Gastrointestinal Microbiota Dysbiosis Associated with SARS-CoV-2 Infection in Colorectal Cancer: The Implication of Probiotics
Abstract
1. Introduction
2. GI Microbiota and CRC
3. GI Microbiota Dysbiosis Associated with SARS-CoV-2 Infection and CRC
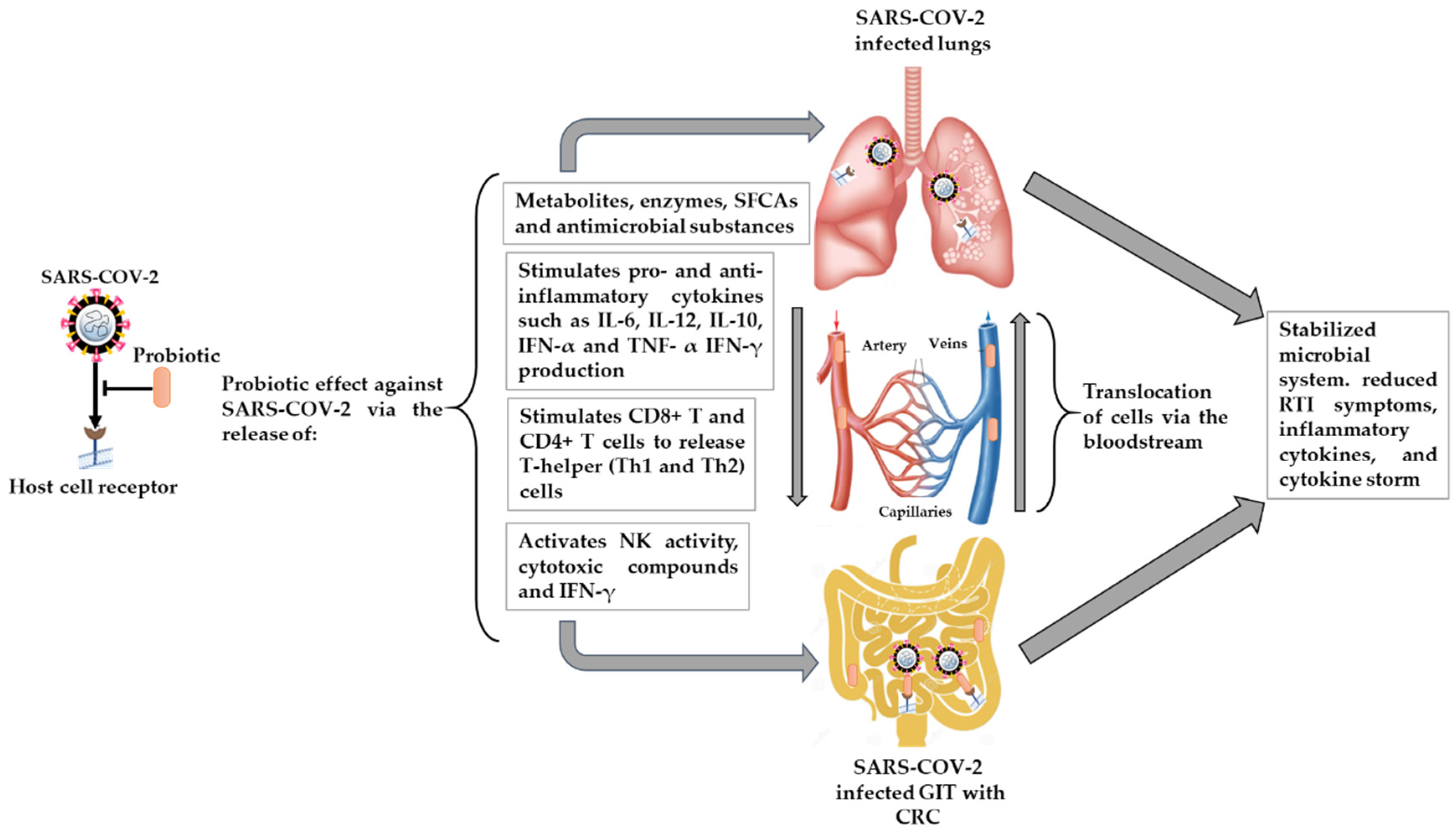
4. Probiotics, GI Microbiota, CRC, and RTIs
4.1. The Effect of Probiotics on CRC and Other GIT Disorders
4.1.1. CRC
4.1.2. IBD
4.1.3. IBS
4.1.4. Diarrhea
4.1.5. Obesity
4.1.6. Possible Mechanism of Probiotics
4.2. The Potential Effect of Probiotics on RTIs, including SARS-CoV-2 Infection, and Possible Mechanisms
4.2.1. Microbial Dysbiosis and GIT–Lung Stability Cross-Talk
4.2.2. Immunomodulatory Effects
4.3. Limitations
5. Conclusions and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO. WHO Coronavirus (COVID-19). Dashboard 2021. Available online: https://covid19.who.int/ (accessed on 23 November 2021).
- Andersen, K.G.; Rambaut, A.; Lipkin, W.I.; Holmes, E.C.; Garry, R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020, 26, 450–452. [Google Scholar] [CrossRef] [PubMed]
- Gorbalenya, A.E.; Baker, S.C.; Baric, R.S.; de Groot, R.J.; Drosten, C.; Gulyaeva, A.A.; Haagmans, B.; Lauber, C.; Leontovich, A.; Neuman, B. Coronaviridae Study Group of the International Committee on Taxonomy of Viruses. The species severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020, 5, 536–544. [Google Scholar]
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N. Genomic characterization and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef]
- Chen, N.; Zhou, M.; Dong, X.; Qu, J.; Gong, F.; Han, Y.; Qiu, Y.; Wang, J.; Liu, Y.; Wei, Y. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet 2020, 395, 507–513. [Google Scholar] [CrossRef]
- Wang, D.; Hu, B.; Hu, C.; Zhu, F.; Liu, X.; Zhang, J.; Wang, B.; Xiang, H.; Cheng, Z.; Xiong, Y. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan, China. JAMA 2020, 323, 1061–1069. [Google Scholar] [CrossRef]
- Zhang, H.; Kang, Z.; Gong, H.; Xu, D.; Wang, J.; Li, Z.; Li, Z.; Cui, X.; Xiao, J.; Zhan, J. Digestive system is a potential route of COVID-19: An analysis of single-cell coexpression pattern of key proteins in the viral entry process. Gut 2020, 69, 1010–1018. [Google Scholar] [CrossRef]
- Lin, L.; Jiang, X.; Zhang, Z.; Huang, S.; Zhang, Z.; Fang, Z.; Gu, Z.; Gao, L.; Shi, H.; Mai, L. Gastrointestinal symptoms of 95 cases with SARS-CoV-2 infection. Gut 2020, 69, 997–1001. [Google Scholar] [CrossRef]
- Hikmet, F.; Méar, L.; Edvinsson, Å.; Micke, P.; Uhlén, M.; Lindskog, C. The protein expression profile of ACE2 in human tissues. Mol. Syst. Biol. 2020, 16, e9610. [Google Scholar] [CrossRef]
- Shang, J.; Wan, Y.; Luo, C.; Ye, G.; Geng, Q.; Auerbach, A.; Li, F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2020, 117, 11727–11734. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020, 181, 271–280.e278. [Google Scholar] [CrossRef]
- Caniglia, J.L.; Asuthkar, S.; Tsung, A.J.; Guda, M.R.; Velpula, K.K. Immunopathology of galectin-3: An increasingly promising target in COVID-19. F1000Research 2020, 9, 1078. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Revilla, J.; Deierborg, T.; Venero, J.L.; Boza-Serrano, A. Hyperinflammation and fibrosis in severe COVID-19 patients: Galectin-3, a target molecule to consider. Front. Immunol. 2020, 11, 2069. [Google Scholar] [CrossRef] [PubMed]
- Baker, A.N.; Richards, S.-J.; Guy, C.S.; Congdon, T.R.; Hasan, M.; Zwetsloot, A.J.; Gallo, A.; Lewandowski, J.R.; Stansfeld, P.J.; Straube, A. The SARS-CoV-2 spike protein binds sialic acids and enables rapid detection in a lateral flow point of care diagnostic device. ACS Cent. Sci. 2020, 6, 2046–2052. [Google Scholar] [CrossRef] [PubMed]
- Helal, M.A.; Shouman, S.; Abdelwaly, A.; Elmehrath, A.O.; Essawy, M.; Sayed, S.M.; Saleh, A.H.; El-Badri, N. Molecular basis of the potential interaction of SARS-CoV-2 spike protein to CD147 in COVID-19 associated-lymphopenia. J. Biomol. Struct. Dyn. 2020, 1–11. [Google Scholar] [CrossRef]
- Reyfman, P.A.; Walter, J.M.; Joshi, N.; Anekalla, K.R.; McQuattie-Pimentel, A.C.; Chiu, S.; Fernandez, R.; Akbarpour, M.; Chen, C.-I.; Ren, Z. Single-cell transcriptomic analysis of human lung provides insights into the pathobiology of pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 2019, 199, 1517–1536. [Google Scholar] [CrossRef]
- Xiao, F.; Tang, M.; Zheng, X.; Liu, Y.; Li, X.; Shan, H. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology 2020, 158, 1831–1833.e3. [Google Scholar] [CrossRef]
- Zang, R.; Castro, M.F.G.; McCune, B.T.; Zeng, Q.; Rothlauf, P.W.; Sonnek, N.M.; Liu, Z.; Brulois, K.F.; Wang, X.; Greenberg, H.B. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci. Immunol. 2020, 5, eabc3582. [Google Scholar] [CrossRef]
- Niu, P.; Lei, F.; Gu, J. Colorectal cancer and COVID-19: Do we need to raise awareness and vigilance? Cancer 2021, 127, 979. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Gagnière, J.; Raisch, J.; Veziant, J.; Barnich, N.; Bonnet, R.; Buc, E.; Bringer, M.-A.; Pezet, D.; Bonnet, M. Gut microbiota imbalance and colorectal cancer. World J. Gastroenterol. 2016, 22, 501. [Google Scholar] [CrossRef]
- Louis, P.; Hold, G.L.; Flint, H.J. The gut microbiota, bacterial metabolites, and colorectal cancer. Nat. Rev. Microbiol. 2014, 12, 661–672. [Google Scholar] [CrossRef]
- Gu, S.; Chen, Y.; Wu, Z.; Chen, Y.; Gao, H.; Lv, L.; Guo, F.; Zhang, X.; Luo, R.; Huang, C. Alterations of the gut microbiota in patients with coronavirus disease 2019 or H1N1 influenza. Clin. Infect. Dis. 2020, 71, 2669–2678. [Google Scholar] [CrossRef]
- Khatiwada, S.; Subedi, A. Lung microbiome and coronavirus disease 2019 (COVID-19): Possible link and implications. Hum. Microbiome J. 2020, 17, 100073. [Google Scholar] [CrossRef] [PubMed]
- Zuo, T.; Zhang, F.; Lui, G.C.; Yeoh, Y.K.; Li, A.Y.; Zhan, H.; Wan, Y.; Chung, A.C.; Cheung, C.P.; Chen, N. Alterations in the gut microbiota of patients with COVID-19 during the time of hospitalization. Gastroenterology 2020, 159, 944–955. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-C.; Lin, W.-C.; Kong, M.-S.; Shi, H.N.; Walker, W.A.; Lin, C.-Y.; Huang, C.-T.; Lin, Y.-C.; Jung, S.-M.; Lin, T.-Y. Oral inoculation of probiotics Lactobacillus acidophilus NCFM suppresses tumor growth both in segmental orthotopic colon cancer and extra-intestinal tissue. Br. J. Nutr. 2012, 107, 1623–1634. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Guo, B.; Gao, R.; Zhu, Q.; Qin, H. Microbiota dysbiosis is associated with colorectal cancer. Front. Microbiol. 2015, 6, 20. [Google Scholar] [CrossRef]
- Beech, A.S.; Lea, S.; Kolsum, U.; Wang, Z.; Miller, B.E.; Donaldson, G.C.; Wedzicha, J.A.; Brightling, C.E.; Singh, D. Bacteria and sputum inflammatory cell counts; a COPD cohort analysis. Respir. Res. 2020, 21, 289. [Google Scholar] [CrossRef] [PubMed]
- Edouard, S.; Million, M.; Bachar, D.; Dubourg, G.; Michelle, C.; Ninove, L.; Charrel, R.; Raoult, D. The nasopharyngeal microbiota in patients with viral respiratory tract infections is enriched in bacterial pathogens. Eur. J. Clin. Microbiol. Infect. Dis. 2018, 37, 1725–1733. [Google Scholar] [CrossRef]
