Biofilm Formation and Phenotypic Detection of ESBL, MBL, KPC and AmpC Enzymes and Their Coexistence in Klebsiella spp. Isolated at the National Reference Laboratory, Kathmandu, Nepal
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design and Study Sample
2.2. Sample Collection and Transportation
2.3. Macroscopic and Microscopic Examinations
2.4. Culture of Specimens and Identification of the Isolates
2.5. Antibiotic Susceptibility Testing
2.6. Screening and Confirmation of ESBL Producers
2.7. Screening and Confirmation of MBL Producers
2.8. Screening and Phenotypic Confirmation of KPC Producers
2.9. Screening and Confirmation of AmpC Producers
2.10. Detection of Biofilm Formation by Micro-Titer Plate Method
2.11. Quality Control
2.12. Data Analysis
3. Results
3.1. Distribution of Growth Pattern of Bacterial Isolates
3.2. Antibiotic Susceptibility Pattern of the Isolates
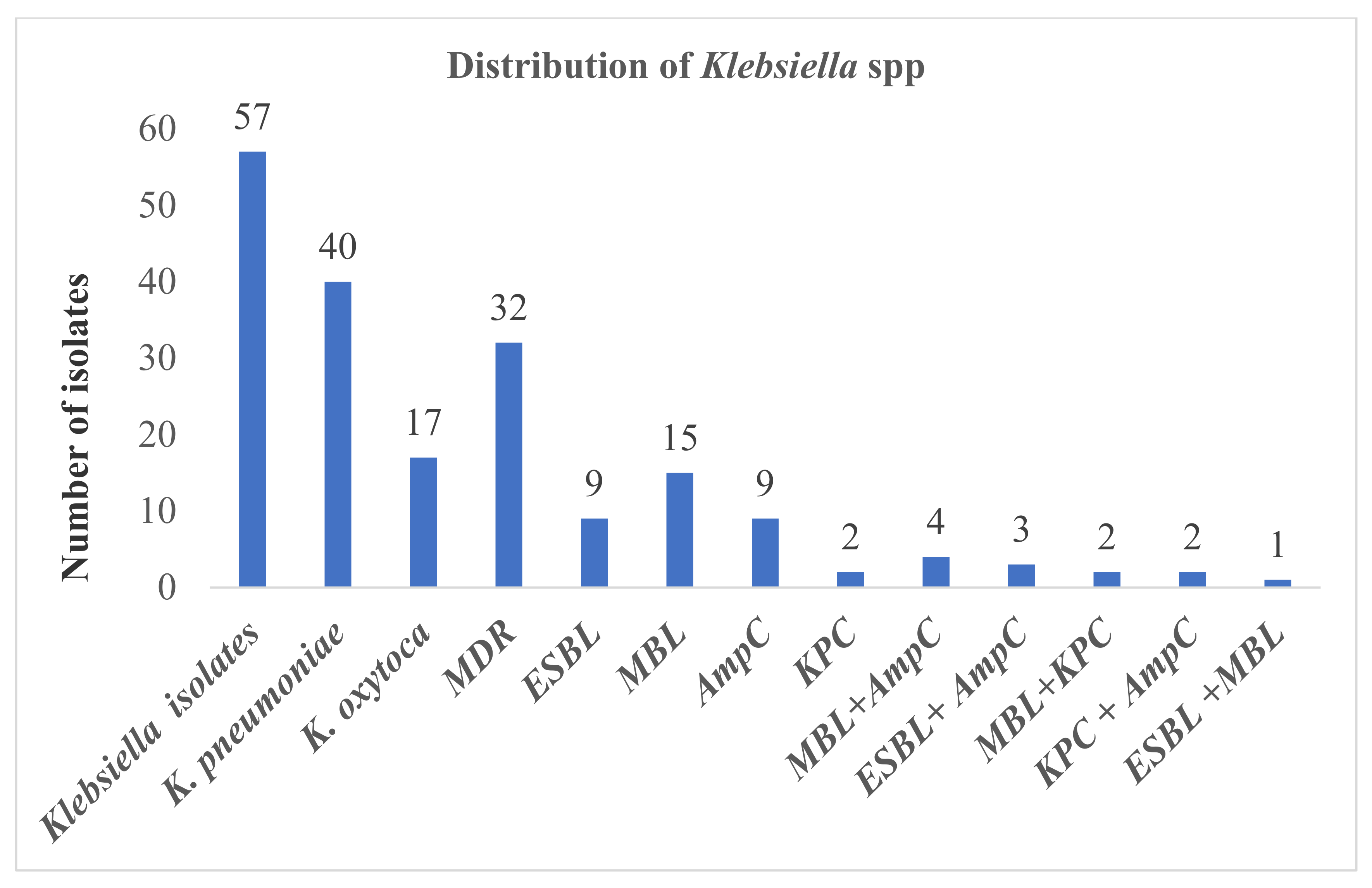
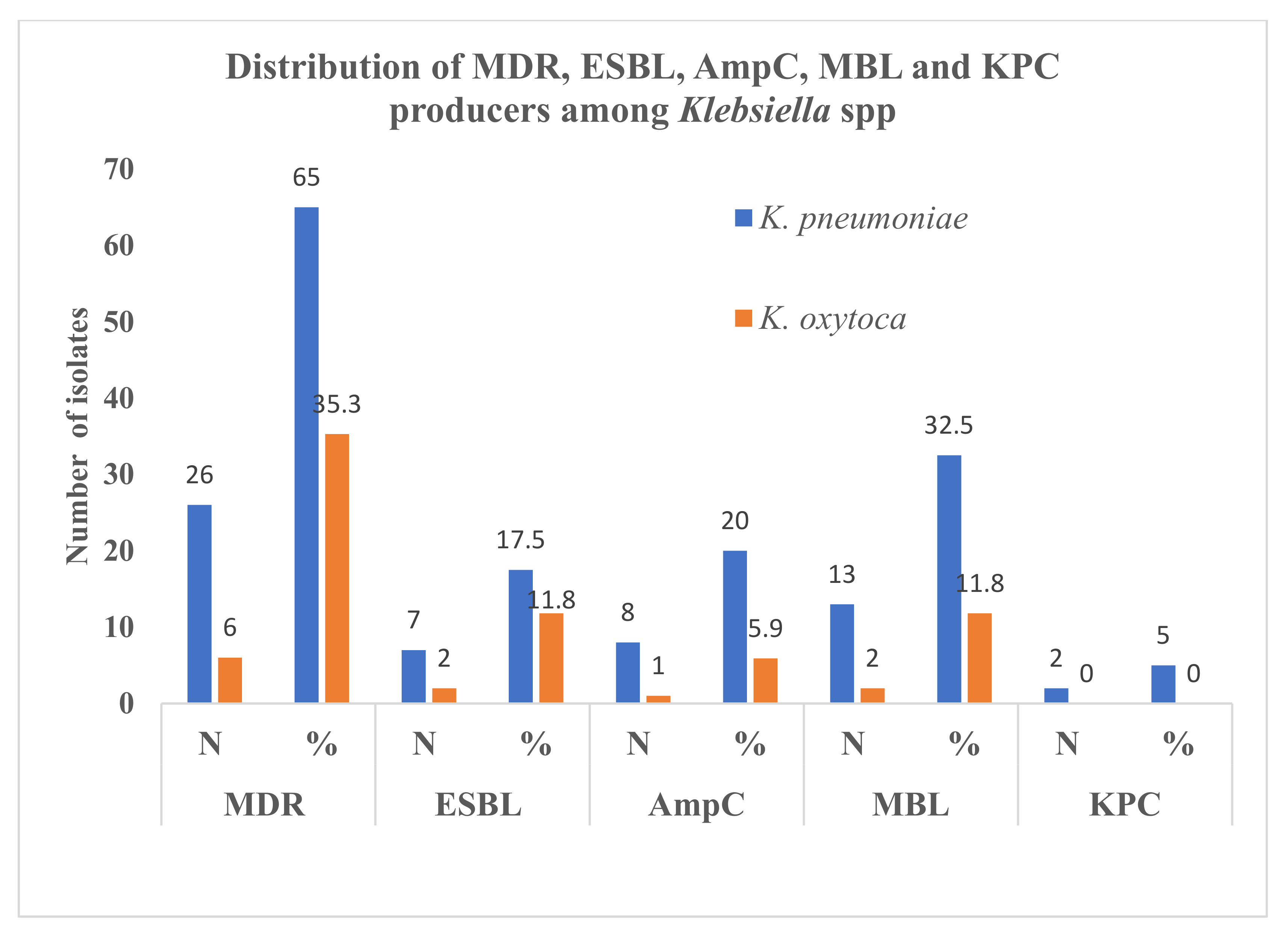
3.3. ESBL, AmpC, MBL, KPC Production among Klebsiella spp.
3.4. Distribution of ESBL, AmpC, MBL and KPC Producers in Differential Specimens and Age Groups
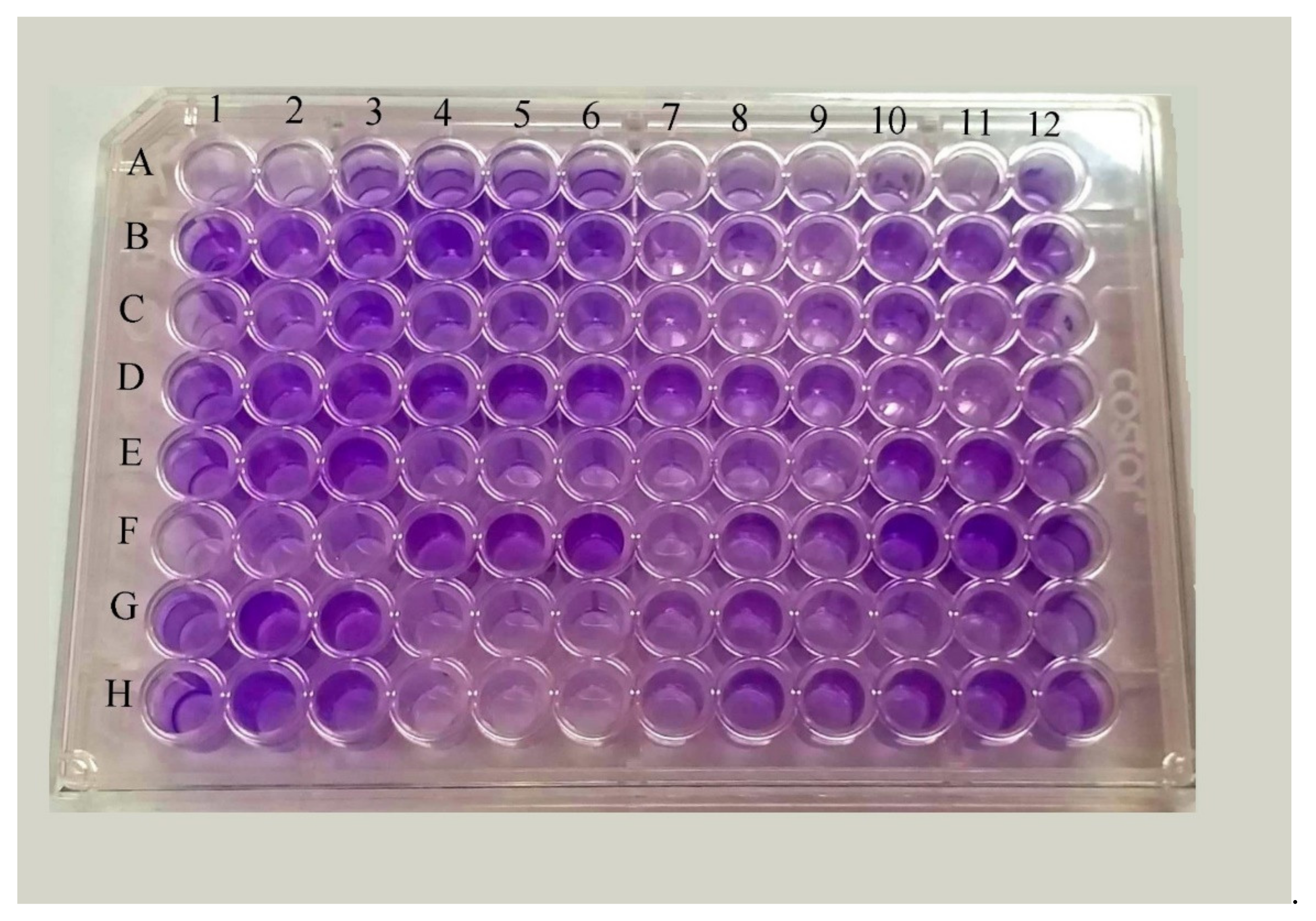
3.5. Detection of Biofilm
3.6. Biofilm Production among ESBL, AmpC, MBL and KPC Producing Klebsiella spp.
3.7. Co-Production of ESBL and AmpC, ESBL and Carbapenemase (MBL and KPC), MBL and KPC, MBL and AmpC and KPC and AmpC
4. Discussion
Strengths and Limitations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Consent for Publication
Abbreviations
References
- Afzal, A.M.S. Antibiotic resistant pattern of E. coli and Klebsiella species in Pakistan: Brief overview. J. Microb. Biochem. Technol. 2017, 9, 277–279. [Google Scholar]
- Martínez, J.; Martínez, L.; Rosenblueth, M.; Silva, J.; Martinez-Romero, E. How are gene sequence analyses modifying bacterial taxonomy? The case of Klebsiella. Int. Microbiol. 2004, 7, 261–268. [Google Scholar] [PubMed]
- Podschun, R.; Ullmann, U. Klebsiella spp. as nosocomial pathogens: Epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin. Microbiol. Rev. 1998, 11, 589–603. [Google Scholar] [CrossRef] [Green Version]
- Namratha, K.G.; Sreeshma, P.; Subbannayya, K.; Dinesh, P.V.; Champa, H. Characterization and antibiogram of Klebsiella spp. isolated from clinical specimen in a rural teaching hospital. Sch. J. App. Med. Sci. 2015, 3, 878–883. [Google Scholar]
- Lederman, E.R.; Crum, N.F. Pyogenic liver abscess with a focus on Klebsiella pneumoniae as a primary pathogen: An emerging disease with unique clinical characteristics. Am. J. Gastroenterol. 2005, 100, 322–331. [Google Scholar] [CrossRef]
- Bonelli, R.R.; Moreira, B.M.; Picao, R.C. Antimicrobial resistance among Enterobacteriaceae in South America: History, current dissemination status and associated socioeconomic factors. Drug Resist. Updates 2014, 17, 24–36. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization (WHO). Antimicrobial Resistance: Global Report on Surveillance. 2014. Geneva. Available online: https://www.who.int/antimicrobial-resistance/publications/surveillancereport/en/ (accessed on 15 March 2020).
- Paczosa, M.K.; Mecsas, J. Klebsiella pneumoniae: Going on the Offense with a Strong Defense. Microbiol. Mol. Biol. Rev. 2016, 80, 629–661. [Google Scholar] [CrossRef] [Green Version]
