Systematic Review of Pharmacogenetics of Immunosuppressants in Heart Transplantation
Abstract
1. Introduction
2. Materials and Methods
3. Results
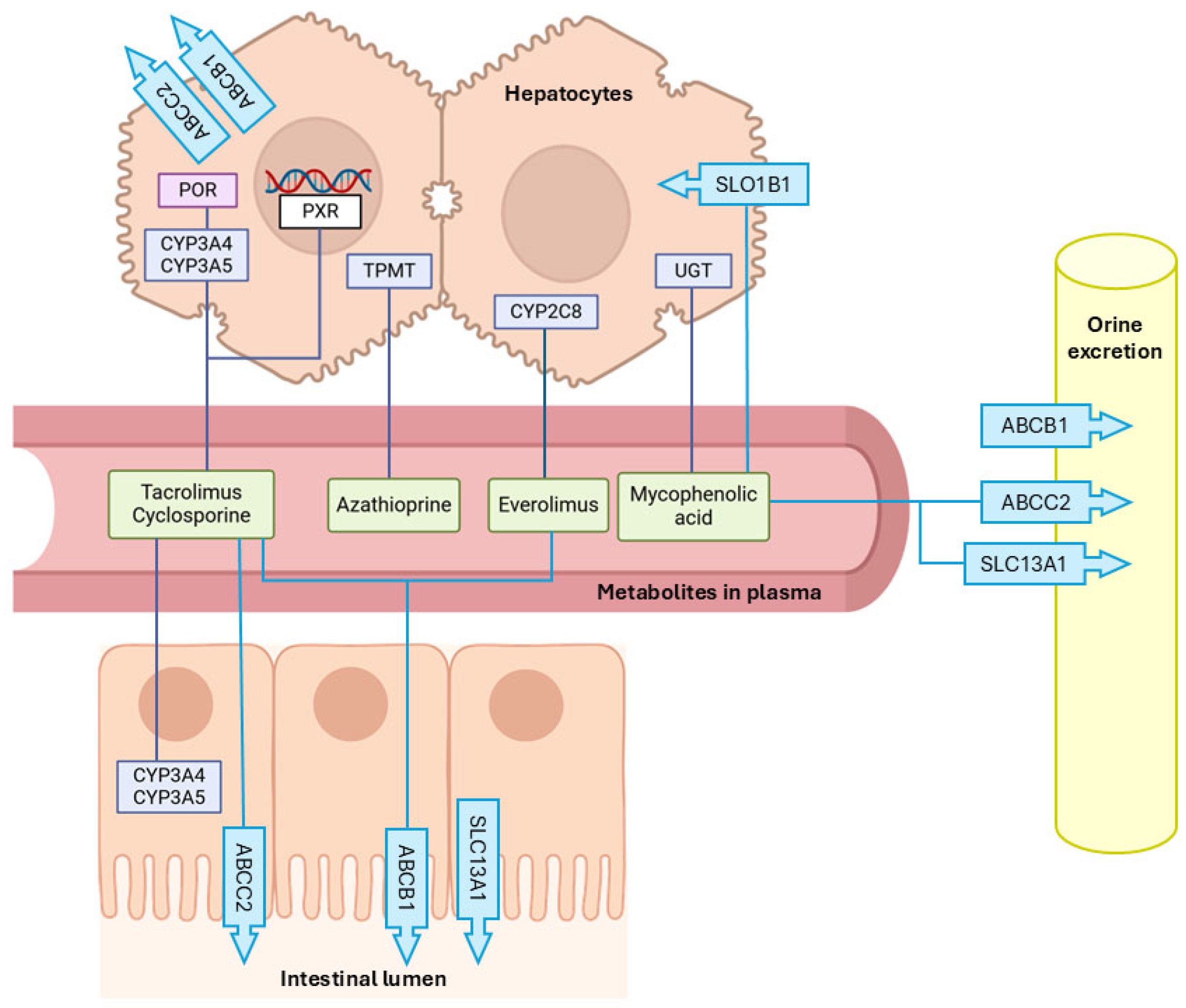
3.1. Immunosuppressant Metabolizing Enzymes
Cytochrome P450 (CYP) and Other Related Genes
- CYP3A4
- CYP3A5
- CYP2C8
- Cytochrome P450 oxidoreductase (POR)
- Nuclear Receptor Subfamily 1 Group I Member 2 (NR1I2)/Pregnane X Receptor (PXR)
- Uridine glycosyltransferase (UGT)
- Inosine monophosphate dehydrogenase (IMPDH)
- Thiopurine methyltransferase (TPMT)
| SNP | Study | n | Age Recipient (Range) | Age Donor (Range) | Ethnia Recipient | Ethnia Donor | HWE | Immunossupresant Scheme | Clinical Outcomes |
|---|---|---|---|---|---|---|---|---|---|
| CYP3A4 | |||||||||
| CYP3A4*18B (C > T) rs2242480 | Cheng 2023 [4] | 53 | 48.45 ± 11.78 | NR | Unknown (China) | NR | Yes | TAC + MMF + CORT |
× (1 + 0.3546 × (rs2242480 − TT)) × (1 + 0.5815 × (rs2242480 − CT))] × exp (CL) |
| CYP3A4 (C > A, G, T) (rs2740574) | Díaz-Molina 2012 [23] | 65 | 54.55 ± 10 | NR | Unknown (Spain) | NR | NR | TAC + MMF + PRED |
|
| 390 A > G (rs2740574) | Isla 2009 [24] | 30 | 43 ± 14 | NR | Caucasian | NR | Yes | CSA ± AZA/MMF/CORT/ SIR ± BSX/ATG |
|
| 390 A > G (rs2740574) | Liu 2024 [5] | 177 | 54 | NR | Unknown (USA) | NR | NR | TAC |
|
| CYP3A4*22 (G > A) (rs35599367) | Deininger 2016 [9] | 76 | 44 ± 14 | NR | Caucasian 81.6% African American 3.9% Asian or Pacific Islander 6.6% American Indian, Eskimo, or Aleutian 1.3% Other 6.6% | NR | Yes | TAC + MMF (77.6%), PRED (23.7%), AZA (13.2%) or SIR (10.5%) |
|
| CYP3A4*22 (G > A) (rs35599367) | Gijsen 2013 [3] | 60 | Median = 4 IQR = 12 | NR | Caucasian 65.0% African American 6.7% Asian 5.0% Native 1.7% Unknown 21.6% | NR | Yes | TAC + MMF + CORT |
|
| CYP3A4*22 (G > A) (rs35599367) | Liu 2024 [5] | 177 | 54 | NR | Unknown (USA) | NR | NR | TAC |
|
| Lesche 2014 [13] | 104 | 47.4 ± 14.3 | NR | White 95% Asian 3% Other 2% | NR | Yes | TAC/CSA/EVE/SIR + AZA/MMF + CORT |
| |
| CYP3A4*1G (20230G > A) (rs2242480) | Liu 2022 [7] | 66 | Median = 10 IQR = 7m–17y | NR | Unknown (China) | NR | Yes | TAC + MMF + PRED |
|
| CYP3A4*1G (20230G > A) (rs2242480) | Liu 2024 [5] | 177 | 54 | NR | Unknown (USA) | NR | NR | TAC |
|
| CYP3A5 | |||||||||
| G > A | Antignac 2010 [25] | 60 | NR | NR | NR | NR | NR | EVE |
|
| CYP3A5*3/*1 6986A > G rs776746 | Lemaitre 2012 [26] | 59 | 50 ± 14 (17–80) | NR | NR (France) | NR (France) | NR | EVE + TAC or CSA + MMF or AZA + CORT |
|
| Feingold 2012 [27] | 453 | 6.2 ± 6.1 | NR | White 60% Black 13% Hispanic 23% | NR | No | CSA or TAC |
| |
| Lesche 2015 [14] | 37 | 53 (16–75) | NR | Caucasian 95% Other 5% | NR | Yes | EVE + MMF or AZA + CORT |
