Serotonin Exposure Improves Stress Resistance, Aggregation, and Biofilm Formation in the Probiotic Enterococcus faecium NCIMB10415
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strain and Growth Conditions
2.2. Growth Kinetics
2.3. Phenotypic Evaluations
2.3.1. Resistance to Bile Salts
2.3.2. Auto-Aggregation
2.3.3. Biofilm Formation
2.3.4. Antibiotic Susceptibility Test
2.4. Whole Cell Proteomic Analyses
2.4.1. Soluble Proteins Extraction
2.4.2. In-Solution Protein Digestion
2.4.3. SWATH-MS Analysis
2.4.4. Protein Data Search
2.4.5. Protein Quantification
2.4.6. Protein Classification
2.5. Immune-Stimulating Activity of Killed E. faecium NCIMB 104145 Cells and Cell Free Supernatants
2.5.1. Preparation of the Bacterial Strain and Cell-Free Culture Supernatant for Dendritic Cells (DC) Maturation Assays
2.5.2. Monocyte-Derived DC Preparation
2.5.3. Monocyte-Derived DC Stimulation
2.5.4. Flow Cytometric Analysis
2.6. Statistical Analyses
3. Results
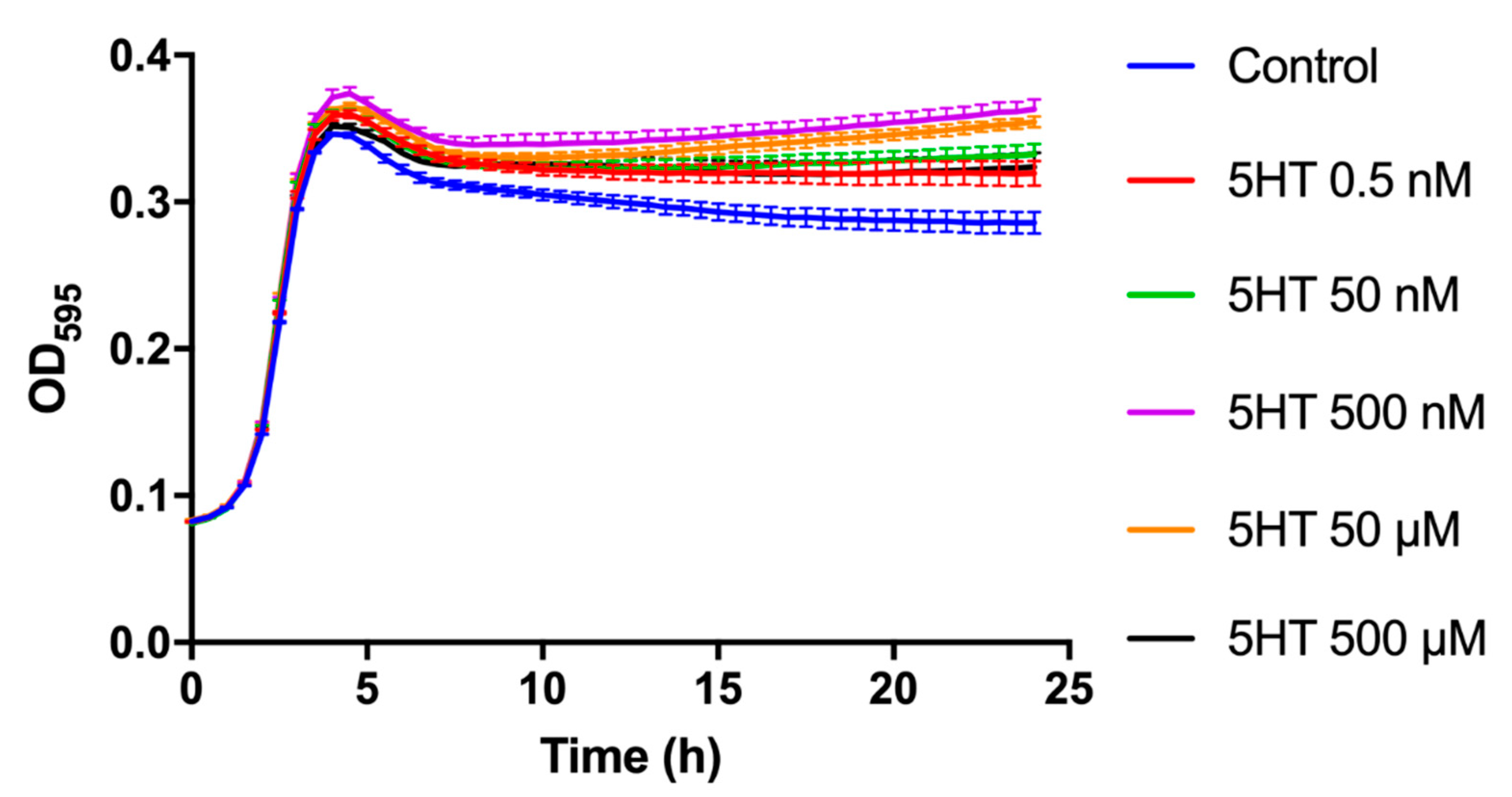
3.1. Growth Kinetics
3.2. Phenotypic Evaluations
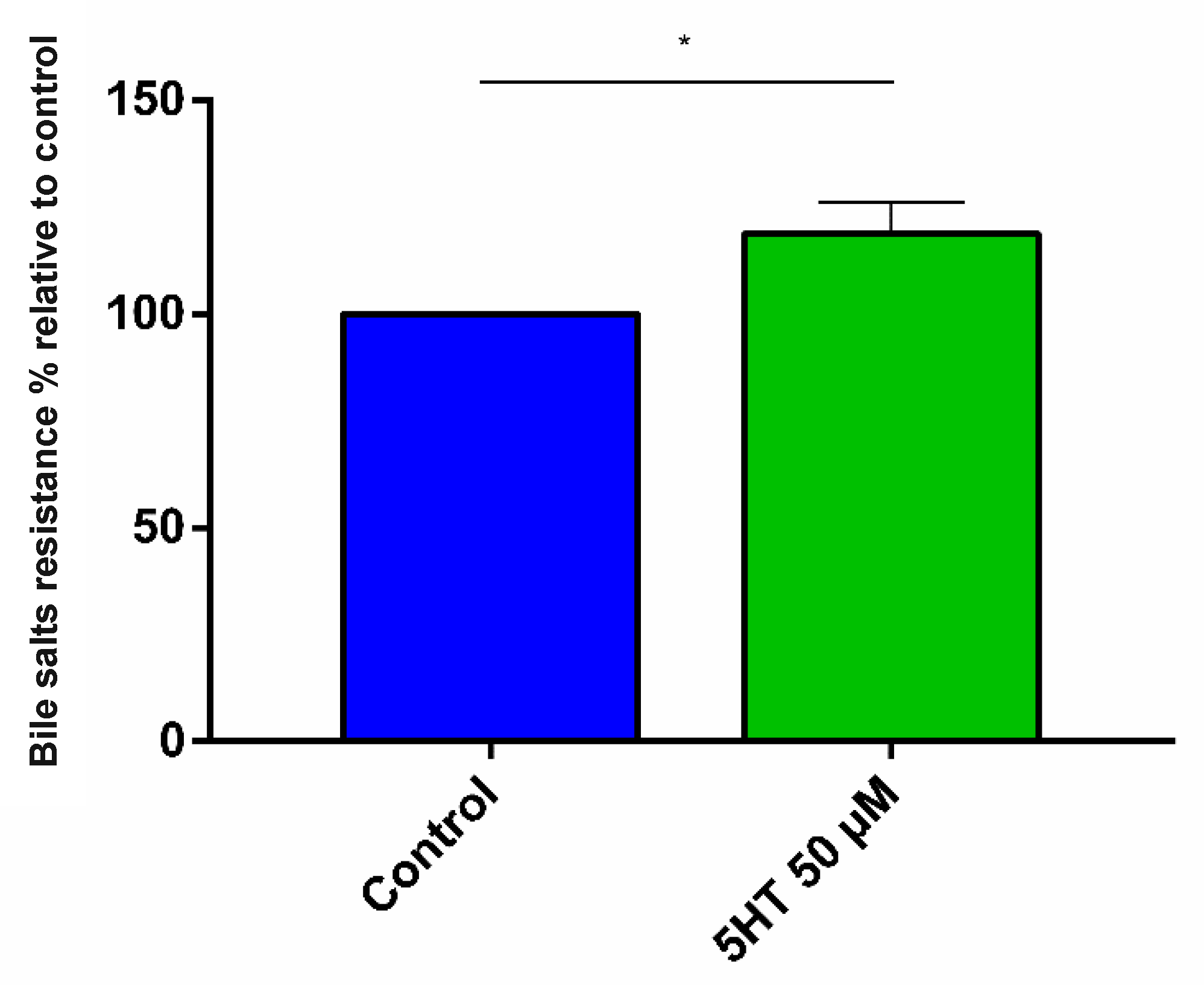
3.2.1. Bile Salt Resistance
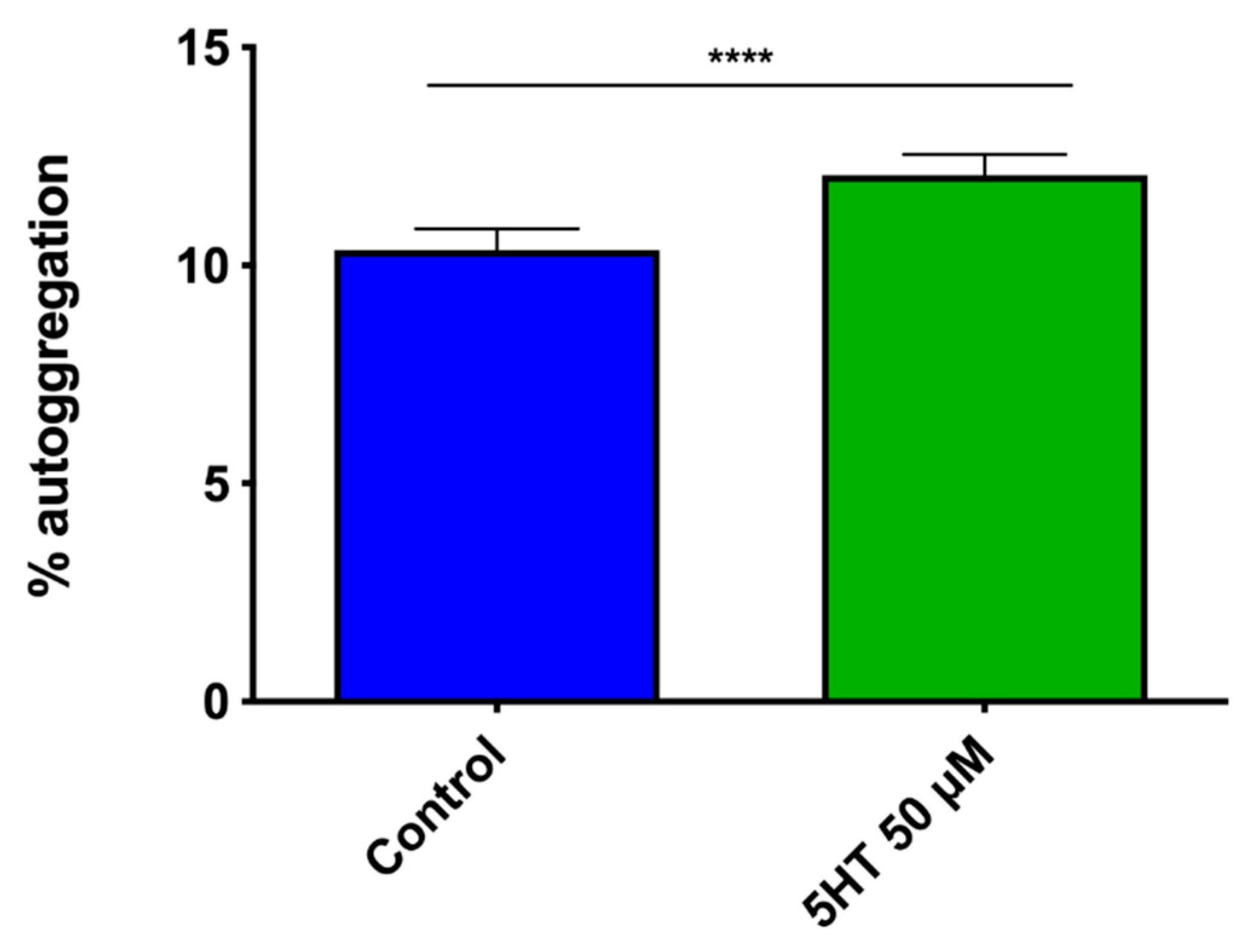
3.2.2. Auto-Aggregation Assay
3.2.3. Biofilm Formation
3.2.4. Antibiotic Susceptibility
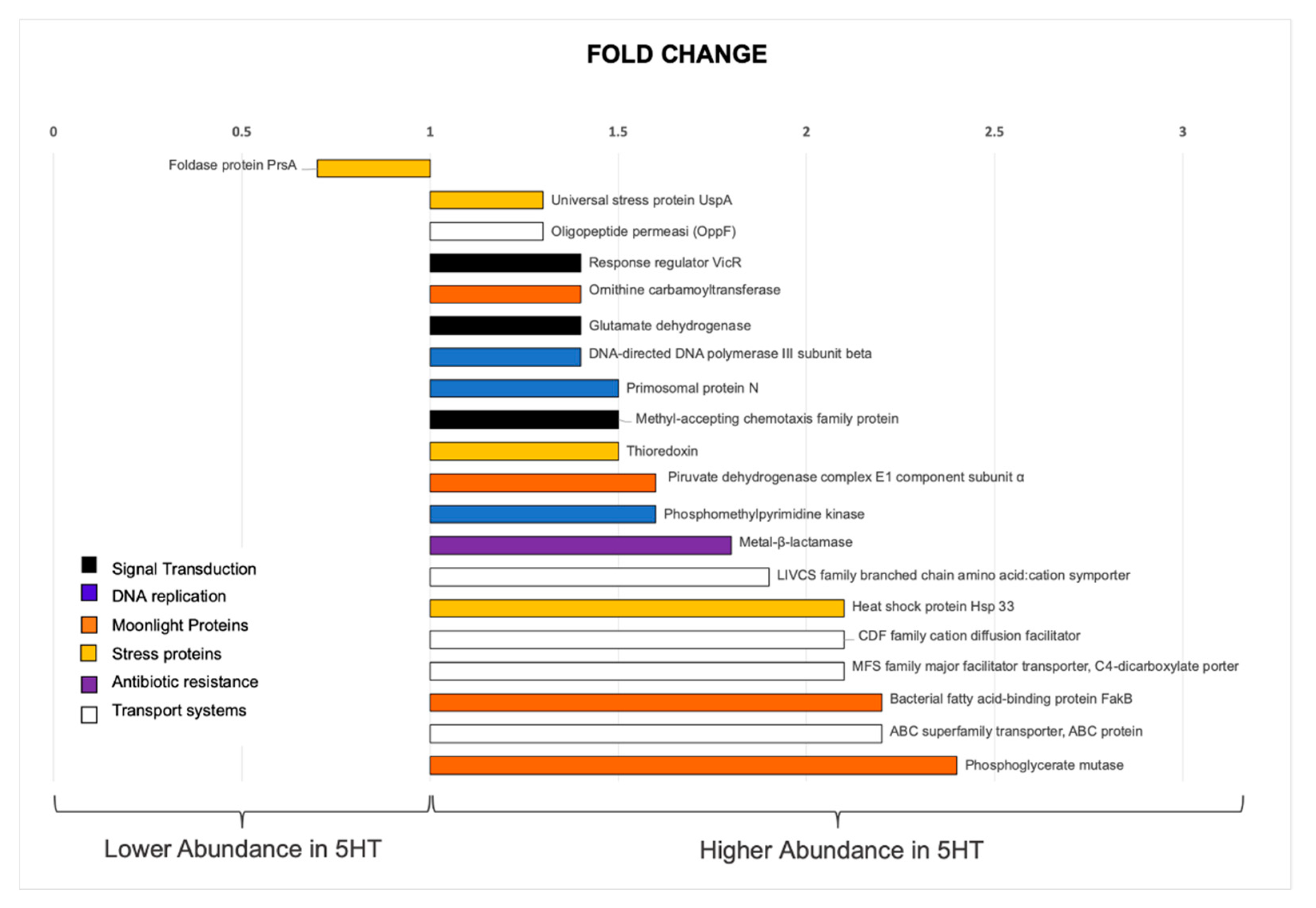
3.3. Whole Cell Proteomic Analyses
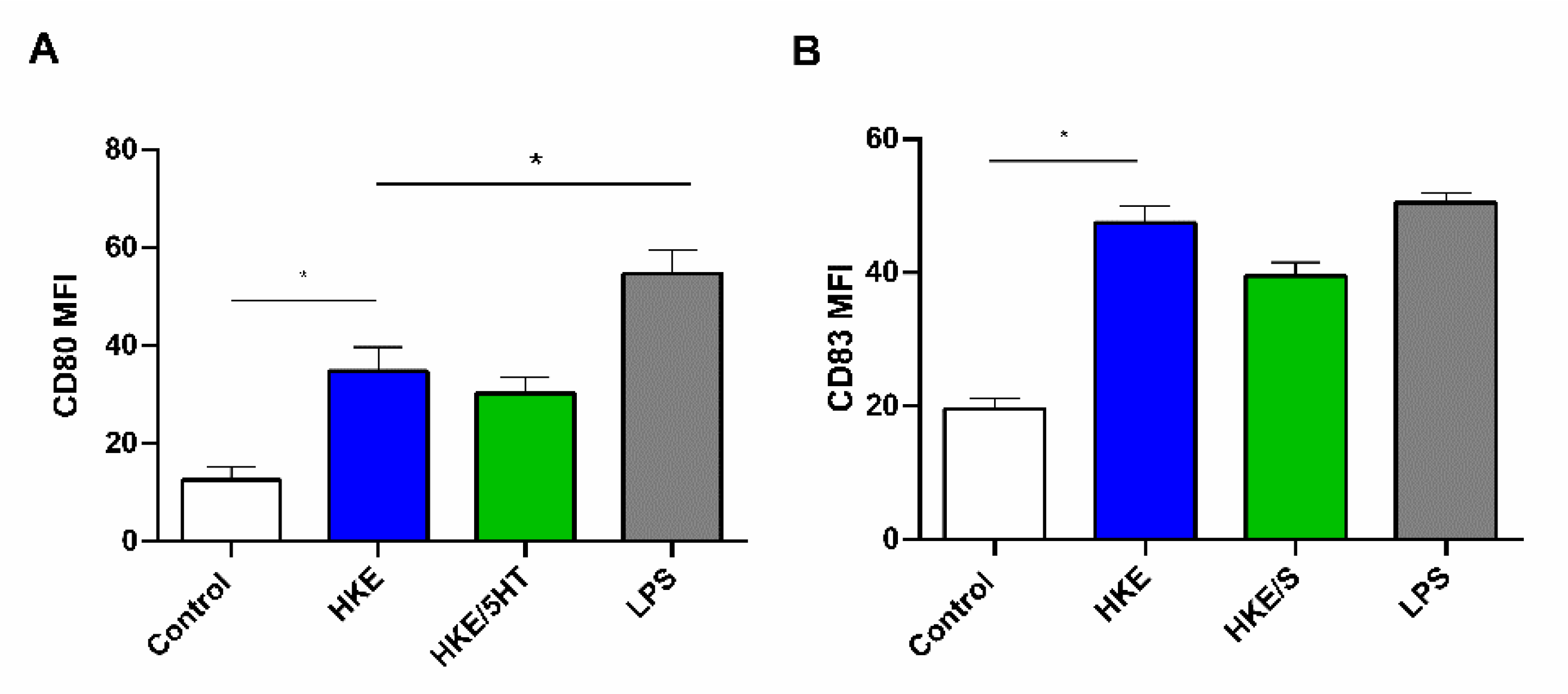
3.4. 5HT Treated and Control E. faecium NCIMB10415 Effects on DC Maturation
4. Discussion
4.1. 5HT Sensing
4.2. Modification of the Growth Profiles
4.3. Enhanced Resistance to Stressors
4.3.1. Bile Stress Resistance
4.3.2. Antibiotic Stress Resistance
4.3.3. Stationary Phase-Related Stress
4.3.4. Oxidative Stress
4.4. Better Interbacterial Interaction: Autoaggregation and Biofilm Formation
4.5. Possible Interaction with the Host
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mazzoli, R.; Pessione, E. The Neuro-endocrinological Role of Microbial Glutamate and GABA Signaling. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef]
- Lyte, M. Microbial Endocrinology: Interkingdom Signaling in Infectious Disease and Health; Springer International Publishing: Cham, Germany, 2016. [Google Scholar] [CrossRef]
- Segain, J.-P. Butyrate inhibits inflammatory responses through NFkappa B inhibition: Implications for Crohn’s disease. Gut 2000, 47, 397–403. [Google Scholar] [CrossRef] [PubMed]
- Cryan, J.F.; Dinan, T.G. Mind-altering microorganisms: The impact of the gut microbiota on brain and behaviour. Nat. Rev. Neurosci. 2012, 13, 701–712. [Google Scholar] [CrossRef] [PubMed]
- Mazzoli, R.; Pessione, E.; Dufour, M.; Laroute, V.; Giuffrida, M.G.; Giunta, C.; Cocaign-Bousquet, M.; Loubière, P. Glutamate-induced metabolic changes in Lactococcus lactis NCDO 2118 during GABA production: Combined transcriptomic and proteomic analysis. Amino Acids 2010, 39, 727–737. [Google Scholar] [CrossRef]
- Richard, H.; Foster, J.W. Escherichia coli Glutamate- and Arginine-Dependent Acid Resistance Systems Increase Internal pH and Reverse Transmembrane Potential. J. Bacteriol. 2004, 186, 6032–6041. [Google Scholar] [CrossRef] [PubMed]
- Lyte, M. Probiotics function mechanistically as delivery vehicles for neuroactive compounds: Microbial endocrinology in the design and use of probiotics. BioEssays 2011, 33, 574–581. [Google Scholar] [CrossRef]
- RWong, K.; Yang, C.; Song, G.-H.; Wong, J.; Ho, K.-Y. Melatonin Regulation as a Possible Mechanism for Probiotic (VSL#3) in Irritable Bowel Syndrome: A Randomized Double-Blinded Placebo Study. Dig. Dis. Sci. 2015, 60, 186–194. [Google Scholar] [CrossRef]
