Construction of Recombinant Escherichia coli with a High L-Phenylalanine Production Yield from Glucose
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains
2.2. Gene Deletion
2.3. Plasmids Construction
2.4. Growth Conditions
2.5. Analytical Methods
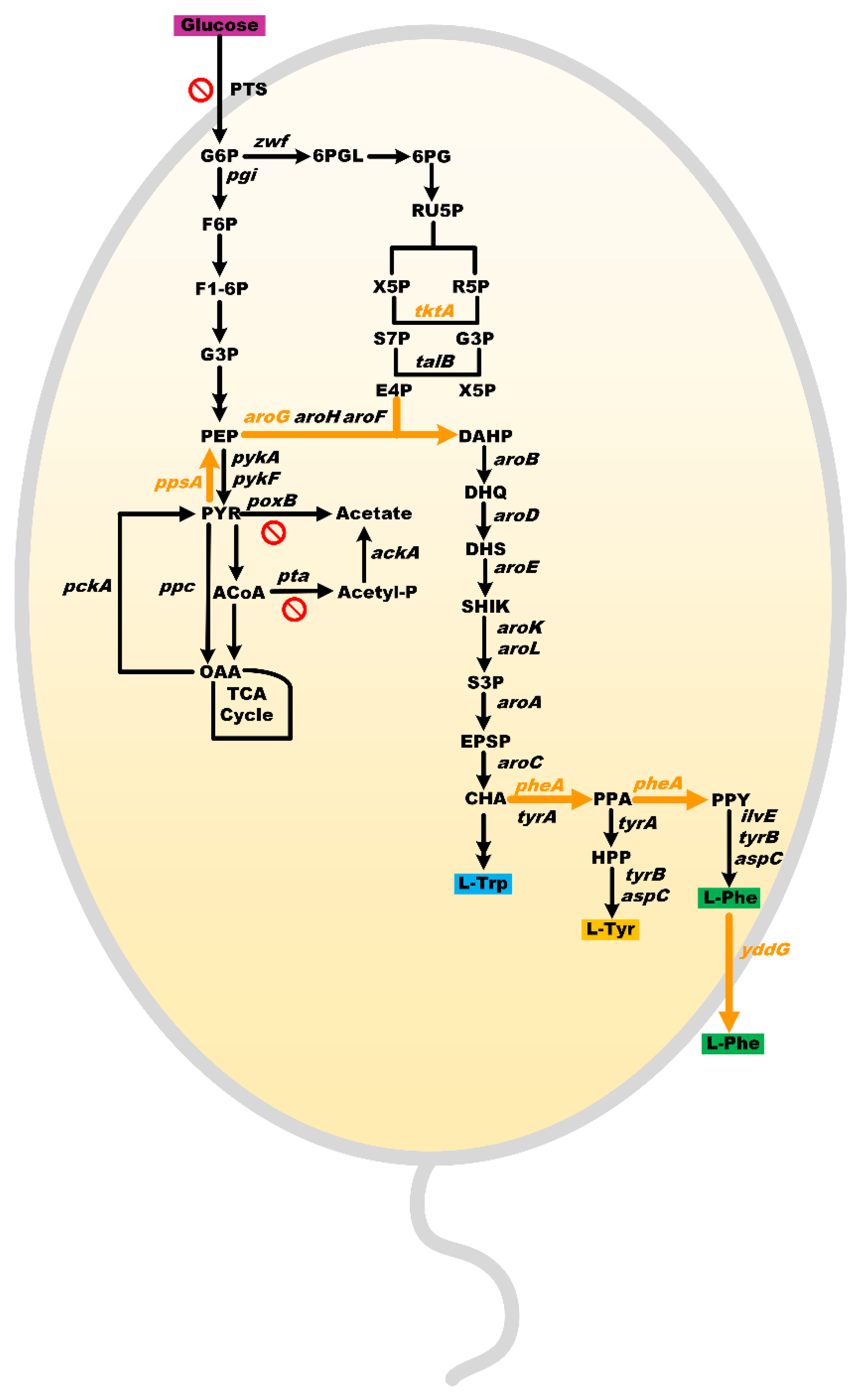
3. Results and Discussion
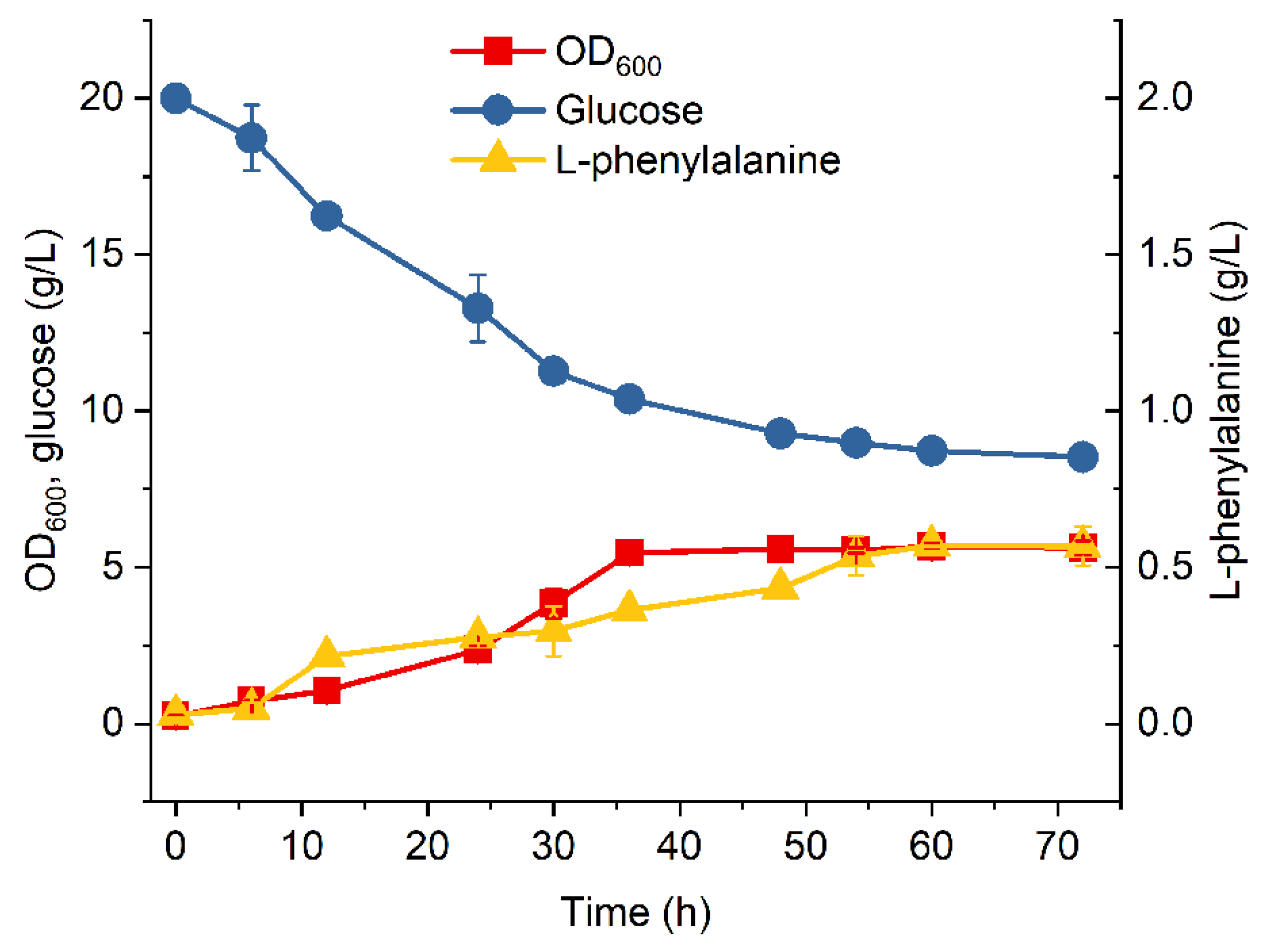
3.1. Engineering a Base E. coli Strain for L-Phenylalanine Production
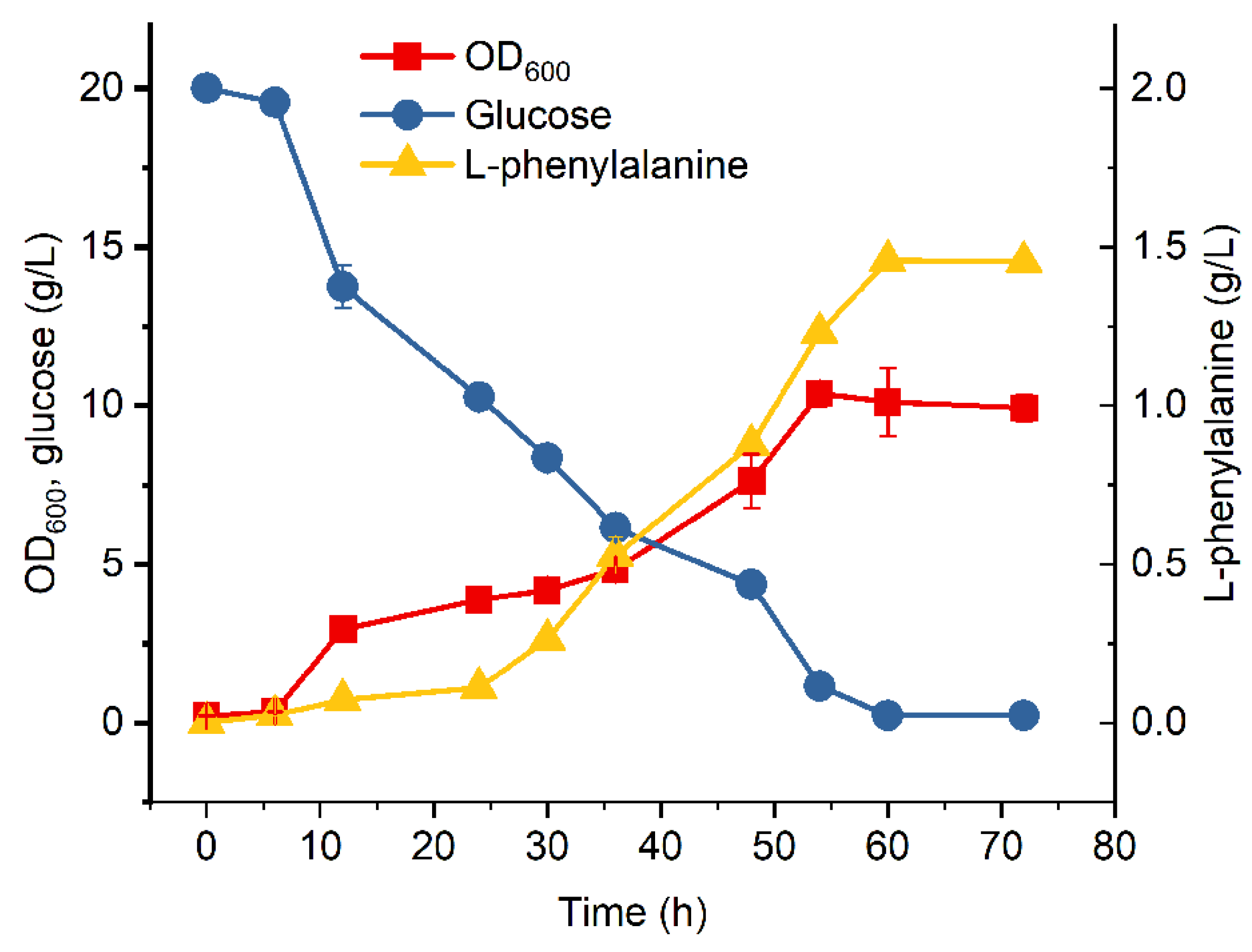
3.2. Introduction of Glf to Improve Glucose Consumption of MPH-2
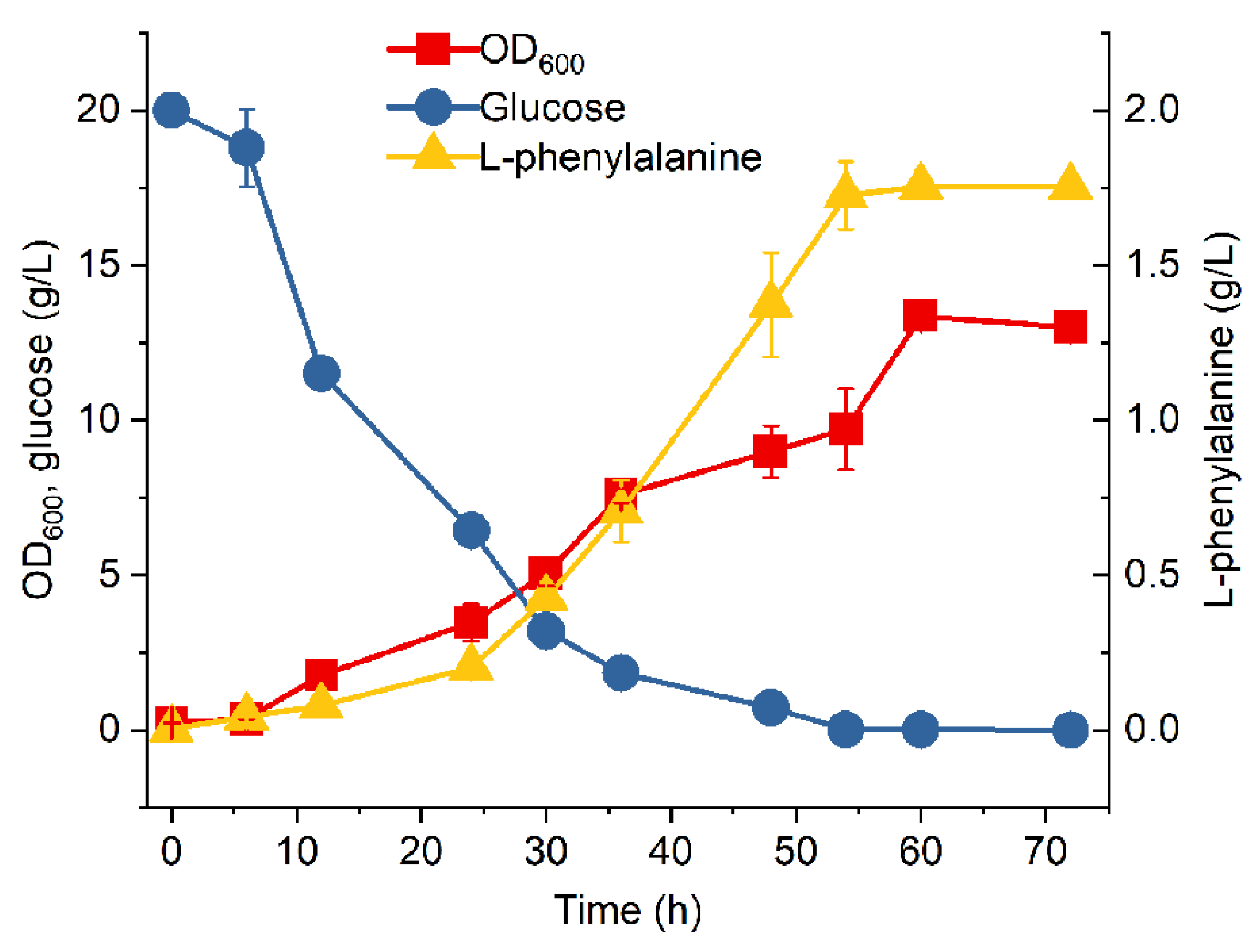
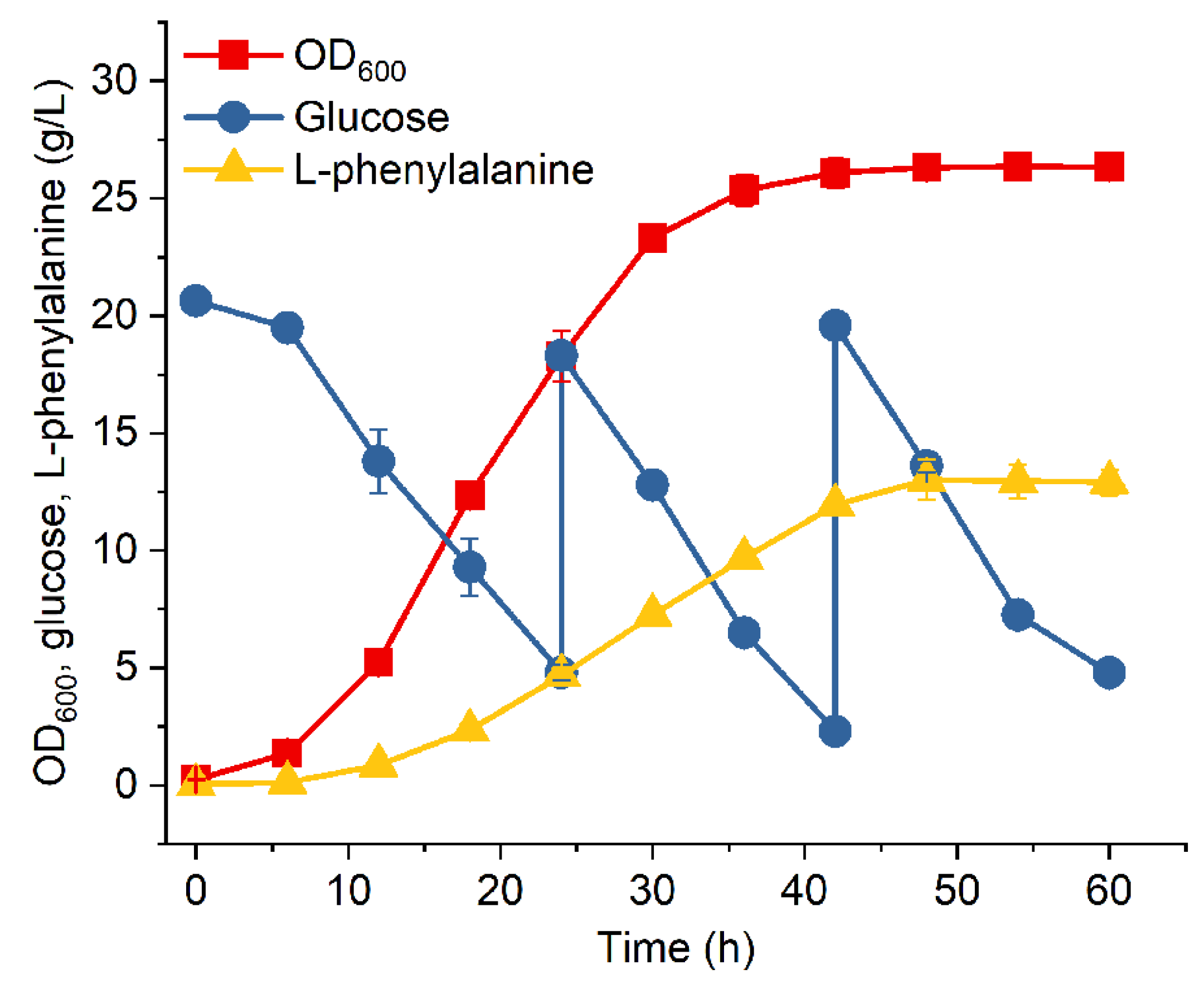
3.3. Fed-Batch Fermentation of MPH-3
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhou, H.; Liao, X.; Wang, T.; Du, G.; Chen, J. Enhanced L-phenylalanine biosynthesis by co-expression of pheA(fbr) and aroF(wt). Bioresour. Technol. 2010, 101, 4151–4156. [Google Scholar] [CrossRef] [PubMed]
- Huccetogullari, D.; Luo, Z.W.; Lee, S.Y. Metabolic engineering of microorganisms for production of aromatic compounds. Microb. Cell. Fact. 2019, 18, 41. [Google Scholar] [CrossRef] [PubMed]
- Kataoka, N.; Matsutani, M.; Matsushita, K.; Yakushi, T. Stepwise metabolic engineering of Corynebacterium glutamicum for the production of phenylalanine. J. Gen. Appl. Microbiol. 2023, 69, 11–23. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Niu, H.; Li, Q.; Gu, P. Metabolic engineering for the production of L-phenylalanine in Escherichia coli. 3 Biotech 2019, 9, 85. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Liao, X.; Liu, L.; Wang, T.; Du, G.; Chen, J. Enhanced L-phenylalanine production by recombinant Escherichia coli BR-42 (pAP-B03) resistant to bacteriophage BP-1 via a two-stage feeding approach. J. Ind. Microbiol. Biotechnol. 2011, 38, 1219–1227. [Google Scholar] [CrossRef] [PubMed]
