Aureimonas altamirensis: First Isolation from a Chicken Slaughterhouse in Italy Followed by Genotype and Phenotype Evaluations
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation and Phenotypical Characterization
2.2. DNA Extraction and Genome Sequencing
2.3. Identification by MALDI-TOF MS and 16S rDNA Analysis
2.4. Bioinformatic Analysis
2.5. Antibiotic Resistance Evaluation
2.6. Biofilm Production Evaluation
2.7. Evaluation of A. altamirensis Colonization on Human Cells
2.8. Statistical Analysis
3. Results and Discussion
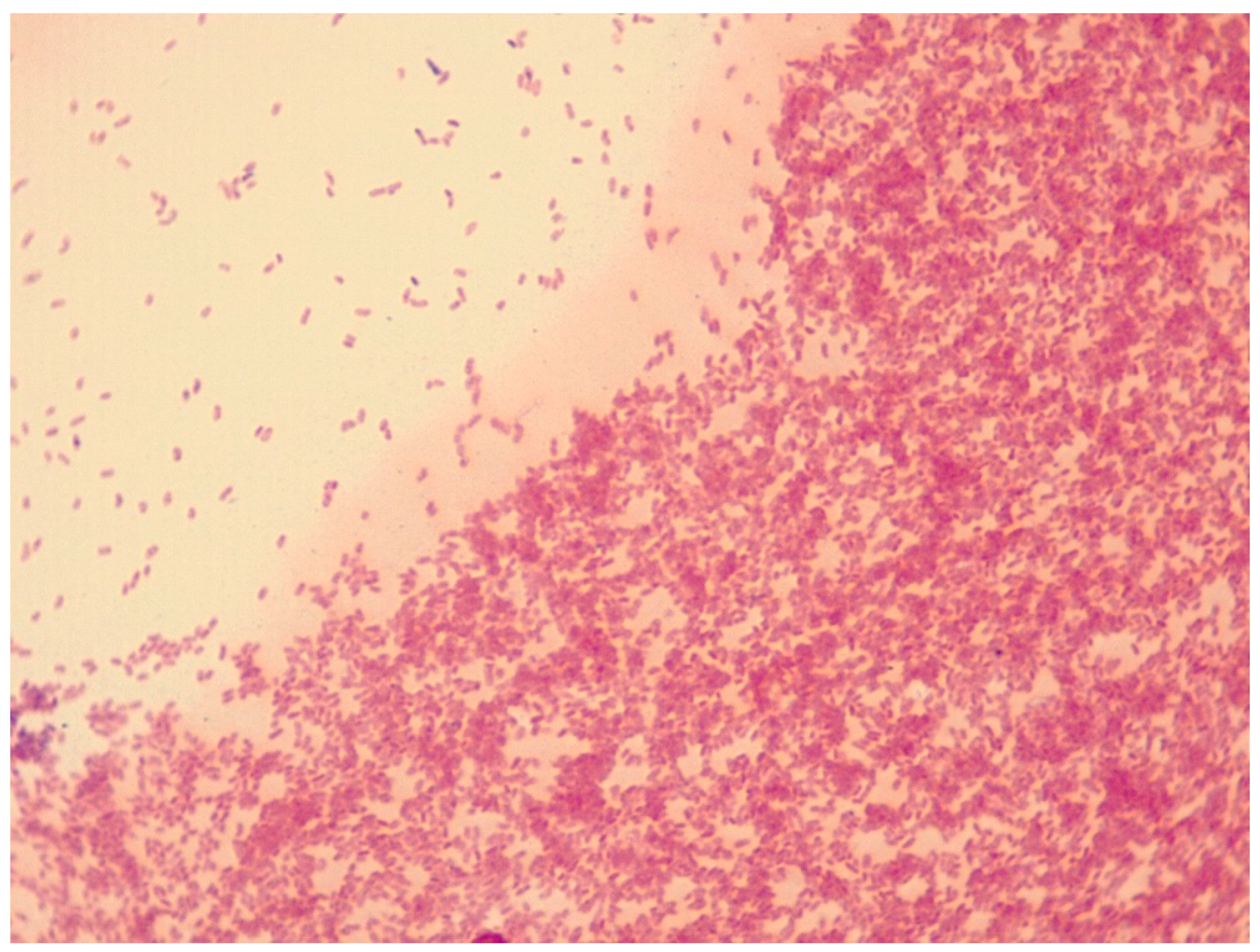
3.1. A. altamirensis Isolation, Identification, and Characterization
3.2. Human Cell Colonization, BFI, and Antibiotic Resistance of A. altamirensis AAI
3.3. Gene Enrichment and Virulence—Antibiotic Resistance Gene Traits
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Bintsis, T. Foodborne pathogens. AIMS Microbiol. 2017, 3, 529–563. [Google Scholar] [CrossRef] [PubMed]
- Rouger, A.; Tresse, O.; Zagorec, M. Bacterial contaminants of poultry meat: Sources, species, and dynamics. Microorganisms 2017, 5, 50. [Google Scholar] [CrossRef] [PubMed]
- Rani, Z.T.; Mhlongo, L.C.; Hugo, A. Microbial profiles of meat at different stages of the distribution chain from the abattoir to retail outlets. Int. J. Environ. Res. Public Health 2023, 20, 1986. [Google Scholar] [CrossRef] [PubMed]
- Rajaei, M.; Moosavy, M.-H.; Gharajalar, S.N.; Khatibi, S.A. Antibiotic resistance in the pathogenic foodborne bacteria isolated from raw kebab and hamburger: Phenotypic and genotypic study. BMC Microbiol. 2021, 21, 272. [Google Scholar] [CrossRef]
- López, A.C.A.; Fernández, J.J.B.; Cortés, E.A.; Gúrpide, B.D.; Suárez, C.R. Aureimonas altamirensis: Primer caso de peritonitis en diálisis peritoneal. Nefrología 2019, 39, 675–677. [Google Scholar] [CrossRef]
- Caniça, M.; Manageiro, V.; Abriouel, H.; Moran-Gilad, J.; Franz, C.M.A.P. Antibiotic resistance in foodborne bacteria. Trends Food Sci. Technol. 2019, 84, 41–44. [Google Scholar] [CrossRef]
- García-Díez, J.; Saraiva, S.; Moura, D.; Grispoldi, L.; Cenci-Goga, B.T.; Saraiva, C. The importance of the slaughterhouse in surveilling animal and public health: A systematic review. Vet. Sci. 2023, 10, 167. [Google Scholar] [CrossRef]
- Marmion, M.; Ferone, M.T.; Whyte, P.; Scannell, A.G.M. The changing microbiome of poultry meat; from farm to fridge. Food Microbiol. 2021, 99, 103823. [Google Scholar] [CrossRef]
- Gonçalves-Tenório, A.; Silva, B.; Rodrigues, V.; Cadavez, V.; Gonzales-Barron, U. Prevalence of pathogens in poultry meat: A meta-analysis of european published surveys. Foods 2018, 7, 69. [Google Scholar] [CrossRef]