- Norris, V.; Molina, F.; Gewirtz, A.T. Hypothesis: Bacteria Control Host Appetites. J. Bacteriol. 2013, 195, 411–416. [Google Scholar] [CrossRef]
- Masek, K.; Kadlec, O. Sleep factor, muramyl peptides, and the serotoninergic system. Lancet 1983, 1, 1277. [Google Scholar] [CrossRef] [PubMed]
- Özoğul, F. Production of biogenic amines by Morganella morganii, Klebsiella pneumoniae and Hafnia alvei using a rapid HPLC method. Eur. Food Res. Technol. 2004, 219, 465–469. [Google Scholar] [CrossRef]
- Sandrini, S.; Alghofaili, F.; Freestone, P.; Yesilkaya, H. Host stress hormone norepinephrine stimulates pneumococcal growth, biofilm formation and virulence gene expression. BMC Microbiol. 2014, 14, 180. [Google Scholar] [CrossRef]
- Scardaci, R.; Varese, F.; Manfredi, M.; Marengo, E.; Mazzoli, R.; Pessione, E. Enterococcus faecium NCIMB10415 responds to norepinephrine by altering protein profiles and phenotypic characters. J. Proteom. 2021, 231, 104003. [Google Scholar] [CrossRef]
- Yang, Q.; Anh, N.D.Q.; Bossier, P.; Defoirdt, T. Norepinephrine and dopamine increase motility, biofilm formation, and virulence of Vibrio harveyi. Front. Microbiol. 2014, 5, 584. [Google Scholar] [CrossRef]
- Cogan, T.A.; Thomas, A.O.; Rees, L.E.N.; Taylor, A.H.; Jepson, M.A.; Williams, P.H.; Ketley, J.; Humphrey, T.J. Norepinephrine increases the pathogenic potential of Campylobacter jejuni. Gut 2007, 56, 1060–1065. [Google Scholar] [CrossRef]
- Hiller, C.C.; Lucca, V.; Carvalho, D.; Borsoi, A.; Borges, K.A.; Furian, T.Q.; do Nascimento, V.P. Influence of catecholamines on biofilm formation by Salmonella Enteritidis. Microb. Pathog. 2019, 130, 54–58. [Google Scholar] [CrossRef] [PubMed]
- Halang, P.; Toulouse, C.; Geißel, B.; Michel, B.; Flauger, B.; Müller, M.; Voegele, R.T.; Stefanski, V.; Steuber, J. Response of Vibrio cholerae to the Catecholamine Hormones Epinephrine and Norepinephrine. J. Bacteriol. 2015, 197, 3769–3778. [Google Scholar] [CrossRef] [PubMed]
- Paulose, J.K.; Wright, J.M.; Patel, A.G.; Cassone, V.M. Human Gut Bacteria Are Sensitive to Melatonin and Express Endogenous Circadian Rhythmicity. PLoS ONE 2016, 11, e0146643. [Google Scholar] [CrossRef]
- Biaggini, K.; Barbey, C.; Borrel, V.; Feuilloley, M.; Déchelotte, P.; Connil, N. The pathogenic potential of Pseudomonas fluorescens MFN1032 on enterocytes can be modulated by serotonin, substance P and epinephrine. Arch. Microbiol. 2015, 197, 983–990. [Google Scholar] [CrossRef] [PubMed]
- O’Mahony, S.M.; Clarke, G.; Borre, Y.E.; Dinan, T.G.; Cryan, J.F. Serotonin, tryptophan metabolism and the brain-gut-microbiome axis. Behav. Brain Res. 2015, 277, 32–48. [Google Scholar] [CrossRef]
- Galligan, J.J. Beneficial actions of microbiota-derived tryptophan metabolites. Neurogastroenterol. Motil. 2018, 30, e13283. [Google Scholar] [CrossRef]
- Yanofsky, C. RNA-based regulation of genes of tryptophan synthesis and degradation, in bacteria. RNA 2007, 13, 1141–1154. [Google Scholar] [CrossRef]
- Raboni, S.; Bettati, S.; Mozzarelli, A. Tryptophan synthase: A mine for enzymologists. Cell. Mol. Life Sci. 2009, 66, 2391–2403. [Google Scholar] [CrossRef]
- Krishnan, S.; Ding, Y.; Saedi, N.; Choi, M.; Sridharan, G.V.; Sherr, D.H.; Yarmush, M.L.; Alaniz, R.C.; Jayaraman, A.; Lee, K. Gut Microbiota-Derived Tryptophan Metabolites Modulate Inflammatory Response in Hepatocytes and Macrophages. Cell Rep. 2018, 23, 1099–1111. [Google Scholar] [CrossRef]