- Umbarger, H.E. Amino acid biosynthesis and its regulation. Annu. Rev. Biochem. 1978, 47, 532–606. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, M. Towards bacterial strains overproducing L-tryptophan and other aromatics by metabolic engineering. Appl. Microbiol. Biotechnol. 2006, 69, 615–626. [Google Scholar] [CrossRef]
- Wallace, B.J.; Pittard, J. Regulator gene controlling enzymes concerned in tyrosine biosynthesis in Escherichia coli. J. Bacteriol. 1969, 97, 1234–1241. [Google Scholar] [CrossRef]
- Rihtar, E.; Zgur Bertok, D.; Podlesek, Z. The Uropathogenic specific protein gene usp from Escherichia coli and Salmonella bongori is a Novel Member of the TyrR and H-NS Regulons. Microorganisms 2020, 8, 330. [Google Scholar] [CrossRef]
- Lawley, B.; Fujita, N.; Ishihama, A.; Pittard, A.J. The TyrR protein of Escherichia coli is a class I transcription activator. J. Bacteriol. 1995, 177, 238–241. [Google Scholar] [CrossRef][Green Version]
- Gunsalus, R.P.; Yanofsky, C. Nucleotide sequence and expression of Escherichia coli trpR, the structural gene for the trp aporepressor. Proc. Natl. Acad. Sci. USA 1980, 77, 7117–7121. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, A.; Martinez, J.A.; Flores, N.; Escalante, A.; Gosset, G.; Bolivar, F. Engineering Escherichia coli to overproduce aromatic amino acids and derived compounds. Microb. Cell. Fact. 2014, 13, 126. [Google Scholar] [CrossRef] [PubMed]
- Gavini, N.; Davidson, B.E. Regulation of pheA expression by the pheR product in Escherichia coli is mediated through attenuation of transcription. J. Biol. Chem. 1991, 266, 7750–7753. [Google Scholar] [CrossRef] [PubMed]
- Postma, P.W.; Lengeler, J.W. Phosphoenolpyruvate:carbohydrate phosphotransferase system of bacteria. Microbiol. Rev. 1985, 49, 232–269. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Niu, H.; Huang, Z.; Li, Q.; Gu, P. Construction of a switchable synthetic Escherichia coli for aromatic amino acids by a tunable switch. J. Ind. Microbiol. Biotechnol. 2020, 47, 233–242. [Google Scholar] [CrossRef] [PubMed]
