The Rise of Pluripotent Stem Cell-Derived Glia Models of Neuroinflammation
Abstract
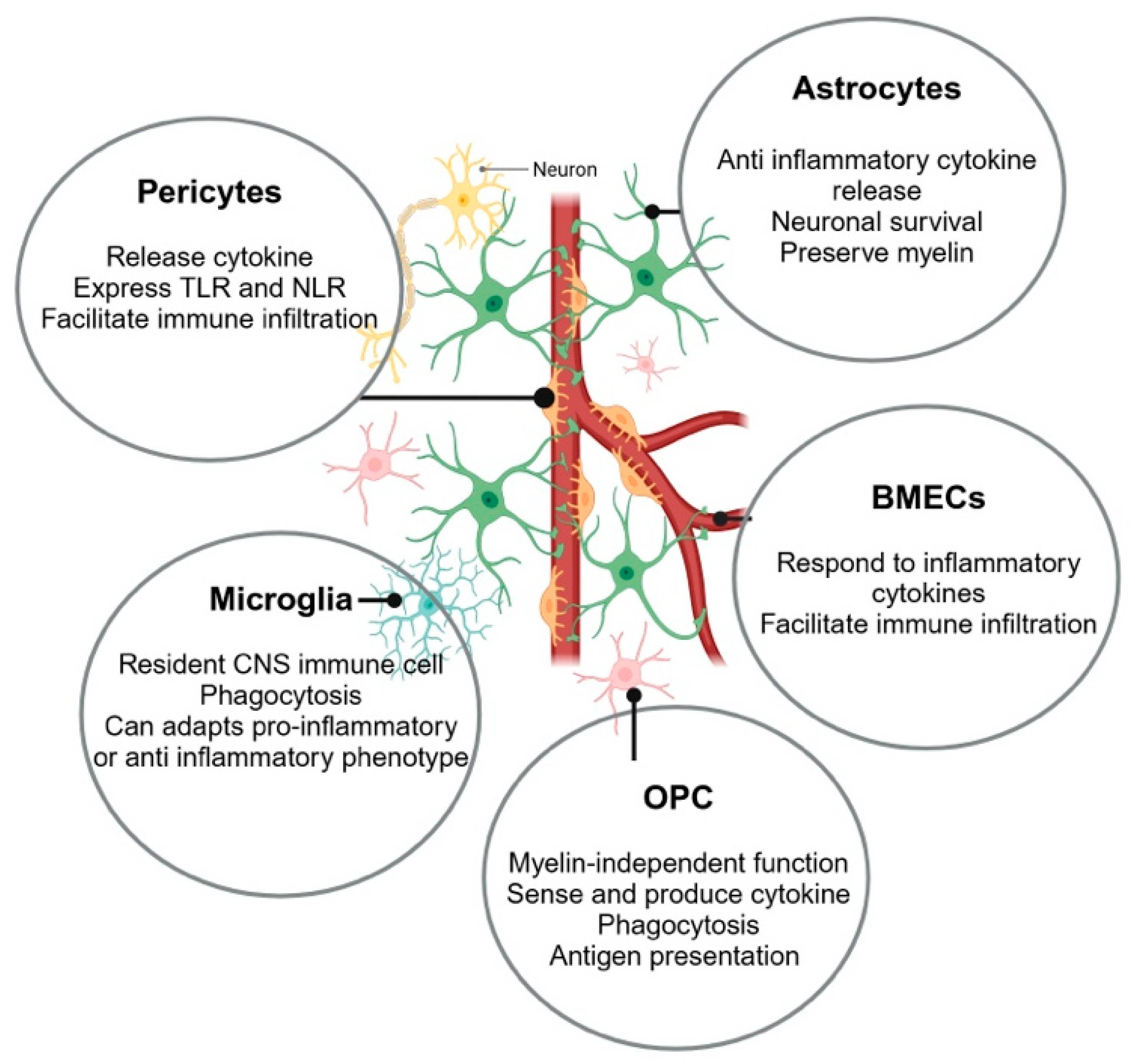
:1. Introduction
2. Microglia
2.1. Differentiation Concepts
2.2. Neuroinflammatory Insights
3. Astrocytes
3.1. Differentiation Concepts
3.2. Neuroinflammatory Insights
| Disease | Key Findings | Source |
|---|---|---|
| NMO |
| [95] |
| AD |
| [59] |
| MS |
| [93] |
| MS |
| [76] |
| General inflammation |
| [79] |
| General inflammation |
| [96] |
| AD |
| [97] |
| AD |
| [98] |
| TDP-43 dementia |
| [99] |
4. Oligodendrocyte Precursor Cells (OPCs)
4.1. Differentiation Concepts
4.2. Neuroinflammatory Insights
| Disease | hiOPC Findings | Source |
|---|---|---|
| AD | hiAs with CLU deletion release proinflammatory cytokines/chemokines resulting in decreased hiOPC proliferation and basic myelin protein production | [137] |
| AD | hiOLs, not hiOPCs, produce higher levels of Aꞵ40 | [138] |
| Secondary progressive MS | hiOPCs have diminished migratory phenotypes and secretomes | [128] |
5. Brain Microvascular Endothelial Cells
5.1. Differentiation Concepts
5.2. Neuroinflammatory Insights
6. Pericytes
6.1. Role in Neuroinflammation
6.2. Differentiation Concepts
7. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CNS | Central nervous system | FGF | Fibroblast growth factor |
| ROS | Reactive oxygen species | BMP | Bone morphogenetic proteins |
| RNS | Reactive nitrogen species | LIF | Leukemia inhibitory factor |
| GWAS | Genome-wide association studies | CNTF | Ciliary neurotrophic factor |
| HIF-1α | Hypoxia-inducible factor 1 alpha | GFAP | Glial fibrillary acidic protein |
| APOE | Apolipoprotein E | GLAST | Glutamate Aspartate Transporter |
| M-CSF | Macrophage colony-stimulating factor | AQP4 | Aquaporin 4 |
| hPSCs | Human pluripotent stem cells | Aβ | Amyloid beta |
| hiPCSs | Human induced pluripotent stem cells | TNF | Tissue necrosis factor |
| MS | Multiple sclerosis | NF-kB | Nuclear factor kappa light chain enhancer of activated B cell |
| PD | Parkinson’s Disease | COVID | Coronavirus disease |
| MHC | Major histocompatibility factor | NMO | Neuromyelitis optica |
| BBB | Blood–brain barrier | TBI | Traumatic brain injury |
| hiMGs | hiPSC-derived microglia-like cells | hiBMECs | hiPSC-derived brain microvascular endothelial cells |
| TMEM119 | Transmembrane protein 119 | VCAM | Vascular cell adhesion protein |
| P2RY12 | Purinergic receptor P2Y G-protein coupled 12 | JAK-STAT | Janus kinase signal transducer and activator of transcription |
| TREM2 | Triggering receptor expressed on myeloid cell 2 | TDP-43 | Transactive response DNA binding protein 43 |
| CD11b | Cluster of differentiation molecule 11b | OPCs | Oligodendrocyte precursor cells |
| CD45 | Cluster of differentiation molecule 45 | NG2 | Neuron glia antigen-2 |
| DAMPs | Damage-associated molecular patterns | hiOPCs | hiPSC-derived OPCs |
| PAMPs | Pathogen-associated molecular patterns | NPC | Neural precursor neuroectoderm-like cell |
| MAF bZIP | Musculoaponeurotic fibrosarcoma oncogene, basic leucine zipper | SOX10 | SRY-box transcription factor 10 |
| AD | Alzheimer’s disease | CLU | Clusterin |
| KO | Knockout | EPCs | Endothelial progenitor cells |
| ATP | Adenosine triphosphate | GO | Gene ontology |
| ADP | Adenosine diphosphate | ICAM | Intercellular Adhesion Molecule |
| aS | Alpha-synuclein | PECAM | Platelet Endothelial Adhesion Molecule |
| IL-1β | Interleukin 1-beta | LPS | Lipopolysaccharide |
| NLRP3 | Nucleotide-binding domain leucine-rich-containing family pyrin domain-containing 3 | PBMCs | Peripheral blood mononuclear cells |
| CRISPR | Clustered regularly interspaced short palindromic repeats | TLRs | Toll-like receptors |
| ALS | Amyotrophic lateral sclerosis | NLRs | Nod-like receptors |
| TBI | Traumatic brain injury | PRRs | Pattern recognition receptors |
| FTD | Frontotemporal dementia | IP-10 | Interferon-inducible protein-10 |
| hiA | hiPSC-astrocyte-like cell | TGF-β1 | Transforming growth factor beta |
| PDGRF β | Platelet-derived growth factor receptor beta |
References
- Leenaars, C.H.C.; Teerenstra, S.; Meijboom, F.L.B.; Bleich, A. Predicting animal to human translation: A proof of concept study using qualitative comparative analysis. medRxiv 2022, medRxiv:2022-01. [Google Scholar] [CrossRef]
- Ritskes-Hoitinga, M.; Leenaars, C.; Beumer, W.; Coenen-de Roo, T.; Stafleu, F.; Meijboom, F.L.B. Improving Translation by Identifying Evidence for More Human-Relevant Preclinical Strategies. Animals 2020, 10, 1170. [Google Scholar] [CrossRef]
- Vasile, F.; Dossi, E.; Rouach, N. Human astrocytes: Structure and functions in the healthy brain. Brain Struct. Funct. 2017, 222, 2017–2029. [Google Scholar] [CrossRef]
- Degl’Innocenti, E.; Dell’Anno, M.T. Human and mouse cortical astrocytes: A comparative view from development to morphological and functional characterization. Front. Neuroanat. 2023, 17, 1130729. [Google Scholar] [CrossRef] [PubMed]
- Padmashri, R.; Ren, B.; Oldham, B.; Jung, Y.; Gough, R.; Dunaevsky, A. Modeling human-specific interlaminar astrocytes in the mouse cerebral cortex. J. Comp. Neurol. 2021, 529, 802–810. [Google Scholar] [CrossRef] [PubMed]
- Geirsdottir, L.; David, E.; Keren-Shaul, H.; Weiner, A.; Bohlen, S.C.; Neuber, J.; Balic, A.; Giladi, A.; Sheban, F.; Dutertre, C.-A.; et al. Cross-Species Single-Cell Analysis Reveals Divergence of the Primate Microglia Program. Cell 2020, 181, 746. [Google Scholar] [CrossRef] [PubMed]
- Patel, T.; Carnwath, T.P.; Wang, X.; Allen, M.; Lincoln, S.J.; Lewis-Tuffin, L.J.; Quicksall, Z.S.; Lin, S.; Tutor-New, F.Q.; Ho, C.C.G.; et al. Transcriptional landscape of human microglia implicates age, sex, and APOE-related immunometabolic pathway perturbations. Aging Cell 2022, 21, e13606. [Google Scholar] [CrossRef]
- Lee, S.; Devanney, N.A.; Golden, L.R.; Smith, C.T.; Schwartz, J.L.; Walsh, A.E.; Clarke, H.A.; Goulding, D.S.; Allenger, E.J.; Morillo-Segovia, G.; et al. APOE modulates microglial immunometabolism in response to age, amyloid pathology, and inflammatory challenge. Cell Rep. 2023, 42, 112196. [Google Scholar] [CrossRef] [PubMed]
- Gastfriend, B.D.; Foreman, K.L.; Katt, M.E.; Palecek, S.P.; Shusta, E.V. Integrative analysis of the human brain mural cell transcriptome. J. Cereb. Blood Flow Metab. 2021, 41, 3052–3068. [Google Scholar] [CrossRef] [PubMed]
- Song, H.W.; Foreman, K.L.; Gastfriend, B.D.; Kuo, J.S.; Palecek, S.P.; Shusta, E.V. Transcriptomic comparison of human and mouse brain microvessels. Sci. Rep. 2020, 10, 12358. [Google Scholar] [CrossRef] [PubMed]
- Goshi, N.; Morgan, R.K.; Lein, P.J.; Seker, E. A primary neural cell culture model to study neuron, astrocyte, and microglia interactions in neuroinflammation. J. Neuroinflamm. 2020, 17, 155. [Google Scholar] [CrossRef] [PubMed]
- van Niekerk, E.A.; Kawaguchi, R.; de Freria, C.M.; Groeniger, K.; Marchetto, M.C.; Dupraz, S.; Bradke, F.; Geschwind, D.H.; Gage, F.H.; Tuszynski, M.H. Methods for culturing adult CNS neurons reveal a CNS conditioning effect. Cell Rep. Methods 2022, 2, 100255. [Google Scholar] [CrossRef]
- Sikora, E.; Bielak-Zmijewska, A.; Dudkowska, M.; Krzystyniak, A.; Mosieniak, G.; Wesierska, M.; Wlodarczyk, J. Cellular Senescence in Brain Aging. Front. Aging Neurosci. 2021, 13, 646924. [Google Scholar] [CrossRef]