- Matur, E.; Eraslan, E. The Impact of Probiotics on the Gastrointestinal Physiology. In New Advances in the Basic and Clinical Gastroenterology; Brzozowski, T., Ed.; IntechOpen: London, UK, 2012. [Google Scholar] [CrossRef]
- Özoğul, F.; Kuley, E.; Özoğul, Y.; Özoğul, İ. The Function of Lactic Acid Bacteria on Biogenic Amines Production by Food-Borne Pathogens in Arginine Decarboxylase Broth. Food Sci. Technol. Res. 2012, 18, 795–804. [Google Scholar] [CrossRef]
- Manocha, M.; Khan, W.I. Serotonin and GI Disorders: An Update on Clinical and Experimental Studies. Clin. Transl. Gastroenterol. 2012, 3, e13. [Google Scholar] [CrossRef]
- Kumar, A.; Russell, R.M.; Pifer, R.; Menezes-Garcia, Z.; Cuesta, S.; Narayanan, S.; MacMillan, J.B.; Sperandio, V. The Serotonin Neurotransmitter Modulates Virulence of Enteric Pathogens. Cell Host Microbe 2020, 28, 41–53.e8. [Google Scholar] [CrossRef] [PubMed]
- Hanchi, H.; Mottawea, W.; Sebei, K.; Hammami, R. The Genus Enterococcus: Between Probiotic Potential and Safety Concerns—An Update. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef]
- Otto, R.; Brink, B.; Veldkamp, H.; Konings, W.N. The relation between growth rate and electrochemical proton gradient of Streptococcus cremoris. FEMS Microbiol. Lett. 1983, 16, 69–74. [Google Scholar] [CrossRef][Green Version]
- Poolman, B.; Konings, W.N. Relation of growth of Streptococcus lactis and Streptococcus cremoris to amino acid transport. J. Bacteriol. 1988, 170, 700–707. [Google Scholar] [CrossRef]
- Zommiti, M.; Cambronel, M.; Maillot, O.; Barreau, M.; Sebei, K.; Feuilloley, M.; Ferchichi, M.; Connil, N. Evaluation of Probiotic Properties and Safety of Enterococcus faecium Isolated From Artisanal Tunisian Meat “Dried Ossban”. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Baccouri, O.; Boukerb, A.M.; Farhat, L.B.; Zébré, A.; Zimmermann, K.; Domann, E.; Cambronel, M.; Barreau, M.; Maillot, O.; Rincé, I.; et al. Probiotic Potential and Safety Evaluation of Enterococcus faecalis OB14 and OB15, Isolated From Traditional Tunisian Testouri Cheese and Rigouta, Using Physiological and Genomic Analysis. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- O’Toole, G.A. Microtiter Dish Biofilm Formation Assay. J. Vis. Exp. 2011. [Google Scholar] [CrossRef] [PubMed]
- Pessione, E.; Mazzoli, R.; Giuffrida, M.G.; Lamberti, C.; Garcia-Moruno, E.; Barello, C.; Conti, A.; Giunta, C. A proteomic approach to studying biogenic amine producing lactic acid bacteria. Proteomics 2005, 5, 687–698. [Google Scholar] [CrossRef]
- Pessione, E.; Pessione, A.; Lamberti, C.; Coïsson, D.J.; Riedel, K.; Mazzoli, R.; Bonetta, S.; Eberl, L.; Giunta, C. First evidence of a membrane-bound, tyramine and β-phenylethylamine producing, tyrosine decarboxylase in Enterococcus faecalis: A two-dimensional electrophoresis proteomic study. Proteomics 2009, 9, 2695–2710. [Google Scholar] [CrossRef]
- Wessel, D.M.; Flügge, U.I. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 1984, 138, 141–143. [Google Scholar] [CrossRef]
- Collins, B.C.; Hunter, C.L.; Liu, Y.; Schilling, B.; Rosenberger, G.; Bader, S.L.; Chan, D.W.; Gibson, B.W.; Gingras, A.-C.; Held, J.M.; et al. Multi-laboratory assessment of reproducibility, qualitative and quantitative performance of SWATH-mass spectrometry. Nat. Commun. 2017, 8, 291. [Google Scholar] [CrossRef]