| |
| CYP3A5 (-A392G) (rs776746) | Díaz-Molina 2012 [23] | 65 | 54.55 ± 10 | NR | Unknown (Spain) | NR | NR | TAC + MMF + PRED |
|
| CYP3A5*1 (T > C) (rs776746) | Mirza 2021 [28] | 66 (Poor: 41 Inter: 21 Exten:4) | Poor (Median = 58 IQR = 55–65) Inter (Median = 63 IQR = 60–67) Exten (Median = 58 IQR = 55–62) | NR | Poor (Caucasian 80.5% Hispanic/Latino 4.9% African American 12.2% Other 2.4%) Inter (Caucasian 43% Hispanic/Latino 24% African American 33%) Exten (African American 100%) | NR | NR | TAC ± MMF ± AZA |
|
| CYP3A5*1 (T > C) (rs776746) | Uno 2018 [8] | 65 (*1/*1: 5 *1/*3: 22 *3/*3: 38) | *1/*1 y *1/*3 Median = 43 IQR = 30–55 *3/*3 Median = 40 IQR = 29–47.8 | NR | Unknown (Japan) | NR | Yes | TAC + MMF + AZA |
|
| CYP3A5*1 (T > C) (rs776746) | Uno 2019 [29] | 26 (*1/*1: 2 *1/*3: 8 *3/*3: 16) | *1/*1 y *1/*3 Median = 45.1 IQR = 14.8 *3/*3 Median = 44.3 IQR = 11.8 | NR | Unknown (Japan) | NR | Yes | TAC + MMF + AZA |
|
| CYP3A5*3 (T > C) (rs776746) | Zheng 2003 [6] | 54 | CYP3A5 *1/*3: 8.7 ± 1.7 CYP3A5 *3/*3: 7.2 ± 1.2 | NR | CYP3A5: African American: 5.6% Other: 94.4% | NR | NR | TAC + PRED ± MMF/AZA |
|
| CYP3A5*3 (T > C) (rs776746) | Deininger 2016 [9] | 76 | 44 ± 14 | NR | Caucasian 81.6% African American 3.9% Asian or Pacific Islander 6.6% American Indian, Eskimo, or Aleutian 1.3% Other 6.6% | NR | Yes | TAC + MMF (77.6%), PRED (23.7%), AZA (13.2%) or SIR (10.5%) |
|
| CYP3A5*3 (6986A > G) (rs776746) | Déri 2021 [30] | 78 | 53.1 (range:19.5–68.7) | NR | Unknown (Hungary) | NR | NR | TAC + MMF + CORT |
|
| CYP3A5*3 (T > C) (rs776746) | Gijsen 2011 [31] | 39 | Median = 6 IQR = 13.75 | NR | White 71.8% African American 5.1% Asian 10.3% Unknown 12.8% | NR | Yes | TAC ± MMF ± CORT | CYP3A5 expressers:
|
| CYP3A5*3 (T > C) (rs776746) | Gijsen 2013 [3] | 60 | Median = 4 IQR = 12 | NR | Caucasian 65.0% African American 6.7% Asian 5.0% Native 1.7% Unknown 21.6% | NR | Yes | TAC + MMF + CORT |
|
| CYP3A5*3 (T > C) (rs776746) | Kniepeiss 2011 [10] | 45 (TAC 15 + EVE 30) | TAC *3*3 54 ± 11 TAC *3*1 37 ± 3 EVE *3*3 47 ± 14 EVE *3*1 60 ± 8 | NR | Caucasian 100% | NR | Yes | TAC/CSA + MMF + PRED EVE + CSA+ PRED | TAC:
|
| CYP3A5*3 (A > G) (rs776746) | Liu 2019 [32] | 55 | *1/*3 (27.3%) 39.20 ± 11.73 *1/*1 (72.7%) 40.92 ± 14.26 | NR | Unknown (China) | NR | Yes | TAC + MMF + PRED |
|
| CYP3A5*3 (6986A > G) (rs776746) | Liu 2022 [7] | 66 | Median = 10 IQR = 7m–17y | NR | Unknown (China) | NR | Yes | TAC + MMF + PRED |
|
| CYP3A5*3 (rs776746) CYP3A5*6 (rs10264272) CYP3A5*7 (rs41303343) | Pasternak 2021 [33] | 37 | 9.2 ± 6.3 | NR | White 81.1% Black/African American 10.8% Other 8.1% | NR | NR | TAC ± UNKNOWN |