- Guo, C.; Yang, L.; Wan, C.-X.; Xia, Y.-Z.; Zhang, C.; Chen, M.-H.; Wang, Z.-D.; Li, Z.-R.; Li, X.-M.; Geng, Y.-D.; et al. Anti-neuroinflammatory effect of Sophoraflavanone G from Sophora alopecuroides in LPS-activated BV2 microglia by MAPK, JAK/STAT and Nrf2/HO-1 signaling pathways. Phytomedicine 2016, 23, 1629–1637. [Google Scholar] [CrossRef]
- Smith, A.M.; Gibbons, H.M.; Oldfield, R.L.; Bergin, P.M.; Mee, E.W.; Curtis, M.A.; Faull, R.L.M.; Dragunow, M. M-CSF increases proliferation and phagocytosis while modulating receptor and transcription factor expression in adult human microglia. J. Neuroinflamm. 2013, 10, 859. [Google Scholar] [CrossRef] [PubMed]
- Pan, C.; Kumar, C.; Bohl, S.; Klingmueller, U.; Mann, M. Comparative Proteomic Phenotyping of Cell Lines and Primary Cells to Assess Preservation of Cell Type-specific Functions. Mol. Cell Proteom. 2009, 8, 443–450. [Google Scholar] [CrossRef]
- Hyvärinen, T.; Hagman, S.; Ristola, M.; Sukki, L.; Veijula, K.; Kreutzer, J.; Kallio, P.; Narkilahti, S. Co-stimulation with IL-1β and TNF-α induces an inflammatory reactive astrocyte phenotype with neurosupportive characteristics in a human pluripotent stem cell model system. Sci. Rep. 2019, 9, 16944. [Google Scholar] [CrossRef] [PubMed]
- Canfield, S.G.; Stebbins, M.J.; Faubion, M.G.; Gastfriend, B.D.; Palecek, S.P.; Shusta, E.V. An isogenic neurovascular unit model comprised of human induced pluripotent stem cell-derived brain microvascular endothelial cells, pericytes, astrocytes, and neurons. Fluids Barriers CNS 2019, 16, 25. [Google Scholar] [CrossRef] [PubMed]
- Gastfriend, B.D.; Palecek, S.P.; Shusta, E.V. Modeling the blood–brain barrier: Beyond the endothelial cells. Curr. Opin. Biomed. Eng. 2018, 5, 6–12. [Google Scholar] [CrossRef] [PubMed]
- Lippmann, E.S.; Azarin, S.M.; Palecek, S.P.; Shusta, E.V. Commentary on human pluripotent stem cell-based blood–brain barrier models. Fluids Barriers CNS 2020, 17, 64. [Google Scholar] [CrossRef] [PubMed]
- Leventoux, N.; Morimoto, S.; Imaizumi, K.; Sato, Y.; Takahashi, S.; Mashima, K.; Ishikawa, M.; Sonn, I.; Kondo, T.; Watanabe, H.; et al. Human Astrocytes Model Derived from Induced Pluripotent Stem Cells. Cells 2020, 9, 2680. [Google Scholar] [CrossRef] [PubMed]
- Williams, L.M.; Fujimoto, T.; Weaver, R.R.; Logsdon, A.F.; Evitts, K.M.; Young, J.E.; Banks, W.A.; Erickson, M.A. Prolonged culturing of iPSC-derived brain endothelial-like cells is associated with quiescence, downregulation of glycolysis, and resistance to disruption by an Alzheimer’s brain milieu. Fluids Barriers CNS 2022, 19, 10. [Google Scholar] [CrossRef]
- Kobolak, J.; Teglasi, A.; Bellak, T.; Janstova, Z.; Molnar, K.; Zana, M.; Bock, I.; Laszlo, L.; Dinnyes, A. Human Induced Pluripotent Stem Cell-Derived 3D-Neurospheres Are Suitable for Neurotoxicity Screening. Cells 2020, 9, 1122. [Google Scholar] [CrossRef] [PubMed]
- Jusop, A.S.; Thanaskody, K.; Tye, G.J.; Dass, S.A.; Wan Kamarul Zaman, W.S.; Nordin, F. Development of brain organoid technology derived from iPSC for the neurodegenerative disease modelling: A glance through. Front. Mol. Neurosci. 2023, 16, 1173433. [Google Scholar] [CrossRef]
- Katt, M.E.; Xu, Z.S.; Gerecht, S.; Searson, P.C. Human Brain Microvascular Endothelial Cells Derived from the BC1 iPS Cell Line Exhibit a Blood-Brain Barrier Phenotype. PLoS ONE 2016, 11, e0152105. [Google Scholar] [CrossRef] [PubMed]
- Nishihara, H.; Perriot, S.; Gastfriend, B.D.; Steinfort, M.; Cibien, C.; Soldati, S.; Matsuo, K.; Guimbal, S.; Mathias, A.; Palecek, S.P.; et al. Intrinsic blood–brain barrier dysfunction contributes to multiple sclerosis pathogenesis. Brain 2022, 145, 4334–4348. [Google Scholar] [CrossRef] [PubMed]
- Haenseler, W.; Zambon, F.; Lee, H.; Vowles, J.; Rinaldi, F.; Duggal, G.; Houlden, H.; Gwinn, K.; Wray, S.; Luk, K.C.; et al. Excess α-synuclein compromises phagocytosis in iPSC-derived macrophages. Sci. Rep. 2017, 7, 9003. [Google Scholar] [CrossRef] [PubMed]
- Dolmetsch, R.; Geschwind, D.H. The Human Brain in a Dish: The Promise of iPSC-Derived Neurons. Cell 2011, 145, 831–834. [Google Scholar] [CrossRef] [PubMed]
- Engle, S.J.; Blaha, L.; Kleiman, R.J. Best Practices for Translational Disease Modeling Using Human iPSC-Derived Neurons. Neuron 2018, 100, 783–797. [Google Scholar] [CrossRef] [PubMed]
- Shirakawa, T.; Suzuki, I. Approach to Neurotoxicity using Human iPSC Neurons: Consortium for Safety Assessment using Human iPS Cells. Curr. Pharm. Biotechnol. 2020, 21, 780–786. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.; Eo, J.-C.; Lee, C.; Yu, J.-W. Distinct Features of Brain-Resident Macrophages: Microglia and Non-Parenchymal Brain Macrophages. Mol. Cells 2021, 44, 281–291. [Google Scholar] [CrossRef] [PubMed]
- Alliot, F.; Godin, I.; Pessac, B. Microglia derive from progenitors, originating from the yolk sac, and which proliferate in the brain. Dev. Brain Res. 1999, 117, 145–152. [Google Scholar] [CrossRef] [PubMed]
- Crapser, J.D.; Arreola, M.A.; Tsourmas, K.I.; Green, K.N. Microglia as hackers of the matrix: Sculpting synapses and the extracellular space. Cell Mol. Immunol. 2021, 18, 2472–2488. [Google Scholar] [CrossRef] [PubMed]
- de Miranda, A.S.; Zhang, C.-J.; Katsumoto, A.; Teixeira, A.L. Hippocampal adult neurogenesis: Does the immune system matter? J. Neurol. Sci. 2017, 372, 482–495. [Google Scholar] [CrossRef] [PubMed]
- Pósfai, B.; Cserép, C.; Orsolits, B.; Dénes, Á. New Insights into Microglia–Neuron Interactions: A Neuron’s Perspective. Neuroscience 2019, 405, 103–117. [Google Scholar] [CrossRef] [PubMed]
- Cockram, T.O.J.; Puigdellívol, M.; Brown, G.C. Calreticulin and Galectin-3 Opsonise Bacteria for Phagocytosis by Microglia. Front. Immunol. 2019, 10, 2647. [Google Scholar] [CrossRef]
- Neumann, H.; Kotter, M.R.; Franklin, R.J.M. Debris clearance by microglia: An essential link between degeneration and regeneration. Brain 2009, 132, 288–295. [Google Scholar] [CrossRef]
- Abud, E.M.; Ramirez, R.N.; Martinez, E.S.; Healy, L.M.; Nguyen, C.H.H.; Newman, S.A.; Yeromin, A.V.; Scarfone, V.M.; Marsh, S.E.; Fimbres, C.; et al. iPSC-Derived Human Microglia-like Cells to Study Neurological Diseases. Neuron 2017, 94, 278–293.e9. [Google Scholar] [CrossRef] [PubMed]
- Choi, I.; Zhang, Y.; Seegobin, S.P.; Pruvost, M.; Wang, Q.; Purtell, K.; Zhang, B.; Yue, Z. Microglia clear neuron-released α-synuclein via selective autophagy and prevent neurodegeneration. Nat. Commun. 2020, 11, 1386. [Google Scholar] [CrossRef] [PubMed]
- Kipnis, J.; Avidan, H.; Caspi, R.R.; Schwartz, M. Dual effect of CD4+CD25+ regulatory T cells in neurodegeneration: A dialogue with microglia. Proc. Natl. Acad. Sci. USA 2004, 101, 14663–14669. [Google Scholar] [CrossRef] [PubMed]
- Louveau, A.; Smirnov, I.; Keyes, T.J.; Eccles, J.D.; Rouhani, S.J.; Peske, J.D.; Derecki, N.C.; Castle, D.; Mandell, J.W.; Lee, K.S.; et al. Structural and functional features of central nervous system lymphatic vessels. Nature 2015, 523, 337–341. [Google Scholar] [CrossRef]
- Paolicelli, R.C.; Sierra, A.; Stevens, B.; Tremblay, M.-E.; Aguzzi, A.; Ajami, B.; Amit, I.; Audinat, E.; Bechmann, I.; Bennett, M.; et al. Microglia states and nomenclature: A field at its crossroads. Neuron 2022, 110, 3458–3483. [Google Scholar] [CrossRef]
- Hedegaard, A.; Stodolak, S.; James, W.S.; Cowley, S.A. Honing the Double-Edged Sword: Improving Human iPSC-Microglia Models. Front. Immunol. 2020, 11, 614972. [Google Scholar] [CrossRef] [PubMed]