- Man, W.H.; van Houten, M.A.; Mérelle, M.E.; Vlieger, A.M.; Chu, M.L.J.; Jansen, N.J.; Sanders, E.A.; Bogaert, D. Bacterial and viral respiratory tract microbiota and host characteristics in children with lower respiratory tract infections: A matched case-control study. Lancet Respir. Med. 2019, 7, 417–426. [Google Scholar] [CrossRef]
- Zuo, T.; Zhan, H.; Zhang, F.; Liu, Q.; Tso, E.Y.; Lui, G.C.; Chen, N.; Li, A.; Lu, W.; Chan, F.K. Alterations in the fecal fungal microbiome of patients with COVID-19 during the time of hospitalization until discharge. Gastroenterology 2020, 159, 1302–1310. [Google Scholar] [CrossRef] [PubMed]
- Ahlawat, S.; Sharma, K.K. Immunological coordination between gut and lungs in SARS-CoV-2 infection. Virus Res. 2020, 286, 198103. [Google Scholar] [CrossRef] [PubMed]
- AKTAŞ, B.; Aslim, B. Gut-lung axis and dysbiosis in COVID-19. Turk. J. Biol. 2020, 44, 265–272. [Google Scholar] [CrossRef] [PubMed]
- Din, A.U.; Mazhar, M.; Waseem, M.; Ahmad, W.; Bibi, A.; Hassan, A.; Ali, N.; Gang, W.; Qian, G.; Ullah, R. SARS-CoV-2 microbiome dysbiosis linked disorders and possible probiotics role. Biomed. Pharmacother. 2021, 133, 110947. [Google Scholar] [CrossRef] [PubMed]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S. Expert consensus document: The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef] [PubMed]
- Hotel, A.C.P.; Cordoba, A. Health and nutritional properties of probiotics in food including powder milk with live lactic acid bacteria. Prevention 2001, 5, 1–10. [Google Scholar]
- Vasiljevic, T.; Shah, N.P. Probiotics—from Metchnikoff to bioactive. Int. Dairy J. 2008, 18, 714–728. [Google Scholar] [CrossRef]
- Sanders, M.E.; Gibson, G.; Gill, H.S.; Guarner, F. Probiotics: Their potential to impact human health. Counc. Agric. Sci. Technol. Issue Pap. 2007, 36, 1–20. [Google Scholar]
- Baud, D.; Dimopoulou Agri, V.; Gibson, G.R.; Reid, G.; Giannoni, E. Using probiotics to flatten the curve of coronavirus disease COVID-2019 pandemic. Front. Public Health 2020, 8, 186. [Google Scholar] [CrossRef]
- Lenoir-Wijnkoop, I.; Gerlier, L.; Roy, D.; Reid, G. The clinical and economic impact of probiotics consumption on respiratory tract infections: Projections for Canada. PLoS ONE 2016, 11, e0166232. [Google Scholar] [CrossRef]
- Szajewska, H.; Kołodziej, M.; Gieruszczak-Białek, D.; Skórka, A.; Ruszczyński, M.; Shamir, R. Systematic review with meta-analysis: Lactobacillus rhamnosus GG for treating acute gastroenteritis in children—A 2019 update. Aliment. Pharmacol. Ther. 2019, 49, 1376–1384. [Google Scholar] [CrossRef]
- Guillemard, E.; Tondu, F.; Lacoin, F.; Schrezenmeir, J. Consumption of a fermented dairy product containing the probiotic Lactobacillus casei DN-114 001 reduces the duration of respiratory infections in the elderly in a randomized controlled trial. Br. J. Nutr. 2010, 103, 58–68. [Google Scholar] [CrossRef] [PubMed]
- Manna, S.; Chowdhury, T.; Chakraborty, R.; Mandal, S.M. Probiotics-derived peptides and their immunomodulatory molecules can play a preventive role against viral diseases including COVID-19. Probiotics Antimicrob. Proteins 2021, 13, 611–623. [Google Scholar] [CrossRef] [PubMed]
- Power, D.; Burton, J.; Chilcott, C.; Dawes, P.; Tagg, J. Preliminary investigations of the colonization of upper respiratory tract tissues of infants using a pediatric formulation of the oral probiotic Streptococcus salivarius K12. Eur. J. Clin. Microbiol. Infect. Dis. 2008, 27, 1261–1263. [Google Scholar] [CrossRef] [PubMed]
- Weiss, G.; Rasmussen, S.; Zeuthen, L.H.; Nielsen, B.N.; Jarmer, H.; Jespersen, L.; Frøkiær, H. Lactobacillus acidophilus induces virus immune defense genes in murine dendritic cells by a Toll-like receptor-2-dependent mechanism. Immunology 2010, 131, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Lehtoranta, L.; Kalima, K.; He, L.; Lappalainen, M.; Roivainen, M.; Närkiö, M.; Mäkelä, M.; Siitonen, S.; Korpela, R.; Pitkäranta, A. Specific probiotics and virological findings in symptomatic conscripts attending military service in Finland. J. Clin. Virol. 2014, 60, 276–281. [Google Scholar] [CrossRef]
- Hendler, R.; Zhang, Y. Probiotics in the treatment of colorectal cancer. Medicines 2018, 5, 101. [Google Scholar] [CrossRef]
- Meng, C.; Bai, C.; Brown, T.D.; Hood, L.E.; Tian, Q. Human gut microbiota and gastrointestinal cancer. Genom. Proteom. Bioinform. 2018, 16, 33–49. [Google Scholar] [CrossRef]
- Raskov, H.; Burcharth, J.; Pommergaard, H.-C. Linking gut microbiota to colorectal cancer. J. Cancer 2017, 8, 3378. [Google Scholar] [CrossRef]
- Sender, R.; Fuchs, S.; Milo, R. Are we really vastly outnumbered? Revisiting the ratio of bacterial to host cells in humans. Cell 2016, 164, 337–340. [Google Scholar] [CrossRef]
- Boleij, A.; Tjalsma, H. Gut bacteria in health and disease: A survey on the interface between intestinal microbiology and colorectal cancer. Biol. Rev. 2012, 87, 701–730. [Google Scholar] [CrossRef]
- Aureli, P.; Capurso, L.; Castellazzi, A.M.; Clerici, M.; Giovannini, M.; Morelli, L.; Poli, A.; Pregliasco, F.; Salvini, F.; Zuccotti, G.V. Probiotics and health: An evidence-based review. Pharmacol. Res. 2011, 63, 366–376. [Google Scholar] [CrossRef] [PubMed]
- Claesson, M.J.; Cusack, S.; O′Sullivan, O.; Greene-Diniz, R.; de Weerd, H.; Flannery, E.; Marchesi, J.R.; Falush, D.; Dinan, T.; Fitzgerald, G. Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. S1), 4586–4591. [Google Scholar] [CrossRef]
- Marchesi, J.R. Human distal gut microbiome. Environ. Microbiol. 2011, 13, 3088–3102. [Google Scholar] [CrossRef] [PubMed]
- Hakansson, A.; Molin, G. Gut microbiota and inflammation. Nutrients 2011, 3, 637–682. [Google Scholar] [CrossRef] [PubMed]
- Sekirov, I.; Russell, S.L.; Antunes, L.C.M.; Finlay, B.B. Gut microbiota in health and disease. Physiol. Rev. 2010, 90, 859–904. [Google Scholar] [CrossRef]
- Quigley, E.M.M. Gut microbiota and the role of probiotics in therapy. Curr. Opin. Pharmacol. 2011, 11, 593–603. [Google Scholar] [CrossRef]
- Swidsinski, A.; Weber, J.; Loening-Baucke, V.; Hale, L.P.; Lochs, H. Spatial organization, and composition of the mucosal flora in patients with inflammatory bowel disease. J. Clin. Microbiol. 2005, 43, 3380–3389. [Google Scholar] [CrossRef]
- Quigley, E.M.; Abu-Shanab, A. Small intestinal bacterial overgrowth. Infect. Dis. Clin. 2010, 24, 943–959. [Google Scholar] [CrossRef]
- Srikanth, C.; McCormick, B.A. Interactions of the intestinal epithelium with the pathogen and the indigenous microbiota: A three-way crosstalk. Interdiscip. Perspect. Infect. Dis. 2008, 2008, 626827. [Google Scholar] [CrossRef]
- Castellarin, M.; Warren, R.L.; Freeman, J.D.; Dreolini, L.; Krzywinski, M.; Strauss, J.; Barnes, R.; Watson, P.; Allen-Vercoe, E.; Moore, R.A. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012, 22, 299–306. [Google Scholar] [CrossRef]
- Marchesi, J.R.; Dutilh, B.E.; Hall, N.; Peters, W.H.; Roelofs, R.; Boleij, A.; Tjalsma, H. Towards the human colorectal cancer microbiome. PLoS ONE 2011, 6, e20447. [Google Scholar] [CrossRef] [PubMed]
- Sobhani, I.; Tap, J.; Roudot-Thoraval, F.; Roperch, J.P.; Letulle, S.; Langella, P.; Corthier, G.; Van Nhieu, J.T.; Furet, J.P. Microbial dysbiosis in colorectal cancer (CRC) patients. PLoS ONE 2011, 6, e16393. [Google Scholar] [CrossRef] [PubMed]