|
| CYP3A5*3 (rs776746) CYP3A5*6 (rs10264272) CYP3A5*7 (rs41303343) | Pasternak 2023 [34] | 38 | 12.2 ± 4.1 | NR | White 76.3% African American 13.2% Other/unknown 10.5% | NR | NR | TAC + MMF/AZA + CORT |
|
| *1 *1/*3 6986 A > G *3 (rs776746) | De Denus 2011 [11] | 160 | 53.2 (IQR: 43.5–58.2) | NR | Caucasian 98.1% | NR | Yes | CSA/TAC/MMF/AZA + CORT |
|
| Klauke 2008 [35] | 106 | RI (n = 53): 50.2 ± 14.6 noRI (n = 53): 51.3 ± 10.7 | RI (n = 53): 35.5 ± 13.7 noRI (n = 53): 34.2 ± 12.1 | NR | NR | NR | CSA/TAC + AZA/MMF + CORT ± mTOR |
| |
| 6986 A > G *3 (rs776746) | Herrero 2010 [2] | 18 | NR | NR | NR | NR | NR | CSA/TAC |
|
| Isla 2009 [24] | 30 | 43 ± 14 | NR | Caucasian | NR | Yes | CSA ± AZA/MMF/CORT/ SIR ± BSX/ATG |
| |
| Jordán 2011 [36] | 41 | NR | NR | NR | NR | NR | CSA or TAC |
| |
| Lesche 2014 [13] | 104 | 47.4 ± 14.3 | NR | White 95% Asian 3% Other 2% | NR | Yes | TAC/CSA/EVE/SIR + AZA/MMF + CORT |
-eGFR: no differences (TAC). | |
| Sigurdardottir 2013 [16] | 107 | NR | NR | NR | NR | NR | TAC or CSA |
| |
| 267871 G > A *6 (rs10264272) | Herrero 2010 [2] | 18 | NR | NR | NR | NR | NR | CSA/TAC |
|
| Jordán 2011 [36] | 41 | NR | NR | NR | NR | NR | CSA or TAC |
| |
| CYP2C8 | |||||||||
| CYP2C8*1, CYP2C8*3 | Kniepeiss 2013 [12] | 30 | *1/*1 47 ± 16 *1/*3 52 ± 8 | NR | NR (Austria) | NR (Austria) | Yes | EVE + CSA + CORT (previously CNI + MMF + CORT) |
|
| Lesche 2015 [14] | 37 | 53 (16–75) | NR | Caucasian 95% Other 5% | NR | Yes | EVE + MMF or AZA + CORT |
| |
| c.-635_-634 delAT rs372775254 | Lesche 2015 [14] | 37 | 53 (16–75) | NR | Caucasian 95% Other 5% | NR | Yes | EVE + MMF or AZA + CORT |
|
| c.-411T > C rs7912549 | Lesche 2015 [14] | 37 | 53 (16–75) | NR | Caucasian 95% Other 5% | NR | Yes | EVE + MMF or AZA + CORT |
|
| c.-370T > G CYP2C8*1C rs17110453 | Lesche 2015 [14] | 37 | 53 (16–75) | NR | Caucasian 95% Other 5% | NR | Yes | EVE + MMF or AZA + CORT |
|
| c.-271C > A CYP2C8*1B rs7909236 | Lesche 2015 [14] | 37 | 53 (16–75) | NR | Caucasian 95% Other 5% | NR | Yes | EVE + MMF or AZA + CORT |
|
| c.332-6_332-5InsT rs11572078 | Lesche 2015 [14] | 37 | 53 (16–75) | NR | Caucasian 95% Other 5% | NR | Yes | EVE + MMF or AZA + CORT |
|
| c.332-64G > A rs2275622 | Lesche 2015 [14] | 37 | 53 (16–75) | NR | Caucasian 95% Other 5% | NR | Yes | EVE + MMF or AZA + CORT |
|
| c.416G > A (p.Arg139Lys) CYP2C8*3 rs11572080 | Lesche 2015 [14] | 37 | 53 (16–75) | NR | Caucasian 95% Other 5% | NR | Yes | EVE + MMF or AZA + CORT |
|
| c.1196A > G (p.Lys399Arg) CYP2C8*3 rs10509681 | Lesche 2015 [14] | 37 | 53 (16–75) | NR | Caucasian 95% Other 5% | NR | Yes | EVE + MMF or AZA + CORT |
|