- Ludwig, C.; Gillet, L.; Rosenberger, G.; Amon, S.; Collins, B.C.; Aebersold, R. Data-independent acquisition-based SWATH—MS for quantitative proteomics: A tutorial. Mol. Syst. Biol. 2018, 14. [Google Scholar] [CrossRef] [PubMed]
- Usai, G.; Cirrincione, S.; Re, A.; Manfredi, M.; Pagnani, A.; Pessione, E.; Mazzoli, R. Clostridium cellulovorans metabolism of cellulose as studied by comparative proteomic approach. J. Proteom. 2020, 216, 103667. [Google Scholar] [CrossRef]
- Pozza, E.D.; Manfredi, M.; Brandi, J.; Buzzi, A.; Conte, E.; Pacchiana, R.; Cecconi, D.; Marengo, E.; Donadelli, M. Trichostatin A alters cytoskeleton and energy metabolism of pancreatic adenocarcinoma cells: An in depth proteomic study. J. Cell. Biochem. 2018, 119, 2696–2707. [Google Scholar] [CrossRef]
- Martinotti, S.; Patrone, M.; Manfredi, M.; Gosetti, F.; Pedrazzi, M.; Marengo, E.; Ranzato, E. HMGB1 Osteo-Modulatory Action on Osteosarcoma SaOS-2 Cell Line: An Integrated Study From Biochemical and -Omics Approaches. J. Cell. Biochem. 2016, 117, 2559–2569. [Google Scholar] [CrossRef] [PubMed]
- Carbonare, L.D.; Manfredi, M.; Caviglia, G.; Conte, E.; Robotti, E.; Marengo, E.; Cheri, S.; Zamboni, F.; Gabbiani, D.; Deiana, M.; et al. Can half-marathon affect overall health? The yin-yang of sport. J. Proteom. 2018, 170, 80–87. [Google Scholar] [CrossRef]
- Albanese, P.; Manfredi, M.; Meneghesso, A.; Marengo, E.; Saracco, G.; Barber, J.; Morosinotto, T.; Pagliano, C. Dynamic reorganization of photosystem II supercomplexes in response to variations in light intensities. Biochim. Biophys. Acta (BBA) Bioenerg. 2016, 1857, 1651–1660. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef] [PubMed]
- Huerta-Cepas, J.; Szklarczyk, D.; Forslund, K.; Cook, H.; Heller, D.; Walter, M.C.; Rattei, T.; Mende, D.R.; Sunagawa, S.; Kuhn, M.; et al. eggNOG 4.5: A hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 2016, 44, D286–D293. [Google Scholar] [CrossRef]
- Tatusov, R.L.; Koonin, E.V. A genomic perspective on protein families.(cover story). Science 1997, 278, 631–637. [Google Scholar] [CrossRef]
- Qin, X.; Galloway-Peña, J.R.; Sillanpaa, J.; Roh, J.H.; Nallapareddy, S.R.; Chowdhury, S.; Bourgogne, A.; Choudhury, T.; Muzny, D.M.; Buhay, C.J.; et al. Complete genome sequence of Enterococcus faecium strain TX16 and comparative genomic analysis of Enterococcus faecium genomes. BMC Microbiol. 2012, 12, 135. [Google Scholar] [CrossRef] [PubMed]
- Cambronel, M.; Tortuel, D.; Biaggini, K.; Maillot, O.; Taupin, L.; Réhel, K.; Rincé, I.; Muller, C.; Hardouin, J.; Feuilloley, M.; et al. Epinephrine affects motility, and increases adhesion, biofilm and virulence of Pseudomonas aeruginosa H103. Sci. Rep. 2019, 9. [Google Scholar] [CrossRef] [PubMed]
- Mohammad-Zadeh, L.F.; Moses, L.; Gwaltney-Brant, S.M. Serotonin: A review. J. Vet. Pharmacol. Ther. 2008, 31, 187–199. [Google Scholar] [CrossRef]
- Hancock, L.; Perego, M. Two-Component Signal Transduction in Enterococcus faecalis. J. Bacteriol. 2002, 184, 5819–5825. [Google Scholar] [CrossRef]
- Cambronel, M.; Nilly, F.; Mesguida, O.; Boukerb, A.M.; Racine, P.-J.; Baccouri, O.; Borrel, V.; Martel, J.; Fécamp, F.; Knowlton, R.; et al. Influence of Catecholamines (Epinephrine/Norepinephrine) on Biofilm Formation and Adhesion in Pathogenic and Probiotic Strains of Enterococcus faecalis. Front. Microbiol. 2020, 11. [Google Scholar] [CrossRef]
