Oda, K.; Sakamura, S. Novel triterpenoids and a steroid from the fungus Ganoderma luicidum. Agric. Biol. Chem. 1988, 52, 211–216. [Google Scholar] [CrossRef]
- Hu, G.S.; Zhai, M.H.; Niu, R.; Xu, X.Q.; Liu, Q.; Jia, J.M. Optimization of culture condition for ganoderic acid production in Ganoderma lucidum liquid static culture and design of a suitable bioreactor. Molecules 2018, 23, 2563–2574. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.W.; Zhao, W.; Zhong, J.J. Biotechnological production and application of ganoderic acids. Appl. Microbiol. Biotechnol. 2010, 87, 457–466. [Google Scholar] [CrossRef] [PubMed]
- Paterson, R.R.M.; Lima, N. Failed PCR of Ganoderma type specimens affects nomenclature. Phytochemistry 2015, 114, 16–17. [Google Scholar] [CrossRef] [PubMed][Green Version]
- USP NSP39-NF34; United States Pharmacopeial Convention Inc.: North Bethesda, MA, USA, 2016; pp. 6641–6647.
- Galappaththi, M.C.A.; Patabendige, N.M.; Premarathne, B.M.; Hapuarachchi, K.K.; Tibpromma, S.; Dai, D.Q.; Suwannarach, N.; Rapior, S.; Karunarathna, S.C. A review of Ganoderma triterpenoids and their bioactivities. Biomolecules 2023, 13, 24. [Google Scholar] [CrossRef]
- Nishitoba, T.; Sato, H.; Oda, K.; Sakamura, S. Novel mycelial components, ganoderic acid Mg, Mh, Mi, Mj and Mk, from the fungus Ganoderma lucidum. Agric. Biol. Chem. 1987, 51, 1149–1153. [Google Scholar] [CrossRef]

| 1 | 2 | 3 | 4 | 5 | |
|---|---|---|---|---|---|
| No. | |||||
| 1 | 30.6 (t) | 37.1 (t) | 31.2 (t) | 37.1 (t) | 30.8 (t) |
| 2 | 23.1 (t) | 28.3 (t) | 26.7 (t) | 28.5 (t) | 27.6 (t) |
| 3 | 78.1 (d) | 79.6 (d) | 76.8 (d) | 79.6 (d) | 217.9 (s) |
| 4 | 36.5 (s) | 38.6 (s) | 38.4 (s) | 38.6 (s) | 36.9 (s) |
| 5 | 44.0 (d) | 50.7 (d) | 44.3 (d) | 50.5 (d) | 51.2 (d) |
| 6 | 22.8 (t) | 24.1 (t) | 24.0 (t) | 24.0 (t) | 26.3 (t) |
| 7 | 121.2 (d) | 121.6 (d) | 122.6 (d) | 122.6 (d) | 19.4 (t) |
| 8 | 140.1 (s) | 143.9 (s) | 142.4 (s) | 141.7 (s) | 133.3 (s) |
| 9 | 145.9 (s) | 147.5 (s) | 147.9 (s) | 147.4 (s) | 135.1 (s) |
| 10 | 37.3 (s) | 39.8 (s) | 38.5 (s) | 39.8 (s) | 44.5 (s) |
| 11 | 115.6 (d) | 117.4 (d) | 116.7 (d) | 117.4 (d) | 21.0 (t) |
| 12 | 38.0 (t) | 39.1 (t) | 39.8 (t) | 39.3 (t) | 31.0 (t) |
| 13 | 43.9 (s) | 44.8 (s) | 45.4 (s) | 45.1 (s) | 47.4 (s) |
| 14 | 51.4 (s) | 51.5 (s) | 53.4 (s) | 52.6 (s) | 49.9 (s) |
| 15 | 77.3 (d) | 32.6 (t) | 75.2 (d) | 78.8 (d) | 36.0 (t) |
| 16 | 36.4 (t) | 28.5 (t) | 39.9 (t) | 37.4 (t) | 35.2 (t) |