| c.1291 + 106G > A rs1934951 | Lesche 2015 [14] | 37 | 53 (16–75) | NR | Caucasian 95% Other 5% | NR | Yes | EVE + MMF or AZA + CORT |
|
| POR | |||||||||
| c.1508C > T (p.Ala503Val) POR*28 rs1057868 | Lesche 2015 [14] | 37 | 53 (16–75) | NR | Caucasian 95% Other 5% | NR | Yes | EVE + MMF or AZA + CORT |
|
| *28 (rs1057868) | Lesche 2014 [13] | 104 | 47.4 ± 14.3 | NR | White 95% Asian 3% Other 2% | NR | Yes | TAC/CSA/EVE/SIR + AZA/MMF + CORT |
|
| Sigurdardottir 2013 [16] | 107 | NR | NR | NR | NR | NR | TAC or CSA |
| |
| POR (6593A > G) (rs2868177) | Liu 2022 [7] | 66 | Median = 10 IQR = 7m–17y | NR | Unknown (China) | NR | Yes | TAC + MMF + PRED |
|
| NR1I2 NR112/PXR | |||||||||
| c.-1135C > T rs3814055 | Lesche 2015 [14] | 37 | 53 (16–75) | NR | Caucasian 95% Other 5% | NR | Yes | EVE + MMF or AZA + CORT |
|
| G25385C > T (rs3814055) | Lesche 2014 [13] | 104 | 47.4 ± 14.3 | NR | White 95% Asian 3% Other 2% | NR | Yes | TAC/CSA/EVE/SIR + AZA/MMF + CORT |
|
| Sigurdardottir 2013 [16] | 107 | NR | NR | NR | NR | NR | TAC or CSA |
| |
| UGT1A9 | |||||||||
| T > G (rs6731242) | Oreschak 2021 [37] | 148 | 49 ± 13 | NR | European American 77.7% African American 10.8% Asian 4.1% Other 7.4% | NR | NR | TAC/CSA ± MMF/AZA + CORT |
|
| UGT2B7 | |||||||||
| C802T (rs749366) | Ting 2010 [17] | 32 | 61.3 ((R: 23.2–77.6) | NR | NR | NR | NR | MMF ± CSA/TAC/SIR ± CORT |
|
| IMPDH2 | |||||||||
| rs11706052 | Ohmann 2010 [19] | 59 | Pediatric | NR | Caucasian 86% African 10% Hispanic 3% | NR | Yes | MMF + TAC or CSA + CORT |
|
| IMPDH1 | |||||||||
| rs2288553 | Ohmann 2010 [19] | 59 | Pediatric | NR | Caucasian 86% African 10% Hispanic 3% | NR | Yes | MMF + TAC or CSA + CORT |
|
| rs2288549 | Ohmann 2010 [19] | 59 | Pediatric | NR | Caucasian 86% African 10% Hispanic 3% | NR | Yes | MMF + TAC or CSA + CORT |
|
| rs2278293 | Ohmann 2010 [19] | 59 | Pediatric | NR | Caucasian 86% African 10% Hispanic 3% | NR | Yes | MMF + TAC or CSA + CORT |
|
| rs2278294 | Ohmann 2010 [19] | 59 | Pediatric | NR | Caucasian 86% African 10% Hispanic 3% | NR | Yes | MMF + TAC or CSA + CORT |
|
| rs2228075 | Ohmann 2010 [19] | 59 | Pediatric | NR | Caucasian 86% African 10% Hispanic 3% | NR | Yes | MMF + TAC or CSA + CORT |
|
| rs2288553, rs2288549, rs2278293, rs2278294, rs2220875 (H: TCCCC, TCTTT, TTCCC, TCTCC, ACCCC) | Ohmann 2010 [19] | 59 | 8.2 (IQR: 1.9–13.5) | NR | White non-Hispanic 86.4% White Hispanic 3.4% Black non-Hispanic 10.2% | NR | NR | MMF + TAC/CSA |
No significant differences for the rest of haplotypes. |
| A > G (rs11761662) | Oreschak 2021 [37] | 148 | 49 ± 13 | NR | European American 77.7% African American 10.8% Asian 4.1% Other 7.4% | NR | NR | TAC/CSA ± MMF/AZA + CORT |
|