- Ray, K. Fusobacterium nucleatum found in colon cancer tissue—Could an infection cause colorectal cancer? Nat. Rev. Gastroenterol. Hepatol. 2011, 8, 662. [Google Scholar] [CrossRef] [PubMed]
- Strauss, J.; Kaplan, G.G.; Beck, P.L.; Rioux, K.; Panaccione, R.; DeVinney, R.; Lynch, T.; Allen-Vercoe, E. Invasive potential of gut mucosa-derived Fusobacterium nucleatum positively correlates with IBD status of the host. Inflamm. Bowel Dis. 2011, 17, 1971–1978. [Google Scholar] [CrossRef]
- Flynn, K.J.; Baxter, N.T.; Schloss, P.D. Metabolic and community synergy of oral bacteria in colorectal cancer. Msphere 2016, 1, e00102–e00116. [Google Scholar] [CrossRef] [PubMed]
- Kostic, A.D.; Chun, E.; Robertson, L.; Glickman, J.N.; Gallini, C.A.; Michaud, M.; Clancy, T.E.; Chung, D.C.; Lochhead, P.; Hold, G.L. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 2013, 14, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Hirayama, A.; Kami, K.; Sugimoto, M.; Sugawara, M.; Toki, N.; Onozuka, H.; Kinoshita, T.; Saito, N.; Ochiai, A.; Tomita, M. Quantitative metabolome profiling of colon and stomach cancer microenvironment by capillary electrophoresis time-of-flight mass spectrometry. Cancer Res. 2009, 69, 4918–4925. [Google Scholar] [CrossRef]
- Tjalsma, H.; Boleij, A.; Marchesi, J.R.; Dutilh, B.E. A bacterial driver–passenger model for colorectal cancer: Beyond the usual suspects. Nat. Rev. Microbiol. 2012, 10, 575–582. [Google Scholar] [CrossRef] [PubMed]
- Sears, C.L.; Garrett, W.S. Microbes, microbiota, and colon cancer. Cell Host Microbe 2014, 15, 317–328. [Google Scholar] [CrossRef] [PubMed]
- Uronis, J.M.; Mühlbauer, M.; Herfarth, H.H.; Rubinas, T.C.; Jones, G.S.; Jobin, C. Modulation of the intestinal microbiota alters colitis-associated colorectal cancer susceptibility. PLoS ONE 2009, 4, e6026. [Google Scholar] [CrossRef]
- Wang, X.; Allen, T.D.; May, R.J.; Lightfoot, S.; Houchen, C.W.; Huycke, M.M. Enterococcus faecalis induces aneuploidy and tetraploidy in colonic epithelial cells through a bystander effect. Cancer Res. 2008, 68, 9909–9917. [Google Scholar] [CrossRef] [PubMed]
- Advani, S.M.; Advani, P.S.; Brown, D.W.; DeSantis, S.M.; Korphaisarn, K.; VonVille, H.M.; Bressler, J.; Lopez, D.S.; Davis, J.S.; Daniel, C.R. Global differences in the prevalence of the CpG island methylator phenotype of colorectal cancer. BMC Cancer 2019, 19, 964. [Google Scholar] [CrossRef]
- Cheriyamundath, S.; Ben-Ze’ev, A. Wnt/β-Catenin target genes in colon cancer metastasis: The special case of L1cam. Cancers 2020, 12, 3444. [Google Scholar] [CrossRef] [PubMed]
- McCrea, P.D.; Gottardi, C.J. Beyond β-catenin: Prospects for a larger catenin network in the nucleus. Nat. Rev. Mol. Cell Biol. 2016, 17, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Boland, C.R.; Goel, A. Microsatellite instability in colorectal cancer. Gastroenterology 2010, 138, 2073–2087. [Google Scholar] [CrossRef] [PubMed]
- Arthur, J.C.; Gharaibeh, R.Z.; Mühlbauer, M.; Perez-Chanona, E.; Uronis, J.M.; McCafferty, J.; Fodor, A.A.; Jobin, C. Microbial genomic analysis reveals the essential role of inflammation in bacteria-induced colorectal cancer. Nat. Commun. 2014, 5, 4724. [Google Scholar] [CrossRef]
- Enaud, R.; Prevel, R.; Ciarlo, E.; Beaufils, F.; Wieërs, G.; Guery, B.; Delhaes, L. The gut-lung axis in health and respiratory diseases: A place for inter-organ and inter-kingdom crosstalks. Front. Cell. Infect. Microbiol. 2020, 10, 9. [Google Scholar] [CrossRef]
- Mulak, A. The impact of probiotics on interactions within the microbiota-gut-lung triad in COVID-19. Int. J. Food Sci. Nutr. 2021, 72, 577–578. [Google Scholar] [CrossRef]
- Zuo, T.; Liu, Q.; Zhang, F.; Lui, G.C.-Y.; Tso, E.Y.; Yeoh, Y.K.; Chen, Z.; Boon, S.S.; Chan, F.K.; Chan, P.K. Depicting SARS-CoV-2 fecal viral activity in association with gut microbiota composition in patients with COVID-19. Gut 2021, 70, 276–284. [Google Scholar]
- Viana, S.D.; Nunes, S.; Reis, F. ACE2 imbalance as a key player for the poor outcomes in COVID-19 patients with age-related comorbidities–Role of gut microbiota dysbiosis. Ageing Res. Rev. 2020, 62, 101123. [Google Scholar] [CrossRef]
- Mönkemüller, K.; Fry, L.C.; Rickes, S. Systemic inflammatory response and thrombosis due to alterations in the gut microbiota in COVID-19. Rev. Esp. Enferm. Dig. Organo Of. Soc. Esp. Patol. Dig. 2020, 112, 584–585. [Google Scholar]
- Tang, L.; Gu, S.; Gong, Y.; Li, B.; Lu, H.; Li, Q.; Zhang, R.; Gao, X.; Wu, Z.; Zhang, J. Clinical significance of the correlation between changes in the major intestinal bacteria species and COVID-19 severity. Engineering 2020, 6, 1178–1184. [Google Scholar] [CrossRef]
- Dhar, D.; Mohanty, A. Gut microbiota and COVID-19—Possible link and implications. Virus Res. 2020, 285, 198018. [Google Scholar] [CrossRef] [PubMed]
- Gou, W.; Fu, Y.; Yue, L.; Chen, G.-d.; Cai, X.; Shuai, M.; Xu, F.; Yi, X.; Chen, H.; Zhu, Y.J. Gut microbiota may underlie the predisposition of healthy individuals to COVID-19. medRxiv 2020. [Google Scholar] [CrossRef]
- Brown, A.; Fernández, I.S.; Gordiyenko, Y.; Ramakrishnan, V. Ribosome-dependent activation of stringent control. Nature 2016, 534, 277–280. [Google Scholar] [CrossRef]
- Brown, M.V.; Reader, J.S.; Tzima, E. Mammalian aminoacyl-tRNA synthetases: Cell signaling functions of the protein translation machinery. Vasc. Pharmacol. 2010, 52, 21–26. [Google Scholar] [CrossRef]
- Kim, Y.; Sundrud, M.S.; Zhou, C.; Edenius, M.; Zocco, D.; Powers, K.; Zhang, M.; Mazitschek, R.; Rao, A.; Yeo, C.-Y. Aminoacyl-tRNA synthetase inhibition activates a pathway that branches from the canonical amino acid response in mammalian cells. Proc. Natl. Acad. Sci. USA 2020, 117, 8900–8911. [Google Scholar] [CrossRef]
- Zhang, S.; Zeng, X.; Ren, M.; Mao, X.; Qiao, S. Novel metabolic and physiological functions of branched-chain amino acids: A review. J. Anim. Sci. Biotechnol. 2017, 8, 10. [Google Scholar] [CrossRef]
- Bao, R.; Hernandez, K.; Huang, L.; Luke, J.J. ACE2 and TMPRSS2 expression by clinical, HLA, immune, and microbial correlates across 34 human cancers and matched normal tissues: Implications for SARS-CoV-2 COVID-19. J. Immunother. Cancer 2020, 8, e001020. [Google Scholar] [CrossRef]
- Lamers, M.M.; Beumer, J.; van der Vaart, J.; Knoops, K.; Puschhof, J.; Breugem, T.I.; Ravelli, R.B.; Van Schayck, J.P.; Mykytyn, A.Z.; Duimel, H.Q. SARS-CoV-2 productively infects human gut enterocytes. Science 2020, 369, 50–54. [Google Scholar] [CrossRef]
- Liu, C.; Wang, K.; Zhang, M.; Hu, X.; Hu, T.; Liu, Y.; Hu, Q.; Wu, S.; Yue, J. High expression of ACE2 and TMPRSS2 and clinical characteristics of COVID-19 in colorectal cancer patients. NPJ Precis. Oncol. 2021, 5, 1. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Li, H.-B.; Lyu, J.-R.; Lei, X.-M.; Li, W.; Wu, G.; Lyu, J.; Dai, Z.-M. Specific ACE2 expression in small intestinal enterocytes may cause gastrointestinal symptoms and injury after 2019-nCoV infection. Int. J. Infect. Dis. 2020, 96, 19–24. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Zhang, H.; Zhong, Y.; Chua, M.L.K.; Xie, C. Reply to colorectal cancer and COVID-19: Do we need to raise awareness and vigilance? Cancer 2021, 127, 980–981. [Google Scholar] [CrossRef] [PubMed]