- Schröttner, P.; Rudolph, W.W.; Taube, F.; Gunzer, F. First report on the isolation of Aureimonas altamirensis from a patient with peritonitis. Int. J. Infect. Dis. 2014, 29, e71–e73. [Google Scholar] [CrossRef][Green Version]
- Andreozzi, F.; Wittnebel, S.; Yvette, M.; Lewalle, P.; Georgala, A. First case of pleural empyema caused by Aureimonas altamirensis: An unusual opportunistic pathogen to be aware of in hemato-oncologic patients. Clin. Lymphoma Myeloma Leuk. 2019, 19, S213–S214. [Google Scholar] [CrossRef]
- Kim, N.; Hwang, J.-H.; Cho, Y.G.; Kim, D.S.; Lee, H.S.; Lee, J. FirstcCase of Aureimonas altamirensis bacteremia in Korea. Ann. Lab. Med. 2019, 39, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Téllez-Castillo, C.J.; Granda, D.G.; Alepuz, M.B.; Lobo, V.J.; Saiz-Jimenez, C.; Juan, J.L.; Soria, J.M. Isolation of Aurantimonas altamirensis from pleural effusions. J. Med. Microbiol. 2010, 59, 1126–1129. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Chen, W.; Lan, Y.; Chen, M.; Li, J.; Guan, Y.; Hu, F.; Li, F.; Tang, X.; Li, L. First case of biliary Aureimonas altamirensis infection in a patient with colon cancer in China. Infect. Dis. Res. 2021, 2, 9. [Google Scholar] [CrossRef]
- Eshaghi, A.; Shahinas, D.; Patel, S.N.; Kus, J.V. First draft genome sequence of Aureimonas altamirensis, isolated from patient blood culture. FEMS Microbiol. Lett. 2015, 362, fnv016. [Google Scholar] [CrossRef]
- Jurado, V.; Gonzalez, J.M.; Laiz, L.; Saiz-Jimenez, C. Aurantimonas altamirensis sp. nov. a member of the order Rhizobiales isolated from Altamira Cave. Int. J. Syst. Evol. Microbiol. 2006, 56, 2583–2585. [Google Scholar] [CrossRef]
- Becker, R.; Ulrich, K.; Behrendt, U.; Schneck, V.; Ulrich, A. Genomic Characterization of Aureimonas altamirensis C2P003—A specific member of the microbiome of Fraxinus excelsior trees tolerant to ash dieback. Plants 2022, 11, 3487. [Google Scholar] [CrossRef]
- Mano, H.; Tanaka, F.; Nakamura, C.; Kaga, H.; Morisaki, H. Culturable endophytic bacterial flora of the maturing leaves and roots of rice plants (Oryza sativa) Cultivated in a paddy field. Microbes Environ. 2007, 22, 175–185. [Google Scholar] [CrossRef]
- Reilly, T.J.; Calcutt, M.J.; Wennerdahl, L.A.; Williams, F.; Evans, T.J.; Ganjam, I.K.; Bowman, J.W.; Fales, W.H. Isolation of Aureimonas altamirensis, a Brucella canis–like bacterium, from an edematous canine testicle. J. Vet. Diagnostic Investig. 2014, 26, 795–798. [Google Scholar] [CrossRef]
- Luong, M.L.; Békal, S.; Vinh, D.C.; Lauzon, D.; Leung, V.; Al-Rawahi, G.N.; Ng, B.; Burdz, T.; Bernard, K. First report of isolation and characterization of Aurantimonas altamirensis from clinical samples. J. Clin. Microbiol. 2008, 46, 2435–2437. [Google Scholar] [CrossRef]