- Azzopardi, E.A.; Ferguson, E.L.; Thomas, D.W. The enhanced permeability retention effect: A new paradigm for drug targeting in infection. J. Antimicrob. Chemother. 2013, 68, 257–274. [Google Scholar] [CrossRef] [Green Version]
- Macia, M.D.; Rojo-Molinero, E.; Oliver, A. Antimicrobial susceptibility testing in biofilm-growing bacteria. Clin. Microbiol. Infect. 2014, 20, 981–990. [Google Scholar] [CrossRef] [Green Version]
- Sadekuzzaman, M.; Yang, S.; Mizan, F.R.; Ha, S.; Sadekuzzaman, M.; Yang, S.; Mizan, F.R.; Ha, S. Current and recent advanced strategies for combating biofilms. Compr. Rev. Food Sci. Food Saf. 2015, 14, 491–509. [Google Scholar] [CrossRef]
- Boddicker, J.D.; Anderson, R.A.; Jagnow, J.; Clegg, S. Signature-tagged mutagenesis of Klebsiella pneumoniae to identify genes that influence biofilm formation on extracellular matrix material. Infect. Immun. 2006, 74, 4590–4597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balestrino, D.; Ghigo, J.M.; Charbonnel, N.; Haagensen, J.A.; Forestier, C. The characterization of functions involved in the establishment and maturation of Klebsiella pneumoniae in vitro biofilm reveals dual roles for surface exopolysaccharides. Environ. Microbiol. 2008, 10, 685–701. [Google Scholar] [CrossRef]
- Raut, S.; Gokhale, S.; Adhikari, B. Prevalence of extended spectrum beta-lactamases among E. coli and Klebsiella spp. isolates in Manipal Teaching Hospital, Pokhara, Nepal. J. Microbiol. Infect. Dis. 2015, 5, 69–75. [Google Scholar] [CrossRef]
- Yusuf, I.; Haruna, M.; Yahaya, H. Prevalence and antibiotic susceptibility of AmpC and ESBL producing clinical isolates at a tertiary health care center in Kano, North west Nigeria. Afr. J. Exp. Microbiol. 2013, 14, 109–119. [Google Scholar] [CrossRef] [Green Version]
- Paterson, D.L. Resistance in gram-negative bacteria: Enterobacteriaceae. Am. J. Med. 2006, 119, S20–S28. [Google Scholar] [CrossRef] [PubMed]
- Kuenzli, E.; Jaeger, V.K.; Frei, R.; Neumayr, A.; DeCrom, S.; Haller, S.; Blum, J.; Widmer, A.F.; Furrer, H.; Battegay, M.; et al. High colonization rates of extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli in Swiss travellers to South Asia- a prospective observational multicentre cohort study looking at epidemiology, microbiology and risk factors. BMC Infect. Dis. 2014, 14, 528. [Google Scholar] [CrossRef] [Green Version]
- Harris, P.N.; Peleg, A.Y.; Iredell, J.; Ingram, P.R.; Miyakis, S.; Stewardson, A.J.; Rogers, B.A.; McBryde, E.S.; Roberts, J.A.; Lipman, J.; et al. Meropenem versus piperacillin-tazobactam for definitive treatment of bloodstream infections due to ceftriaxone non-susceptible Escherichia coli and Klebsiella spp (the MERINO trial): Study protocol for a randomised controlled trial. Trials 2015, 16, 24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azimi, L.; Rastegar-Lari, A.; Talebi, M.; Ebrahimzadeh-Namvar, A.; Soleymanzadeh-Moghadam, S. Evaluation of phenotypic methods for detection of Klebsiella pneumoniae carbapenemase-producing K. pneumoniae in Tehran. J. Med. Bacteriol. 2013, 2, 26–31. [Google Scholar]
- Chaudhary, A.K.; Bhandari, D.; Amatya, J.; Chaudhary, P.; Acharya, B. Metallo-beta-lactamase producing Gram negative bacteria among patients visiting Shahid Gangalal National Heart Central. Austin J. Microbiol. 2016, 2, 1010. [Google Scholar]