| TPMT | |||||||||
| A719G G460A G238C | Liang 2013 [22] | 93 | 49.4 | 33.7 | Caucasian | NR | NR | CSA or TAC + AZA |
|
3.2. Membrane Transporters
3.2.1. Efflux Transporters
- ATP-binding cassette transporter B family member 1 (ABCB1)
- ATP-binding cassette transporter B family member 2 (ABCB2)
3.2.2. Efflux Transporters
- Solute Carrier Organic Anion Transporter Family Member 1B1 (SLCO1B1)
- Solute Carrier Family 13 Member 1 (SLC13A1)
| SNP | Study | n | Age Recipient (Range) | Age Donor (Range) | Ethnia Recipient | Ethnia Donor | HWE | Immunossupresant Scheme | Clinical Outcomes |
|---|---|---|---|---|---|---|---|---|---|
| ABCB1 | |||||||||
| C3435T T/T. C/C. C/T (exon 26; rs1045642) | Barnard 2006 [38] | 170 | T/T 50.5 ± 1.6 C/C 46.6 ± 2.8 C/T 47.2 ± 1.2 | T/T 31.4 ± 1.9 C/C 29.2 ± 2.6 C/T 32 ± 1.2 | NR | NR | Yes | CSA + AZA + CORT |
|
| Chowbay 2003 [45] | 14 | 47.9 ± 9.8 | NR | Chinese 71.4% Indian 28.6% | NR | No | CSA + AZA + CORT |
| |
| De Denus 2011 [11] | 160 | 53.2 (RIC: 43.5–58.2) | NR | Caucasian 98.1% | NR | Yes | CSA/TAC/MMF/AZA + CORT |
| |
| Díaz-Molina 2012 [23] | 65 | 54.55 ± 10 | NR | Unknown (Spain) | NR | NR | TAC + MMF + PRED |
| |
| Feingold 2012 [27] | 453 | 6.2 ± 6.1 | NR | White 60% Black 13% Hispanic 23% | NR | Yes | CSA or TAC |
| |
| De Iudicibus 2008 [39] | 50: Heart 21 Renal 26 Lung 4 | Overgrowth (mix) < 30% 56.3 (34–73) ≥ 30% 57.9 (35–75) | NR | NR | NR | NR | CSA |
| |
| Herrero 2010 [2] | 18 | NR | NR | NR | NR | NR | CSA/TAC |
| |
| Isla 2009 [24] | 30 | 43 ± 14 | NR | Caucasian | NR | Yes | CSA ± AZA/MMF/CORT/ SIR ± BSX/ATG |
| |
| Lesche 2014 [13] | 104 | 47.4 ± 14.3 | NR | White 95% Asian 3% Other 2% | NR | Yes | TAC/CSA/EVE/SIR + AZA/MMF + CORT |
| |
| Taegtmeyer 2010 [40] | 337 | 48.1 ± 11.1 | 32.5 ± 13.2 | White 90.5% | NR | Yes | CSA ± AZA ± CORT ± ATG |
| |
| Liu 2022 [7] | 66 | Median = 10 IQR = 7m–17y | NR | Unknown (China) | NR | Yes | TAC + MMF + PRED |
| |
| C3435T rs1045642 | Antignac 2010 [25] | 60 | NR | NR | NR | NR | NR | EVE |
|
| Lemaitre 2012 [26] | 59 | 50 ± 14 (17–80) | NR | NR (France) | NR (France) | NR | EVE + TAC or CSA + MMF or AZA + CORT |
| |
| Lesche 2015 [14] | 37 | 53 (16–75) | NR | Caucasian 95% Other 5% | NR | Yes | EVE + MMF or AZA + CORT |
| |
| Kunicki 2016 [41] | 31 | 55 (24–76) | NR | NR (Poland) | NR (Poland) | NR | EVE + CNI or MMF |
| |
| G2677T A/G. A/T. G/G. G/T. T/T (exon 21; rs2032582) | Barnard 2006 [38] | 170 | A/G 42.5 ± 5.9 A/T 52.6 ± 4.5 G/G 48.1 ± 2.0 G/T 48.9 ±1.3 T/T 45.8 ± 2.0 | A/G 36 ± 4.4 A/T 42.3 ± 9.6 G/G 29.2 ± 1.9 G/T 33 ± 1.4 T/T 29.3 ± 2.0 | NR | NR | Yes | CSA + AZA + CORT |
|