- Sprenger, G.A. From scratch to value: Engineering Escherichia coli wild type cells to the production of L-phenylalanine and other fine chemicals derived from chorismate. Appl. Microbiol. Biotechnol. 2007, 75, 739–749. [Google Scholar] [CrossRef] [PubMed]
- Flores, S.; Gosset, G.; Flores, N.; de Graaf, A.A.; Bolivar, F. Analysis of carbon metabolism in Escherichia coli strains with an inactive phosphotransferase system by (13)C labeling and NMR spectroscopy. Metab. Eng. 2002, 4, 124–137. [Google Scholar] [CrossRef]
- Guo, L.; Ding, S.; Liu, Y.; Gao, C.; Hu, G.; Song, W.; Liu, J.; Chen, X.; Liu, L. Enhancing tryptophan production by balancing precursors in Escherichia coli. Biotechnol. Bioeng. 2022, 119, 983–993. [Google Scholar] [CrossRef]
- Liu, Y.; Xu, Y.; Ding, D.; Wen, J.; Zhu, B.; Zhang, D. Genetic engineering of Escherichia coli to improve L-phenylalanine production. BMC Biotechnol. 2018, 18, 5. [Google Scholar] [CrossRef]
- Datsenko, K.A.; Wanner, B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 2000, 97, 6640–6645. [Google Scholar] [CrossRef]
- Cherepanov, P.P.; Wackernagel, W. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 1995, 158, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Kuhlman, T.E.; Cox, E.C. Site-specific chromosomal integration of large synthetic constructs. Nucleic Acids Res. 2010, 38, e92. [Google Scholar] [CrossRef] [PubMed]
- Lerner, C.G.; Inouye, M. Low copy number plasmids for regulated low-level expression of cloned genes in Escherichia coli with blue/white insert screening capability. Nucleic Acids Res. 1990, 18, 4631. [Google Scholar] [CrossRef] [PubMed]
- Gu, P.; Yang, F.; Li, F.; Liang, Q.; Qi, Q. Knocking out analysis of tryptophan permeases in Escherichia coli for improving L-tryptophan production. Appl. Microbiol. Biotechnol. 2013, 97, 6677–6683. [Google Scholar] [CrossRef] [PubMed]
- Gu, P.; Yang, F.; Kang, J.; Wang, Q.; Qi, Q. One-step of tryptophan attenuator inactivation and promoter swapping to improve the production of L-tryptophan in Escherichia coli. Microb. Cell Fact. 2012, 11, 30. [Google Scholar] [CrossRef] [PubMed]
