Abstract
Background/Objectives: Legionella micdadei is a clinically significant species within the Legionella genus, requiring accurate detection methods, surveillance, and precise clinical diagnosis. Our objective was to develop a sensitive polymerase chain reaction (PCR) assay specific for L. micdadei to detect its presence in environmental specimens. Methods: We targeted the 23S–5S intergenic spacer region, which can differentiate Legionella spp. We tested the detection of L. micdadei with 20 strains and determined the limit of detection with 2 strains. We verified assay specificity with 17 strains of other Legionella spp., 62 strains of other bacterial and fungal genera, and three human DNA specimens. We evaluated intra- and inter-run precision. We tested 15 environmental specimens (water, swabs of water faucets, mulch, and soil) by PCR. Results: The PCR assay demonstrated 100% analytical specificity (no cross-reactivity with non-targeted species), 100% inclusivity (detection of all L. micdadei strains), and high precision, with a coefficient of variation ≤ 2% across replicates. The limit of detection was estimated at 5 genomic DNA copies per reaction. We detected L. micdadei in environmental specimens. Conclusions: This PCR assay enables accurate detection of L. micdadei and is not subject to competition with other Legionella spp., thereby addressing limitations of current broad-spectrum Legionella approaches. The evaluation supports its application in environmental detection for surveillance.
1. Introduction
Legionella spp. are Gram-negative mesophilic bacteria that are ubiquitous in freshwater ecosystems and soils [1]. Of the more than 60 species described, about half have been reported to infect humans [2]. These pathogenic bacteria are responsible for Legionnaires’ disease (LD) and Pontiac fever, both acquired through the inhalation of contaminated aerosols [3]. They can colonize water systems, including cooling towers, hot water tanks, and water distribution systems, posing significant public health risks [3]. In addition, certain Legionella spp. have also been detected in soils, potting mixes, and composts. These can serve as additional environmental reservoirs and sources of infection, particularly for individuals exposed to contaminated dust or aerosols during gardening or agricultural work [4,5,6]. From 2000 to 2019, the national incidence rate of Legionnaires’ disease in the United States increased 6.5-fold, from 0.42 to 2.71 cases per 100,000 persons [2]. While L. pneumophila causes approximately 90% of these infections, other species also contribute to the burden of disease [1]. Of these, the most frequently isolated from infected patients are L. bozemanae, L. dumoffii, L. longbeachae, and L. micdadei [7]. Based on 2018–2019 data from the Supplemental Legionnaires’ Disease Surveillance System (SLDSS), L. micdadei accounted for 1.3% of all clinical Legionella spp. cases diagnosed in the United States [2]. While L. pneumophila typically causes lobar consolidation or patchy pulmonary infiltrates, L. micdadei more frequently causes nodular lung lesions that may cavitate or enlarge over time [8,9]. This radiological pattern can closely mimic invasive fungal infections, potentially misleading clinicians toward a fungal diagnosis and prompt empiric antifungal therapy before the correct etiology is identified [10,11,12].
Standard detection methods currently rely on culture using buffered charcoal yeast extract (BCYE) agar media. Though specific for the Legionella genus, certain media formulations favor the rapid growth of L. pneumophila, while species like L. micdadei may require 1–2 weeks to form visible colonies due to their inherently slower growth rates, or fail to grow entirely [13]. This delay can hinder a timely response in water and cooling tower monitoring, when rapid intervention is critical to prevent and contain outbreaks. To improve selectivity, antibiotic supplements are added to BCYE agar to suppress bacteria and fungi other than Legionella spp. Some media, such as BCYE-MWY and BCYE-GVPC, support L. micdadei growth, whereas BCYE-BMPA inhibits it due to the presence of cefamandole [14]. Clinical laboratories usually use two to three culture media in parallel to detect a broader diversity of Legionella spp. However, this approach does not always include a medium suited for L. micdadei, which may lead to underdetection of this species [15]. Finally, Legionella spp. can be found co-occurring in environmental and clinical specimens, which can make their epidemiological correlation particularly complex [16,17,18].
Polymerase chain reaction (PCR)-based methods address these limitations by targeting conserved Legionella spp. DNA sequences, delivering results within 6 to 8 h [19,20,21]. They also detect low bacterial loads that can be missed by culture and allow for efficient monitoring of water systems [19,22]. While PCR cannot distinguish viable from non-viable bacteria, its speed and sensitivity support water and cooling tower monitoring, enable the detection of viable but nonculturable (VBNC) bacteria, and identify Legionella spp. contained within protozoa [22,23,24]. Multiple PCR-based methods have been developed for the detection and identification of Legionella spp., including both genus-level and species-specific assays targeting genetic regions such as 16S rRNA, mip, and various intergenic spacer regions [13,25,26,27,28,29,30]. While these approaches have improved diagnostic capabilities, the detection of Legionella spp. like L. micdadei remains challenging, especially in environmental specimens where specificity and sensitivity may be compromised by potential inhibitors and competing bacterial flora [30,31]. Applications that can discriminate between closely related Legionella spp. with high specificity in environmental matrices are limited. In this study, we present a PCR assay that specifically detects L. micdadei [25].