- Roo-Brand, G.; Schuurs, T.A.; van Zeijl, J.H. Cutaneous Brucellosis unmasked as Aureimonas altamirensis in a wound culture. IDCases 2022, 29, e01532. [Google Scholar] [CrossRef] [PubMed]
- Cosseau, C.; Romano-Bertrand, S.; Duplan, H.; Lucas, O.; Ingrassia, I.; Pigasse, C.; Roques, C.; Jumas-Bilak, E. Proteobacteria from the human skin microbiota: Species-level diversity and hypotheses. One Health 2016, 2, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Andrade López, A.C.; Bande Fernández, J.J.; Cortés, E.A.; Gúrpide, D.; Suárez, C.R. Aureimonas altamirensis: The first case of peritonitis on peritoneal dialysis. Nefrologia 2019, 39, 675–677. [Google Scholar] [CrossRef] [PubMed]
- Houf, K.; Devriese, L.A.; De Zutter, L.; Van Hoof, J.; Vandamme, P. Development of a new protocol for the isolation and quantification of Arcobacter species from poultry products. Int. J. Food Microbiol. 2001, 71, 189–196. [Google Scholar] [CrossRef] [PubMed]
- Buzzanca, D.; Kerkhof, P.; Alessandria, V.; Rantsiou, K.; Houf, K. Arcobacteraceae comparative genome analysis demonstrates genome heterogeneity and reduction in species isolated from animals and associated with human illness. Heliyon 2023, 9, e17652. [Google Scholar] [CrossRef]
- Ramees, T.P.; Dhama, K.; Karthik, K.; Rathore, R.S.; Kumar, A.; Saminathan, M.; Tiwari, R.; Malik, Y.S.; Singh, R.K. Arcobacter: An emerging food-borne zoonotic pathogen, its public health concerns and advances in diagnosis and control—A comprehensive review. Vet. Q. 2017, 37, 136–161. [Google Scholar] [CrossRef]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef]
- Olson, R.D.; Assaf, R.; Brettin, T.; Conrad, N.; Cucinell, C.; Davis, J.J.; Dempsey, D.M.; Dickerman, A.; Dietrich, E.M.; Kenyon, R.W.; et al. Introducing the Bacterial and Viral Bioinformatics Resource Center (BV-BRC): A resource combining PATRIC, IRD and ViPR. Nucleic Acids Res. 2023, 51, D678–D689. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Tian, L.; Huang, C.; Mazloom, R.; Heath, L.S.; Vinatzer, B.A. LINbase: A web server for genome-based identification of prokaryotes as members of crowdsourced taxa. Nucleic Acids Res. 2020, 48, W529–W537. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Brettin, T.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Olsen, G.J.; Olson, R.; Overbeek, R.; Parrello, B.; Pusch, G.D.; et al. RASTtk: A modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 2015, 5, 8365. [Google Scholar] [CrossRef] [PubMed]