- Doroshenko, V.; Airich, L.; Vitushkina, M.; Kolokolova, A.; Livshits, V.; Mashko, S. YddG from Escherichia coli promotes export of aromatic amino acids. FEMS Microbiol. Lett. 2007, 275, 312–318. [Google Scholar] [CrossRef] [PubMed]
- Niu, H.; Li, R.; Gao, J.; Fan, X.; Li, Q.; Gu, P. Different performance of Escherichia coli mutants with defects in the phosphoenolpyruvate: Carbohydrate phosphotransferase system under low glucose condition. 3 Biotech 2019, 9, 50. [Google Scholar] [CrossRef]
- Gosset, G. Improvement of Escherichia coli production strains by modification of the phosphoenolpyruvate:sugar phosphotransferase system. Microb. Cell Fact. 2005, 4, 14. [Google Scholar] [CrossRef]
- Cases, I.; Velazquez, F.; de Lorenzo, V. The ancestral role of the phosphoenolpyruvate-carbohydrate phosphotransferase system (PTS) as exposed by comparative genomics. Res. Microbiol. 2007, 158, 666–670. [Google Scholar] [CrossRef]
- Yang, M.; Zhang, X. Construction of pyruvate producing strain with intact pyruvate dehydrogenase and genome-wide transcription analysis. World J. Microbiol. Biotechnol. 2017, 33, 59. [Google Scholar] [CrossRef]
- Hernandez-Montalvo, V.; Martinez, A.; Hernandez-Chavez, G.; Bolivar, F.; Valle, F.; Gosset, G. Expression of galP and glk in a Escherichia coli PTS mutant restores glucose transport and increases glycolytic flux to fermentation products. Biotechnol. Bioeng. 2003, 83, 687–694. [Google Scholar] [CrossRef] [PubMed]
- Kurgan, G.; Onyeabor, M.; Holland, S.C.; Taylor, E.; Schneider, A.; Kurgan, L.; Billings, T.; Wang, X. Directed evolution of Zymomonas mobilis sugar facilitator Glf to overcome glucose inhibition. J. Ind. Microbiol. Biotechnol. 2022, 49, kuab066. [Google Scholar] [CrossRef]
- Shimada, T.; Nakazawa, K.; Tachikawa, T.; Saito, N.; Niwa, T.; Taguchi, H.; Tanaka, K. Acetate overflow metabolism regulates a major metabolic shift after glucose depletion in Escherichia coli. FEBS Lett. 2021, 595, 2047–2056. [Google Scholar] [CrossRef] [PubMed]