2. Materials and Methods
2.1. Bacterial and Fungal Strains
Twenty L. micdadei strains (Table 1), 17 strains from other Legionella spp., and 62 bacterial and fungal strains from other genera (Table S1) were obtained from the American Type Culture Collection (Manassas, VA, USA) (ATCC), the Collection de Culture du Centre de recherche en Infectiologie (Québec, QC, Canada) (CCRI), and the Laboratoire de Santé Publique du Québec (Québec, QC, Canada) (L00 and ID). All Legionella spp. strains were grown at 35–40 °C under aerobic conditions on BCYE for 72 to 96 h. Other bacterial and yeast strains were cultured on tryptic soy agar with 5% sheep blood or Sabouraud dextrose agar for 24–48 h. Filamentous fungi were grown on potato dextrose agar for 7–21 days.

Table 1.
PCR results of Legionella micdadei and other Legionella spp. strains used in this study.
2.2. DNA Isolation
Bacterial DNA (both purified and crude genomic DNA (gDNA)) was isolated from cultures using one of the following methods: eMAG (Cat. No. 418591, bioMérieux, Marcy-l’Étoile, France), BioSprint 15 DNA Blood Kit (Cat. No. 940014, Qiagen, Mississauga, ON, Canada) automated with a KingFisher system (Cat. No. 5400610, Thermo Fisher Scientific, Waltham, MA, USA) [32], or crude lysate prepared using a standardized glass bead-beating lysis technique as previously described [33]. DNA concentration was measured using a NanoDrop spectrophotometer (Cat. No. ND-ONE-W, Thermo Fisher Scientific), a Qubit fluorometer (Cat. No. Q33226, Thermo Fisher Scientific), or a BioTek Synergy H1 (Cat. No. SYS-BT-SYNH1, Agilent, Santa Clara, CA, USA).
2.3. Primer and Probe Design
We targeted the three genome copies of 23S-5S rRNA intergenic spacers of L. micdadei, thereby increasing the number of detectable templates and improving assay sensitivity [34]. This spacer exhibits highly conserved extremities across the genus, while its central section varies between species, enabling species-specific discrimination. We retrieved sequences from GenBank and aligned them using MAFFT (version 7.520, RIMD, Osaka, Japan): ten sequences from four L. micdadei strains and 504 sequences from 22 other Legionella spp. (L. anisa (n = 9), L. birminghamensis (n = 1), L. bozemanae (n = 2), L. cardiaca (n = 1), L. cherrii (n = 4), L. cincinnatiensis (n = 1), L. clemsonensis (n = 1), L. dumoffii (n = 1), L. feeleii (n = 2), L. gormanii (n = 1), L. hackeliae (n = 5), L. jordanis (n = 5), L. lansingensis (n = 5), L. longbeachae (n = 30), L. maceachernii (n = 2), L. oakridgensis (n = 7), L. parisiensis (n = 1), L. pneumophila (n = 408), L. rubrilucens (n = 2), L. sainthelensi (n = 14), L. tucsonensis (n = 1), L. wadsworthii (n = 1)). We designed a forward primer, which we combined with the reverse primer and probe from Cross et al. (2016) to obtain specific amplification of L. micdadei (Table 2) [25]. The fluorophore tag of the probe was modified to CalRed610-BHQ2 to suit our detection system. Additionally, we designed two primers and a probe targeting B. subtilis strain CCRI-21428 as an internal control.

Table 2.
Primer and probe characteristics.
To assess inclusivity and specificity in silico, we analyzed primer and probe sequence cross-reactivity using Primer-BLAST (accessed August 2024, NCBI, Bethesda, MD, USA), querying the GenBank nucleotide collection (core_nt, National Center for Biotechnology Information, Bethesda, MD, USA). The search included all available complete and partial genome sequences of L. micdadei as well as sequences from 51 non-target species, selected based on phylogenetic relatedness and/or their presence in respiratory microbiota.