- Tanizawa, Y.; Fujisawa, T.; Nakamura, Y. Genome analysis DFAST: A flexible prokaryotic genome annotation pipeline for faster genome publication. Genome Anal. DFAST 2017, 34, 1037–1039. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Szklarczyk, D.; Heller, D.; Hernández-Plaza, A.; Forslund, S.K.; Cook, H.; Mende, D.R.; Letunic, I.; Rattei, T.; Jensen, L.J.; et al. eggNOG 5.0: A hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019, 47, D309–D314. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef]
- Couvin, D.; Bernheim, A.; Toffano-Nioche, C.; Touchon, M.; Michalik, J.; Néron, B.; Rocha, E.P.C.; Vergnaud, G.; Gautheret, D.; Pourcel, C. CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Res. 2018, 46, W246–W251. [Google Scholar] [CrossRef]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Edalatmand, A.; Huynh, W.; Nguyen, A.V.; Cheng, A.A.; Liu, S.; Min, S.Y.; et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020, 48, 517–525. [Google Scholar] [CrossRef]
- Teh, K.H.; Flint, S.; French, N. Biofilm formation by Campylobacter jejuni in controlled mixed-microbial populations. Int. J. Food Microbiol. 2010, 143, 118–124. [Google Scholar] [CrossRef]
- Buzzanca, D.; Botta, C.; Ferrocino, I.; Alessandria, V.; Houf, K.; Rantsiou, K. Functional pangenome analysis reveals high virulence plasticity of Aliarcobacter butzleri and affinity to human mucus. Genomics 2021, 113, 2065–2076. [Google Scholar] [CrossRef]
- Buzzanca, D.; Alessandria, V.; Botta, C.; Zadeh, N.S.; Ferrocino, I.; Houf, K.; Cocolin, L.; Rantsiou, K. Transcriptome analysis of Arcobacter butzleri infection in a mucus-producing human intestinal in vitro model. Microbiol. Spectr. 2023, 11, e02071-22. [Google Scholar] [CrossRef]
- Moran, A.P.; Gupta, A.; Joshi, L. Sweet-talk: Role of host glycosylation in bacterial pathogenesis of the gastrointestinal tract. Gut 2011, 60, 1412–1425. [Google Scholar] [CrossRef]
- Garneau-Tsodikova, S.; Labby, K.J. Mechanisms of resistance to aminoglycoside antibiotics: Overview and perspectives. Medchemcomm 2016, 7, 11–27. [Google Scholar] [CrossRef]
- Sultan, I.; Rahman, S.; Jan, A.T.; Siddiqui, M.T. Antibiotics, resistome and resistance mechanisms: A bacterial perspective. Front. Microbiol. 2018, 9, 2066. [Google Scholar] [CrossRef] [PubMed]
- Rovetto, F.; Carlier, A.; Van Den Abeele, A.M.; Illeghems, K.; Van Nieuwerburgh, F.; Cocolin, L.; Houf, K. Characterization of the emerging zoonotic pathogen Arcobacter thereius by whole genome sequencing and comparative genomics. PLoS ONE 2017, 12, e0180493. [Google Scholar] [CrossRef] [PubMed]
- Kunin, V.; Sorek, R.; Hugenholtz, P. Evolutionary conservation of sequence and secondary structures in CRISPR repeats. Genome Biol. 2007, 8, R61. [Google Scholar] [CrossRef] [PubMed]