- Pinto, D.; Mascher, T. (Actino)Bacterial “intelligence”: Using comparative genomics to unravel the information processing capacities of microbes. Curr. Genet. 2016, 62, 487–498. [Google Scholar] [CrossRef] [PubMed]
- Lyon, P. The cognitive cell: Bacterial behavior reconsidered. Front. Microbiol. 2015, 6. [Google Scholar] [CrossRef]
- Stock, A.M.; Robinson, V.L.; Goudreau, P.N. Two-Component Signal Transduction. Ann. Rev. Biochem. 2000, 69, 183–215. [Google Scholar] [CrossRef]
- Senadheera, M.D.; Guggenheim, B.; Spatafora, G.A.; Huang, Y.-C.C.; Choi, J.; Hung, D.C.I.; Treglown, J.S.; Goodman, S.D.; Ellen, R.P.; Cvitkovitch, D.G. A VicRK Signal Transduction System in Streptococcus mutans Affects gtfBCD, gbpB, and ftf Expression, Biofilm Formation, and Genetic Competence Development. J. Bacteriol. 2005, 187, 4064–4076. [Google Scholar] [CrossRef]
- Clarke, M.B.; Hughes, D.T.; Zhu, C.; Boedeker, E.C.; Sperandio, V. The QseC sensor kinase: A bacterial adrenergic receptor. Proc. Natl. Acad. Sci. USA 2006, 103, 10420–10425. [Google Scholar] [CrossRef]
- Lange, R.; Wagner, C.; de Saizieu, A.; Flint, N.; Molnos, J.; Stieger, M.; Caspers, P.; Kamber, M.; Keck, W.; Amrein, K.E. Domain organization and molecular characterization of 13 two-component systems identified by genome sequencing of Streptococcus pneumoniae. Gene 1999, 237, 223–234. [Google Scholar] [CrossRef]
- Gunka, K.; Newman, J.A.; Commichau, F.M.; Herzberg, C.; Rodrigues, C.; Hewitt, L.; Lewis, R.J.; Stülke, J. Functional Dissection of a Trigger Enzyme: Mutations of the Bacillus subtilis Glutamate Dehydrogenase RocG That Affect Differentially Its Catalytic Activity and Regulatory Properties. J. Mol. Biol. 2010, 400, 815–827. [Google Scholar] [CrossRef]
- Kwon, Y.H.; Wang, H.; Denou, E.; Ghia, J.-E.; Rossi, L.; Fontes, M.E.; Bernier, S.P.; Shajib, M.S.; Banskota, S.; Collins, S.M.; et al. Modulation of Gut Microbiota Composition by Serotonin Signaling Influences Intestinal Immune Response and Susceptibility to Colitis. Cell. Mole. Gastroenterol. Hepatol. 2019, 7, 709–728. [Google Scholar] [CrossRef]
- Ganong, W.F. Review of Medical Physiology; Mcgraw-Hill: New York, NY, USA, 1995. [Google Scholar]
- Alvarado, A.; Behrens, W.; Josenhans, C. Protein Activity Sensing in Bacteria in Regulating Metabolism and Motility. Front. Microbiol. 2020, 10. [Google Scholar] [CrossRef] [PubMed]
- Saier, M.H., Jr. Families of transmembrane transporters selective for amino acids and their derivatives The information presented in this review was initially prepared for presentation at the FASEB meeting on amino acid transport held in Copper Mountain, Colorado, June 26–July 1, 1999 and was updated in January 2000 following the meeting of the Transport Nomenclature Panel of the International Union of Biochemistry and Molecular Biology (IUBMB) in Geneva, November 28–30, 1999. The system of classification described in this review reflects the recommendations of that panel. Microbiology 2000, 146, 1775–1795. [Google Scholar] [CrossRef] [PubMed]
- Mills, O.E.; Thomas, T.D. Nitrogen sources for growth of lactic streptococci in milk. N. Z. J. Dairy Sci. Technol. 1981, 16, 43–45. [Google Scholar]
- Garault, P.; Letort, C.; Juillard, V.; Monnet, V. Branched-Chain Amino Acid Biosynthesis Is Essential for Optimal Growth of Streptococcus thermophilus in Milk. Appl. Environ. Microbiol. 2000, 66, 5128–5133. [Google Scholar] [CrossRef] [PubMed]
- Pessione, E. Lactic acid bacteria contribution to gut microbiota complexity: Lights and shadows. Front. Cell. Infect. Microbiol. 2012, 2. [Google Scholar] [CrossRef]