- Enjalbert, B.; Cocaign-Bousquet, M.; Portais, J.C.; Letisse, F. Acetate exposure determines the diauxic behavior of Escherichia coli during the glucose-acetate transition. J. Bacteriol. 2015, 197, 3173–3181. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tyagi, N.; Saini, D.; Guleria, R.; Mukherjee, K.J. Designing an Escherichia coli strain for phenylalanine overproduction by metabolic engineering. Mol. Biotechnol. 2017, 59, 168–178. [Google Scholar] [CrossRef] [PubMed]
- Baez-Viveros, J.L.; Osuna, J.; Hernandez-Chavez, G.; Soberon, X.; Bolivar, F.; Gosset, G. Metabolic engineering and protein directed evolution increase the yield of L-phenylalanine synthesized from glucose in Escherichia coli. Biotechnol. Bioeng. 2004, 87, 516–524. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.B.; Guo, X.L.; Zhang, M.L.; Huang, Q.G.; Qi, F.; Huang, J.Z. Enhancement of l-phenylalanine production in Escherichia coli by heterologous expression of Vitreoscilla hemoglobin. Biotechnol. Appl. Biochem. 2018, 65, 476–483. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.P.; Liu, R.X.; Xiao, M.R.; Zhang, L.; Ding, Z.Y.; Gu, Z.H.; Shi, G.Y. A systems level engineered E. coli capable of efficiently producing L-phenylalanine. Process Biochem. 2014, 49, 751–757. [Google Scholar] [CrossRef]
- Liu, S.P.; Xiao, M.R.; Zhang, L.; Xu, J.; Ding, Z.Y.; Gu, Z.H.; Shi, G.Y. Production of l-phenylalanine from glucose by metabolic engineering of wild type Escherichia coli W3110. Process Biochem. 2013, 48, 413–419. [Google Scholar] [CrossRef]
- Ding, R.; Liu, L.; Chen, X.; Cui, Z.; Ao, Z.; Ren, D.; Zhang, L. Introduction of two mutations into AroG increases phenylalanine production in Escherichia coli. Biotechnol. Lett. 2014, 36, 2103–2108. [Google Scholar] [CrossRef]