2.4. PCR Conditions
The PCR mixture (total 20 µL) contained four primers and two probes (Table 2), X FastStart Essential DNA Probes Master Mix (Roche Life Science, Basel, Switzerland), gDNA of B. subtilis (1000 genome copies) as an internal control, and 5 µL of specimens (Supplementary Table S1). PCR amplification was performed on a CFX96 thermal cycler (Bio-Rad, Hercules, CA, USA) as follows: 95 °C for 10 min, followed by 45 cycles of 95 °C for 10 s (ramp rate 1 °C/s) and 60 °C for 30 s, and finalized with a hold at 25 °C for 5 s. Each PCR was performed in duplicate (unless otherwise specified).
2.5. In Vitro Performance Characteristics of the PCR Assay
Analytical specificity was evaluated using purified gDNA and crude lysates from 17 Legionella strains other than L. micdadei, 56 other bacterial strains, six fungal strains, and three human gDNA (Supplementary Table S1) [35]. The inclusivity of the test for the target species was evaluated using 20 L. micdadei strains. Binomial 95% confidence intervals for analytical specificity and inclusivity were calculated using the exact Clopper–Pearson interval method on RStudio (v2025.05.1+513; Posit PBC, Boston, MA, USA) [36].
The limit of detection (LOD) was defined as the lowest genome copy number at which amplification was detected in at least 95% of replicates [37]. LOD evaluation involved testing two L. micdadei strains in eight replicates across five serial dilutions (20, 10, 5, 2, and 1 genome copies/reaction; Table 3) [37].

Table 3.
Limit of detection of the L. micdadei PCR assay.
PCR efficiency and linearity were assessed using duplicate reactions of L. micdadei gDNA (10–105 genome copies per reaction; Figure 1). To evaluate the precision of the PCR assay, we tested two strains of L. micdadei (Table 4). Mean Ct values and standard deviations were calculated for each strain, and the coefficient of variation (CV) was calculated for both repeatability (intra-assay) and reproducibility (inter-assay). Competition evaluation for the internal control. We evaluated the competition between B. subtilis internal control and L. micdadei detection. We mixed gDNA from B. subtilis at concentrations of 1000, 5000, 10,000, 50,000, and 220,000 genome copies with 10 genome copies of L. micdadei gDNA per reaction.
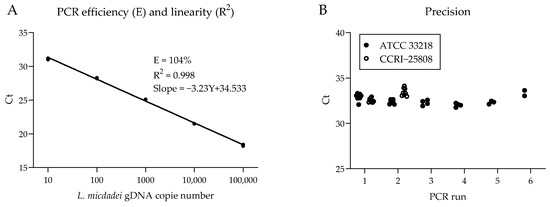
Figure 1.
PCR amplification efficiency, repeatability, and reproducibility. (A) Standard curve for PCR efficiency and linearity assessment of Ct values against gDNA dilutions of L. micdadei ATCC 33218 strain. (B) Variability of Ct values across eight independent PCR runs using 10 genome copies of gDNA from L. micdadei strains ATCC 33218 and CCRI-25808.

Table 4.
PCR repeatability and reproducibility for precision evaluation.
2.6. Competition Evaluation from Other Legionella spp.
To assess interference from other Legionella spp., purified gDNA or crude lysates from multiple Legionella spp. were tested in duplicates. Each reaction contained a fixed 10 copies of L. micdadei gDNA and one of 17 other Legionella spp. strains, introduced as either purified gDNA (1 ng) or crude lysate (1:100 dilution).
2.7. Environmental Specimen Preparation
One-liter tap water specimens were collected from hospital faucets in sterile bottles and processed on the same day, following ISO 12869:2019 guidelines. However, sodium thiosulfate was not added to the bottles to neutralize chlorine. Water specimens were filtered using a 0.4 µm membrane (PALL, Port Washington, NY, USA). The filters were folded and placed in a 2 mL plastic tube (Sarstedt, Nümbrecht, Germany). Sterile molecular-grade water (0.6 mL) was added to each tube. The tubes were vortexed three times for 30 s. Swabs of water faucets were collected using Roche Cobas flocked swab (Roche Diagnostics, Laval, QC, Canada). The swab was broken in a 15 mL conical plastic tube (Sarstedt, Nümbrecht, Germany) containing 1 mL of sterile molecular-grade water. The tubes were vortexed three times for 30 s. A volume of 50 µL of the suspensions was plated onto BCYE-MWY agar for culture, and 300 µL were transferred to 1.5 mL conical tubes with standardized glass beads for crude lysate DNA extraction [33]. The suspensions were centrifuged at 15,800× g for 5 min. The supernatants were discarded and the resulting pellets were resuspended in 50 uL of 5× TE buffer, (50 mM Tris-HCl, 5 mM disodium EDTA, pH 8.0; Thermo Fisher Scientific, Waltham, MA, USA), followed by 5 min bead-beating lysis and a 2 min heating step at 95 °C [33].