- KBarrett, E.; Ganong, W.F. Ganong’s Review of Medical Physiology, 24th ed.; McGraw-Hill Med: New York, NY, USA, 2012. [Google Scholar]
- Merritt, M.E.; Donaldson, J.R. Effect of bile salts on the DNA and membrane integrity of enteric bacteria. J. Med. Microbiol. 2009, 58, 1533–1541. [Google Scholar] [CrossRef]
- Breton, Y.L.; Mazé, A.; Hartke, A.; Lemarinier, S.; Auffray, Y.; Rincé, A. Isolation and Characterization of Bile Salts-Sensitive Mutants of Enterococcus faecalis. Curr. Microbiol. 2002, 45, 0434–0439. [Google Scholar] [CrossRef] [PubMed]
- HSaito, E.; Harp, J.R.; Fozo, E.M. Incorporation of Exogenous Fatty Acids Protects Enterococcus faecalis from Membrane-Damaging Agents. Appl. Environ. Microbiol. 2014, 80, 6527–6538. [Google Scholar] [CrossRef]
- Andersen, B.M. Bacterial resistance against beta-lactam antibiotics. Tidsskr. Nor. Laegeforen 1990, 110, 3233–3239. [Google Scholar]
- Sacha, P.; Wieczorek, P.; Hauschild, T.; Zórawski, M.; Olszańska, D.; Tryniszewska, E. Metallo-beta-lactamases of Pseudomonas aeruginosa--a novel mechanism resistance to beta-lactam antibiotics. Folia Histochem. Cytobiol. 2008, 46. [Google Scholar] [CrossRef] [PubMed]
- Anton, A.; Weltrowski, A.; Haney, C.J.; Franke, S.; Grass, G.; Rensing, C.; Nies, D.H. Characteristics of Zinc Transport by Two Bacterial Cation Diffusion Facilitators from Ralstonia metallidurans CH34 and Escherichia coli. J. Bacteriol. 2004, 186, 7499–7507. [Google Scholar] [CrossRef]
- Schiene, C.; Fischer, G. Enzymes that catalyse the restructuring of proteins. Curr. Opin. Struct. Biol. 2000, 10, 40–45. [Google Scholar] [CrossRef]
- Kontinen, V.P.; Sarvas, M. The PrsA lipoprotein is essential for protein secretion in Bacillus subtilis and sets a limit for high-level secretion. Mol. Microbiol. 1993, 8, 727–737. [Google Scholar] [CrossRef] [PubMed]
- Lindholm, A.; Ellmén, U.; Tolonen-Martikainen, M.; Palva, A. Heterologous protein secretion in Lactococcus lactis is enhanced by the Bacillus subtilis chaperone-like protein PrsA. Appl. Microbiol. Biotechnol. 2006, 73, 904–914. [Google Scholar] [CrossRef] [PubMed]
- Hyyryläinen, H.-L.; Marciniak, B.C.; Dahncke, K.; Pietiäinen, M.; Courtin, P.; Vitikainen, M.; Seppala, R.; Otto, A.; Becher, D.; Chapot-Chartier, M.-P.; et al. Penicillin-binding protein folding is dependent on the PrsA peptidyl-prolyl cis-trans isomerase in Bacillus subtilis. Mol. Microbiol. 2010, 77, 108–127. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.H.; Kingston, A.W.; Helmann, J.D. Glutamate Dehydrogenase Affects Resistance to Cell Wall Antibiotics in Bacillus subtilis. J. Bacteriol. 2012, 194, 993–1001. [Google Scholar] [CrossRef] [PubMed]
- Freestone, P.; Nyström, T.; Trinei, M.; Norris, V. The universal stress protein, UspA, of Escherichia coli is phosphorylated in response to stasis. J. Mol. Biol. 1997, 274, 318–324. [Google Scholar] [CrossRef]
- Goo, E.; Majerczyk, C.D.; An, J.H.; Chandler, J.R.; Seo, Y.-S.; Ham, H.; Lim, J.Y.; Kim, H.; Lee, B.; Jang, M.S.; et al. Bacterial quorum sensing, cooperativity, and anticipation of stationary-phase stress. Proc. Natl. Acad. Sci. USA 2012, 109, 19775–19780. [Google Scholar] [CrossRef]
- Mitchell, A.; Romano, G.H.; Groisman, B.; Yona, A.; Dekel, E.; Kupiec, M.; Dahan, O.; Pilpel, Y. Adaptive prediction of environmental changes by microorganisms. Nature 2009, 460, 220–224. [Google Scholar] [CrossRef]