- Mishra, S.K.; Acharya, J.; Kattel, H.P.; Koirala, J.; Rijal, B.P.; Pokhrel, B.M.; Mishra, S.K.; Acharya, J.; Kattel, H.P.; Koirala, J.; et al. Metallo-beta-lactamase producing gram-negative bacterial isolates. J. Nepal Health Res. Counc. 2012, 10, 208–213. [Google Scholar] [PubMed]
- Marchaim, D.; Navon-Venezia, S.; Schwaber, M.J.; Carmeli, Y. Isolation of imipenem-resistant Enterobacter species: Emergence of KPC-2 carbapenemase, molecular characterization, epidemiology, and outcomes. Antimicrob. Agents Chemother. 2008, 52, 1413–1418. [Google Scholar] [CrossRef] [Green Version]
- Fisher, M.A.; Stamper, P.D.; Hujer, K.M.; Love, Z.; Croft, A.; Cohen, S.; Bonomo, R.A.; Carroll, K.C.; Petti, C.A. Performance of the Phoenix bacterial identification system compared with disc diffusion methods for identifying extended-spectrum beta-lactamase, AmpC and KPC producers. J. Med. Microbiol. 2009, 58, 774–778. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanchez, G.V.; Master, R.N.; Clark, R.B.; Fyyaz, M.; Duvvuri, P.; Ekta, G.; Bordon, J. Klebsiella pneumoniae antimicrobial drug resistance, United States, 1998–2010. Emerg. Infect. Dis. 2013, 19, 133–136. [Google Scholar] [CrossRef] [PubMed]
- Kayastha, K.; Dhungel, B.; Karki, S.; Adhikari, B.; Banjara, M.R.; Rijal, K.R.; Ghimire, P. Extended-Spectrum beta-Lactamase-Producing Escherichia coli and Klebsiella species in pediatric patients visiting International Friendship Children’s Hospital, Kathmandu, Nepal. Infect. Dis. (Auckl.) 2020, 13, 1178633720909798. [Google Scholar] [PubMed] [Green Version]
- Dhungana, K.; Awal, B.K.; Dhungel, B.; Sharma, S.; Banjara, M.R.; Rijal, K.R. Detection of Klebsiella pneumoniae carbapenemase (KPC) and metallo betalactamae (MBL) producing Gram negative bacteria isolated from different clinical samples in a Transplant Center, Kathmandu, Nepal. ASMI 2019, 2, 60–69. [Google Scholar]
- Guragain, N.; Pradhan, A.; Dhungel, B.; Banjara, M.R.; Rijal, K.R.; Ghimire, P. Extended spectrum B-lactamase producing Gram negative bacterial isolates from urine of patients visiting Everest Hospital, Kathmandu, Nepal. TUJM 2019, 6, 26–31. [Google Scholar] [CrossRef]
- Raut, S.; Rijal, K.R.; Khatiwada, S.; Karna, S.; Khanal, R.; Adhikari, J.; Adhikari, B. Trend and characteristics of Acinetobacter baumannii infections in patients attending Universal College of Medical Sciences, Bhairahawa, Western Nepal: A longitudinal study of 2018. Infect. Drug Resist. 2020, 13, 1631–1641. [Google Scholar] [CrossRef]
- Gurung, S.; Kafle, S.; Dhungel, B.; Adhikari, N.; Shrestha, U.T.; Adhikari, B.; Banjara, M.R.; Rijal, K.R.; Ghimire, P. Detection of OXA-48 gene in carbapenem-resistant Escherichia coli and Klebsiella pneumoniae from urine samples. Infect. Drug Resist. 2020, 13, 2311–2321. [Google Scholar] [CrossRef] [PubMed]
- Parajuli, N.P.; Acharya, S.P.; Mishra, S.K.; Parajuli, K.; Rijal, B.P.; Pokhrel, B.M. High burden of antimicrobial resistance among Gram negative bacteria causing healthcare associated infections in a critical care unit of Nepal. Antimicrob. Resist. Infect. Control 2017, 6, 67. [Google Scholar] [CrossRef] [Green Version]