| 17 | 45.2 (d) | 48.6 (d) | 46.4 (d) | 46.5 (d) | 46.8 (d) |
| 18 | 15.9 (q) | 16.2 (q) | 16.5 (q) | 16.4 (q) | 15.8 (q) |
| 19 | 22.6 (q) | 23.3 (q) | 23.3 (q) | 23.3 (q) | 24.2 (q) |
| 20 | 40.8 (d) | 42.4 (d) | 42.3 (d) | 42.1 (d) | 41.4 (d) |
| 21 | 11.5 (q) | 12.1 (q) | 12.2 (q) | 12.1 (q) | 11.7 (q) |
| 22 | 72.5 (d) | 73.4 (d) | 73.4 (d) | 73.2 (d) | 72.8 (d) |
| 23 | 35.2 (t) | 35.8 (t) | 35.8 (t) | 35.7 (t) | 34.6 (t) |
| 24 | 141.0 (d) | 141.0 (d) | 141.0 (d) | 140.8 (d) | 151.3 (d) |
| 25 | 128.9 (s) | 130.1 (s) | 130.1 (s) | 130.2 (s) | 140.6 (s) |
| 26 | 171.3 (s) | 171.7 (s) | 171.5 (s) | 173.1 (s) | 195.2 (d) |
| 27 | 12.4 (q) | 12.8 (q) | 12.7 (q) | 12.8 (q) | 9.5 (q) |
| 28 | 18.5 (q) | 26.2 (q) | 18.0 (q) | 18.9 (q) | 18.7 (q) |
| 29 | 27.8 (q) | 28.8 (q) | 28.9 (q) | 28.8 (q) | 26.2 (q) |
| 30 | 22.4 (q) | 16.5 (q) | 23.4 (q) | 16.5 (q) | 21.3 (q) |
| OAc | 170.8 (s) | 171.6 (s) | |||
| OAc | 170.8 (s) | ||||
| OCOCH3 | 21.3 (q) | 21.2 (q) | |||
| OCOCH3 | 21.4 (q) |
| 1 | 2 | 3 | 4 | 5 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| δH | HMBC (H to C) | δH | HMBC (H to C) | δH | HMBC (H to C) | δH | HMBC (H to C) | δH | HMBC (H to C) | |
| 1 | 1.64 (a, br.t, J = 14.0 Hz) | 10,19 | 1.42 (a, m) | 4 | 1.69 (a, dt, J = 13.3, 3.5 Hz) | 1.43 (a, ddd, J = 14.7, 10.5, 4.2 Hz) | 1.26 (a, t, J = 7.5 Hz) | |||
| 1.75 (b, dt, J = 13.3, 3.5 Hz) | 10,19 | 2.04 (b, m) | 3,4,5 | 1.81 (b, br.d. J = 10.5, 3.5 Hz) | 2.04 (b, ddd, J = 13.5, 7.7, 3.5 Hz) | 5 | 1.61 (b, m) | 19 | ||
| 2 | 1.73 (m) | 1.35 (m) | 2.03 (m) | 1.68 (m) | 3 | 1.38 (a, dd, J = 7.5, 5.5, 2.0 Hz) | ||||
| 1.95 (br.t, J = 12.6, 14.2 Hz) | 2.09 (m) | 1.98 (b, m) | ||||||||
| 3 | 4.69 (s) | 1,2,4,5 | 3.16 (dd, J = 11.2, 4.2 Hz) | 3.38 (s) | 1,5 | 3.16 (dd, J = 11.2, 4.9 Hz) | 29,30 | |||
| 5 | 1.50 (dd, J = 11.9, 3.56 Hz) | 4,6,10 | 1.10 (dd, J = 11.2, 4.2 Hz) | 6 | 1.57 (dd, J = 11.2, 4.9 Hz) | 9,10 | 1.08 (dd, J = 11.9, 4.2 Hz) | 2,4,6,10,29 | 1.60 (m) | |
| 6 | 2.03 (m) | 7,8,10 | 2.10 (m) | 5 | 2.08 (m) | 2.10 (m) | 2.08 (m) | |||
| 7 | 5.50 (d, J = 4.9 Hz) | 5,6,9,14 | 5.51 (br.d, J = 5.6 Hz) | 6,9 | 5.91 (d, J = 5.6 Hz) | 8 | 5.52 (d, J = 7.0 Hz) | 5,6,9,14 | 1.64 (m) | |
| 11 | 5.34 (d, J = 5.6 Hz) | 8,12,13 | 5.37 (br.d, J = 6.3 Hz) | 8,12,13 | 5.37 (d, J = 6.3 Hz) | 9,10,13 | 5.40 (d, J = 6.3 Hz) | 8,12,13 | 2.05 (m) | |
| 12 | 2.05 (a, m) | 8,9,11,13,18 | 2.13 (a,m) | 11,13,14 | 2.09 (a, m) | 9,11,14 | 2.13 (a,m) | 9,11,13,14,18 | 1.70 (a, m) | |