- Louwen, R.; Horst-Kreft, D.; Boer, A.G.; Graaf, L.; Knegt, G.; Hamersma, M.; Heikema, A.P.; Timms, A.R.; Jacobs, B.C.; Wagenaar, J.A.; et al. A novel link between Campylobacter jejuni bacteriophage defence, virulence and Guillain–Barré syndrome. Eur. J. Clin. Microbiol. Infect. Dis. 2013, 32, 207–226. [Google Scholar] [CrossRef]
- Torres, A.G.; Redford, P.; Welch, R.A.; Payne, S.M. TonB-Dependent Systems of Uropathogenic Escherichia coli: Aerobactin and heme transport and TonB are required for virulence in the mouse. Infect. Immun. 2001, 69, 6179–6185. [Google Scholar] [CrossRef]
- Jefferson, K. What drives bacteria to produce a biofilm? FEMS Microbiol. Lett. 2004, 236, 163–173. [Google Scholar] [CrossRef] [PubMed]
- Wang, R. Biofilms and meat safety: A Mini-Review. J. Food Prot. 2019, 82, 120–127. [Google Scholar] [CrossRef] [PubMed]
- Haider, A.; Ringer, M.; Kotroczó, Z.; Mohácsi-Farkas, C.; Kocsis, T. The current level of MALDI-TOF MS applications in the detection of microorganisms: A short review of benefits and limitations. Microbiol. Res. 2023, 14, 80–90. [Google Scholar] [CrossRef]
- Lahti, E.T.; Karamehmedovic, N.; Riedel, H.; Blom, L.; Boel, J.; Delibato, E.; Denis, M.; van Essen-Zandbergen, A.; Garcia-Fernandez, A.; Hendriksen, R.; et al. One Health surveillance—A cross-sectoral detection, characterization, and notification of foodborne pathogens. Front. Public Health 2023, 11, 1129083. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Buzzanca, D.; Chiarini, E.; Mania, I.; Chiesa, F.; Alessandria, V. Aureimonas altamirensis: First Isolation from a Chicken Slaughterhouse in Italy Followed by Genotype and Phenotype Evaluations. Microbiol. Res. 2023, 14, 1319-1330. https://doi.org/10.3390/microbiolres14030089
Buzzanca D, Chiarini E, Mania I, Chiesa F, Alessandria V. Aureimonas altamirensis: First Isolation from a Chicken Slaughterhouse in Italy Followed by Genotype and Phenotype Evaluations. Microbiology Research. 2023; 14(3):1319-1330. https://doi.org/10.3390/microbiolres14030089
Chicago/Turabian StyleBuzzanca, Davide, Elisabetta Chiarini, Ilaria Mania, Francesco Chiesa, and Valentina Alessandria. 2023. "Aureimonas altamirensis: First Isolation from a Chicken Slaughterhouse in Italy Followed by Genotype and Phenotype Evaluations" Microbiology Research 14, no. 3: 1319-1330. https://doi.org/10.3390/microbiolres14030089
APA StyleBuzzanca, D., Chiarini, E., Mania, I., Chiesa, F., & Alessandria, V. (2023). Aureimonas altamirensis: First Isolation from a Chicken Slaughterhouse in Italy Followed by Genotype and Phenotype Evaluations. Microbiology Research, 14(3), 1319-1330. https://doi.org/10.3390/microbiolres14030089