- Isenberg, H.D. Clinical Microbiology Procedures Handbook, 2nd ed.; ASM Press: Washington, DC, USA, 2004. [Google Scholar]
- Cheesbrough, M. District Laboratory Practice in Tropical Countries, 2nd ed.; Cambridge University Press: Cambridge, UK, 2005. [Google Scholar]
- Forbes, B.A.; Sahm, D.F.; Weissfelt, S.A. Bailey and Scott’s Diagnostic Microbiology; Mosby Publication: St. Louis, MO, USA, 2007. [Google Scholar]
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing, 28th ed.; Informational supplement M100-S28; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018. [Google Scholar]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [Green Version]
- Panchal, C.A.; Oza, S.S.; Mehta, S.J. Comparison of four phenotypic methods for detection of metallo-beta-lactamase-producing Gram-negative bacteria in rural teaching hospital. J. Lab. Physicians 2017, 9, 81–83. [Google Scholar]
- Tsakris, A.; Poulou, A.; Pournaras, S.; Voulgari, E.; Vrioni, G.; Themeli-Digalaki, K.; Petropoulou, D.; Sofianou, D. A simple phenotypic method for the differentiation of metallo-beta-lactamases and class A KPC carbapenemases in Enterobacteriaceae clinical isolates. J. Antimicrob. Chemother. 2010, 65, 1664–1671. [Google Scholar] [CrossRef] [Green Version]
- Coudron, P.E. Inhibitor-based methods for detection of plasmid-mediated AmpC beta-lactamases in Klebsiella spp., Escherichia coli, and Proteus mirabilis. J. Clin. Microbiol. 2005, 43, 4163–4167. [Google Scholar] [CrossRef] [Green Version]
- Stepanović, S.; Vuković, D.; Dakić, I.; Savić, B.; Švabić-Vlahović, M. A modified microtiter-plate test for quantification of Staphylococcal biofilm formation. J. Microbiol. Methods 2000, 40, 175–179. [Google Scholar] [CrossRef]
- Lamichhane, K.; Adhikari, N.; Bastola, A.; Devkota, L.; Bhandari, P.; Dhungel, B.; Shrestha, U.T.; Adhikari, B.; Banjara, M.R.; Rijal, K.R.; et al. Biofilm-Producing Candida Species Causing Oropharyngeal Candidiasis in HIV Patients Attending Sukraraj Tropical and Infectious Diseases Hospital in Kathmandu, Nepal. HIV AIDS (Auckl.) 2020, 12, 211–220. [Google Scholar] [CrossRef] [PubMed]
- Poudyal, S.; Bhatta, D.R.; Shakya, G.; Upadhyaya, B.; Dumre, S.P.; Buda, G.; Kandel, B.P. Extended spectrum a-lactamase producing multidrug resistant clinical bacterial isolates at National Public Health Laboratory, Nepal. Nepal Med. Coll. J. 2011, 13, 34–38. [Google Scholar] [PubMed]
- Bina, M.; Pournajaf, A.; Mirkalantari, S.; Talebi, M.; Irajian, G. Detection of the Klebsiella pneumoniae carbapenemase (KPC) in K. pneumoniae isolated from the clinical samples by the phenotypic and genotypic methods. Iran. J. Pathol. 2015, 10, 199–205. [Google Scholar] [PubMed]
- Sah, R.S.P.; Dhungel, B.; Yadav, B.K.; Adhikari, N.; Thapa Shrestha, U.; Lekhak, B.; Banjara, M.R.; Adhikari, B.; Ghimire, P.; Rijal, K.R. Detection of TEM and CTX-M Genes in Escherichia coli Isolated from Clinical Specimens at Tertiary Care Heart Hospital, Kathmandu, Nepal. Diseases 2021, 9, 15. [Google Scholar] [CrossRef]
- Tamang, K.; Shrestha, P.; Koirala, A.; Khadka, J.; Gautam, N.; Rijal, K.R. Prevalence of bacterial uropathogens among diabetic patients attending Padma Nursing Hospital of Western Nepal. HiJOST 2017, 1, 15–19. [Google Scholar] [CrossRef]