- Cheng, L.K.; Wang, J.; Xu, Q.Y.; Zhao, C.G.; Shen, Z.Q.; Xie, X.X.; Chen, N. Strategy for pH control and pH feedback-controlled substrate feeding for high-level production of L-tryptophan by Escherichia coli. World J. Microbiol. Biotechnol. 2013, 29, 883–890. [Google Scholar] [CrossRef] [PubMed]
- Jing, K.; Tang, Y.; Yao, C.; Del Rio-Chanona, E.A.; Ling, X.; Zhang, D. Overproduction of L-tryptophan via simultaneous feed of glucose and anthranilic acid from recombinant Escherichia coli W3110: Kinetic modeling and process scale-up. Biotechnol. Bioeng. 2018, 115, 371–381. [Google Scholar] [CrossRef] [PubMed]
- Gerigk, M.; Bujnicki, R.; Ganpo-Nkwenkwa, E.; Bongaerts, J.; Sprenger, G.; Takors, R. Process control for enhanced L-phenylalanine production using different recombinant Escherichia coli strains. Biotechnol. Bioeng. 2002, 80, 746–754. [Google Scholar] [CrossRef] [PubMed]
- Weiner, M.; Trondle, J.; Albermann, C.; Sprenger, G.A.; Weuster-Botz, D. Improvement of constraint-based flux estimation during L-phenylalanine production with Escherichia coli using targeted knock-out mutants. Biotechnol. Bioeng. 2014, 111, 1406–1416. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Ding, D.; Bai, D.; Lin, Y.; Zhu, Y.; Zhang, C.; Zhang, D. Transcriptomics and metabolomics analysis of L-phenylalanine overproduction in Escherichia coli. Microb. Cell Fact. 2023, 22, 65. [Google Scholar] [CrossRef] [PubMed]
- Mahr, R.; von Boeselager, R.F.; Wiechert, J.; Frunzke, J. Screening of an Escherichia coli promoter library for a phenylalanine biosensor. Appl. Microbiol. Biotechnol. 2016, 100, 6739–6753. [Google Scholar] [CrossRef]
- Yakandawala, N.; Romeo, T.; Friesen, A.D.; Madhyastha, S. Metabolic engineering of Escherichia coli to enhance phenylalanine production. Appl. Microbiol. Biotechnol. 2008, 78, 283–291. [Google Scholar] [CrossRef]
- Ojima, Y.; Komaki, M.; Nishioka, M.; Iwatani, S.; Tsujimoto, N.; Taya, M. Introduction of a stress-responsive gene, yggG, enhances the yield of L-phenylalanine with decreased acetic acid production in a recombinant Escherichia coli. Biotechnol. Lett. 2009, 31, 525–530. [Google Scholar] [CrossRef]