For gardening soil specimens, approximately 100 mg were suspended in 5 mL of sterile molecular-grade water and vortexed for 5 min. Specimens were then left to settle for 30 min. The clarified supernatants were collected and centrifuged at 500× g for 2 min to remove large debris. The resulting supernatants were filtered through a 5 µm syringe filter, then centrifuged at 10,000× g for 15 min to concentrate bacterial cells. After discarding the supernatant, the pellets were resuspended in 200 µL of sterile molecular-grade water by vortexing. 100 µL were then transferred into a glass-bead lysis tube, mixed with 900 µL of 1× TE buffer, and centrifuged at 21,000× g for 5 min. The supernatants were discarded. The resulting pellets were processed by glass-bead crude lysate DNA extraction as described above.
For dust and wood mulch mixed with soil, DNA was extracted and purified using the DNeasy PowerSoil Pro Kit (Cat. No. 47014, Qiagen, Hilden, Germany). Specimens were homogenized by manual mixing, then 250 mg were transferred to bead-beating tubes. Lysis and purification were then performed following the manufacturer’s instructions.
3. Results
3.1. In Silico Analysis of Primers and Probes
The in silico evaluation confirmed 100% identity for the primer and probe target regions across ten L. micdadei gene sequences available in GenBank, corresponding to four distinct strains (accession numbers: CP020615.1, Z24694.1, LN614830.1, and CP020614.1). Specificity analysis revealed between 7 and 17 mismatches in the forward primer compared to other Legionella spp., supporting assay specificity in silico.
3.2. In Vitro Performance Characteristics of the PCR Assay
We established the LOD at 5 genome copies of L. micdadei gDNA per reaction. With 5, 2, and 1 genome copies per reaction, the detection rates were 100% (32/32), 90.63% (29/32), and 68.75% (22/32), respectively (Table 3). The assay showed 100% inclusivity (20/20, 95% CI: 83.2–100%) for L. micdadei strains (Table 1). Analytical specificity was 100% (0/102 false positives; 95% CI: 96.5–100%), with no amplification detected in 17 strains of Legionella spp. other than L. micdadei, 56 bacterial strains from other genera, 6 fungal strains, and 3 human DNA specimens (Supplementary Table S1).
L. micdadei PCR showed linear amplification with an efficiency of 104% and a correlation coefficient (R2) of 0.998 (Figure 1A). We also tested the PCR precision on two strains of L. micdadei. Mean PCR cycle threshold (Ct) was similar, with average Ct values of 32.7 and 33.0, respectively. In all cases, the CV for repeatability and reproducibility was ≤2% (Figure 1B, Table 4).
3.3. Competition and Co-Amplification Analysis
We evaluated the potential for amplification competition within the assay by testing L. micdadei gDNA in the presence of high concentrations of internal control (B. subtilis) gDNA. The L. micdadei amplification exhibited no evidence of competition with the internal control gDNA, with a similar Ct value (31.38 ± 0.36) in all conditions (Figure 2A). Similarly, we observed no competition with other Legionella spp. during amplification, as indicated by comparable Ct values of 32.95 ± 0.25 for L. micdadei and 32.65 ± 0.27 for the internal control (B. subtilis) (Figure 2B).
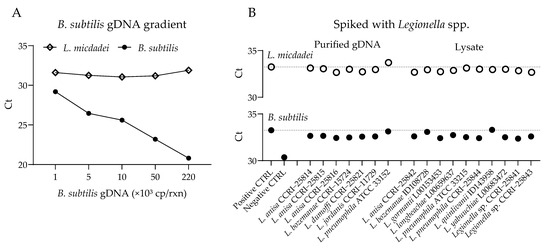
Figure 2.
Competition assays for PCR amplification of L. micdadei and B. subtilis in the presence of Legionella spp. (A) Amplification with L. micdadei (10 genome copies (cp) gDNA per reaction) and increasing amounts of internal control B. subtilis gDNA. (B) Amplification of L. micdadei (10 cp gDNA) and B. subtilis (1000 cp gDNA) with the addition of another Legionella sp., introduced as either purified gDNA (1 ng) or lysate (1:100 dilution). Seventeen Legionella spp. were tested.
3.4. Environmental Specimens
We evaluated the assay’s capacity to detect L. micdadei in environmental specimens: eight tap water, two swabs from water faucets, one dust, one wood mulch mixed with soil, and three gardening soil (Table 5). In parallel, tap water and swab specimens were also analyzed by culture. We detected L. micdadei by PCR in the wood mulch mixed with soil specimen and in one gardening soil specimen. However, we observed no amplification in any of the tap water, swab from water faucet, or dust specimens. Culture results identified the presence of L. anisa, L. feeleii, and L. pneumophila in five specimens. However, L. micdadei did not grow in cultures of environmental specimens tested.