| G2677T A/G. G/G. G/T. T/T (exon 21; rs2032582) | De Denus 2011 [11] | 160 | 53.2 (IQR: 43.5–58.2) | NR | Caucasian 98.1% | NR | Yes | CSA/TAC/MMF/AZA+ CORT |
|
| G2677T G/G. G/T. T/T (exon 21; rs2032582) | Chowbay 2003 [45] | 14 | 47.9 ± 9.8 | NR | Chinese 71.4% Indian 28.6% | NR | No | CSA + AZA + CORT |
|
| De Iudicibus 2008 [39] | 50: H21 R26 L4 | Overgrowth (mix) < 30% 56.3 (34–73) ≥ 30% 57.9 (35–75) | NR | NR | NR | NR | CSA |
| |
| Herrero 2010 [2] | 18 | NR | NR | NR | NR | NR | CSA/TAC |
| |
| Klauke 2008 [35] | 106 | RI (n = 53): 50.2 ± 14.6 noRI (n = 53): 51.3 ± 10.7 | RI (n = 53): 35.5 ± 13.7 noRI (n = 53): 34.2 ± 12.1 | NR | NR | NR | CSA/TAC + AZA/MMF + CORT ± mTOR |
| |
| Taegtmeyer 2010 [40] | 337 | 48.1 ± 11.1 | 32.5 ± 13.2 | White 90.5% | NR | Yes | CSA ± AZA ± CORT ± ATG |
| |
| Feingold 2012 [27] | 453 | 6.2 ± 6.1 | NR | White 60% Black 13% Hispanic 23% | NR | Yes | CSA or TAC |
| |
| Lesche 2014 [13] | 104 | 47.4 ± 14.3 | NR | White 95% Asian 3% Other 2% | NR | Yes | TAC/CSA/EVE/SIR + AZA/MMF + CORT |
| |
| Lesche 2015 [14] | 37 | 53 (16–75) | NR | Caucasian 95% Other 5% | NR | Yes | EVE + MMF or AZA + CORT |
| |
| C1236T C/C. C/T. T/T (exon 12; rs1128503) | Chowbay 2003 [45] | 14 | 47.9 ± 9.8 | NR | Chinese 71.4% Indian 28.6% | NR | No | CSA + AZA + CORT |
|
| Herrero 2010 [2] | 18 | NR | NR | NR | NR | NR | CSA/TAC |
| |
| Jordán 2012 [46] | 60 | NR | NR | NR | NR | NR | CSA or TAC |
| |
| Lesche 2014 [13] | 104 | 47.4 ± 14.3 | NR | White 95% Asian 3% Other 2% | NR | Yes | TAC/CSA/EVE/SIR + AZA/MMF + CORT |
| |
| Lesche 2015 [14] | 37 | 53 (16–75) | NR | Caucasian 95% Other 5% | NR | Yes | EVE + MMF or AZA + CORT |
| |
| Sánchez-Lázaro 2015 [15] | 60 | CSA (n = 36): 53 ± 11 TAC (n = 24): 52 ± 10 | CSA (n = 36): 42 ± 12 TAC (n = 24): 43 ± 9 | NR | NR | NR | CSA or TAC + MMF + CORT |
| |
| Taegtmeyer 2010 [40] | 337 | 48.1 ± 11.1 | 32.5 ± 13.2 | White 90.5% | NR | Yes | CSA ± AZA ± CORT ± ATG |
| |
| C3435T/G2677T GG/CC, GT/CT, TT/TT (H) | Chowbay 2003 [45] | 14 | 47.9 ± 9.8 | NR | Chinese 71.4% Indian 28.6% | NR | No | CSA + AZA + CORT |
|
| Taegtmeyer 2010 [40] | 337 | 48.1 ± 11.1 | 32.5 ± 13.2 | White 90.5% | NR | Yes | CSA ± AZA ± CORT ± ATG |
| |
| Zheng 2003 [6] | C3435T: 63 G2677T: 63 | C3435T CC: 6.3 ± 6.0 C3435T CT/TT: 7.9 ± 7.4 G2677T GG: 6.1 ± 5.8 G2677T GT/TT: 8.2 ± 7.5 | NR | C3435T: African American: 6.3% Other: 93.7% G2677T: African American: 6.3% Other: 93.7% | NR | NR | TAC + PRED ± MMF/AZA |
| |
| C3435T T/T, C/C, C/T G2677T G/G, G/T, T/T C1236T C/C, C/T, T/T (H) | Chowbay 2003 [45] | 14 | 47.9 ± 9.8 | NR | Chinese 71.4% Indian 2.6% | NR | No | CSA + AZA + CORT |
|
| Jordán 2011 [36] | 41 | NR | NR | NR | NR | NR | CSA or TAC |
| |