- Maleki, F.; Khosravi, A.; Nasser, A.; Taghinejad, H.; Azizian, M. Bacterial Heat Shock Protein Activity. J. Clin. Diagn Res. 2016, 10, BE01–BE03. [Google Scholar] [CrossRef]
- Sugimoto, S.; Abdullah-Al-Mahin; Sonomoto, K. Molecular Chaperones in Lactic Acid Bacteria: Physiological Consequences and Biochemical Properties. J. Biosci. Bioeng. 2008, 106, 324–336. [Google Scholar] [CrossRef] [PubMed]
- Graumann, J.; Lilie, H.; Tang, X.; Tucker, K.A.; Hoffmann, J.H.; Vijayalakshmi, J.; Saper, M.; Bardwell, J.C.A.; Jakob, U. Activation of the Redox-Regulated Molecular Chaperone Hsp33—A Two-Step Mechanism. Structure 2001, 9, 377–387. [Google Scholar] [CrossRef]
- Holmgren, A.; Bjornstedt, M. Thioredoxin and thioredoxin reductase. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 1995; pp. 199–208. [Google Scholar] [CrossRef]
- MCollado, C.; Meriluoto, J.; Salminen, S. Adhesion and aggregation properties of probiotic and pathogen strains. Eur. Food Res. Technol. 2008, 226, 1065–1073. [Google Scholar] [CrossRef]
- Trunk, T.; Khalil, H.S.; Leo, J.C. Bacterial Cell Surface Group, Section for Genetics and Evolutionary Biology, Department of Biosciences, University of Oslo, Oslo, Norway, Bacterial autoaggregation. AIMS Microbiol. 2018, 4, 140–164. [Google Scholar] [CrossRef]
- Guo, L.; Wu, T.; Hu, W.; He, X.; Sharma, S.; Webster, P.; Gimzewski, J.K.; Zhou, X.; Lux, R.; Shi, W. Phenotypic characterization of the foldase homologue PrsA in Streptococcus mutans. Mol. Oral Microbiol. 2013, 28, 154–165. [Google Scholar] [CrossRef]
- Costerton, J. Overview of microbial biofilms. J. Ind. Microbiol. 1995, 15, 137–140. [Google Scholar] [CrossRef]
- Monnet, V.; Gardan, R. Quorum-sensing regulators in Gram-positive bacteria: ‘cherchez le peptide’: Quorum-sensing regulators in Gram-positive bacteria. Mol. Microbiol. 2015, 97, 181–184. [Google Scholar] [CrossRef] [PubMed]
- Visick, K.L.; Fuqua, C. Decoding Microbial Chatter: Cell-Cell Communication in Bacteria. J. Bacteriol. 2005, 187, 5507–5519. [Google Scholar] [CrossRef] [PubMed]
- Leonard, B.A.; Podbielski, A.; Hedberg, P.J.; Dunny, G.M. Enterococcus faecalis pheromone binding protein, PrgZ, recruits a chromosomal oligopeptide permease system to import sex pheromone cCF10 for induction of conjugation. Proc. Natl. Acad. Sci. USA 1996, 93, 260–264. [Google Scholar] [CrossRef]
- Salas-Jara, M.; Ilabaca, A.; Vega, M.; García, A. Biofilm Forming Lactobacillus: New Challenges for the Development of Probiotics. Microorganisms 2016, 4, 35. [Google Scholar] [CrossRef] [PubMed]
- Meijerink, M.; van Hemert, S.; Taverne, N.; Wels, M.; de Vos, P.; Bron, P.A.; Savelkoul, H.F.; van Bilsen, J.; Kleerebezem, M.; Wells, J.M. Identification of Genetic Loci in Lactobacillus plantarum That Modulate the Immune Response of Dendritic Cells Using Comparative Genome Hybridization. PLoS ONE 2010, 5, e10632. [Google Scholar] [CrossRef] [PubMed]
- Molina, M.A.; Díaz, A.M.; Hesse, C.; Ginter, W.; Gentilini, M.V.; Nuñez, G.G.; Canellada, A.M.; Sparwasser, T.; Berod, L.; Castro, M.S.; et al. Immunostimulatory Effects Triggered by Enterococcus faecalis CECT7121 Probiotic Strain Involve Activation of Dendritic Cells and Interferon-Gamma Production. PLoS ONE 2015, 10, e0127262. [Google Scholar] [CrossRef]
- Khalkhali, S.; Mojgani, N. Enterococcus faecium; a Suitable Probiotic Candidate for Modulation of Immune Responses against Pathogens. Int. J. Basic Sci. Med. 2017, 2, 77–82. [Google Scholar] [CrossRef]