- Thakur, P.; Ghimire, P.; Rijal, K.R.; Singh, G.K. Antimicrobial resistance pattern of Escherichia coli isolated from urine samples in patients visiting tertiary health care centre in eastern Nepal. Sunsari Tech. Coll. J. 2012, 1, 22–26. [Google Scholar] [CrossRef] [Green Version]
- Adhikari, S.; Khadka, S.; Rana, J.C.; Baniya, S.; Poudel, S.; Chapagain, A.; Regmi, R. Prevalence of β-Lactamase producing carbapenem-resistant Enterobacteriaceae among the patients attending Bharatpur Hospital. Biosci. Dis. 2019, 10, 64–71. [Google Scholar]
- Pérez-Vazquez, M.; Oteo-Iglesias, J.; Sola-Campoy, P.J.; Carrizo-Manzoni, H.; Bautista, V.; Lara, N.; Aracil, B.; Alhambra, A.; Martínez-Martínez, L.; Campos, J.; et al. Characterization of carbapenemase-producing Klebsiella oxytoca in Spain, 2016–2017. Antimicrob. Agents Chemother. 2019, 63, e02529-18. [Google Scholar] [CrossRef] [Green Version]
- Munoz-Davila, M.J. Role of old antibiotics in the era of antibiotic resistance. Highlighted nitrofurantoin for the treatment of lower urinary tract infections. Antibiotics 2014, 3, 39–48. [Google Scholar] [CrossRef] [Green Version]
- Rijal, K.R.; Banjara, M.R.; Dhungel, B.; Kafle, S.; Gautam, K.; Ghimire, B.; Ghimire, P.; Dhungel, S.; Adhikari, N.; Shrestha, U.T.; et al. Use of antimicrobials and antimicrobial resistance in Nepal: A nationwide survey. Sci. Rep. 2021, 11, 11554. [Google Scholar] [CrossRef]
- Nirwati, H.; Sinanjung, K.; Fahrunissa, F.; Wijaya, F.; Napitupulu, S.; Hati, V.P.; Hakim, M.S.; Meliala, A.; Aman, A.T.; Nuryastuti, T. Biofilm formation and antibiotic resistance of Klebsiella pneumoniae isolated from clinical samples in a tertiary care hospital, Klaten, Indonesia. BMC Proc. 2019, 13, 20. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, R.L.; da Silva, B.; Rezende, G.S.; Nakamura-Silva, R.; Pitondo-Silva, A.; Campanini, E.B.; Brito, M.C.; da Silva, E.M.; Freire, C.C.; Cunha, A.F.; et al. High prevalence of multidrug-resistant Klebsiella pneumoniae harboring several virulence and beta-lactamase encoding genes in a Brazilian Intensive Care Unit. Front. Microbiol. 2018, 9, 3198. [Google Scholar] [CrossRef] [Green Version]
- Upadhyay, A.K.; Parajuli, P. Extended spectrum β-lactamases producing Klebsiella species isolated at National Medical College and Teaching Hospital, Nepal. Asian J. Pharma Clin. Res. 2013, 1, 161–164. [Google Scholar]
- Shrestha, U.T.; Shrestha, S.; Adhikari, N.; Rijal, K.R.; Shrestha, B.; Adhikari, B.; Banjara, M.R.; Ghimire, P. Plasmid profiling and occurrence of β-lactamase enzymesin multidrug-resistant uropathogenic Escherichia coli in Kathmandu, Nepal. Infect. Drug Resist. 2020, 13, 1905–1917. [Google Scholar]
- Shu, J.C.; Chia, J.H.; Kuo, A.J.; Su, L.H.; Wu, T.L. A 7-year surveillance for ESBL-producing Escherichia coli and Klebsiella pneumoniae at a university hospital in Taiwan: The increase of CTX-M-15 in the ICU. Epidemiol. Infect. 2010, 138, 253–263. [Google Scholar] [CrossRef]
- Jain, A.; Roy, I.; Gupta, M.K.; Kumar, M.; Agarwal, S.K. Prevalence of extended-spectrum beta-lactamase-producing Gram-negative bacteria in septicaemic neonates in a tertiary care hospital. J. Med. Microbiol. 2003, 52, 421–425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baral, P.; Neupanea, S.; Shresthac, B.; Ghimirea, K.R.; Marasinia, B.P.; Lekhaka, B. Clinical and microbiological observational study on AmpC beta-lactamase-producing Enterobacteriaceae in a hospital of Nepal. Braz. J. Infect. Dis. 2013, 17, 256–259. [Google Scholar] [CrossRef] [Green Version]