Table 5.
Detection of L. micdadei in environmental specimens.
4. Discussion
In this study, we developed a PCR assay targeting L. micdadei’s multi-copy 23S–5S rRNA intergenic region, including a B. subtilis internal control. The assay achieved high specificity for L. micdadei, low LOD, and high inclusivity with excellent repeatability and reproducibility. Even though detection of L. micdadei is rare in the environment, we found two sources (soil and mulch). Environmental matrices such as soil can harbor L. micdadei and serve as potential sources of exposure, as evidenced by our results [30]. Exposure to soil contaminated by Legionella spp. can occur through aerosolization during soil-disturbing activities and handling [37,38,39]. Thus, soil testing is important for outbreak investigation and surveillance.
The presence of multiple copies of the 23S–5S target region within the L. micdadei genome likely plays a role in efficiently detecting small amounts [34]. The present L. micdadei-specific PCR assay offers distinct advantages over previously published studies [25]. The Cross et al. assay detected L. micdadei at 10 genome copies per reaction with 50% positivity. Our assay demonstrated improved sensitivity with a lower LOD of 5 genome copies per reaction in ≥95% of replicates. Environmental matrices frequently contain substances like humic acids, which may co-extract with DNA and act as potent PCR inhibitors, interfering with nucleic acid amplification and potentially leading to false negatives or reduced assay sensitivity [38,39]. Thus, we incorporated an internal amplification control (B. subtilis DNA) to monitor that each reaction worked correctly. This internal control is especially important to detect false negatives when testing environmental specimens containing PCR inhibitors. Finally, our assay tolerates competition with more than 1000 fold excess concentration of other Legionella spp. strains [34]. In addition to these technical strengths, the species-specific PCR assay for L. micdadei offers operational advantages for public health and environmental monitoring. The rapid turnaround time (3–4 h) and low LOD facilitate L. micdadei detection and support surveillance of water and soil sources, potentially enabling more effective outbreak response compared to culture-based methods. Clinical presentation of L. micdadei infections can differ from other Legionella spp. cases and could be missed without a high level of suspicion and access to environmental detection and diagnostic testing. In its current form, the PCR can help public health investigate L. micdadei infections and enable heightened surveillance of suspected or confirmed environmental sources. After clinical validation with patient specimens, this PCR could also be used to diagnose patients faster.
As a limitation, this study relied on PCR assays developed and evaluated with specimens collected from a limited number of sources. While a range of environmental matrices (tap water, water faucet swabs, soil and dust) was included, few (soil and mulch) naturally contain L. micdadei. Thus, the detection of the target organism from water specimens was confirmed by simulation (i.e., spiking bacteria). This evaluation does not fully represent the complexity and variability encountered in the environment.
5. Conclusions
Our L. micdadei-specific PCR assay has the potential to provide rapid and accurate detection of a rare opportunistic pathogen, enhancing environmental surveillance. Legionella species-specific assays like the one presented in this report can provide public health authorities with a better understanding of environmental sources and transmission dynamics, thus enable better responses to outbreaks. Such advances are required as epidemiological surveillance indicates an increase in cases of Legionnaires’ disease and Pontiac fever [40]. The utility of this PCR assay could be expanded by validating it in respiratory specimens from infected patients, thereby enhancing diagnosis and facilitating targeted therapy, particularly in immunocompromised patients at an increased risk of severe disease.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/idr17050131/s1, Supplemental Table S1: Bacterial strains and other specimens used in this study.
Author Contributions
Conceptualization, methodology, software, formal analysis, investigation, resources, data curation, W.N.B., M.B., E.B., M.G. and S.I.; writing—original draft preparation, visualization, W.N.B., M.B., M.T. and S.I.; validation, writing—review and editing, M.G., Y.G.S., V.D., K.B., C.L., C.R.-R., M.J.-W., S.T., D.B.-P., A.R. and I.T.; supervision, project administration, funding acquisition, S.I. All authors have read and agreed to the published version of the manuscript.
Funding
This project was funded by institutional funds and FRQS. S.I. is a recipient of an FRQS Clinical Research Scholars—Junior 1 award.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and approved by the research ethics committee of CHU de Québec—Université Laval (registration number 2025-7714); CHU de Québec—Université Laval (protocol code CRI-2025-09-Sains, approved on 28 October 2024).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
All relevant data supporting the findings of this study are included within the article. No additional datasets were generated or analyzed.