- Hasselmann, J.; Blurton-Jones, M. Human iPSC-derived microglia: A growing toolset to study the brain’s innate immune cells. Glia 2020, 68, 721–739. [Google Scholar] [CrossRef] [PubMed]
- Nikolakopoulou, P.; Rauti, R.; Voulgaris, D.; Shlomy, I.; Maoz, B.M.; Herland, A. Recent progress in translational engineered in vitro models of the central nervous system. Brain 2020, 143, 3181–3213. [Google Scholar] [CrossRef]
- Loewa, A.; Feng, J.J.; Hedtrich, S. Human disease models in drug development. Nat. Rev. Bioeng. 2023, 1, 545–559. [Google Scholar] [CrossRef] [PubMed]
- Bierman-Duquette, R.D.; Safarians, G.; Huang, J.; Rajput, B.; Chen, J.Y.; Wang, Z.Z.; Seidlits, S.K. Engineering Tissues of the Central Nervous System: Interfacing Conductive Biomaterials with Neural Stem/Progenitor Cells. Adv. Healthc. Mater. 2022, 11, 2101577. [Google Scholar] [CrossRef]
- Delbridge, A.R.D.; Huh, D.; Brickelmaier, M.; Burns, J.C.; Roberts, C.; Challa, R.; Raymond, N.; Cullen, P.; Carlile, T.M.; Ennis, K.A.; et al. Organotypic Brain Slice Culture Microglia Exhibit Molecular Similarity to Acutely-Isolated Adult Microglia and Provide a Platform to Study Neuroinflammation. Front. Cell Neurosci. 2020, 14, 592005. [Google Scholar] [CrossRef]
- Speicher, A.M.; Wiendl, H.; Meuth, S.G.; Pawlowski, M. Generating microglia from human pluripotent stem cells: Novel in vitro models for the study of neurodegeneration. Mol. Neurodegener. 2019, 14, 46. [Google Scholar] [CrossRef]
- Vankriekelsvenne, E.; Chrzanowski, U.; Manzhula, K.; Greiner, T.; Wree, A.; Hawlitschka, A.; Llovera, G.; Zhan, J.; Joost, S.; Schmitz, C.; et al. Transmembrane protein 119 is neither a specific nor a reliable marker for microglia. Glia 2022, 70, 1170–1190. [Google Scholar] [CrossRef] [PubMed]
- Van Wageningen, T.A.; Vlaar, E.; Kooij, G.; Jongenelen, C.A.M.; Geurts, J.J.G.; Van Dam, A.-M. Regulation of microglial TMEM119 and P2RY12 immunoreactivity in multiple sclerosis white and grey matter lesions is dependent on their inflammatory environment. Acta Neuropathol. Commun. 2019, 7, 206. [Google Scholar] [CrossRef] [PubMed]
- Tagliatti, E.; Desiato, G.; Mancinelli, S.; Bizzotto, M.; Gagliani, M.C.; Faggiani, E.; Hernández-Soto, R.; Cugurra, A.; Poliseno, P.; Miotto, M.; et al. Trem2 expression in microglia is required to maintain normal neuronal bioenergetics during development. Immunity 2024, 57, 86–105.e9. [Google Scholar] [CrossRef]
- Honarpisheh, P.; Lee, J.; Banerjee, A.; Blasco-Conesa, M.P.; Honarpisheh, P.; d’Aigle, J.; Mamun, A.A.; Ritzel, R.M.; Chauhan, A.; Ganesh, B.P.; et al. Potential caveats of putative microglia-specific markers for assessment of age-related cerebrovascular neuroinflammation. J. Neuroinflamm. 2020, 17, 366. [Google Scholar] [CrossRef]
- Muffat, J.; Li, Y.; Yuan, B.; Mitalipova, M.; Omer, A.; Corcoran, S.; Bakiasi, G.; Tsai, L.-H.; Aubourg, P.; Ransohoff, R.M.; et al. Efficient derivation of microglia-like cells from human pluripotent stem cells. Nat. Med. 2016, 22, 1358–1367. [Google Scholar] [CrossRef] [PubMed]
- Buchrieser, J.; James, W.; Moore, M.D. Human Induced Pluripotent Stem Cell-Derived Macrophages Share Ontogeny with MYB-Independent Tissue-Resident Macrophages. Stem Cell Rep. 2017, 8, 334–345. [Google Scholar] [CrossRef]
- Kierdorf, K.; Erny, D.; Goldmann, T.; Sander, V.; Schulz, C.; Perdiguero, E.G.; Wieghofer, P.; Heinrich, A.; Riemke, P.; Hölscher, C.; et al. Microglia emerge from erythromyeloid precursors via Pu.1- and Irf8-dependent pathways. Nat. Neurosci. 2013, 16, 273–280. [Google Scholar] [CrossRef] [PubMed]
- Dräger, N.M.; Sattler, S.M.; Huang, C.T.-L.; Teter, O.M.; Leng, K.; Hashemi, S.H.; Hong, J.; Aviles, G.; Clelland, C.D.; Zhan, L.; et al. A CRISPRi/a platform in human iPSC-derived microglia uncovers regulators of disease states. Nat. Neurosci. 2022, 25, 1149–1162. [Google Scholar] [CrossRef] [PubMed]
- Douvaras, P.; Sun, B.; Wang, M.; Kruglikov, I.; Lallos, G.; Zimmer, M.; Terrenoire, C.; Zhang, B.; Gandy, S.; Schadt, E.; et al. Directed Differentiation of Human Pluripotent Stem Cells to Microglia. Stem Cell Rep. 2017, 8, 1516–1524. [Google Scholar] [CrossRef] [PubMed]
- Tcw, J.; Qian, L.; Pipalia, N.H.; Chao, M.J.; Liang, S.A.; Shi, Y.; Jain, B.R.; Bertelsen, S.E.; Kapoor, M.; Marcora, E.; et al. Cholesterol and matrisome pathways dysregulated in astrocytes and microglia. Cell 2022, 185, 2213–2233.e25. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.-T.; Seo, J.; Gao, F.; Feldman, H.M.; Wen, H.-L.; Penney, J.; Cam, H.P.; Gjoneska, E.; Raja, W.K.; Cheng, J.; et al. APOE4 Causes Widespread Molecular and Cellular Alterations Associated with Alzheimer’s Disease Phenotypes in Human iPSC-Derived Brain Cell Types. Neuron 2018, 98, 1141–1154.e7. [Google Scholar] [CrossRef] [PubMed]
- Gutbier, S.; Wanke, F.; Dahm, N.; Rümmelin, A.; Zimmermann, S.; Christensen, K.; Köchl, F.; Rautanen, A.; Hatje, K.; Geering, B.; et al. Large-Scale Production of Human iPSC-Derived Macrophages for Drug Screening. Int. J. Mol. Sci. 2020, 21, 4808. [Google Scholar] [CrossRef] [PubMed]
- van Wilgenburg, B.; Browne, C.; Vowles, J.; Cowley, S.A. Efficient, Long Term Production of Monocyte-Derived Macrophages from Human Pluripotent Stem Cells under Partly-Defined and Fully-Defined Conditions. PLoS ONE 2013, 8, e71098. [Google Scholar] [CrossRef] [PubMed]
- Reich, M.; Paris, I.; Ebeling, M.; Dahm, N.; Schweitzer, C.; Reinhardt, D.; Schmucki, R.; Prasad, M.; Köchl, F.; Leist, M.; et al. Alzheimer’s Risk Gene TREM2 Determines Functional Properties of New Type of Human iPSC-Derived Microglia. Front. Immunol. 2021, 11, 617860. [Google Scholar] [CrossRef]
- Brownjohn, P.W.; Smith, J.; Solanki, R.; Lohmann, E.; Houlden, H.; Hardy, J.; Dietmann, S.; Livesey, F.J. Functional Studies of Missense TREM2 Mutations in Human Stem Cell-Derived Microglia. Stem Cell Rep. 2018, 10, 1294. [Google Scholar] [CrossRef] [PubMed]
- Badanjak, K.; Mulica, P.; Smajic, S.; Delcambre, S.; Tranchevent, L.-C.; Diederich, N.; Rauen, T.; Schwamborn, J.C.; Glaab, E.; Cowley, S.A.; et al. iPSC-Derived Microglia as a Model to Study Inflammation in Idiopathic Parkinson’s Disease. Front. Cell Dev. Biol. 2021, 9, 740758. [Google Scholar] [CrossRef]
- Díaz-Castro, B.; Robel, S.; Mishra, A. Astrocyte Endfeet in Brain Function and Pathology: Open Questions. Annu. Rev. Neurosci. 2023, 46, 101–121. [Google Scholar] [CrossRef]
- Matias, I.; Morgado, J.; Gomes, F.C.A. Astrocyte Heterogeneity: Impact to Brain Aging and Disease. Front. Aging Neurosci. 2019, 11, 59. [Google Scholar] [CrossRef] [PubMed]
- Kwon, H.S.; Koh, S.-H. Neuroinflammation in neurodegenerative disorders: The roles of microglia and astrocytes. Transl. Neurodegener. 2020, 9, 42. [Google Scholar] [CrossRef] [PubMed]
- Colombo, E.; Farina, C. Astrocytes: Key Regulators of Neuroinflammation. Trends Immunol. 2016, 37, 608–620. [Google Scholar] [CrossRef] [PubMed]
- Cekanaviciute, E.; Buckwalter, M.S. Astrocytes: Integrative Regulators of Neuroinflammation in Stroke and Other Neurological Diseases. Neurotherapeutics 2016, 13, 685–701. [Google Scholar] [CrossRef]
- Kaur, D.; Sharma, V.; Deshmukh, R. Activation of microglia and astrocytes: A roadway to neuroinflammation and Alzheimer’s disease. Inflammopharmacology 2019, 27, 663–677. [Google Scholar] [CrossRef] [PubMed]