- Deriu, E.; Boxx, G.M.; He, X.; Pan, C.; Benavidez, S.D.; Cen, L.; Rozengurt, N.; Shi, W.; Cheng, G. Influenza virus affects intestinal microbiota and secondary Salmonella infection in the gut through type I Interferons. PLoS Pathog. 2016, 12, e1005572. [Google Scholar] [CrossRef]
- Zha, L.; Garrett, S.; Sun, J. Salmonella infection in chronic inflammation and gastrointestinal cancer. Diseases 2019, 7, 28. [Google Scholar] [CrossRef]
- Ma, W.-T.; Yao, X.-T.; Peng, Q.; Chen, D.-K. The protective and pathogenic roles of IL-17 in viral infections: Friend or foe? Open Biol. 2019, 9, 190109. [Google Scholar] [CrossRef]
- Wang, X.; Ma, K.; Chen, M.; Ko, K.-H.; Zheng, B.-J.; Lu, L. IL-17A Promotes Pulmonary B-1a Cell Differentiation via Induction of Blimp-1 Expression during Influenza Virus Infection. PLoS Pathog. 2016, 12, e1005367. [Google Scholar] [CrossRef]
- Ivanov, I.I.; de Llanos Frutos, R.; Manel, N.; Yoshinaga, K.; Rifkin, D.B.; Sartor, R.B.; Finlay, B.B.; Littman, D.R. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host. Microb. 2008, 4, 337–349. [Google Scholar] [CrossRef]
- Mazmanian, S.K.; Round, J.L.; Kasper, D.L. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 2008, 453, 620–625. [Google Scholar] [CrossRef]
- Sobhani, I.; Le Gouvello, S. Critical role for CD8+ FoxP3+ regulatory T cells in colon cancer immune response in humans. Gut 2009, 58, 743–744. [Google Scholar] [CrossRef]
- Wu, S.; Rhee, K.-J.; Albesiano, E.; Rabizadeh, S.; Wu, X.; Yen, H.-R.; Huso, D.L.; Brancati, F.L.; Wick, E.; McAllister, F. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat. Med. 2009, 15, 1016–1022. [Google Scholar] [CrossRef]
- van Dam, P.A.; Huizing, M.; Mestach, G.; Dierckxsens, S.; Tjalma, W.; Trinh, X.B.; Papadimitriou, K.; Altintas, S.; Vermorken, J.; Vulsteke, C.; et al. SARS-CoV-2 and cancer: Are they really partners in crime? Cancer Treat. Rev. 2020, 89, 102068. [Google Scholar] [CrossRef]
- McGill, A.R.; Kahlil, R.; Dutta, R.; Green, R.; Howell, M.; Mohapatra, S.; Mohapatra, S.S. SARS-CoV-2 Immuno-pathogenesis and potential for diverse vaccines and therapies: Opportunities and challenges. Infect. Dis. Rep. 2021, 13, 102–125. [Google Scholar] [CrossRef] [PubMed]
- Howell, M.C.; Green, R.; McGill, A.R.; Dutta, R.; Mohapatra, S.; Mohapatra, S.S. SARS-CoV-2-Induced Gut Microbiome Dysbiosis: Implications for Colorectal Cancer. Cancers 2021, 13, 2676. [Google Scholar] [CrossRef] [PubMed]
- Vandenplas, Y.; Salvatore, S.; Viera, M.; Devreker, T.; Hauser, B. Probiotics in infectious diarrhea in children: Are they indicated? Eur. J. Pediatr. 2007, 166, 1211–1218. [Google Scholar] [CrossRef] [PubMed]
- Meurman, J.H.; Stamatova, I. Probiotics: Contributions to oral health. Oral Dis. 2007, 13, 443–451. [Google Scholar] [CrossRef]
- Canani, R.B.; Cirillo, P.; Terrin, G.; Cesarano, L.; Spagnuolo, M.I.; De Vincenzo, A.; Albano, F.; Passariello, A.; De Marco, G.; Manguso, F. Probiotics for treatment of acute diarrhea in children: A randomized clinical trial of five different preparations. BMJ 2007, 335, 340. [Google Scholar] [CrossRef]
- Krasaekoopt, W.; Bhandari, B.; Deeth, H. Evaluation of encapsulation techniques of probiotics for yogurt. Int. Dairy J. 2003, 13, 3–13. [Google Scholar] [CrossRef]
- Food and Agriculture Organisation. WHO Working Group Report on Drafting Guidelines for the Evaluation of Probiotics in Food; Food and Agriculture Organization of the United Nations, World Health Organization: London, ON, Canada, 2002; p. 30. [Google Scholar]
- Quigley, E.M. Gut bacteria in health and disease. Gastroenterol. Hepatol. 2013, 9, 560. [Google Scholar]
- Saad, N.; Delattre, C.; Urdaci, M.; Schmitter, J.-M.; Bressollier, P. An overview of the last advances in the probiotic and prebiotic field. LWT Food. Sci. Technol. 2013, 50, 1–16. [Google Scholar] [CrossRef]
- Baldwin, C.; Millette, M.; Oth, D.; Ruiz, M.T.; Luquet, F.-M.; Lacroix, M. Probiotic Lactobacillus acidophilus and Lactobacillus casei mix sensitize colorectal tumoral cells to 5-fluorouracil-induced apoptosis. Nutr. Cancer 2010, 62, 371–378. [Google Scholar] [CrossRef]
- An, J.; Ha, E.-M. Combination therapy of Lactobacillus plantarum supernatant and 5-fluouracil increases chemosensitivity in colorectal cancer cells. J. Microbiol. Biotechnol. 2016, 26, 1490–1503. [Google Scholar] [CrossRef]
- Escamilla, J.; Lane, M.A.; Maitin, V. Cell-free supernatants from probiotic Lactobacillus casei and Lactobacillus rhamnosus GG decrease colon cancer cell invasion in vitro. Nutr. Cancer 2012, 64, 871–878. [Google Scholar] [CrossRef] [PubMed]
- Dallal, M.M.S.; Mojarrad, M.; Baghbani, F.; Raoofian, R.; Mardaneh, J.; Salehipour, Z. Effects of probiotic Lactobacillus acidophilus and Lactobacillus casei on colorectal tumor cells activity (CaCo-2). Arch. Iran. Med. 2015, 18, 167–172. [Google Scholar]
- Cousin, F.J.; Jouan-Lanhouet, S.; Théret, N.; Brenner, C.; Jouan, E.; Le Moigne-Muller, G.; Dimanche-Boitrel, M.-T.; Jan, G. The probiotic Propionibacterium freudenreichii as a new adjuvant for TRAIL-based therapy in colorectal cancer. Oncotarget 2016, 7, 7161. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.-Y.; Hsieh, Y.-M.; Huang, C.-C.; Tsai, C.-C. Inhibitory effects of probiotic Lactobacillus on the growth of human colonic carcinoma cell line HT-29. Molecules 2017, 22, 107. [Google Scholar] [CrossRef]
- Kahouli, I.; Malhotra, M.; Westfall, S.; Alaoui-Jamali, M.A.; Prakash, S. Design and validation of an orally administrated active L. fermentum-L. acidophilus probiotic formulation using colorectal cancer Apc Min/+ mouse model. Appl. Microbiol. Biotechnol. 2017, 101, 1999–2019. [Google Scholar] [CrossRef]
- Appleyard, C.B.; Cruz, M.L.; Isidro, A.A.; Arthur, J.C.; Jobin, C.; De Simone, C. Pretreatment with the probiotic VSL# 3 delays transition from inflammation to dysplasia in a rat model of colitis-associated cancer. Am. J. Physiol. Gastrointest. 2011, 301, G1004–G1013. [Google Scholar]
- Yan, F.; Cao, H.; Cover, T.L.; Washington, M.K.; Shi, Y.; Liu, L.; Chaturvedi, R.; Peek, R.M.; Wilson, K.T.; Polk, D.B. Colon-specific delivery of a probiotic-derived soluble protein ameliorates intestinal inflammation in mice through an EGFR-dependent mechanism. J. Clin. Investig. 2011, 121, 2242–2253. [Google Scholar] [CrossRef]
- Hu, J.; Wang, C.; Ye, L.; Yang, W.; Huang, H.; Meng, F.; Shi, S.; Ding, Z. Anti-tumour immune effect of oral administration of Lactobacillus plantarum to CT26 tumor-bearing mice. J. Biosci. 2015, 40, 269–279. [Google Scholar] [CrossRef]
- Odun-Ayo, F.; Mellem, J.; Reddy, L. Improving the survival of probiotics in simulated conditions and azoxymethane-induced colon tumor-bearing mice using modified citrus pectin-alginate microencapsulation. Afr. J. Tradit. Complement. Altern. Med. 2016, 13, 101–109. [Google Scholar] [CrossRef][Green Version]
- Odun-Ayo, F.; Mellem, J.; Reddy, L. The effect of modified citrus pectin-probiotic on fecal Lactobacilli in Balb/c mice. Food. Sci. Technol. 2017, 37, 478–482. [Google Scholar] [CrossRef]
- Song, H.; Wang, W.; Shen, B.; Jia, H.; Hou, Z.; Chen, P.; Sun, Y. Pretreatment with probiotic Bifico ameliorates colitis-associated cancer in mice: Transcriptome and gut flora profiling. Cancer Sci. 2018, 109, 666–677. [Google Scholar] [CrossRef] [PubMed]