Acknowledgments
This paper is part of the Special Issue entitled “Prevention, Diagnosis, and Treatment of Infectious Diseases”, dedicated to the 50th anniversary of the Research Center in Infectious Diseases founded by Michel G. Bergeron at the CHU de Québec–Université Laval. We would like to extend our sincere gratitude to Michel G. Bergeron for his invaluable contributions and for creating such an exceptional research environment. During the preparation of this manuscript, the authors used Perplexity (Sonar, Perplexity AI) and ChatGPT (GPT4o, OpenAI) AI tools to assist with literature searches and English language editing. The authors have reviewed and edited the output and take full responsibility for the content of this publication.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Winn, W.C., Jr. Legionella. In Bergey’s Manual of Systematics of Archaea and Bacteria; American Cancer Society: Hoboken, NJ, USA, 2015; pp. 1–44. ISBN 978-1-118-96060-8. [Google Scholar]
- Barskey, A.; Lee, S.; Hannapel, E.; Smith, J.; Edens, C. Disease Surveillance Summary Report, United States; Division of Bacterial Diseases, National Center for Immunization and Respiratory Diseases, CDC ; Center for Disease Control and Prevention: Atlanta, GA, USA, 2022; p. 49. [Google Scholar]
- Mercante, J.W.; Winchell, J.M. Current and Emerging Legionella Diagnostics for Laboratory and Outbreak Investigations. Clin. Microbiol. Rev. 2015, 28, 95–133. [Google Scholar] [CrossRef]
- van Heijnsbergen, E.; van Deursen, A.; Bouwknegt, M.; Bruin, J.P.; de Roda Husman, A.M.; Schalk, J.A.C. Presence and Persistence of Viable, Clinically Relevant Legionella pneumophila Bacteria in Garden Soil in the Netherlands. Appl. Environ. Microbiol. 2016, 82, 5125–5131. [Google Scholar] [CrossRef]
- Currie, S.L.; Beattie, T.K.; Knapp, C.W.; Lindsay, D.S.J. Legionella Spp. in UK Composts—A Potential Public Health Issue? Clin. Microbiol. Infect. 2014, 20, O224–O229. [Google Scholar] [CrossRef] [PubMed]
- Conza, L.; Pagani, S.C.; Gaia, V. Presence of Legionella and Free-Living Amoebae in Composts and Bioaerosols from Composting Facilities. PLoS ONE 2013, 8, e68244. [Google Scholar] [CrossRef]
- Bennett, J.E.; Dolin, R.; Blaser, M.J. (Eds.) Mandell, Douglas, and Bennett’s Principles and Practice of Infectious Diseases, 9th ed.; Elsevier: Philadelphia, PA, USA, 2020; Volume 2, ISBN 978-0-323-48255-4. [Google Scholar]
- Del Castillo, M.; Lucca, A.; Plodkowski, A.; Huang, Y.-T.; Kaplan, J.; Gilhuley, K.; Babady, N.E.; Seo, S.K.; Kamboj, M. Atypical Presentation of Legionella pneumonia among Patients with Underlying Cancer: A Fifteen-Year Review. J. Infect. 2016, 72, 45–51. [Google Scholar] [CrossRef]
- Lachant, D.; Prasad, P. Legionella micdadei: A Forgotten Etiology of Growing Cavitary Nodules: A Case Report and Literature Review. Case Rep. Pulmonol. 2015, 2015, 535012. [Google Scholar] [CrossRef] [PubMed]
- Foissac, M.; Bergon, L.; Vidal, J.; Cauquil, P.; Mainar, A.; Mourguet, M. Pneumonia and Pulmonary Abscess Due to Legionella micdadei in an Immunocompromised Patient. Germs 2019, 9, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Waldron, P.R.; Martin, B.A.; Ho, D.Y. Mistaken Identity: Legionella micdadei Appearing as Acid-Fast Bacilli on Lung Biopsy of a Hematopoietic Stem Cell Transplant Patient. Transpl. Infect. Dis. 2015, 17, 89–93. [Google Scholar] [CrossRef]
- Palusińska-Szysz, M.; Jurak, M.; Gisch, N.; Waldow, F.; Zehethofer, N.; Nehls, C.; Schwudke, D.; Koper, P.; Mazur, A. The Human LL-37 Peptide Exerts Antimicrobial Activity against Legionella micdadei Interacting with Membrane Phospholipids. Biochim. Biophys. Acta BBA Mol. Cell Biol. Lipids 2022, 1867, 159138. [Google Scholar] [CrossRef]