- Stöberl, N.; Maguire, E.; Salis, E.; Shaw, B.; Hall-Roberts, H. Human iPSC-derived glia models for the study of neuroinflammation. J. Neuroinflamm. 2023, 20, 231. [Google Scholar] [CrossRef]
- Tcw, J.; Wang, M.; Pimenova, A.A.; Bowles, K.R.; Hartley, B.J.; Lacin, E.; Machlovi, S.I.; Abdelaal, R.; Karch, C.M.; Phatnani, H.; et al. An Efficient Platform for Astrocyte Differentiation from Human Induced Pluripotent Stem Cells. Stem Cell Rep. 2017, 9, 600–614. [Google Scholar] [CrossRef] [PubMed]
- Roybon, L.; Lamas, N.J.; Garcia-Diaz, A.; Yang, E.J.; Sattler, R.; Jackson-Lewis, V.; Kim, Y.A.; Kachel, C.A.; Rothstein, J.D.; Przedborski, S.; et al. Human Stem Cell-Derived Spinal Cord Astrocytes with Defined Mature or Reactive Phenotypes. Cell Rep. 2013, 4, 1035–1048. [Google Scholar] [CrossRef] [PubMed]
- Perriot, S.; Canales, M.; Mathias, A.; Du Pasquier, R. Differentiation of functional astrocytes from human-induced pluripotent stem cells in chemically defined media. STAR Protoc. 2021, 2, 100902. [Google Scholar] [CrossRef] [PubMed]
- Perriot, S.; Mathias, A.; Perriard, G.; Canales, M.; Jonkmans, N.; Merienne, N.; Meunier, C.; El Kassar, L.; Perrier, A.L.; Laplaud, D.-A.; et al. Human Induced Pluripotent Stem Cell-Derived Astrocytes Are Differentially Activated by Multiple Sclerosis-Associated Cytokines. Stem Cell Rep. 2018, 11, 1199–1210. [Google Scholar] [CrossRef]
- Mulica, P.; Venegas, C.; Landoulsi, Z.; Badanjak, K.; Delcambre, S.; Tziortziou, M.; Hezzaz, S.; Ghelfi, J.; Smajic, S.; Schwamborn, J.; et al. Comparison of two protocols for the generation of iPSC-derived human astrocytes. Biol. Proced. Online 2023, 25, 26. [Google Scholar] [CrossRef] [PubMed]
- Santos, R.; Vadodaria, K.C.; Jaeger, B.N.; Mei, A.; Lefcochilos-Fogelquist, S.; Mendes, A.P.D.; Erikson, G.; Shokhirev, M.; Randolph-Moore, L.; Fredlender, C.; et al. Differentiation of Inflammation-Responsive Astrocytes from Glial Progenitors Generated from Human Induced Pluripotent Stem Cells. Stem Cell Rep. 2017, 8, 1757–1769. [Google Scholar] [CrossRef]
- Barbar, L.; Jain, T.; Zimmer, M.; Kruglikov, I.; Sadick, J.S.; Wang, M.; Kalpana, K.; Rose, I.V.L.; Burstein, S.R.; Rusielewicz, T.; et al. CD49f Is a Novel Marker of Functional and Reactive Human iPSC-Derived Astrocytes. Neuron 2020, 107, 436–453.e12. [Google Scholar] [CrossRef] [PubMed]
- Vatine, G.D.; Barrile, R.; Workman, M.J.; Sances, S.; Barriga, B.K.; Rahnama, M.; Barthakur, S.; Kasendra, M.; Lucchesi, C.; Kerns, J.; et al. Human iPSC-Derived Blood-Brain Barrier Chips Enable Disease Modeling and Personalized Medicine Applications. Cell Stem Cell 2019, 24, 995–1005.e6. [Google Scholar] [CrossRef]
- Konstantinidis, E.; Dakhel, A.; Beretta, C.; Erlandsson, A. Long-term effects of amyloid-beta deposits in human iPSC-derived astrocytes. Mol. Cell Neurosci. 2023, 125, 103839. [Google Scholar] [CrossRef] [PubMed]
- Mansur, F.; Teles e Silva, A.L.; Gomes, A.K.S.; Magdalon, J.; De Souza, J.S.; Griesi-Oliveira, K.; Passos-Bueno, M.R.; Sertié, A.L. Complement C4 Is Reduced in iPSC-Derived Astrocytes of Autism Spectrum Disorder Subjects. Int. J. Mol. Sci. 2021, 22, 7579. [Google Scholar] [CrossRef] [PubMed]
- Lundin, A.; Delsing, L.; Clausen, M.; Ricchiuto, P.; Sanchez, J.; Sabirsh, A.; Ding, M.; Synnergren, J.; Zetterberg, H.; Brolén, G.; et al. Human iPS-Derived Astroglia from a Stable Neural Precursor State Show Improved Functionality Compared with Conventional Astrocytic Models. Stem Cell Rep. 2018, 10, 1030–1045. [Google Scholar] [CrossRef]
- Dang, J.; Tiwari, S.K.; Agrawal, K.; Hui, H.; Qin, Y.; Rana, T.M. Glial cell diversity and methamphetamine-induced neuroinflammation in human cerebral organoids. Mol. Psychiatry 2021, 26, 1194–1207. [Google Scholar] [CrossRef] [PubMed]
- Mantle, J.L.; Lee, K.H. Immunoglobulin G transport increases in an in vitro blood–brain barrier model with amyloid-β and with neuroinflammatory cytokines. Biotechnol. Bioeng. 2019, 116, 1752–1761. [Google Scholar] [CrossRef]
- Deivasigamani, S.; Miteva, M.T.; Natale, S.; Gutierrez-Barragan, D.; Basilico, B.; Di Angelantonio, S.; Weinhard, L.; Molotkov, D.; Deb, S.; Pape, C.; et al. Microglia complement signaling promotes neuronal elimination and normal brain functional connectivity. Cereb. Cortex 2023, 33, 10750–10760. [Google Scholar] [CrossRef] [PubMed]
- Oechtering, J.; Stein, K.; Schaedelin, S.A.; Maceski, A.M.; Orleth, A.; Meier, S.; Willemse, E.; Qureshi, F.; Heijnen, I.; Regeniter, A.; et al. Complement Activation Is Associated With Disease Severity in Multiple Sclerosis. Neurol. Neuroimmunol. Neuroinflamm. 2024, 11, e200212. [Google Scholar] [CrossRef] [PubMed]
- Carpanini, S.M.; Torvell, M.; Bevan, R.J.; Byrne, R.A.J.; Daskoulidou, N.; Saito, T.; Saido, T.C.; Taylor, P.R.; Hughes, T.R.; Zelek, W.M.; et al. Terminal complement pathway activation drives synaptic loss in Alzheimer’s disease models. Acta Neuropathol. Commun. 2022, 10, 99. [Google Scholar] [CrossRef] [PubMed]
- Cervia-Hasler, C.; Brüningk, S.C.; Hoch, T.; Fan, B.; Muzio, G.; Thompson, R.C.; Ceglarek, L.; Meledin, R.; Westermann, P.; Emmenegger, M.; et al. Persistent complement dysregulation with signs of thromboinflammation in active Long Covid. Science 2024, 383, eadg7942. [Google Scholar] [CrossRef] [PubMed]
- An, X.; Xi, W.; Gu, C.; Huang, X. Complement protein C5a enhances the β-amyloid-induced neuro-inflammatory response in microglia in Alzheimer’s disease. Med./Sci. 2018, 34, 116–120. [Google Scholar] [CrossRef] [PubMed]
- Thomas, A.; Gasque, P.; Vaudry, D.; Gonzalez, B.; Fontaine, M. Expression of a complete and functional complement system by human neuronal cells in vitro. Int. Immunol. 2000, 12, 1015–1023. [Google Scholar] [CrossRef]
- Lian, H.; Litvinchuk, A.; Chiang, A.C.-A.; Aithmitti, N.; Jankowsky, J.L.; Zheng, H. Astrocyte-Microglia Cross Talk through Complement Activation Modulates Amyloid Pathology in Mouse Models of Alzheimer’s Disease. J. Neurosci. 2016, 36, 577–589. [Google Scholar] [CrossRef]
- Kerkering, J.; Muinjonov, B.; Rosiewicz, K.S.; Diecke, S.; Biese, C.; Schiweck, J.; Chien, C.; Zocholl, D.; Conrad, T.; Paul, F.; et al. iPSC-derived reactive astrocytes from patients with multiple sclerosis protect cocultured neurons in inflammatory conditions. J. Clin. Investig. 2023, 133, e164637. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Viollet, C.; Efthymiou, A.; Khayrullina, G.; Moritz, K.E.; Wilkerson, M.D.; Sukumar, G.; Dalgard, C.L.; Doughty, M.L. Neuroinflammatory astrocytes generated from cord blood-derived human induced pluripotent stem cells. J. Neuroinflamm. 2019, 16, 164. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.; Lee, H.; Jung, M.; Hong, K.; Woo, S.-H.; Lee, Y.-S.; Kim, B.J.; Jeon, M.Y.; Seo, J.; Mun, J.Y. Neuromyelitis optica (NMO)-IgG-driven organelle reorganization in human iPSC-derived astrocytes. FASEB J. 2021, 35, e21894. [Google Scholar] [CrossRef]
- Kim, H.; Leng, K.; Park, J.; Sorets, A.G.; Kim, S.; Shostak, A.; Embalabala, R.J.; Mlouk, K.; Katdare, K.A.; Rose, I.V.L.; et al. Reactive astrocytes transduce inflammation in a blood-brain barrier model through a TNF-STAT3 signaling axis and secretion of alpha 1-antichymotrypsin. Nat. Commun. 2022, 13, 6581. [Google Scholar] [CrossRef] [PubMed]