- Heydari, Z.; Rahaie, M.; Alizadeh, A.M.; Agah, S.; Khalighfard, S.; Bahmani, S. Effects of Lactobacillus acidophilus and Bifidobacterium bifidum probiotics on the expression of microRNAs 135b, 26b, 18a and 155, and their involving genes in mice colon cancer. Probiotics Antimicrob. 2019, 11, 1155–1162. [Google Scholar] [CrossRef]
- Mego, M.; Chovanec, J.; Vochyanova-Andrezalova, I.; Konkolovsky, P.; Mikulova, M.; Reckova, M.; Miskovska, V.; Bystricky, B.; Beniak, J.; Medvecova, L. Prevention of irinotecan induced diarrhea by probiotics: A randomized double blind, placebo controlled pilot study. Complement. Ther. Med. 2015, 23, 356–362. [Google Scholar] [CrossRef] [PubMed]
- Kotzampassi, K.; Stavrou, G.; Damoraki, G.; Georgitsi, M.; Basdanis, G.; Tsaousi, G.; Giamarellos-Bourboulis, E.J. A four-probiotics regimen reduces postoperative complications after colorectal surgery: A randomized, double-blind, placebo-controlled study. World J. Surg. 2015, 39, 2776–2783. [Google Scholar] [CrossRef]
- Theodoropoulos, G.E.; Memos, N.A.; Peitsidou, K.; Karantanos, T.; Spyropoulos, B.G.; Zografos, G. Synbiotics and gastrointestinal function-related quality of life after elective colorectal cancer resection. Ann. Gastroenterol. 2016, 29, 56. [Google Scholar] [PubMed]
- Hibberd, A.A.; Lyra, A.; Ouwehand, A.C.; Rolny, P.; Lindegren, H.; Cedgård, L.; Wettergren, Y. Intestinal microbiota is altered in patients with colon cancer and modified by probiotic intervention. BMJ Open Gastroenterol. 2017, 4, e000145. [Google Scholar] [CrossRef]
- Kahouli, I.; Tomaro-Duchesneau, C.; Prakash, S. Probiotics in colorectal cancer (CRC) with emphasis on mechanisms of action and current perspectives. J. Med. Microbiol. 2013, 62, 1107–1123. [Google Scholar] [CrossRef]
- Malago, J.J.; Tooten, P.C.; Koninkx, J.F. Anti-inflammatory properties of probiotic bacteria on Salmonella-induced IL-8 synthesis in enterocyte-like Caco-2 cells. Benef. Microbes 2010, 1, 121–130. [Google Scholar] [CrossRef]
- Odun-Ayo, F.; Mellem, J.; Naicker, T.; Reddy, L. Chemoprevention of azoxymethane-induced colonic carcinogenesis in Balb/c mice using a modified pectin alginate probiotic. Anticancer Res. 2015, 35, 4765–4775. [Google Scholar]
- Liu, D.; Jiang, X.-Y.; Zhou, L.-S.; Song, J.-H.; Zhang, X. Effects of probiotics on intestinal mucosa barrier in patients with colorectal cancer after operation: A meta-analysis of randomized controlled trials. Medicine 2016, 95, e3342. [Google Scholar] [CrossRef] [PubMed]
- Arora, T.; Singh, S.; Sharma, R.K. Probiotics: Interaction with gut microbiome and antiobesity potential. Nutrition 2013, 29, 591–596. [Google Scholar] [CrossRef] [PubMed]
- Gholizadeh, P.; Mahallei, M.; Pormohammad, A.; Varshochi, M.; Ganbarov, K.; Zeinalzadeh, E.; Yousefi, B.; Bastami, M.; Tanomand, A.; Mahmood, S.S. Microbial balance in the intestinal microbiota and its association with diabetes, obesity and allergic disease. Microb. Pathog. 2019, 127, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Gregoret, V.; Perezlindo, M.; Vinderola, G.; Reinheimer, J.; Binetti, A. A comprehensive approach to determine the probiotic potential of human-derived Lactobacillus for industrial use. Food Microbiol. 2013, 34, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Yadav, H.; Jain, S.; Yadav, M. Probiotics and Diabetes/Obesity. In Bioactive Food as Dietary Interventions for Diabetes; Academic Press: Cambridge, MA, USA, 2012; pp. 307–317. [Google Scholar]
- Fernandez, E.M.; Valenti, V.; Rockel, C.; Hermann, C.; Pot, B.; Boneca, I.G.; Grangette, C. Anti-inflammatory capacity of selected lactobacilli in experimental colitis is driven by NOD2-mediated recognition of a specific peptidoglycan-derived muropeptide. Gut 2011, 60, 1050–1059. [Google Scholar] [CrossRef]
- Doherty, G.; Bennett, G.; Cheifetz, A.; Moss, A. Meta-analysis: Targeting the intestinal microbiota in prophylaxis for postoperative Crohn’s disease. Aliment. Pharmacol. Ther. 2010, 31, 802–809. [Google Scholar] [CrossRef]
- Niedzielin, K.; Kordecki, H.; ena Birkenfeld, B. A controlled, double-blind, randomized study on the efficacy of Lactobacillus plantarum 299V in patients with irritable bowel syndrome. Eur. J. Gastroenterol. Hepatol. 2001, 13, 1143–1147. [Google Scholar] [CrossRef]
- Kajander, K.; Myllyluoma, E.; Rajilić-Stojanović, M.; Kyrönpalo, S.; Rasmussen, M.; Järvenpää, S.; Zoetendal, E.G.; de Vos, W.M.; Vapaatalo, H.; Korpela, R. Clinical trial: Multispecies probiotic supplementation alleviates the symptoms of irritable bowel syndrome and stabilizes intestinal microbiota. Aliment. Pharmacol. Ther. 2008, 27, 48–57. [Google Scholar] [CrossRef]
- McFarland, L.V.; Dublin, S. Meta-analysis of probiotics for the treatment of irritable bowel syndrome. World J. Gastroenterol. 2008, 14, 2650. [Google Scholar] [CrossRef]
- Fernandez-Duarte, K.P.; Olaya-Galán, N.N.; Salas-Cárdenas, S.P.; Lopez-Rozo, J.; Gutierrez-Fernandez, M.F. Bifidobacterium adolescentis (DSM 20083) and Lactobacillus casei (Lafti L26-DSL): Probiotics able to block the in vitro adherence of rotavirus in MA104 cells. Probiotics Antimicrob. 2018, 10, 56–63. [Google Scholar] [CrossRef]
- Lee, D.K.; Park, J.E.; Kim, M.J.; Seo, J.G.; Lee, J.H.; Ha, N.J. Probiotic bacteria, B. longum, and L. acidophilus inhibit infection by rotavirus in vitro and decrease the duration of diarrhea in pediatric patients. Clin. Res. Hepatol. Gastroenterol. 2015, 39, 237–244. [Google Scholar] [CrossRef] [PubMed]
- Olaya Galán, N.; Ulloa Rubiano, J.; Velez Reyes, F.; Fernandez Duarte, K.; Salas Cardenas, S.; Gutierrez Fernandez, M. In vitro antiviral activity of Lactobacillus casei and Bifidobacterium adolescentis against rotavirus infection monitored by NSP 4 protein production. J. Appl. Microbiol. 2016, 120, 1041–1051. [Google Scholar] [CrossRef] [PubMed]
- Hojsak, I.; Abdović, S.; Szajewska, H.; Milošević, M.; Krznarić, Ž.; Kolaček, S. Lactobacillus rhamnosus GG in the prevention of nosocomial gastrointestinal and respiratory tract infections. Pediatrics 2010, 125, e1171–e1177. [Google Scholar] [CrossRef] [PubMed]
- Preidis, G.A.; Hill, C.; Guerrant, R.L.; Ramakrishna, B.; Tannock, G.W.; Versalovic, J. Probiotics, enteric and diarrheal diseases, and global health. Gastroenterology 2011, 140, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Sarker, S.A.; Sultana, S.; Fuchs, G.J.; Alam, N.H.; Azim, T.; Brüssow, H.; Hammarström, L. Lactobacillus paracasei strain ST11 has no effect on rotavirus but ameliorates the outcome of nonrotavirus diarrhea in children from Bangladesh. Pediatrics 2005, 116, e221–e228. [Google Scholar] [CrossRef]
- Turnbaugh, P.J.; Bäckhed, F.; Fulton, L.; Gordon, J.I. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe 2008, 3, 213–223. [Google Scholar] [CrossRef]
- Kadooka, Y.; Sato, M.; Imaizumi, K.; Ogawa, A.; Ikuyama, K.; Akai, Y.; Okano, M.; Kagoshima, M.; Tsuchida, T. Regulation of abdominal adiposity by probiotics (Lactobacillus gasseri SBT2055) in adults with obese tendencies in a randomized controlled trial. Eur. J. Clin. Nutr. 2010, 64, 636–643. [Google Scholar] [CrossRef]
- Rafter, J. Probiotics and colon cancer. Best Pract. Res. Clin. Gastroenterol. 2003, 17, 849–859. [Google Scholar] [CrossRef]
- Sivamaruthi, B.S.; Kesika, P.; Chaiyasut, C. The role of probiotics in colorectal cancer management. Evid. Based Complement. Alternat. Med. 2020, 2020, 3535982. [Google Scholar] [CrossRef]