- Bhandari, P.; Thapa, G.; Pokhrel, B.M.; Bhatta, D.R.; Devkota, U. Nosocomial isolates and their drug resistant pattern in ICU patients at National Institute of Neurological and Allied Sciences, Nepal. Int. J. Microbiol. 2015, 2015, 572163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meletis, G. Carbapenem resistance: Overview of the problem and future perspectives. Adv. Infect. Dis 2016, 3, 15–21. [Google Scholar] [CrossRef] [Green Version]
- Gupta, V.; Bansal, N.; Singla, N.; Chander, J. Occurrence and phenotypic detection of class A carbapenemases among Escherichia coli and Klebsiella pneumoniae blood isolates at a tertiary care center. J. Microbiol. Immunol. Infect. 2013, 46, 104–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chauhan, K.; Pandey, A.; Asthana, A.K.; Madan, M. Evaluation of phenotypic tests for detection of Klebsiella pneumoniae carbapenemase and metallo-beta-lactamase in clinical isolates of Escherichia coli and Klebsiella species. Indian J. Pathol. Microbiol. 2015, 58, 31–35. [Google Scholar]
- Mohamudha Parveen, R.; Harish, B.N.; Parija, S.C. AmpC Beta lactamases among gram negative clinical isolates from a tertiary hospital, South India. Braz. J. Microbiol. 2010, 41, 596–602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khanal, S.; Joshi, D.R.; Bhatta, D.R.; Devkota, U.; Pokhrel, B.M. Beta-lactamase-producing multidrug-resistant bacterial pathogens from tracheal aspirates of Intensive Care Unit patients at National Institute of Neurological and Allied Sciences, Nepal. ISRN Microbiol. 2013, 2013, 847569. [Google Scholar] [CrossRef] [Green Version]
- Rahdar, H.A.; Malekabad, E.S.; Dadashi, A.-R.; Takei, E.; Keikha, M.; Kazemian, H.; Karami-Zarandi, M. Correlation between biofilm formation and carbapenem resistance among clinical isolates of Klebsiella pneumoniae. Ethiop. J. Health Sci. 2019, 29, 745–750. [Google Scholar] [CrossRef]
- Sanchez, C.J., Jr.; Mende, K.; Beckius, M.L.; Akers, K.S.; Romano, D.R.; Wenke, J.C.; Murray, C.K. Biofilm formation by clinical isolates and the implications in chronic infections. BMC Infect. Dis. 2013, 13, 47. [Google Scholar] [CrossRef] [Green Version]
- Thapa, S.; Neopane, P.; Shrestha, R. Biofilm formation and antimicrobial resistance in Klebsiella pneumoniae isolated from patient visiting a tertiary care center of Nepal. Asian Pac. J. Trop. Dis. 2017, 7, 347–351. [Google Scholar]
| Specimens/Character | Culture Positive | |
|---|---|---|
| Number | Percentage | |
| Clinical specimens | ||
| Urine (n = 2032) | 140 | 80 |
| Sputum (n = 150) | 28 | 16 |
| Pus (n = 15) | 7 | 4 |
| Gram type | ||
| Gram-negative bacteria | 151 | 86.3 |
| Gram-positive bacteria | 24 | 13.7 |
| Different type of Gram-positive bacteria (n = 24) | ||
| S. aureus | 15 | 62.5 |
| Enterococcus spp. | 8 | 33.3 |
| S. saprophyticus | 1 | 4.2 |
| Different type of Gram-negative bacteria (n = 151) | ||
| E. coli | 62 | 41.1 |
| K. pneumoniae | 40 | 26.5 |
| K. oxytoca | 17 | 11.3 |