- Bassil, R.; Shields, K.; Granger, K.; Zein, I.; Ng, S.; Chih, B. Improved modeling of human AD with an automated culturing platform for iPSC neurons, astrocytes and microglia. Nat. Commun. 2021, 12, 5220. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, S.E.; Liao, M.; Smith, R.V.; White, C.; Lagomarsino, V.N.; Xu, J.; Taga, M.; Bennett, D.A.; De Jager, P.L.; Young-Pearse, T.L. Candidate-based screening via gene modulation in human neurons and astrocytes implicates FERMT2 in Aβ and TAU proteostasis. Hum. Mol. Genet. 2019, 28, 718–735. [Google Scholar] [CrossRef]
- Serio, A.; Bilican, B.; Barmada, S.J.; Ando, D.M.; Zhao, C.; Siller, R.; Burr, K.; Haghi, G.; Story, D.; Nishimura, A.L.; et al. Astrocyte pathology and the absence of non-cell autonomy in an induced pluripotent stem cell model of TDP-43 proteinopathy. Proc. Natl. Acad. Sci. USA 2013, 110, 4697–4702. [Google Scholar] [CrossRef]
- Qin, C.; Yang, S.; Chu, Y.-H.; Zhang, H.; Pang, X.-W.; Chen, L.; Zhou, L.-Q.; Chen, M.; Tian, D.-S.; Wang, W. Signaling pathways involved in ischemic stroke: Molecular mechanisms and therapeutic interventions. Signal Transduct. Target. Ther. 2022, 7, 215. [Google Scholar] [CrossRef] [PubMed]
- Lai, J.D.; Berlind, J.E.; Fricklas, G.; Lie, C.; Urenda, J.-P.; Lam, K.; Sta Maria, N.; Jacobs, R.; Yu, V.; Zhao, Z.; et al. KCNJ2 inhibition mitigates mechanical injury in a human brain organoid model of traumatic brain injury. Cell Stem Cell 2024, 31, 519–536.e8. [Google Scholar] [CrossRef] [PubMed]
- Abdelhak, A.; Foschi, M.; Abu-Rumeileh, S.; Yue, J.K.; D’Anna, L.; Huss, A.; Oeckl, P.; Ludolph, A.C.; Kuhle, J.; Petzold, A.; et al. Blood GFAP as an emerging biomarker in brain and spinal cord disorders. Nat. Rev. Neurol. 2022, 18, 158–172. [Google Scholar] [CrossRef] [PubMed]
- Phatnani, H.; Maniatis, T. Astrocytes in neurodegenerative disease. Cold Spring Harb. Perspect. Biol. 2015, 7, a020628. [Google Scholar] [CrossRef] [PubMed]
- Dai, D.L.; Li, M.; Lee, E.B. Human Alzheimer’s disease reactive astrocytes exhibit a loss of homeostastic gene expression. Acta Neuropathol. Commun. 2023, 11, 127. [Google Scholar] [CrossRef]
- Rodríguez-Giraldo, M.; González-Reyes, R.E.; Ramírez-Guerrero, S.; Bonilla-Trilleras, C.E.; Guardo-Maya, S.; Nava-Mesa, M.O. Astrocytes as a Therapeutic Target in Alzheimer’s Disease-Comprehensive Review and Recent Developments. Int. J. Mol. Sci. 2022, 23, 13630. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Yoo, I.D.; Lim, J.; Moon, J.-S. Pathological phenotypes of astrocytes in Alzheimer’s disease. Exp. Mol. Med. 2024, 56, 95–99. [Google Scholar] [CrossRef] [PubMed]
- Zou, P.; Wu, C.; Liu, T.C.-Y.; Duan, R.; Yang, L. Oligodendrocyte progenitor cells in Alzheimer’s disease: From physiology to pathology. Transl. Neurodegener. 2023, 12, 52. [Google Scholar] [CrossRef]
- Leenders, F.; Koole, L.; Slaets, H.; Tiane, A.; van den Hove, D.; Vanmierlo, T. Navigating oligodendrocyte precursor cell aging in brain health. Mech. Ageing Dev. 2024, 220, 111959. [Google Scholar] [CrossRef]
- Haroon, A.; Seerapu, H.; Fang, L.-P.; Weß, J.H.; Bai, X. Unlocking the Potential: Immune functions of oligodendrocyte precursor cells. Front. Immunol. 2024, 15, 1425706. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.-C.; Ge, B.; Duncan, I.D. Adult brain retains the potential to generate oligodendroglial progenitors with extensive myelination capacity. Proc. Natl. Acad. Sci. USA 1999, 96, 4089–4094. [Google Scholar] [CrossRef]
- Huang, W.; Bhaduri, A.; Velmeshev, D.; Wang, S.; Wang, L.; Rottkamp, C.A.; Alvarez-Buylla, A.; Rowitch, D.H.; Kriegstein, A.R. Origins and Proliferative States of Human Oligodendrocyte Precursor Cells. Cell 2020, 182, 594–608.e11. [Google Scholar] [CrossRef] [PubMed]
- Beiter, R.M.; Rivet-Noor, C.; Merchak, A.R.; Bai, R.; Johanson, D.M.; Slogar, E.; Sol-Church, K.; Overall, C.C.; Gaultier, A. Evidence for oligodendrocyte progenitor cell heterogeneity in the adult mouse brain. Sci. Rep. 2022, 12, 12921. [Google Scholar] [CrossRef] [PubMed]
- Skaper, S.D. Chapter 2—Oligodendrocyte precursor cells as a therapeutic target for demyelinating diseases. In Progress in Brain Research; Sharma, A., Sharma, H.S., Eds.; Elsevier: Amsterdam, The Netherlands, 2019; Volume 245, pp. 33–58. [Google Scholar]
- Hamanaka, G.; Hernández, I.C.; Takase, H.; Ishikawa, H.; Benboujja, F.; Kimura, S.; Fukuda, N.; Guo, S.; Lok, J.; Lo, E.H.; et al. Myelination-and migration-associated genes are downregulated after phagocytosis in cultured oligodendrocyte precursor cells. J. Neurochem. 2023, 167, 571–581. [Google Scholar] [CrossRef] [PubMed]
- Saraswat, D.; Shayya, H.J.; Polanco, J.J.; Tripathi, A.; Welliver, R.R.; Pol, S.U.; Seidman, R.A.; Broome, J.E.; O’Bara, M.A.; van Kuppervelt, T.H.; et al. Overcoming the inhibitory microenvironment surrounding oligodendrocyte progenitor cells following experimental demyelination. Nat. Commun. 2021, 12, 1923. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Yang, Z.; Wang, R.; He, F.; Yan, R.; Zhang, Y.; Yu, L.; Deng, W.; Nie, Y. Rapid differentiation of hiPSCs into functional oligodendrocytes using an OLIG2 synthetic modified messenger RNA. Commun. Biol. 2022, 5, 1095. [Google Scholar] [CrossRef]
- Li, N.; Leung, G.K.K. Oligodendrocyte Precursor Cells in Spinal Cord Injury: A Review and Update. BioMed Res. Int. 2015, 2015, 235195. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Qiu, M.; Fu, H. Oligodendrocytes in central nervous system diseases: The effect of cytokine regulation. Neural Regen. Res. 2024, 19, 2132. [Google Scholar] [CrossRef]
- Sanchez-Petidier, M.; Guerri, C.; Moreno-Manzano, V. Toll-like receptors 2 and 4 differentially regulate the self-renewal and differentiation of spinal cord neural precursor cells. Stem Cell Res. Ther. 2022, 13, 117. [Google Scholar] [CrossRef] [PubMed]
- Falcão, A.M.; van Bruggen, D.; Marques, S.; Meijer, M.; Jäkel, S.; Agirre, E.; Samudyata; Floriddia, E.M.; Vanichkina, D.P.; ffrench-Constant, C.; et al. Disease-specific oligodendrocyte lineage cells arise in multiple sclerosis. Nat. Med. 2018, 24, 1837–1844. [Google Scholar] [CrossRef]
- Kirby, L.; Jin, J.; Cardona, J.G.; Smith, M.D.; Martin, K.A.; Wang, J.; Strasburger, H.; Herbst, L.; Alexis, M.; Karnell, J.; et al. Oligodendrocyte precursor cells present antigen and are cytotoxic targets in inflammatory demyelination. Nat. Commun. 2019, 10, 3887. [Google Scholar] [CrossRef] [PubMed]
- Gorter, R.P.; Baron, W. Matrix metalloproteinases shape the oligodendrocyte (niche) during development and upon demyelination. Neurosci. Lett. 2020, 729, 134980. [Google Scholar] [CrossRef] [PubMed]
- Marangon, D.; Castro e Silva, J.H.; Cerrato, V.; Boda, E.; Lecca, D. Oligodendrocyte Progenitors in Glial Scar: A Bet on Remyelination. Cells 2024, 13, 1024. [Google Scholar] [CrossRef] [PubMed]
- Chamling, X.; Kallman, A.; Fang, W.; Berlinicke, C.A.; Mertz, J.L.; Devkota, P.; Pantoja, I.E.M.; Smith, M.D.; Ji, Z.; Chang, C.; et al. Single-cell transcriptomic reveals molecular diversity and developmental heterogeneity of human stem cell-derived oligodendrocyte lineage cells. Nat. Commun. 2021, 12, 652. [Google Scholar] [CrossRef] [PubMed]
- Ornelas, I.M.; McLane, L.E.; Saliu, A.; Evangelou, A.V.; Khandker, L.; Wood, T.L. Heterogeneity in oligodendroglia: Is it relevant to mouse models and human disease? J. Neurosci. Res. 2016, 94, 1421–1433. [Google Scholar] [CrossRef] [PubMed]