- Zhu, Y.; Luo, T.M.; Jobin, C.; Young, H.A. Gut microbiota and probiotics in colon tumorigenesis. Cancer Lett. 2011, 309, 119–127. [Google Scholar] [CrossRef]
- Chong, E.S.L. A potential role of probiotics in colorectal cancer prevention: A review of possible mechanisms of action. Best Pract. Res. Clin. Gastroenterol. 2014, 30, 351–374. [Google Scholar] [CrossRef] [PubMed]
- Faghfoori, Z.; Gargari, B.P.; Gharamaleki, A.S.; Bagherpour, H.; Khosroushahi, A.Y. Cellular and molecular mechanisms of probiotics effects on colorectal cancer. J. Funct. Foods 2015, 18, 463–472. [Google Scholar] [CrossRef]
- Pagnini, C.; Corleto, V.D.; Hoang, S.B.; Saeed, R.; Cominelli, F.; Delle Fave, G. Commensal bacteria and “oncologic surveillance”: Suggestions from an experimental model. J. Clin. Gastroenterol. 2008, 42, S193–S196. [Google Scholar] [CrossRef]
- Kumar, R.S.; Kanmani, P.; Yuvaraj, N.; Paari, K.; Pattukumar, V.; Thirunavukkarasu, C.; Arul, V. Lactobacillus plantarum AS1 isolated from south Indian fermented food Kallappam suppress 1,2-dimethyl hydrazine (DMH)-induced colorectal cancer in male Wistar rats. Appl. Biochem. Biotechnol. 2012, 166, 620–631. [Google Scholar] [CrossRef]
- Lin, P.W.; Myers, L.E.; Ray, L.; Song, S.-C.; Nasr, T.R.; Berardinelli, A.J.; Kundu, K.; Murthy, N.; Hansen, J.M.; Neish, A.S. Lactobacillus rhamnosus blocks inflammatory signaling in vivo via reactive oxygen species generation. Free Radic. Biol. Med. 2009, 47, 1205–1211. [Google Scholar] [CrossRef]
- Mohania, D.; Kansal, V.K.; Sagwal, R.; Shah, D. Anticarcinogenic effect of probiotic Dahi and piroxicam on DMH-induced colorectal carcinogenesis in Wistar rats. Am. J. Phys. 2013, 1, 8–24. [Google Scholar]
- Shida, K.; Kiyoshima-Shibata, J.; Nagaoka, M.; Watanabe, K.; Nanno, M. Induction of interleukin-12 by Lactobacillus strains having a rigid cell wall resistant to intracellular digestion. J. Dairy Sci. 2006, 89, 3306–3317. [Google Scholar] [CrossRef]
- Jahani-Sherafat, S.; Alebouyeh, M.; Moghim, S.; Amoli, H.A.; Ghasemian-Safaei, H. Role of gut microbiota in the pathogenesis of colorectal cancer; a review article. Gastroenterol. Hepatol. Bed Bench 2018, 11, 101. [Google Scholar] [PubMed]
- Zhu, Q.; Gao, R.; Wu, W.; Qin, H. The role of gut microbiota in the pathogenesis of colorectal cancer. Tumor Biol. 2013, 34, 1285–1300. [Google Scholar] [CrossRef] [PubMed]
- Den Besten, G.; Van Eunen, K.; Groen, A.K.; Venema, K.; Reijngoud, D.-J.; Bakker, B.M. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J. Lipid Res. 2013, 54, 2325–2340. [Google Scholar] [CrossRef]
- Macfarlane, G.T.; Macfarlane, S. Bacteria, colonic fermentation, and gastrointestinal health. J. AOAC Int. 2012, 95, 50–60. [Google Scholar] [CrossRef]
- Nogal, A.; Valdes, A.M.; Menni, C. The role of short-chain fatty acids in the interplay between gut microbiota and diet in cardio-metabolic health. Gut Microbes 2021, 13, 1897212. [Google Scholar] [CrossRef]
- Ooi, L.-G.; Liong, M.-T. Cholesterol-lowering effects of probiotics and prebiotics: A review of in vivo and in vitro findings. Int. J. Mol. Sci. 2010, 11, 2499–2522. [Google Scholar] [CrossRef]
- De Preter, V.; Raemen, H.; Cloetens, L.; Houben, E.; Rutgeerts, P.; Verbeke, K. Effect of dietary intervention with different pre-and probiotics on intestinal bacterial enzyme activities. Eur. J. Clin. Nutr. 2008, 62, 225–231. [Google Scholar] [CrossRef]
- Nakamura, J.; Kubota, Y.; Miyaoka, M.; Saitoh, T.; Mizuno, F.; Benno, Y. Comparison of four microbial enzymes in Clostridia and Bacteroides isolated from human feces. Microbiol. Immunol. 2002, 46, 487–490. [Google Scholar] [CrossRef]
- Hijová, E.; Kuzma, J.; Strojný, L.; Bomba, A.; Bertková, I.; Chmelárová, A.; Hertelyová, Z.; Kuliková, L.; Štofilová, J.; Ambro, Ľ. Effect of Lactobacillus plantarum LS/07 on intestinal bacterial enzyme activities in the prevention of cancer, atherosclerosis, and dysbiosis. Acta Vet. Beogr. 2016, 66, 294–303. [Google Scholar] [CrossRef]
- Mroczynska, M.; Libudzisz, Z. Beta-glucuronidase and beta-glucosidase activity of Lactobacillus and Enterococcus isolated from human feces. Pol. J. Microbiol. 2010, 59, 265–269. [Google Scholar] [CrossRef]
- Sadeghi-Aliabadi, H.; Mohammadi, F.; Fazeli, H.; Mirlohi, M. Effects of Lactobacillus plantarum A7 with probiotic potential on colon cancer and normal cells proliferation in comparison with a commercial strain. Iran. J. Basic Med. Sci. 2014, 17, 815. [Google Scholar]
- Kim, S.-J.; Cho, S.Y.; Kim, S.H.; Song, O.-J.; Shin, I.-S.; Cha, D.S.; Park, H.J. Effect of microencapsulation on viability and other characteristics in Lactobacillus acidophilus ATCC 43121. LWT-Food Sci. Technol. 2008, 41, 493–500. [Google Scholar] [CrossRef]
- Rowland, I.; Rumney, C.; Coutts, J.; Lievense, L. Effect of Bifidobacterium longum and inulin on gut bacterial metabolism and carcinogen-induced aberrant crypt foci in rats. Carcinogenesis 1998, 19, 281–285. [Google Scholar] [CrossRef] [PubMed]
- Hatakka, K.; Holma, R.; El-Nezami, H.; Suomalainen, T.; Kuisma, M.; Saxelin, M.; Poussa, T.; Mykkänen, H.; Korpela, R. The influence of Lactobacillus rhamnosus LC705 together with Propionibacterium freudenreichii ssp. shermanii JS on potentially carcinogenic bacterial activity in the human colon. Int. J. Food Microbiol. 2008, 128, 406–410. [Google Scholar] [CrossRef]
- Bruce, W.R. Recent hypotheses for the origin of colon cancer. Cancer Res. 1987, 47, 4237–4242. [Google Scholar] [PubMed]
- Lidbeck, A.; Allinger, U.G.; Orrhage, K.; Ottova, L.; Brismar, B.; Gustafsson, J.-Å.; Rafter, J.; Nord, C. Impact of Lactobacillus acidophilus supplements on the fecal microflora and soluble fecal bile acids in colon cancer patients. Microb. Ecol. Health Dis. 1991, 4, 81–88. [Google Scholar]
- De Vrese, M.; Winkler, P.; Rautenberg, P.; Harder, T.; Noah, C.; Laue, C.; Ott, S.; Hampe, J.; Schreiber, S.; Heller, K.; et al. Effect of Lactobacillus gasseri PA 16/8, Bifidobacterium longum SP 07/3, B. bifidum MF 20/5 on common cold episodes: A double-blind, randomized, controlled trial. Clin. Nutr. 2005, 24, 481–491. [Google Scholar] [CrossRef]
- Kawahara, T.; Takahashi, T.; Oishi, K.; Tanaka, H.; Masuda, M.; Takahashi, S.; Takano, M.; Kawakami, T.; Fukushima, K.; Kanazawa, H. Consecutive oral administration of Bifidobacterium longum MM-2 improves the defense system against influenza virus infection by enhancing natural killer cell activity in a murine model. Microbiol. Immunol. 2015, 59, 1–12. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, Q.; Jiang, Y.; Yang, W.; Huang, H.; Shi, C.; Yang, G.; Wang, C. Surface-displayed porcine IFN-λ3 in Lactobacillus plantarum inhibits porcine enteric coronavirus infection of porcine intestinal epithelial cells. J. Microbiol. Biotechnol. 2020, 30, 515–525. [Google Scholar] [CrossRef]
- Namba, K.; Hatano, M.; Yaeshima, T.; Takase, M.; Suzuki, K. Effects of Bifidobacterium longum BB536 administration on influenza infection, influenza vaccine antibody titer, and cell-mediated immunity in the elderly. Biosci. Biotechnol. Biochem. 2010, 74, 939–945. [Google Scholar] [CrossRef]