| P. aeruginosa | 16 | 10.5 |
| Enterobacter aerogenes | 5 | 3.3 |
| Citrobacter fruendii | 4 | 2.6 |
| Morganella morganii | 2 | 1.3 |
| Providencia spp. | 2 | 1.3 |
| Proteus spp. | 1 | 0.7 |
| Haemophilus spp. | 1 | 0.7 |
| Acinetobacter baumannii | 1 | 0.7 |
| Gender | ||
| Male | 92 | 52.5 |
| Female | 83 | 47.5 |
| Age groups (Years) | ||
| 0–15 | 10 | 5.7 |
| 16–45 | 80 | 45.7 |
| 46–60 | 45 | 25.7 |
| >60 | 40 | 22.9 |
| Character | K. pneumoniae (n = 40) | K. oxytoca (n = 17) | Total |
|---|---|---|---|
| N (%) | N (%) | N (%) | |
| Clinical specimens | |||
| Urine | 25 (62.5) | 12 (70.6) | 37 (64.9) |
| Sputum | 13 (32.5) | 5 (29.4) | 18 (31.6) |
| Pus | 2 (5.0) | 0 | 2 (3.5) |
| Gender | |||
| Male | 23 (57.5) | 9 (52.9) | 32 (56.1) |
| Female | 17 (42.5) | 8 (47.1) | 25 (43.9) |
| Age groups (in years) | |||
| 0–15 | 1 (2.5) | 0 | 1 (1.8) |
| 15–45 | 13 (32.5) | 7 (41.2) | 20 (35.1) |
| 45–60 | 10 (25) | 5 (29.4) | 15 (26.3) |
| >60 | 16 (40) | 5 (29.4) | 21 (36.8) |
| Mode of Action | Antimicrobial Category | Antibiotics | K. pneumoniae (n = 57) | ||
|---|---|---|---|---|---|
| Sensitive | Intermediate | Resistant | |||
| N (%) | N (%) | N (%) | |||
| Cell wall synthesis inhibitors | Aminopenicillin | Amoxiclav (30 µg) | 19 (33.3) | 4 (7.1) | 34 (59.6) |
| Extended spectrum cephalosporins | Ceftriaxone (30 µg) | 26 (45.6) | 2 (3.5) | 29 (50.9) | |
| Cefoxitin (30 µg) | 21 (36.8) | 4 (7.0) | 32 (56.2) | ||
| Cefotaxime (30 µg) | 26(45.6) | 1(1.8) | 30 (52.6) | ||
| Carbapenems | Imipenem (10 µg) | 20 (35.0) | 5 (8.8) | 32 (56.2) | |
| Meropenem (10 µg) | 20 (35.0) | 5 (8.8) | 32 (56.2) | ||
| Penicillins and beta-lactamase inhibitors | Piperacillin-tazobactam (100/10 µg) | 20 (35.0) | 6 (10.5) | 31 (54.5) | |
| Protein synthesis inhibitors | Aminoglycosides | Amikacin (30 µg) | 25 (43.9) | 3 (5.3) | 29 (50.8) |
| Gentamicin (10 µg) | 39 (68.4) | 1 (1.8) | 17 (29.8) | ||
| Tetracyclines | Tetracycline (30 µg) | 26 (45.6) | 5 (8.8) | 26 (45.6) | |
| Nucleic acid synthesis inhibitors | Fluoroquinolones | Ciprofloxacin (5 µg) | 24 (42.1) | 3 (5.3) | 30 (52.6) |
| Norfloxacin (10 µg) | 20 (35.1) | 2 (3.5) | 35 (61.4) | ||
| Levofloxacin (5 µg) | 21 (36.8) | 4 (7.0) | 32 (56.2) | ||
| Nitrofurans | Nitrofurantoin (300 µg) | 26 (45.6) | 6 (10.5) | 25 (43.8) | |
| Folate pathway inhibitors | Trimethoprim and Sulfamethoxazole | Cotrimoxazole (25 µg) | 27 (47.4) | 1 (1.8) | 29 (50.9) |
| Character | MDR | ESBL | AmpC | MBL | KPC |
|---|---|---|---|---|---|
| N (%) | N (%) | N (%) | N (%) | N (%) | |
| Clinical specimens | |||||
| Urine | 20 (62.5) | 7 (77.8) | 5 (55.6) | 9 (60) | 1 (50) |
| Sputum | 10 (31.3) | 0 (0) | 3 (33.3) | 6 (40) | 1 (50) |
| Pus | 2 (6.3) | 2 (22.2) | 1 (11.1) | 0 (0) | 0 (0) |
| p value | 0.44 | 0.07 | 0.74 | 0.28 | 0.8 |
| Gender | |||||
| Male | 21 (65.6) | 8 (88.9) | 4 (44.4) | 9 (60) | 0 |
| Female | 11 (34.4) | 1 (11.1) | 5 (55.6 | 6 (40) | 2 (100) |
| p value | 0.11 | 0.7 | 0.11 | 0.52 | 0.04 |
| Age groups (in years) | |||||
| 0–15 | 1 (3.1) | 0 (0) | 0 (0) | 1 (6.7) | 0 (0) |
| 16–45 | 12 (37.5) | 4 (44.4) | 5 (55.5) | 6 (40) | 1 (50) |
| 46–60 | 8 (25) | 2 (22.2) | 3 (33.4) | 3 (20) | 1 (50) |
| >60 | 11 (34.4) | 3 (33.3) | 1 (11.1) | 5 (33.3) | 0 (0) |