- Windrem, M.S.; Nunes, M.C.; Rashbaum, W.K.; Schwartz, T.H.; Goodman, R.A.; McKhann, G.; Roy, N.S.; Goldman, S.A. Fetal and adult human oligodendrocyte progenitor cell isolates myelinate the congenitally dysmyelinated brain. Nat. Med. 2004, 10, 93–97. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; You, H.; Hu, X.; Luo, Y.; Zhang, Z.; Song, Y.; An, J.; Lu, H. Microglia–Astrocyte Interaction in Neural Development and Neural Pathogenesis. Cells 2023, 12, 1942. [Google Scholar] [CrossRef]
- Lopez-Caraballo, L.; Martorell-Marugan, J.; Carmona-Sáez, P.; Gonzalez-Munoz, E. iPS-Derived Early Oligodendrocyte Progenitor Cells from SPMS Patients Reveal Deficient In Vitro Cell Migration Stimulation. Cells 2020, 9, 1803. [Google Scholar] [CrossRef]
- Zveik, O.; Rechtman, A.; Brill, L.; Vaknin-Dembinsky, A. Anti- and pro-inflammatory milieu differentially regulate differentiation and immune functions of oligodendrocyte progenitor cells. Immunology 2024, 171, 618–633. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Castañeda, A.; Chappell, M.S.; Rosen, D.A.; Seki, S.M.; Beiter, R.M.; Johanson, D.M.; Liskey, D.; Farber, E.; Onengut-Gumuscu, S.; Overall, C.C.; et al. The active contribution of OPCs to neuroinflammation is mediated by LRP1. Acta Neuropathol. 2020, 139, 365–382. [Google Scholar] [CrossRef]
- Tepavčević, V.; Lubetzki, C. Oligodendrocyte progenitor cell recruitment and remyelination in multiple sclerosis: The more, the merrier? Brain 2022, 145, 4178–4192. [Google Scholar] [CrossRef]
- Luo, W.; Xu, H.; Xu, L.; Jiang, W.; Chen, C.; Chang, Y.; Liu, C.; Tian, Z.; Qiu, X.; Xie, C.; et al. Remyelination in neuromyelitis optica spectrum disorder is promoted by edaravone through mTORC1 signaling activation. Glia 2023, 71, 284–304. [Google Scholar] [CrossRef]
- Paolilo, R.B.; Deiva, K.; Neuteboom, R.; Rostásy, K.; Lim, M. Acute Disseminated Encephalomyelitis: Current Perspectives. Children 2020, 7, 210. [Google Scholar] [CrossRef]
- Sasmita, A.O.; Depp, C.; Nazarenko, T.; Sun, T.; Siems, S.B.; Ong, E.C.; Nkeh, Y.B.; Böhler, C.; Yu, X.; Bues, B.; et al. Oligodendrocytes produce amyloid-β and contribute to plaque formation alongside neurons in Alzheimer’s disease model mice. Nat. Neurosci. 2024, 27, 1668–1674. [Google Scholar] [CrossRef] [PubMed]
- Lim, R.G.; Al-Dalahmah, O.; Wu, J.; Gold, M.P.; Reidling, J.C.; Tang, G.; Adam, M.; Dansu, D.K.; Park, H.-J.; Casaccia, P.; et al. Huntington disease oligodendrocyte maturation deficits revealed by single-nucleus RNAseq are rescued by thiamine-biotin supplementation. Nat. Commun. 2022, 13, 7791. [Google Scholar] [CrossRef]
- Li, W.; He, T.; Shi, R.; Song, Y.; Wang, L.; Zhang, Z.; Tang, Y.; Yang, G.-Y.; Wang, Y. Oligodendrocyte Precursor Cells Transplantation Improves Stroke Recovery via Oligodendrogenesis, Neurite Growth and Synaptogenesis. Aging Dis. 2022, 12, 2096–2112. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Chao, J.; Wang, C.; Sun, G.; Roeth, D.; Liu, W.; Chen, X.; Li, L.; Tian, E.; Feng, L.; et al. Astrocytic response mediated by the CLU risk allele inhibits OPC proliferation and myelination in a human iPSC model. Cell Rep. 2023, 42, 112841. [Google Scholar] [CrossRef] [PubMed]
- Rajani, R.M.; Ellingford, R.; Hellmuth, M.; Harris, S.S.; Taso, O.S.; Graykowski, D.; Lam, F.K.W.; Arber, C.; Fertan, E.; Danial, J.S.H.; et al. Selective suppression of oligodendrocyte-derived amyloid beta rescues neuronal dysfunction in Alzheimer’s disease. PLoS Biol. 2024, 22, e3002727. [Google Scholar] [CrossRef]
- Lohrasbi, F.; Ghasemi-Kasman, M.; Soghli, N.; Ghazvini, S.; Vaziri, Z.; Abdi, S.; Darban, Y.M. The Journey of iPSC-derived OPCs in Demyelinating Disorders: From In vitro Generation to In vivo Transplantation. Curr. Neuropharmacol. 2023, 21, 1980–1991. [Google Scholar] [CrossRef] [PubMed]
- Koike, H.; Katsuno, M. Macrophages and Autoantibodies in Demyelinating Diseases. Cells 2021, 10, 844. [Google Scholar] [CrossRef] [PubMed]
- Chanoumidou, K.; Mozafari, S.; Baron-Van Evercooren, A.; Kuhlmann, T. Stem cell derived oligodendrocytes to study myelin diseases. Glia 2020, 68, 705–720. [Google Scholar] [CrossRef] [PubMed]
- Ferrari Bardile, C.; Garcia-Miralles, M.; Caron, N.S.; Rayan, N.A.; Langley, S.R.; Harmston, N.; Rondelli, A.M.; Teo, R.T.Y.; Waltl, S.; Anderson, L.M.; et al. Intrinsic mutant HTT-mediated defects in oligodendroglia cause myelination deficits and behavioral abnormalities in Huntington disease. Proc. Natl. Acad. Sci. USA 2019, 116, 9622–9627. [Google Scholar] [CrossRef] [PubMed]
- Vanzulli, I.; Papanikolaou, M.; De-La-Rocha, I.C.; Pieropan, F.; Rivera, A.D.; Gomez-Nicola, D.; Verkhratsky, A.; Rodríguez, J.J.; Butt, A.M. Disruption of oligodendrocyte progenitor cells is an early sign of pathology in the triple transgenic mouse model of Alzheimer’s disease. Neurobiol. Aging 2020, 94, 130–139. [Google Scholar] [CrossRef]
- Bonfanti, E.; Gelosa, P.; Fumagalli, M.; Dimou, L.; Viganò, F.; Tremoli, E.; Cimino, M.; Sironi, L.; Abbracchio, M.P. The role of oligodendrocyte precursor cells expressing the GPR17 receptor in brain remodeling after stroke. Cell Death Dis. 2017, 8, e2871. [Google Scholar] [CrossRef] [PubMed]
- Garza, R.; Sharma, Y.; Atacho, D.A.M.; Thiruvalluvan, A.; Abu Hamdeh, S.; Jönsson, M.E.; Horvath, V.; Adami, A.; Ingelsson, M.; Jern, P.; et al. Single-cell transcriptomics of human traumatic brain injury reveals activation of endogenous retroviruses in oligodendroglia. Cell Rep. 2023, 42, 113395. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, L.; Chow, B.W.; Gu, C. Neuronal regulation of the blood–brain barrier and neurovascular coupling. Nat. Rev. Neurosci. 2020, 21, 416–432. [Google Scholar] [CrossRef]
- Workman, M.J.; Svendsen, C.N. Recent advances in human iPSC-derived models of the blood–brain barrier. Fluids Barriers CNS 2020, 17, 30. [Google Scholar] [CrossRef] [PubMed]
- Al-Ahmad, A.J. Human-Induced Pluripotent Stem Cell-Based Model of the Blood-Brain at 10 Years: A Retrospective on Past and Current Disease Models. In Human iPSC-Derived Disease Models for Drug Discovery; Springer: Berlin/Heidelberg, Germany, 2023; pp. 1–16. [Google Scholar]
- Lu, T.M.; Houghton, S.; Magdeldin, T.; Durán, J.G.B.; Minotti, A.P.; Snead, A.; Sproul, A.; Nguyen, D.-H.T.; Xiang, J.; Fine, H.A.; et al. Pluripotent stem cell-derived epithelium misidentified as brain microvascular endothelium requires ETS factors to acquire vascular fate. Proc. Natl. Acad. Sci. USA 2021, 118, e2016950118. [Google Scholar] [CrossRef] [PubMed]
- Lippmann, E.S.; Al-Ahmad, A.; Azarin, S.M.; Palecek, S.P.; Shusta, E.V. A retinoic acid-enhanced, multicellular human blood-brain barrier model derived from stem cell sources. Sci. Rep. 2014, 4, 4160. [Google Scholar] [CrossRef] [PubMed]
- Neal, E.H.; Marinelli, N.A.; Shi, Y.; McClatchey, P.M.; Balotin, K.M.; Gullett, D.R.; Hagerla, K.A.; Bowman, A.B.; Ess, K.C.; Wikswo, J.P.; et al. A Simplified, Fully Defined Differentiation Scheme for Producing Blood-Brain Barrier Endothelial Cells from Human iPSCs. Stem Cell Rep. 2019, 12, 1380–1388. [Google Scholar] [CrossRef]
- Nishihara, H.; Gastfriend, B.D.; Kasap, P.; Palecek, S.P.; Shusta, E.V.; Engelhardt, B. Differentiation of human pluripotent stem cells to brain microvascular endothelial cell-like cells suitable to study immune cell interactions. STAR Protoc. 2021, 2, 100563. [Google Scholar] [CrossRef] [PubMed]