- Panigrahi, P.; Parida, S.; Nanda, N.C.; Satpathy, R.; Pradhan, L.; Chandel, D.S.; Baccaglini, L.; Mohapatra, A.; Mohapatra, S.S.; Misra, P.R. A randomized synbiotic trial to prevent sepsis among infants in rural India. Nature 2017, 548, 407–412. [Google Scholar] [CrossRef]
- Turner, R.B.; Woodfolk, J.A.; Borish, L.; Steinke, J.W.; Patrie, J.T.; Muehling, L.M.; Lahtinen, S.; Lehtinen, M.J. Effect of probiotic on innate inflammatory response and viral shedding in experimental rhinovirus infection—A randomized controlled trial. Benef. Microbes 2017, 8, 207–215. [Google Scholar] [CrossRef]
- Waki, N.; Matsumoto, M.; Fukui, Y.; Suganuma, H. Effects of probiotic Lactobacillus brevis KB 290 on the incidence of influenza infection among schoolchildren: An open-label pilot study. Lett. Appl. Microbiol. 2014, 59, 565–571. [Google Scholar] [CrossRef]
- Zelaya, H.; Alvarez, S.; Kitazawa, H.; Villena, J. Respiratory antiviral immunity and immunobiotics: Beneficial effects on inflammation-coagulation interaction during influenza virus infection. Front. Immunol. 2016, 7, 633. [Google Scholar] [CrossRef]
- Hao, Q.; Dong, B.R.; Wu, T. Probiotics for preventing acute upper respiratory tract infections. Cochrane Database Syst. Rev. 2015, 21, Cd006895. [Google Scholar] [CrossRef]
- Sugimura, T.; Takahashi, H.; Jounai, K.; Ohshio, K.; Kanayama, M.; Tazumi, K.; Tanihata, Y.; Miura, Y.; Fujiwara, D.; Yamamoto, N. Effects of oral intake of plasmacytoid dendritic cells-stimulative lactic acid bacterial strain on the pathogenesis of influenza-like illness and immunological response to influenza virus. Br. J. Nutr. 2015, 114, 727–733. [Google Scholar] [CrossRef]
- Wang, B.; Hylwka, T.; Smieja, M.; Surrette, M.; Bowdish, D.M.; Loeb, M. Probiotics to prevent respiratory infections in nursing homes: A pilot randomized controlled trial. J. Am. Geriatr. Soc. 2018, 66, 1346–1352. [Google Scholar] [CrossRef]
- Izumo, T.; Maekawa, T.; Ida, M.; Noguchi, A.; Kitagawa, Y.; Shibata, H.; Yasui, H.; Kiso, Y. Effect of intranasal administration of Lactobacillus pentosus S-PT84 on influenza virus infection in mice. Int. Immunopharmacol. 2010, 10, 1101–1106. [Google Scholar] [CrossRef]
- Makino, S.; Sato, A.; Goto, A.; Nakamura, M.; Ogawa, M.; Chiba, Y.; Hemmi, J.; Kano, H.; Takeda, K.; Okumura, K. Enhanced natural killer cell activation by exopolysaccharides derived from yogurt fermented with Lactobacillus delbrueckii ssp. bulgaricus OLL1073R-1. J. Dairy Sci. 2016, 99, 915–923. [Google Scholar] [CrossRef]
- Luoto, R.; Ruuskanen, O.; Waris, M.; Kalliomäki, M.; Salminen, S.; Isolauri, E. Prebiotic and probiotic supplementation prevents rhinovirus infections in preterm infants: A randomized, placebo-controlled trial. J. Allergy Clin. Immunol. 2014, 133, 405–413. [Google Scholar] [CrossRef]
- Jespersen, L.; Tarnow, I.; Eskesen, D.; Morberg, C.M.; Michelsen, B.; Bügel, S.; Dragsted, L.O.; Rijkers, G.T.; Calder, P.C. Effect of Lactobacillus paracasei subsp. paracasei, L. casei 431 on the immune response to influenza vaccination and upper respiratory tract infections in healthy adult volunteers: A randomized, double-blind, placebo-controlled, parallel-group study. Am. J. Clin. Nutr. 2015, 101, 1188–1196. [Google Scholar] [CrossRef]
- Rodríguez-Díaz, J.; Monedero, V. Probiotics against digestive tract viral infections. Bioact. Food Diet Int. Liver Gastrointest. Dis. 2013, 271–284. [Google Scholar]
- Al Kassaa, I.; Hober, D.; Hamze, M.; Chihib, N.E.; Drider, D. Antiviral potential of lactic acid bacteria and their bacteriocins. Probiotics Antimicrob. Proteins 2014, 6, 177–185s. [Google Scholar] [CrossRef]
- Wan, L.; Chen, Z.; Shah, N.; El-Nezami, H. Modulation of intestinal epithelial defense responses by probiotic bacteria. Crit. Rev. Food Sci. Nutr. 2016, 56, 2628–2641. [Google Scholar] [CrossRef]
- Iwabuchi, N.; Xiao, J.-Z.; Yaeshima, T.; Iwatsuki, K. Oral administration of Bifidobacterium longum ameliorates influenza virus infection in mice. Biol. Pharm. Bull. 2011, 34, 1352–1355. [Google Scholar] [CrossRef]
- Chen, Y.-J.; Wang, S.-F.; Weng, I.-C.; Hong, M.-H.; Lo, T.-H.; Jan, J.-T.; Hsu, L.-C.; Chen, H.-Y.; Liu, F.-T. Galectin-3 enhances avian H5N1 influenza a virus-induced pulmonary inflammation by promoting NLRP3 inflammasome activation. Am. J. Pathol. 2018, 188, 1031–1042. [Google Scholar] [CrossRef]
- Woodward, A.; Mauris, J.; Argüeso, P. Binding of transmembrane mucins to galectin-3 limits herpesvirus 1 infection of human corneal keratinocytes. J. Virol. 2013, 87, 5841–5847. [Google Scholar] [CrossRef]
- Odun-Ayo, F.; Reddy, L. Potential roles of modified pectin targeting galectin-3 against Severe Acute Respiratory Syndrome Coronavirus-2. Multidiscip. Sci. J. 2021, 4, 824–837. [Google Scholar] [CrossRef]
- Chong, H.-X.; Yusoff, N.A.A.; Hor, Y.-Y.; Lew, L.-C.; Jaafar, M.H.; Choi, S.-B.; Yusoff, M.S.; Wahid, N.; Abdullah, M.F.I.; Zakaria, N. Lactobacillus plantarum DR7 improved upper respiratory tract infections via enhancing immune and inflammatory parameters: A randomized, double-blind, placebo-controlled study. J. Dairy Sci. 2019, 102, 4783–4797. [Google Scholar] [CrossRef]
- Garcia-Crespo, K.E.; Chan, C.C.; Gabryszewski, S.J.; Percopo, C.M.; Rigaux, P.; Dyer, K.D.; Domachowske, J.B.; Rosenberg, H.F. Lactobacillus priming of the respiratory tract: Heterologous immunity and protection against lethal pneumovirus infection. Antivir. Res. 2013, 97, 270–279. [Google Scholar] [CrossRef]
- Paolillo, R.; Carratelli, C.R.; Sorrentino, S.; Mazzola, N.; Rizzo, A. Immunomodulatory effects of Lactobacillus plantarum on human colon cancer cells. Int. Immunopharmacol. 2009, 9, 1265–1271. [Google Scholar] [CrossRef]
- Gao, Q.Y.; Chen, Y.X.; Fang, J.Y. 2019 Novel coronavirus infection and gastrointestinal tract. J. Dig. Dis. 2020, 21, 125. [Google Scholar] [CrossRef]
- Baindara, P.; Chakraborty, R.; Holliday, Z.M.; Mandal, S.M.; Schrum, A.G. Oral probiotics in coronavirus disease 2019: Connecting the gut–lung axis to viral pathogenesis, inflammation, secondary infection and clinical trials. New Microbes New Infect. 2021, 40, 4783–4797. [Google Scholar] [CrossRef]

| Disease | Model | Dose of Probiotic and Duration | Probiotic Used and Outcome | References |
|---|---|---|---|---|
| In vitro | ||||
| CRC | LS513 cell + 5-FU | 106–109 cfu/mL; 48 h | Lactobacillus acidophilus CL1285 and Lactobacillus casei LBC80R activate caspase-3 protein, downregulate p21 protein, and increase the apoptosis induction capacity of 5-FU. | [113] |
| CRC | HT-29 and HCT-116 cells | 10 μg (≥107 cfu/mL); 72 h | Lactobacillus plantarum supernatant inhibits 5-FU-resistant colorectal cancer cell lines, inhibits the expression of CD44, 133, 166 markers, and the ALDH1 of cancer stem cells. | [114] |
| CRC | Human HCT-116 cells | 109 cfu/mL; 24–72 h | L. casei ATCC 4356 and L. rhamnosus ATCC 39,392 lower MMP-9 activity and increase ZO-1 protein in cultured metastatic CRC cells. | [115] |
| CRC | CaCo-2 cell | 5 × 106 cfu/mL; 24 h | L. acidophilus and L. casei reduce cell proliferation and cell migration and invasion and increase cell apoptosis. | [116] |
| CRC | HT29 cell | 0–100 ng/mL; 24 h | P. freudenreichii (supernatant or metabolites) induced HT29 cell apoptosis and enhanced TRAIL cytotoxic activity. | [117] |