| 2.38 (b, d, J = 17.5 Hz) | 8,9,11,13,18 | 2.27 (b, br.d, J = 17.5 Hz) | 11,13,18 | 2.35 (b, d, J = 17.5 Hz) | 9,11,13,18 | 2.38 (b, d, J = 17.5 Hz) | 9,11,13,18 | 1.80 (b, m) | ||
| 15 | 5.08 (dd, J = 9.1, 5.6 Hz) | 28 | 1.44 (b,m) | 13 | 4.24 (dd, J = 9.1, 5.6 Hz) | 5.07 (dd, J = 9.8, 5.6 Hz) | 28 | 1.63 (a, m) | ||
| 1.67 (a, m) | 28 | 1.98 (b, m) | ||||||||
| 16 | 1.79 (b, ddd, J = 9.8, 5.6, 4.2 Hz) | 13,15 | 1.68 (m) | 1.88 (m) | 1.82 (b, ddd, J = 9.8, 8.4, 4.2 Hz) | 13,15 | 2.42 (b, m) | |||
| 2.17 (a, m) | 14,15,17,OAc | 2.10 (a, m) | 13,17 | 2.61 (a, m) | ||||||
| 17 | 2.08 (m) | 13,14,16,20,18 | 2.03 (m) | 16 | 2.08 (m) | 2.11 (m) | 16,18 | 1.87 (dd, J = 23.5, 7.0 Hz) | 13,18,20 | |
| 18 | 0.68 (s) | 12,13,14,17 | 0.61 (s) | 8,12,13,14,17 | 0.66 (s) | 12,13,14,17 | 0.69 (s) | 12,13,14,17 | 0.73 (s) | 12,13,14,17 |
| 19 | 0.99 (s) | 1,5,9,10 | 1.01 (s) | 1,5,9,10 | 1.03 (s) | 1,5,9,10 | 1.01 (s) | 1,4,5,9,10 | 0.93 (s) | 1,3,9,10 |
| 20 | 1.44 (m) | 17,21,22 | 1.44 (m) | 1.38 (m) | 1.40 (m) | 17 | 1.48(m) | |||
| 21 | 0.93 (d, J = 6.3Hz) | 17,20,22 | 0.92 (d, J = 7.0 Hz) | 17,20,22 | 0.91 (d, J = 6.3 Hz) | 17,20,22 | 0.92 (d, J = 7.0 Hz) | 17,20,22 | 0.96 (d, J = 5.0 Hz) | 17,20,22 |
| 22 | 3.80 (dd, J = 8.4, 4.2 Hz) | 3.80 (dd, J = 7.0, 6.3 Hz) | 23 | 3.74 (t, J = 7.7 Hz) | 3.69 (t, J = 7.0 Hz) | 17,21,24 | 3.90 (dd, J = 6.5 3.5 Hz) | |||
| 23 | 2.24 (m) | 20,22,24,25 | 2.29 (ddd, J = 14.0, 7.7, 7.0 Hz) | 22 | 2.28(ddd, J = 14.0, 7.7, 7.0 Hz) | 2.29 (ddd, J = 14.0, 7.7, 7.0 Hz) | 22,25 | 2.42 (m) | 24 | |
| 2.49 (ddd, J = 15.4, 8.4, 7.0 Hz) | 20,22,24,25 | 2.43 (ddd, J = 14.7, 7.7, 7.0 Hz); | 22,24,26 | 2.43(ddd, J = 14.7, 7.7, 7.0 Hz); | 24 | 2.43 (ddd, J = 14.7, 7.7, 7.0 Hz) | 22,25 | 2.60 (m) | 22,24,25 | |
| 24 | 6.92 (t, J = 7.0 Hz) | 22,23,25,27 | 6.83 (dd, J = 7.0, 6.3 Hz) | 23,26 | 6.84 (t, J = 7.0 Hz) | 6.82 (t, J = 7.7 Hz) | 23,25,27 | 6.59 (t, J = 5.0 Hz) | ||
| 26 | 9.44 (s) | 25,27 | ||||||||
| 27 | 1.87 (s) | 24,25,26 | 1.83 (s) | 24,25,26 | 1.84 (s) | 24,25,26 | 1.83 (s) | 24,25 | 1.78 (s) | 24,25,26 |
| 28 | 1.10 (s) | 8,13,14,15 | 0.94 (s) | 8,13,14,15 | 0.98 (s) | 7,8,13,14,15 | 1.07 (s) | 8,13,14,15 | 1.12 (s) | 8,15 |
| 29 | 0.90 (s) | 3,4,5,30 | 0.99 (s) | 3,5,10,30 | 0.96 (s) | 3,4,5,30 | 0.99 (s) | 3,5,10,30 | 1.10 (s) | 3,5,30 |
| 30 | 1.00 (s) | 3,4,5,29 | 0.88 (s) | 3,5,10,29 | 0.94 (s) | 3,4,29 | 0.87 (s) | 3,5,10,29 | 1.07 (s) | 3,5,29 |
| OCOCH3 | 2.06 (s) | 2.09 (s) | ||||||||