| Taegtmeyer 2010 [40] | 337 | 48.1 ± 11.1 | 32.5 ± 13.2 | White 90.0% | NR | Yes | CSA ± AZA ± CORT ± ATG |
| |
| ABCB1 (C3435T, G2677T, C1236T) | Gijsen 2011 [31] | 39 | Median = 6 IQR = 13.75 | NR | White 71.8% African American 5.1% Asian 10.3% Unknown 12.8% | NR | Yes | TAC ± MMF ± CORT |
|
| ABCB1 (c.1236 C > T) (rs1128503) (c.2677 G > T/A) (rs2032582) (c.3435 C > T) (rs1045642) | Oreschak 2017 [47] | 76 | 53 ± 15 | NR | Caucasian 82% Other 18% | NR | NR | TAC ± UNKNOWN |
|
| rs9282564 | Jordán 2012 [46] | 60 | NR | NR | NR | NR | NR | CSA or TAC |
|
| Jordán 2011 [36] | 41 | NR | NR | NR | NR | NR | CSA or TAC |
| |
| Sánchez-Lázaro 2015 [15] | 60 | CSA (n = 36): 53 ± 11 TAC (n = 24): 52 ± 10 | CSA (n = 36): 42 ± 12 TAC (n = 24): 43 ± 9 | NR | NR | NR | CSA or TAC + MMF + CORT |
| |
| rs2235013 | Jordán 2011 [36] | 41 | NR | NR | NR | NR | NR | CSA or TAC |
|
| rs2235033 | Jordán 2011 [36] | 41 | NR | NR | NR | NR | NR | CSA or TAC |
|
| ABCC2 | |||||||||
| rs717620 (G > A) | Ohmann 2010 [20] | 59 | Pediatric | NR | Caucasian 86% African 10% Hispanic 3% | NR | Yes | MMF + TAC or CSA + CORT |
|
| Burckart 2014 [43] | 290 | Pediatric | NR | Caucasian 74% African 19% Other 7% | NR | Yes | MMF ± UNKNOWN |
| |
| ABCC2 (c.-24 C > T) (rs717620) (c.1249 G > A) (rs2273697) (c.3972 C > T) (rs3740066) | Oreschak 2018 [42] | 89 | 54 ± 15 | NR | Caucasian 83% Other 17% | NR | NR | TAC ± UNKNOWN |
|
| SLCO1B1 | |||||||||
| rs4149056 | Sánchez-Lázaro 2015 [15] | 60 | CSA (n = 36): 53 ± 11 TAC (n = 24): 52 ± 10 | CSA (n = 36): 42 ± 12 TAC (n = 24): 43 ± 9 | NR | NR | NR | CSA or TAC + MMF + CORT |
|
| rs2306283 | Sánchez-Lázaro 2015 [15] | 60 | CSA (n = 36): 53 ± 11 TAC (n = 24): 52 ± 10 | CSA (n = 36): 42 ± 12 TAC (n = 24): 43 ± 9 | NR | NR | NR | CSA or TAC + MMF + CORT |
|
| SLCO1A2 | |||||||||
| rs11568564 | Sánchez-Lázaro 2015 [15] | 60 | CSA (n = 36): 53 ± 11 TAC (n = 24): 52 ± 10 | CSA (n = 36): 42 ± 12 TAC (n = 24): 43 ± 9 | NR | NR | NR | CSA or TAC + MMF + CORT |
|
| rs72559749 | Sánchez-Lázaro 2015 [15] | 60 | CSA (n = 36): 53 ± 11 TAC (n = 24): 52 ± 10 | CSA (n = 36): 42 ± 12 TAC (n = 24): 43 ± 9 | NR | NR | NR | CSA or TAC + MMF + CORT |
|
| rs11568563 | Sánchez-Lázaro 2015 [15] | 60 | CSA (n = 36): 53 ± 11 TAC (n = 24): 52 ± 10 | CSA (n = 36): 42 ± 12 TAC (n = 24): 43 ± 9 | NR | NR | NR | CSA or TAC + MMF + C |
|
| SLC13A1 | |||||||||
| T > C (rs2140516) | Oreschak 2021 [37] | 148 | 49 ± 13 | NR | European American 77.7% African American 10.8% Asian 4.1% Other 7.4% | NR | NR | TAC/CSA ± MMF/AZA + CORT |
|
3.3. Immunomodulatory Pathway