| p value | 0.63 | 0.83 | 0.15 | 0.55 | 0.52 |
| Character | Biofilm Producers | Biofilm Non-Producers | |||
|---|---|---|---|---|---|
| Strong | Moderate | Weak | Total Producer | Non-Producer | |
| N (%) | N (%) | N (%) | N (%) | N (%) | |
| Klebsiella spp. (K. pneumoniae and K. oxytoca) | 4 (7%) | 34 (59.7%) | 16 (28.1%) | 54 (94.8%) | 3 (5.2%) |
| K. pneumoniae | 2 (50) | 24 (70.6) | 13 (81.3) | 39 (72.2) | 1 (33.3) |
| K. oxytoca | 2 (50) | 10 (29.4) | 3 (18.8) | 17 (27.8) | 2 (66.7) |
| Clinical specimens | |||||
| Urine | 4 (100) | 19 (55.9) | 12 (75) | 35 (64.8) | 2 (66.7) |
| Sputum | 0 (0) | 13 (38.2) | 4 (24) | 17 (31.5) | 1 (33.3) |
| Pus | 0 (0) | 2 (5.9) | 0 (0) | 2 (3.7) | 0 (0) |
| Gender | |||||
| Male | 1 (25) | 20 (58.8) | 9 (56.3) | 30 (55.6) | 2 (66.7) |
| Female | 3 (75) | 14 (41.2) | 7 (43.8) | 24 (44.4) | 1 (33.3) |
| Age group (in years) | |||||
| 0–15 | 0 (0) | 0 (0) | 1 (6.3) | 1 (1.9) | 0 (0) |
| 16–45 | 2 (50) | 12 (35.3) | 5 (31.3) | 19 (35.2) | 1 (33.3) |
| 46–60 | 1 (25) | 10 (29.4) | 4 (25) | 15 (27.7) | 0 (0) |
| >60 | 1 (25) | 12 (35.3) | 6 (37.5) | 19 (35.2) | 2 (66.7) |
| Relation of biofilm production according to MDR, ESBL, MBL, AmpC and KPC production | |||||
| MDR producers | 1 (3.1) | 21 (65.6) | 8 (25) | 30 (93.7) | 2 (6.3) |
| ESBL producers | 1 (11.1) | 5 (55.6) | 3 (33.3) | 9 (100) | 0 (0) |
| MBL producers | 1 (6.7) | 8 (53.3) | 5 (33.3) | 14 (93.3) | 1 (6.7) |
| AmpC producers | 0 (0) | 7 (77.8) | 2 (22.2) | 9 (100) | 0 (0) |
| KPC producers | 0 (0) | 2 (100) | 0 (0) | 2 (100) | 0 (0) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kuinkel, S.; Acharya, J.; Dhungel, B.; Adhikari, S.; Adhikari, N.; Shrestha, U.T.; Banjara, M.R.; Rijal, K.R.; Ghimire, P. Biofilm Formation and Phenotypic Detection of ESBL, MBL, KPC and AmpC Enzymes and Their Coexistence in Klebsiella spp. Isolated at the National Reference Laboratory, Kathmandu, Nepal. Microbiol. Res. 2021, 12, 683-697. https://doi.org/10.3390/microbiolres12030049
Kuinkel S, Acharya J, Dhungel B, Adhikari S, Adhikari N, Shrestha UT, Banjara MR, Rijal KR, Ghimire P. Biofilm Formation and Phenotypic Detection of ESBL, MBL, KPC and AmpC Enzymes and Their Coexistence in Klebsiella spp. Isolated at the National Reference Laboratory, Kathmandu, Nepal. Microbiology Research. 2021; 12(3):683-697. https://doi.org/10.3390/microbiolres12030049
Chicago/Turabian StyleKuinkel, Susmita, Jyoti Acharya, Binod Dhungel, Sanjib Adhikari, Nabaraj Adhikari, Upendra Thapa Shrestha, Megha Raj Banjara, Komal Raj Rijal, and Prakash Ghimire. 2021. "Biofilm Formation and Phenotypic Detection of ESBL, MBL, KPC and AmpC Enzymes and Their Coexistence in Klebsiella spp. Isolated at the National Reference Laboratory, Kathmandu, Nepal" Microbiology Research 12, no. 3: 683-697. https://doi.org/10.3390/microbiolres12030049
APA StyleKuinkel, S., Acharya, J., Dhungel, B., Adhikari, S., Adhikari, N., Shrestha, U. T., Banjara, M. R., Rijal, K. R., & Ghimire, P. (2021). Biofilm Formation and Phenotypic Detection of ESBL, MBL, KPC and AmpC Enzymes and Their Coexistence in Klebsiella spp. Isolated at the National Reference Laboratory, Kathmandu, Nepal. Microbiology Research, 12(3), 683-697. https://doi.org/10.3390/microbiolres12030049