| OCOCH3 | 2.10 (s) | |||||||||
| Linear Regression Calibration Curves | ||||||
|---|---|---|---|---|---|---|
| 10 | 12 | |||||
| R2 | 0.998 | 1.000 | ||||
| Linear range (μg/mL) | 25–3000 | 25–1000 | ||||
| LOD (Limit of Detection, μg/mL) | 2.2 | 2.1 | ||||
| LOQ (Limit of Quantitation, μg/mL) | 6.4 | 6.4 | ||||
| Intraday and interday precision | ||||||
| Concentration (μg/mL) | Mean ± SD (RSD, %) | |||||
| 10 | 12 | |||||
| Intraday | Interday | Intraday | Interday | |||
| 100.0 | 101.8 ± 0.2 (0.15) | 100.7 ± 0.1 (0.06) | 101.8 ± 0.4 (0.41) | 100.9 ± 0.1 (0.11) | ||
| 400.0 | 406.9 ± 1.6 (0.40) | 403.6 ± 0.3 (0.06) | 407.4 ± 0.5 (0.12) | 401.9 ± 0.4 (0.09) | ||
| 800.0 | 813.8 ± 0.4 (0.04) | 808.0 ± 0.7 (0.09) | 814.7 ± 1.7 (0.21) | 803.9 ± 3.6 (0.44) | ||
| Recovery tests | ||||||
| Amount added (μg/mL) | Amount measured (μg/mL), mean ± SD | Recovery (%) mean ± SD | RSD (%) | |||
| 10 | 12 | 10 | 12 | 10 | 12 | |
| 0 (blank) | 1216.0 ± 9.60 a | 368.1 ± 3.6 a | ||||
| 100.0 | 1322.3 ± 2.39 b | 473.6 ± 1.6 b | 106.3 ± 2.4 | 105.5 ± 1.6 | 2.25 | 1.50 |
| 400.0 | 1636.7 ± 6.78 b | 777.0 ± 11.4 b | 105.2 ± 1.7 | 102.2 ± 2.8 | 1.61 | 2.78 |
| 800.0 | 2067.3 ± 13.14 b | 1139.3 ± 5.91 b | 106.4 ± 1.6 | 96.4 ± 0.7 | 1.54 | 0.77 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, D.-H.; Wang, J.-Y.; Chen, M.-T.; Liu, Y.-C.; Chen, K.-D. The Triterpenoid High-Performance Liquid Chromatography Analytical Profiles of the Mycelia of Ganoderma lucidum (lingzhi). Microbiol. Res. 2023, 14, 1353-1363. https://doi.org/10.3390/microbiolres14030092
Chen D-H, Wang J-Y, Chen M-T, Liu Y-C, Chen K-D. The Triterpenoid High-Performance Liquid Chromatography Analytical Profiles of the Mycelia of Ganoderma lucidum (lingzhi). Microbiology Research. 2023; 14(3):1353-1363. https://doi.org/10.3390/microbiolres14030092
Chicago/Turabian StyleChen, Deng-Hai, Jian-Yuan Wang, Mon-Tarng Chen, Yen-Chun Liu, and Kuang-Dee Chen. 2023. "The Triterpenoid High-Performance Liquid Chromatography Analytical Profiles of the Mycelia of Ganoderma lucidum (lingzhi)" Microbiology Research 14, no. 3: 1353-1363. https://doi.org/10.3390/microbiolres14030092
APA StyleChen, D.-H., Wang, J.-Y., Chen, M.-T., Liu, Y.-C., & Chen, K.-D. (2023). The Triterpenoid High-Performance Liquid Chromatography Analytical Profiles of the Mycelia of Ganoderma lucidum (lingzhi). Microbiology Research, 14(3), 1353-1363. https://doi.org/10.3390/microbiolres14030092