- Praça, C.; Rosa, S.C.; Sevin, E.; Cecchelli, R.; Dehouck, M.-P.; Ferreira, L.S. Derivation of Brain Capillary-like Endothelial Cells from Human Pluripotent Stem Cell-Derived Endothelial Progenitor Cells. Stem Cell Rep. 2019, 13, 599–611. [Google Scholar] [CrossRef] [PubMed]
- Foreman, K.L.; Shusta, E.V.; Palecek, S.P. Defined Differentiation of Human Pluripotent Stem Cells to Brain Microvascular Endothelial-Like Cells for Modeling the Blood-Brain Barrier. In Stem Cell-Based Neural Model Systems for Brain Disorders; Huang, Y.-W.A., Pak, C., Eds.; Springer: New York, NY, USA, 2023; pp. 113–133. ISBN 978-1-07-163287-1. [Google Scholar]
- Appelt-Menzel, A.; Oerter, S.; Mathew, S.; Haferkamp, U.; Hartmann, C.; Jung, M.; Neuhaus, W.; Pless, O. Human iPSC-Derived Blood-Brain Barrier Models: Valuable Tools for Preclinical Drug Discovery and Development? Curr. Protoc. Stem Cell Biol. 2020, 55, e122. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.-C.; Sonninen, T.-M.; Peltonen, S.; Koistinaho, J.; Lehtonen, Š. Blood–Brain Barrier and Neurodegenerative Diseases—Modeling with iPSC-Derived Brain Cells. Int. J. Mol. Sci. 2021, 22, 7710. [Google Scholar] [CrossRef] [PubMed]
- Katt, M.E.; Mayo, L.N.; Ellis, S.E.; Mahairaki, V.; Rothstein, J.D.; Cheng, L.; Searson, P.C. The role of mutations associated with familial neurodegenerative disorders on blood–brain barrier function in an iPSC model. Fluids Barriers CNS 2019, 16, 20. [Google Scholar] [CrossRef] [PubMed]
- Ohshima, M.; Kamei, S.; Fushimi, H.; Mima, S.; Yamada, T.; Yamamoto, T. Prediction of Drug Permeability Using In Vitro Blood–Brain Barrier Models with Human Induced Pluripotent Stem Cell-Derived Brain Microvascular Endothelial Cells. BioRes. Open Access 2019, 8, 200–209. [Google Scholar] [CrossRef] [PubMed]
- Nishihara, H.; Gastfriend, B.D.; Soldati, S.; Perriot, S.; Mathias, A.; Sano, Y.; Shimizu, F.; Gosselet, F.; Kanda, T.; Palecek, S.P.; et al. Advancing human induced pluripotent stem cell-derived blood-brain barrier models for studying immune cell interactions. FASEB J. 2020, 34, 16693–16715. [Google Scholar] [CrossRef] [PubMed]
- Linville, R.M.; Sklar, M.B.; Grifno, G.N.; Nerenberg, R.F.; Zhou, J.; Ye, R.; DeStefano, J.G.; Guo, Z.; Jha, R.; Jamieson, J.J.; et al. Three-dimensional microenvironment regulates gene expression, function, and tight junction dynamics of iPSC-derived blood-brain barrier microvessels. Fluids Barriers CNS 2022, 19, 87. [Google Scholar] [CrossRef]
- Linville, R.M.; DeStefano, J.G.; Sklar, M.B.; Xu, Z.; Farrell, A.M.; Bogorad, M.I.; Chu, C.; Walczak, P.; Cheng, L.; Mahairaki, V.; et al. Human iPSC-derived blood-brain barrier microvessels: Validation of barrier function and endothelial cell behavior. Biomaterials 2019, 190–191, 24–37. [Google Scholar] [CrossRef] [PubMed]
- Lian, X.; Bao, X.; Al-Ahmad, A.; Liu, J.; Wu, Y.; Dong, W.; Dunn, K.K.; Shusta, E.V.; Palecek, S.P. Efficient differentiation of human pluripotent stem cells to endothelial progenitors via small-molecule activation of WNT signaling. Stem Cell Rep. 2014, 3, 804–816. [Google Scholar] [CrossRef] [PubMed]
- Rosa, S.; Praça, C.; Pitrez, P.R.; Gouveia, P.J.; Aranguren, X.L.; Ricotti, L.; Ferreira, L.S. Functional characterization of iPSC-derived arterial- and venous-like endothelial cells. Sci. Rep. 2019, 9, 3826. [Google Scholar] [CrossRef] [PubMed]
- Fengler, S.; Kurkowsky, B.; Kaushalya, S.K.; Roth, W.; Fava, E.; Denner, P. Human iPSC-derived brain endothelial microvessels in a multi-well format enable permeability screens of anti-inflammatory drugs. Biomaterials 2022, 286, 121525. [Google Scholar] [CrossRef] [PubMed]
- Brown, L.S.; Foster, C.G.; Courtney, J.-M.; King, N.E.; Howells, D.W.; Sutherland, B.A. Pericytes and Neurovascular Function in the Healthy and Diseased Brain. Front. Cell Neurosci. 2019, 13, 282. [Google Scholar] [CrossRef] [PubMed]
- Alarcon-Martinez, L.; Yilmaz-Ozcan, S.; Yemisci, M.; Schallek, J.; Kılıç, K.; Can, A.; Di Polo, A.; Dalkara, T. Capillary pericytes express α-smooth muscle actin, which requires prevention of filamentous-actin depolymerization for detection. eLife 2018, 7, e34861. [Google Scholar] [CrossRef]
- Fu, J.; Liang, H.; Yuan, P.; Wei, Z.; Zhong, P. Brain pericyte biology: From physiopathological mechanisms to potential therapeutic applications in ischemic stroke. Front. Cell Neurosci. 2023, 17, 1267785. [Google Scholar] [CrossRef] [PubMed]
- Bergers, G.; Song, S. The role of pericytes in blood-vessel formation and maintenance. Neuro-Oncol. 2005, 7, 452–464. [Google Scholar] [CrossRef]
- Bell, R.D.; Winkler, E.A.; Sagare, A.P.; Singh, I.; LaRue, B.; Deane, R.; Zlokovic, B.V. Pericytes control key neurovascular functions and neuronal phenotype in the adult brain and during brain aging. Neuron 2010, 68, 409–427. [Google Scholar] [CrossRef] [PubMed]
- Armulik, A.; Genové, G.; Mäe, M.; Nisancioglu, M.H.; Wallgard, E.; Niaudet, C.; He, L.; Norlin, J.; Lindblom, P.; Strittmatter, K.; et al. Pericytes regulate the blood–brain barrier. Nature 2010, 468, 557–561. [Google Scholar] [CrossRef] [PubMed]
- Winkler, E.A.; Sengillo, J.D.; Sullivan, J.S.; Henkel, J.S.; Appel, S.H.; Zlokovic, B.V. Blood–spinal cord barrier breakdown and pericyte reductions in amyotrophic lateral sclerosis. Acta Neuropathol. 2013, 125, 111–120. [Google Scholar] [CrossRef]
- Zlokovic, B.V. Neurovascular pathways to neurodegeneration in Alzheimer’s disease and other disorders. Nat. Rev. Neurosci. 2011, 12, 723–738. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Zhao, Z.; Sagare, A.P.; Wu, Y.; Wang, M.; Owens, N.C.; Verghese, P.B.; Herz, J.; Holtzman, D.M.; Zlokovic, B.V. Blood-brain barrier-associated pericytes internalize and clear aggregated amyloid-β42 by LRP1-dependent apolipoprotein E isoform-specific mechanism. Mol. Neurodegener. 2018, 13, 57. [Google Scholar] [CrossRef] [PubMed]
- Fabry, Z.; Fitzsimmons, K.M.; Herlein, J.A.; Moninger, T.O.; Dobbs, M.B.; Hart, M.N. Product ion of the cytokines interleukin 1 and 6 by murine brain microvessel endothelium and smooth muscle pericytes. J. Neuroimmunol. 1993, 47, 23–34. [Google Scholar] [CrossRef] [PubMed]
- Nyúl-Tóth, Á.; Kozma, M.; Nagyőszi, P.; Nagy, K.; Fazakas, C.; Haskó, J.; Molnár, K.; Farkas, A.E.; Végh, A.G.; Váró, G.; et al. Expression of pattern recognition receptors and activation of the non-canonical inflammasome pathway in brain pericytes. Brain. Behav. Immun. 2017, 64, 220–231. [Google Scholar] [CrossRef] [PubMed]
- Broz, P.; Dixit, V.M. Inflammasomes: Mechanism of assembly, regulation and signalling. Nat. Rev. Immunol. 2016, 16, 407–420. [Google Scholar] [CrossRef] [PubMed]
- Guijarro-Muñoz, I.; Compte, M.; Álvarez-Cienfuegos, A.; Álvarez-Vallina, L.; Sanz, L. Lipopolysaccharide Activates Toll-like Receptor 4 (TLR4)-mediated NF-κB Signaling Pathway and Proinflammatory Response in Human Pericytes. J. Biol. Chem. 2014, 289, 2457–2468. [Google Scholar] [CrossRef] [PubMed]
- Stark, K.; Eckart, A.; Haidari, S.; Tirniceriu, A.; Lorenz, M.; von Brühl, M.-L.; Gärtner, F.; Khandoga, A.G.; Legate, K.R.; Pless, R.; et al. Capillary and arteriolar pericytes attract innate leukocytes exiting through venules and “instruct” them with pattern-recognition and motility programs. Nat. Immunol. 2013, 14, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Kaushik, D.K.; Bhattacharya, A.; Lozinski, B.M.; Wee Yong, V. Pericytes as mediators of infiltration of macrophages in multiple sclerosis. J. Neuroinflamm. 2021, 18, 301. [Google Scholar] [CrossRef]