- Baez, A.; Cho, K.M.; Liao, J.C. High-flux isobutanol production using engineered Escherichia coli: A bioreactor study with in situ product removal. Appl. Microbiol. Biotechnol. 2011, 90, 1681–1690. [Google Scholar] [CrossRef]
| Name | Relevant Genotype | Reference |
|---|---|---|
| DH5α | F−, endA1, hsdR17 (rK−, mK+), supE44, thi-l, λ−, recA1, gyrA96, ΔlacU169 (Φ80lacZ ΔM15) | Lab stock |
| MG1655 | F−, λ−, rph-1 | Lab stock |
| MG-1 | MG1655 (ΔpoxB) | This study |
| MG-2 | MG1655 (ΔpoxBΔpta) | This study |
| MG-3 | MG1655/pT1 | |
| MG-4 | MG-1/pT1 | This study |
| MG-5 | MG-2/pT1 | This study |
| MPH-1 | MG1655 (ΔpoxBΔptaΔptsI) | This study |
| MPH-2 | MPH-1/pT1 | This study |
| MPH-3 | MPH-1/pT1/pT2 | This study |
| MPH-4 | MPH-1 with tyrA replacement by glf containing plasmid pT1 | This study |
| Name | Relevant Genotype | Reference |
|---|---|---|
| pKD3 | bla, FRT-cat-FRT | [20] |
| pCP20 | bla and cat, helper plasmid | [21] |
| pTKRed | SpcR, IPTG induced λRed enzymes | [22] |
| pCL1920 | SpcR | [23] |
| pTrc99a | bla | Lab stock |
| pT1 | pCL1920-pheAfbr-tktA-aroGfbr-ppsA-yddG | Synthesized by TSINGKE Biological Technology |
| pT2 | pTrc99a-glf | Synthesized by TSINGKE Biological Technology |
| E. coli Strains | Genes Overexpressed in Plasmids | Promoters and Replicon in Plasmids | Host Engineering | Culture Methods | L-Phenylalanine Production | References | |
|---|---|---|---|---|---|---|---|
| Titer (g/L) | Yields (g/g) | ||||||
| Xllp21 | pheA (Thr326Pro), aroF, galP, glk, and aroD | pBR322 replicon and BBa_J23106 promoter | W3110 mutant with L-tyrosine auxotrophic (ΔptsH and tyrR (T495I)) | 5 L fed-batch fermentation | 72.9 | 0.26 | [19] |
| PAPV | aroF, pheAfbr, and vgb from Vitreoscilla | PLPRpromoters and replicon was not indicated | Derived from Escherichia coli K-12, Hfr (PO1), λ−, el4-, tyrA4, relA1, spoT1, thiE1 | 3 L fed-batch fermentation | 44.21 | 0.071 | [37] |