- Yang, G.; Benson, R.; Pelish, T.; Brown, E.; Winchell, J.M.; Fields, B. Dual Detection of Legionella pneumophila and Legionella Species by Real-Time PCR Targeting the 23S-5S rRNA Gene Spacer Region. Clin. Microbiol. Infect. 2010, 16, 255–261. [Google Scholar] [CrossRef]
- Lee, T.C.; Vickers, R.M.; Yu, V.L.; Wagener, M.M. Growth of 28 Legionella Species on Selective Culture Media: A Comparative Study. J. Clin. Microbiol. 1993, 31, 2764–2768. [Google Scholar] [CrossRef]
- Leber, A.L. (Ed.) Clinical Microbiology Procedures Handbook; ASM Press: Washington, DC, USA, 2016; ISBN 978-1-68367-076-6. [Google Scholar]
- Buchbinder, S.; Leitritz, L.; Trebesius, K.; Banas, B.; Heesemann, J. Mixed Lung Infection by Legionella pneumophila and Legionella gormanii Detected by Fluorescent in Situ Hybridization. Infection 2004, 32, 242–245. [Google Scholar] [CrossRef]
- Coscollá, M.; Fernández, C.; Colomina, J.; Sánchez-Busó, L.; González-Candelas, F. Mixed Infection by Legionella pneumophila in Outbreak Patients. Int. J. Med. Microbiol. 2014, 304, 307–313. [Google Scholar] [CrossRef]
- Wewalka, G.; Schmid, D.; Harrison, T.G.; Uldum, S.A.; Lück, C. Dual Infections with Different Legionella Strains. Clin. Microbiol. Infect. 2014, 20, O13–O19. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Trigui, H.; Matthews, S.; Bedard, E.; Charron, D.; Chea, S.; Fleury, C.; Maldonado, J.F.G.; Rivard, M.; Faucher, S.P.; Prevost, M. Assessment of Monitoring Approaches to Control Legionella pneumophila within a Complex Cooling Tower System. Sci. Total Environ. 2024, 950, 175136. [Google Scholar] [CrossRef] [PubMed]
- Bai, L.; Yang, W.; Li, Y. Clinical and Laboratory Diagnosis of Legionella pneumonia. Diagnostics 2023, 13, 280. [Google Scholar] [CrossRef] [PubMed]
- Jahan, R.; Tarafder, S.; Saleh, A.A.; Miah, R.A. Identification of Legionella from Clinically Diagnosed Pneumonia Patients and Environmental Samples. Bangladesh Med. Res. Counc. Bull. 2016, 41, 24–28. [Google Scholar] [CrossRef][Green Version]
- Young, C.; Smith, D.; Wafer, T.; Crook, B. Rapid Testing and Interventions to Control Legionella Proliferation Following a Legionnaires’ Disease Outbreak Associated with Cooling Towers. Microorganisms 2021, 9, 615. [Google Scholar] [CrossRef]
- Josephson, K.L.; Gerba, C.P.; Pepper, I.L. Polymerase Chain Reaction Detection of Nonviable Bacterial Pathogens. Appl. Environ. Microbiol. 1993, 59, 3513–3515. [Google Scholar] [CrossRef]
- Guo, L.; Wan, K.; Zhu, J.; Ye, C.; Chabi, K.; Yu, X. Detection and Distribution of Vbnc/Viable Pathogenic Bacteria in Full-Scale Drinking Water Treatment Plants. J. Hazard. Mater. 2021, 406, 124335. [Google Scholar] [CrossRef]
- Cross, K.E.; Mercante, J.W.; Benitez, A.J.; Brown, E.W.; Diaz, M.H.; Winchell, J.M. Simultaneous Detection of Legionella Species and L. anisa, L. bozemanii, L. longbeachae and L. micdadei Using Conserved Primers and Multiple Probes in a Multiplex Real-Time PCR Assay. Diagn. Microbiol. Infect. Dis. 2016, 85, 295–301. [Google Scholar] [CrossRef][Green Version]
- Chang, B.; Sugiyama, K.; Taguri, T.; Amemura-Maekawa, J.; Kura, F.; Watanabe, H. Specific Detection of Viable Legionella Cells by Combined Use of Photoactivated Ethidium Monoazide and PCR/Real-Time PCR. Appl. Environ. Microbiol. 2009, 75, 147–153. [Google Scholar] [CrossRef]