- Jansson, D.; Rustenhoven, J.; Feng, S.; Hurley, D.; Oldfield, R.L.; Bergin, P.S.; Mee, E.W.; Faull, R.L.; Dragunow, M. A role for human brain pericytes in neuroinflammation. J. Neuroinflamm. 2014, 11, 104. [Google Scholar] [CrossRef]
- Rustenhoven, J.; Aalderink, M.; Scotter, E.L.; Oldfield, R.L.; Bergin, P.S.; Mee, E.W.; Graham, E.S.; Faull, R.L.M.; Curtis, M.A.; Park, T.I.-H.; et al. TGF-beta1 regulates human brain pericyte inflammatory processes involved in neurovasculature function. J. Neuroinflamm. 2016, 13, 37. [Google Scholar] [CrossRef] [PubMed]
- Mäe, M.A.; He, L.; Nordling, S.; Vazquez-Liebanas, E.; Nahar, K.; Jung, B.; Li, X.; Tan, B.C.; Foo, J.C.; Cazenave-Gassiot, A.; et al. Single-Cell Analysis of Blood-Brain Barrier Response to Pericyte Loss. Circ. Res. 2021, 128, e46–e62. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; D’Souza, S.S.; Moskvin, O.V.; Toh, H.; Wang, B.; Zhang, J.; Swanson, S.; Guo, L.-W.; Thomson, J.A.; Slukvin, I.I. Specification and Diversification of Pericytes and Smooth Muscle Cells from Mesenchymoangioblasts. Cell Rep. 2017, 19, 1902–1916. [Google Scholar] [CrossRef] [PubMed]
- Faal, T.; Phan, D.T.T.; Davtyan, H.; Scarfone, V.M.; Varady, E.; Blurton-Jones, M.; Hughes, C.C.W.; Inlay, M.A. Induction of Mesoderm and Neural Crest-Derived Pericytes from Human Pluripotent Stem Cells to Study Blood-Brain Barrier Interactions. Stem Cell Rep. 2019, 12, 451–460. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Schwartz, M.P.; Hou, Z.; Bai, Y.; Ardalani, H.; Swanson, S.; Steill, J.; Ruotti, V.; Elwell, A.; Nguyen, B.K.; et al. A Genome-wide Analysis of Human Pluripotent Stem Cell-Derived Endothelial Cells in 2D or 3D Culture. Stem Cell Rep. 2017, 8, 907–918. [Google Scholar] [CrossRef] [PubMed]
- Stebbins, M.J.; Gastfriend, B.D.; Canfield, S.G.; Lee, M.S.; Richards, D.; Faubion, M.G.; Li, W.J.; Daneman, R.; Palecek, S.P.; Shusta, E.V. Human pluripotent stem cell–derived brain pericyte–like cells induce blood-brain barrier properties. Sci. Adv. 2019, 5, eaau7375. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Huang, Y.; Gong, J.; Wang, J.; Fan, Y.; Cai, J.; Wang, Y.; Qiu, Y.; Wei, Y.; Xiong, C.; et al. Transplantation of hPSC-derived pericyte-like cells promotes functional recovery in ischemic stroke mice. Nat. Commun. 2020, 11, 5196. [Google Scholar] [CrossRef] [PubMed]
- Kelleher, J.; Dickinson, A.; Cain, S.; Hu, Y.; Bates, N.; Harvey, A.; Ren, J.; Zhang, W.; Moreton, F.C.; Muir, K.W.; et al. Patient-Specific iPSC Model of a Genetic Vascular Dementia Syndrome Reveals Failure of Mural Cells to Stabilize Capillary Structures. Stem Cell Rep. 2019, 13, 817–831. [Google Scholar] [CrossRef] [PubMed]
- Payne, L.B.; Hoque, M.; Houk, C.; Darden, J.; Chappell, J.C. Pericytes in Vascular Development. Curr. Tissue Microenviron. Rep. 2020, 1, 143–154. [Google Scholar] [CrossRef] [PubMed]
- Wanjare, M.; Kusuma, S.; Gerecht, S. Defining differences among perivascular cells derived from human pluripotent stem cells. Stem Cell Rep. 2014, 2, 561–575. [Google Scholar] [CrossRef]
- Orlova, V.V.; Drabsch, Y.; Freund, C.; Petrus-Reurer, S.; van den Hil, F.E.; Muenthaisong, S.; Dijke, P.T.; Mummery, C.L. Functionality of Endothelial Cells and Pericytes From Human Pluripotent Stem Cells Demonstrated in Cultured Vascular Plexus and Zebrafish Xenografts. Arterioscler. Thromb. Vasc. Biol. 2014, 34, 177–186. [Google Scholar] [CrossRef] [PubMed]
- Orlova, V.V.; van den Hil, F.E.; Petrus-Reurer, S.; Drabsch, Y.; ten Dijke, P.; Mummery, C.L. Generation, expansion and functional analysis of endothelial cells and pericytes derived from human pluripotent stem cells. Nat. Protoc. 2014, 9, 1514–1531. [Google Scholar] [CrossRef]
- Fortune, A.J.; Fletcher, J.L.; Blackburn, N.B.; Young, K.M. Using MS induced pluripotent stem cells to investigate MS aetiology. Mult. Scler. Relat. Disord. 2022, 63, 103839. [Google Scholar] [CrossRef]
- Tachibana, M.; Yamazaki, Y.; Liu, C.-C.; Bu, G.; Kanekiyo, T. Pericyte implantation in the brain enhances cerebral blood flow and reduces amyloid-β pathology in amyloid model mice. Exp. Neurol. 2018, 300, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Smyth, L.C.D.; Rustenhoven, J.; Scotter, E.L.; Schweder, P.; Faull, R.L.M.; Park, T.I.H.; Dragunow, M. Markers for human brain pericytes and smooth muscle cells. J. Chem. Neuroanat. 2018, 92, 48–60. [Google Scholar] [CrossRef] [PubMed]
- Katt, M.E.; Shusta, E.V. In vitro models of the blood-brain barrier: Building in physiological complexity. Curr. Opin. Chem. Eng. 2020, 30, 42–52. [Google Scholar] [CrossRef] [PubMed]
- Han, J.J. FDA Modernization Act 2.0 allows for alternatives to animal testing. Artif. Organs 2023, 47, 449–450. [Google Scholar] [CrossRef]
- Doss, M.X.; Sachinidis, A. Current Challenges of iPSC-Based Disease Modeling and Therapeutic Implications. Cells 2019, 8, 403. [Google Scholar] [CrossRef] [PubMed]
- Kilpatrick, S.; Irwin, C.; Singh, K.K. Human pluripotent stem cell (hPSC) and organoid models of autism: Opportunities and limitations. Transl. Psychiatry 2023, 13, 217. [Google Scholar] [CrossRef] [PubMed]
- Volpato, V.; Webber, C. Addressing variability in iPSC-derived models of human disease: Guidelines to promote reproducibility. Dis. Model. Mech. 2020, 13, dmm042317. [Google Scholar] [CrossRef] [PubMed]
| Disease | Differentiation | Key Findings | Source |
|---|---|---|---|
| Abud et al. 2017 [38] |
| [58] | |
| AD | Abud et al. 2017 [38] |
| [38] |
| AD | Abud et al. 2017 [38] |
| [59] |
| AD | Muffat et al. 2016 [54] |
| [60] |
| AD | Gutbier et al. 2020 [61] adapted from Wilgenburg et al. 2013 [62] |
| [63] |
| AD | Transcription factor enhanced |
| [57] |
| PD | Wilgenburg et al. 2013 [62] |
| [27] |
| Disease | Differentiation | Key Findings | Source |
|---|---|---|---|
| AD | Lippmann et al. 2014 [150] |
| [85] |
| General Inflammation | Lippmann et al. 2014 [150] |
| [161] |
| General Inflammation | Lippmann et al. 2014 [150] |
| [160] |
| General Inflammation | Lippmann et al. 2014 [150] |
| [80] |
| General Inflammation | Comparison between Lippmann et al. 2014 [150] Lian et al. 2014 [162]. Nishihara et al. 2021 [152] |
| [159] |
| MS | Nishihara et al. 2021 [152] |
| [26] |
| General Inflammation | Rosa et al. 2019 [163] |
| [153] |
| Differentiation Progenitor | Key Findings | Source |
|---|---|---|
| Mesenchymal progenitor |
| [183] |
| Mesoderm/neural crest |
| [184] |
| Mesodermal lineage |
| [185] |
| Neural crest |
| [186] |
| [187] | |
| [188] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kala, S.; Strutz, A.G.; Katt, M.E. The Rise of Pluripotent Stem Cell-Derived Glia Models of Neuroinflammation. Neurol. Int. 2025, 17, 6. https://doi.org/10.3390/neurolint17010006
Kala S, Strutz AG, Katt ME. The Rise of Pluripotent Stem Cell-Derived Glia Models of Neuroinflammation. Neurology International. 2025; 17(1):6. https://doi.org/10.3390/neurolint17010006
Chicago/Turabian StyleKala, Srishti, Andrew G. Strutz, and Moriah E. Katt. 2025. "The Rise of Pluripotent Stem Cell-Derived Glia Models of Neuroinflammation" Neurology International 17, no. 1: 6. https://doi.org/10.3390/neurolint17010006
APA StyleKala, S., Strutz, A. G., & Katt, M. E. (2025). The Rise of Pluripotent Stem Cell-Derived Glia Models of Neuroinflammation. Neurology International, 17(1), 6. https://doi.org/10.3390/neurolint17010006