| W14 (pR15BABKG) | aroG15, pheAfbr, aroK, ydiB, yddG, and tyrB | λcIts857 replicon and PLPR promoters | W3110 mutant with L-tyrosine auxotrophic (Δcrr) | 15 L fed-batch fermentation | 47 | 0.252 | [38] |
| W3110 (pNpheABK15) | pheAfbr and aroG15 | pBR322 replicon and PN25 promoter | Wild W3110 | 15 L fed-batch fermentation | 23.8 | 0.154 | [39] |
| W3110 (pQPTABG8/15) | AroG (A202T and M147I) | pBR322 replicon and Ptacpromoter | Wild W3110 | 3 L fed-batch fermentation | 26.78 | 0.231 | [40] |
| WSH-Z06 (pAP-B03) | pheAfbr and aroFwt | p15A replicon and PLPR promoters | W3110 mutant with L-tyrosine auxotrophic | 3 L fed-batch fermentation | 35.38 | 0.238 | [1] |
| MPH-3 | pheAfbr, tktA, aroGfbr, ppsA, and yddG | pBR322 replicon with Ptrc promoter and pSC101 replicon with Plac promoter | MG1655 (ΔpoxBΔptaΔptsI) | 5 L fed-batch fermentation | 19.24 | 0.279 | This study |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gu, P.; Zhao, S.; Li, C.; Jiang, S.; Zhou, H.; Li, Q. Construction of Recombinant Escherichia coli with a High L-Phenylalanine Production Yield from Glucose. Microbiol. Res. 2023, 14, 1185-1198. https://doi.org/10.3390/microbiolres14030079
Gu P, Zhao S, Li C, Jiang S, Zhou H, Li Q. Construction of Recombinant Escherichia coli with a High L-Phenylalanine Production Yield from Glucose. Microbiology Research. 2023; 14(3):1185-1198. https://doi.org/10.3390/microbiolres14030079
Chicago/Turabian StyleGu, Pengfei, Shuo Zhao, Chengwei Li, Shuixing Jiang, Hao Zhou, and Qiang Li. 2023. "Construction of Recombinant Escherichia coli with a High L-Phenylalanine Production Yield from Glucose" Microbiology Research 14, no. 3: 1185-1198. https://doi.org/10.3390/microbiolres14030079
APA StyleGu, P., Zhao, S., Li, C., Jiang, S., Zhou, H., & Li, Q. (2023). Construction of Recombinant Escherichia coli with a High L-Phenylalanine Production Yield from Glucose. Microbiology Research, 14(3), 1185-1198. https://doi.org/10.3390/microbiolres14030079