- Bernander, S.; Hanson, H.-S.; Johansson, B.; Von Stedingk, L.-V. A Nested Polymerase Chain Reaction for Detection of Legionella pneumophila in Clinical Specimens. Clin. Microbiol. Infect. 1997, 3, 95–101. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhan, X.-Y.; Li, L.-Q.; Hu, C.-H.; Zhu, Q.-Y. Two-Step Scheme for Rapid Identification and Differentiation of Legionella pneumophila and Non-Legionella pneumophila Species. J. Clin. Microbiol. 2010, 48, 433–439. [Google Scholar] [CrossRef] [PubMed]
- Yong, S.F.Y.; Tan, S.H.; Wee, J.; Tee, J.J.; Sansom, F.M.; Newton, H.J.; Hartland, E.L. Molecular Detection of Legionella: Moving on From mip. Front. Microbiol. 2010, 1, 7584. [Google Scholar] [CrossRef] [PubMed]
- Zhan, X.-Y.; Yang, J.-L.; Sun, H.; Zhou, X.; Qian, Y.-C.; Huang, K.; Leng, Y.; Huang, B.; He, Y. Presence of Viable, Clinically Relevant Legionella Bacteria in Environmental Water and Soil Sources of China. Microbiol. Spectr. 2022, 10, e01140-21. [Google Scholar] [CrossRef]
- Toplitsch, D.; Platzer, S.; Zehner, R.; Maitz, S.; Mascher, F.; Kittinger, C. Comparison of Updated Methods for Legionella Detection in Environmental Water Samples. Int. J. Environ. Res. Public. Health 2021, 18, 5436. [Google Scholar] [CrossRef]
- Isabel, S.; Leblanc, É.; Boissinot, M.; Boudreau, D.K.; Grondin, M.; Picard, F.J.; Martel, E.A.; Parham, N.J.; Chain, P.S.; Bader, D.E.; et al. Divergence among Genes Encoding the Elongation Factor Tu of Yersinia Species. J. Bacteriol. 2008, 190, 7548–7558. [Google Scholar] [CrossRef]
- Isabel, S.; Boissinot, M.; Charlebois, I.; Fauvel, C.M.; Shi, L.-E.; Lévesque, J.-C.; Paquin, A.T.; Bastien, M.; Stewart, G.; Leblanc, É.; et al. Rapid Filtration Separation-Based Sample Preparation Method for Bacillus Spores in Powdery and Environmental Matrices. Appl. Environ. Microbiol. 2012, 78, 1505–1512. [Google Scholar] [CrossRef]
- Parks, D.H.; Chuvochina, M.; Rinke, C.; Mussig, A.J.; Chaumeil, P.-A.; Hugenholtz, P. GTDB: An Ongoing Census of Bacterial and Archaeal Diversity through a Phylogenetically Consistent, Rank Normalized and Complete Genome-Based Taxonomy. Nucleic Acids Res. 2022, 50, D785–D794. [Google Scholar] [CrossRef]
- ISO 12869:2019; Water Quality—Detection and Quantification of Legionella spp. and/or Legionella pneumophila by Concentration and Genic Amplification by Quantitative Polymerase Chain Reaction (qPCR). ISO: Geneva, Switzerland, 2019.
- Yin, X.; Chen, Y.-Z.; Ye, Q.-Q.; Liao, L.-J.; Cai, Z.-R.; Lin, M.; Li, J.-N.; Zhang, G.-B.; Peng, X.-L.; Shi, W.-F.; et al. Detection Performance of PCR for Legionella pneumophila in Environmental Samples: A Systematic Review and Meta-Analysis. Ann. Clin. Microbiol. Antimicrob. 2022, 21, 12. [Google Scholar] [CrossRef]
- Kralik, P.; Ricchi, M. A Basic Guide to Real Time PCR in Microbial Diagnostics: Definitions, Parameters, and Everything. Front. Microbiol. 2017, 8, 108. [Google Scholar] [CrossRef]
- van Doorn, R.; Klerks, M.M.; van Gent-Pelzer, M.P.E.; Speksnijder, A.G.C.L.; Kowalchuk, G.A.; Schoen, C.D. Accurate Quantification of Microorganisms in PCR-Inhibiting Environmental DNA Extracts by a Novel Internal Amplification Control Approach Using Biotrove OpenArrays. Appl. Environ. Microbiol. 2009, 75, 7253–7260. [Google Scholar] [CrossRef][Green Version]
- Trombley Hall, A.; McKay Zovanyi, A.; Christensen, D.R.; Koehler, J.W.; Devins Minogue, T. Evaluation of Inhibitor-Resistant Real-Time PCR Methods for Diagnostics in Clinical and Environmental Samples. PLoS ONE 2013, 8, e73845. [Google Scholar] [CrossRef]
- Han, X.Y. Effects of Climate Changes and Road Exposure on the Rapidly Rising Legionellosis Incidence Rates in the United States. PLoS ONE 2021, 16, e0250364. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).