- Jeffery, C.J. Moonlighting proteins—An update. Mol. BioSyst. 2009, 5, 345. [Google Scholar] [CrossRef] [PubMed]
- Vastano, V.; Salzillo, M.; Siciliano, R.A.; Muscariello, L.; Sacco, M.; Marasco, R. The E1 beta-subunit of pyruvate dehydrogenase is surface-expressed in Lactobacillus plantarum and binds fibronectin. Microbiol. Res. 2014, 169, 121–127. [Google Scholar] [CrossRef]
- Qi, J.; Zhang, F.; Wang, Y.; Liu, T.; Tan, L.; Wang, S.; Tian, M.; Li, T.; Wang, X.; Ding, C.; et al. Characterization of Mycoplasma gallisepticum pyruvate dehydrogenase alpha and beta subunits and their roles in cytoadherence. PLoS ONE 2018, 13, e0208745. [Google Scholar] [CrossRef]
- Sheng, X.; Liu, M.; Liu, H.; Tang, X.; Xing, J.; Zhan, W. Identification of immunogenic proteins and evaluation of recombinant PDHA1 and GAPDH as potential vaccine candidates against Streptococcus iniae infection in flounder (Paralichthys olivaceus). PLoS ONE 2018, 13, e0195450. [Google Scholar] [CrossRef]
- Candela, M.; Bergmann, S.; Vici, M.; Vitali, B.; Turroni, S.; Eikmanns, B.J.; Hammerschmidt, S.; Brigidi, P. Binding of Human Plasminogen to Bifidobacterium. J. Bacteriol. 2007, 189, 5929–5936. [Google Scholar] [CrossRef]
- Genovese, F.; Coïsson, J.D.; Majumder, A.; Pessione, A.; Svensson, B.; Jacobsen, S.; Pessione, E. An exoproteome approach to monitor safety of a cheese-isolated Lactococcus lactis. Food Res. Int. 2013, 54, 1072–1079. [Google Scholar] [CrossRef]
- Hussain, M.; Peters, G.; Chhatwal, G.S.; Herrmann, M. A lithium chloride-extracted, broad-spectrum-adhesive 42-kilodalton protein of Staphylococcus epidermidis is ornithine carbamoyltransferase. Infect. Immun. 1999, 67, 6688–6690. [Google Scholar] [CrossRef] [PubMed]
- Alam, S.; Bansod, S.; Kumar, R.; Sengupta, N.; Singh, L. Differential proteomic analysis of Clostridium perfringens ATCC13124; identification of dominant, surface and structure associated proteins. BMC Microbiol. 2009, 9, 162. [Google Scholar] [CrossRef] [PubMed]

| Treatment | MIC Amp (µg/mL) | MIC Van (µg/mL) |
|---|---|---|
| Control | 0.38 ± 0.0 | 1.1 ± 0.3 |
| 5HT 50 μM | 0.56 ± 0.1** | 1.0 ± 0.3 ns |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Scardaci, R.; Manfredi, M.; Barberis, E.; Scutera, S.; Marengo, E.; Pessione, E. Serotonin Exposure Improves Stress Resistance, Aggregation, and Biofilm Formation in the Probiotic Enterococcus faecium NCIMB10415. Microbiol. Res. 2021, 12, 606-625. https://doi.org/10.3390/microbiolres12030043
Scardaci R, Manfredi M, Barberis E, Scutera S, Marengo E, Pessione E. Serotonin Exposure Improves Stress Resistance, Aggregation, and Biofilm Formation in the Probiotic Enterococcus faecium NCIMB10415. Microbiology Research. 2021; 12(3):606-625. https://doi.org/10.3390/microbiolres12030043
Chicago/Turabian StyleScardaci, Rossella, Marcello Manfredi, Elettra Barberis, Sara Scutera, Emilio Marengo, and Enrica Pessione. 2021. "Serotonin Exposure Improves Stress Resistance, Aggregation, and Biofilm Formation in the Probiotic Enterococcus faecium NCIMB10415" Microbiology Research 12, no. 3: 606-625. https://doi.org/10.3390/microbiolres12030043
APA StyleScardaci, R., Manfredi, M., Barberis, E., Scutera, S., Marengo, E., & Pessione, E. (2021). Serotonin Exposure Improves Stress Resistance, Aggregation, and Biofilm Formation in the Probiotic Enterococcus faecium NCIMB10415. Microbiology Research, 12(3), 606-625. https://doi.org/10.3390/microbiolres12030043