| CRC | HT29 cell | 109 cfu/mL; 8 h | L. johnsonii BCRC17010 and L. reuteri BCRC14625 show an increase in lactate dehydrogenase (LDH) activity inhibiting HT29 cells. L. plantarum PM153 and L. johnsonii BCRC17010 show good adhesion ability while the latter display potential apoptotic effects. | [118] |
| CRC | Caco-2 cell | 6 × 103–5 × 106 cells; 24–48 h | L. acidophilus ATCC 314 and L. fermentum NCIMB 5221 reduce cell proliferation and increase apoptosis. | [119] |
| In vivo | ||||
| Colitis-associated cancer | 6 weeks old male Sprague-Dawley rats | 5 × 109 cfu per 100 g body weight; 1 week | The probiotic VSL#3 enhances the antiangiogenic factor angiostatin, VDR expression, and alkaline sphingomyelinase. | [120] |
| Acute colitis | 8–12 weeks old WT C57BL/6 mice | 10 μg in pectin/zein beads/mouse/day; 5 days | L. rhamnosus GG inhibits cytokine-induced apoptosis in intestinal epithelial cells and the impairment of barrier function in the colon epithelium, in an EGFR-dependent manner. | [121] |
| CRC | 6–8 weeks old Balb/c CT26 induced adenocarcinoma mice | 1 × 109 cfu/day; 14 weeks | L. plantarum A and L. rhamnosus B up-regulates IFN and promotes Th1-type CD4+ T differentiation. | [122] |
| CRC | 7 weeks old male AOM-induced Balb/c mice | 10 × 109 cfu/mL, intragastric; 4 weeks | L. acidophilus ATCC 4356 significantly increases the number of fecal lactobacilli and intestinal microbiota in treated mice. | [123,124] |
| CRC | 4 weeks old C57BL/6J–APCMin mice | 0.5 × 1010 cfu/intragastric; 12 weeks | L. acidophilus ATCC 314 and L. fermentum NCIMB 5221 reduces intestinal tumor multiplicity and cellular marker downregulation. | [119] |
| Colitis-associated cancer | AOM-induced C57BL/6 mice | 1.2 × 107 cfu/day; 9 weeks | Probiotic Bifico reduces the abundance of Desulfovibrio, Mucispirillum, Odoribacter, and Lactobacillus, which are associated with the expression of CXCR2 ligand genes. | [125] |
| CRC | AOM-induced Balb/c mice | 1.5 g powders of 1 × 109 cfu/g; 5 months | Lactobacillus acidophilus and Bifidobacterium bifidum influence the expression of the tumor suppressor miRNAs and their target genes. | [126] |
| Human | ||||
| CRC | Clinical | 10 × 109 cfu/mL; 3 times daily for 12 weeks | The probiotic formula, Colon DophilusTM, reduces the occurrence of diarrhea and enterocolitis. | [127] |
| CRC | Post-operative treatment | 0.5–1.75 × 109 cfu/capsule twice daily; 14 days | L. acidophilus, L. plantarum, B. lactis, and Saccharomyces boulardii reduce the rate of all main complications after surgery, post-operative pneumonia, and surgical site infections. | [128] |
| CRC | Post-operative treatment | 12 g of sachets/ 10 × 1011 cfu each probiotic + prebiotics; 15 days | Pediococcus pentosaceus, Leuconostoc mesenteroides, Lactobacillus paracasei ssp. paracasei 19, and Lactobacillus plantarum improve postcolectomy gastrointestinal function. | [129] |
| CRC | Pre-operative treatment | 2 daily tablets of 1.4 × 1010 cfu B. lactis and 7 × 109 cfu L. acidophilus | Reduced levels of Fusobacterium and Peptostreptococcus in fecal microbiota, increased number of Faecalibacterium and Clostridiales spp. in the tumor microbiota of CRC patients treated with probiotics. | [130] |
| Disease | Study | Infection | Dose and Duration | Mode of Action and Outcome | References |
|---|---|---|---|---|---|
| Influenza | Clinical | Viral | 5 × 107 cfu/one tablet per day; 14 days | Lactobacillus gasseri PA 16/8, Bifidobacterium longum SP 07/3, and Bifidobacterium bifidum MF 20/5 significantly enhance cytotoxic plus T suppressor (CD8+) and T helper (CD4+) cells, thus reducing the symptoms and duration of common cold episodes and fever, | [178] |
| RTI | Clinical | Viral and bacterial | 1010 cfu/100 g of the probiotic; 3 months | Lactobacillus casei DN-114 001 significantly reduces RTI duration and rhinopharyngitis. | [42] |
| Influenza | Clinical | Viral | 1 × 1011 cfu; 5 weeks | B. longum BB536 reduces the occurrence of influenza and fever, most likely by enhancing innate immunity. | [181] |
| Influenza | Clinical | Viral | 6 × 109 cfu; 8 weeks | Lactobacillus brevis KB290 reduces influenza infection among children. | [184] |
| Influenza | In vivo (6 week-old female Balb/c mice) | Viral | 2.0 × 109 cfu per day; 17 days | B. longum MM-2 decreases virus titers, cell death, and pro-inflammatory cytokines such as IL-6 and TNF-α. | [179] |
| Influenza | Clinical | Viral | 1 × 1011 cfu per day; 10 weeks | Lactococcus lactis ssp. lactis JCM5805 enhances activation and increases IFN-α mediated response. | [187] |
| RTI | Clinical | Viral | 109 cfu per capsule/2 capsules per day; 6 months | Lactobacillus rhamnosus GG significantly reduces RTI duration. | [188] |
| Influenza | In vivo (7 week-old female Balb/c mice) | Viral | 20 μL at a concentration of 10 mg/mL−1 (200 μg per mouse) once daily; 3 days | L. rhamnosus GG improves the level of IFN-γ, proinflammatory factors, and immunoregulatory cytokines such as IL-12, which allow the clearance of virus with minimal inflammatory lung tissue damage. | [185,189] |
| Influenza | Clinical | Viral | 1 × 109 cfu/mL once daily; 3 weeks | Lactobacillus delbrueckii ssp. bulgaricus OLL1073R-1 and Streptococcus thermophilus OLS3059 activate natural killer (NK) cell activity and induce IFN-γ production. | [190] |
| LRTI and rhinovirus infection | Clinical | Viral | 109 cfu/capsule/seven daily doses; 60 days | L. rhamnosus GG reduces LRTI and the incidence of rhinovirus-induced episodes in children. | [191] |
| rhinovirus infection | Clinical | Viral | ≥2 × 109 cfu/satchet daily dose; 28 days | B. animalis subspecies lactis Bl-04 reduces nasal lavage virus titer and influences the innate immune response in the nasal cavity. | [183] |
| URTI | Clinical | Viral | 1 × 109 cfu/subjects once daily; 6 weeks | L. paracasei subsp. Paracasei and L. casei 431 reduce the frequency of RTI episodes. | [192] |
| Coronavirus | In vitro (IPEC-J2 cell line) | Viral | 2 × 106 recombinant cells; 2 h | L. plantarum enhances the expression levels of IFN stimulated genes, thus suppressing the viral infection. | [180] |
| COVID-19 | In silico docking | Viral | Probiotics-derived peptides were docked targeting viral proteins | Probiotics-derived polypeptides show strong affinity binding to the S1-protein receptor-binding domain of SARS-CoV-2 | [43] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Odun-Ayo, F.; Reddy, L. Gastrointestinal Microbiota Dysbiosis Associated with SARS-CoV-2 Infection in Colorectal Cancer: The Implication of Probiotics. Gastroenterol. Insights 2022, 13, 35-59. https://doi.org/10.3390/gastroent13010006
Odun-Ayo F, Reddy L. Gastrointestinal Microbiota Dysbiosis Associated with SARS-CoV-2 Infection in Colorectal Cancer: The Implication of Probiotics. Gastroenterology Insights. 2022; 13(1):35-59. https://doi.org/10.3390/gastroent13010006
Chicago/Turabian StyleOdun-Ayo, Frederick, and Lalini Reddy. 2022. "Gastrointestinal Microbiota Dysbiosis Associated with SARS-CoV-2 Infection in Colorectal Cancer: The Implication of Probiotics" Gastroenterology Insights 13, no. 1: 35-59. https://doi.org/10.3390/gastroent13010006
APA StyleOdun-Ayo, F., & Reddy, L. (2022). Gastrointestinal Microbiota Dysbiosis Associated with SARS-CoV-2 Infection in Colorectal Cancer: The Implication of Probiotics. Gastroenterology Insights, 13(1), 35-59. https://doi.org/10.3390/gastroent13010006