- Cytokines and growth factors may play a significant role in modulating the immune response and triggering acute transplant rejection; therefore, controlling cytokine production represents a potential strategy to reduce the risk of rejection. Human Leukocyte Antigen G (HLA-G)
- Tumor Necrosis Factor alfa (TNF-α)
- Transforming Growth Factor- ß1 (TGF-ß1)
- Other cytokines and growth factors
3.4. Other Genes
| SNP | Study | n | Age Recipient (Range) | Age Donor (Range) | Ethnia Recipient | Ethnia Donor | HWE | Immunossupresant Scheme | Clinical Outcomes |
|---|---|---|---|---|---|---|---|---|---|
| HLA-G | |||||||||
| 14-bp indel rs371194629 | Torres 2009 [48] | 37 | 44 ± 12 | NR | NR (Spain) | NR (Spain) | Yes | CSA + MMF or EVE + CORT |
|
| Adamson 2020 [49] | 123 | 48 ± 12 | 36 ± 14 | Caucasian 23% Black 2% Other 3% Unknown 72% | NR | Yes | TAC or CSA + MMF or AZA + CORT |
| |
| −201 G/A rs1233333 | Adamson 2020 [49] | 123 | 48 ± 12 | 36 ± 14 | Caucasian 23% Black 2% Other 3% Unknown 72% | NR | Yes | TAC or CSA + MMF or AZA + CORT |
|
| −725C/G/T rs1233334 | Adamson 2020 [49] | 123 | 48 ± 12 | 36 ± 14 | Caucasian 23% Black 2% Other 3% Unknown 72% | NR | No | TAC or CSA + MMF or AZA + CORT |
|
| +3196C/G rs1610696 | Adamson 2020 [49] | 123 | 48 ± 12 | 36 ± 14 | Caucasian 23% Black 2% Other 3% Unknown 72% | NR | Yes | TAC or CSA + MMF or AZA + CORT |
|
| +3187A/G rs9380142 | Adamson 2020 [49] | 123 | 48 ± 12 | 36 ± 14 | Caucasian 23% Black 2% Other 3% Unknown 72% | NR | Yes | TAC or CSA + MMF or AZA + CORT |
|
| +3142C/G rs1063320 | Adamson 2020 [49] | 123 | 48 ± 12 | 36 ± 14 | Caucasian 23% Black 2% Other 3% Unknown 72% | NR | Yes | TAC or CSA + MMF or AZA + CORT |
|
| TNF-A | Tumor necrosis factor-alpha | ||||||||
| −308 G > A rs1800629 | Azzawi 2001 [50] | 119 | NR | NR | NR (United Kingdom) | NR (United Kingdom) | NR | CSA + AZA + CORT |
|
| Ternstrom 2005 [51] | 70 | 48 ± 2 | 36 ± 2 | NR (Sweden) | NR (Sweden) | NR | CSA + AZA + CORT |
| |
| Girnita 2008 [52] | 323 | NR (Pediatric) | NR | White 63.5% Black 13.3% Hispanic 23.2% | NR | Yes | TAC or CSA + MMF or AZA + CORT |
| |
| Feingold 2012 [27] | 453 | 6.2 ± 6.1 | NR | White 60% Black 13% Hispanic 23% | NR | Yes | CSA or TAC |
| |
| TNF-B | |||||||||
| +252 G > A (rs909253) | Ternstrom 2005 [51] | 70 | 48 ± 2 | 36 ± 2 | NR (Sweden) | NR (Sweden) | NR | CSA + AZA + CORT |
|
| TGF-B1 | |||||||||
| 914 G > C (syn. Arg25 > Pro, codon 25) rs1800471 | Densem 1999 [61] | 121 | NR | NR | NR (United Kingdom) | NR (United Kingdom) | NR | CSA based schemes |
|
| Baan 2000 [53] | 168 | 44 ± 12 or 48 ± 9 (Cr < or > 250 µmol/L ) | NR | Caucasian 95% Unknown 5% | NR | NR | CSA based schemes |
| |
| Lacha 2001 [62] | 298 | NR | NR | NR (Czech Republic) | NR (Czech Republic) | NR | CSA + AZA + CORT |
| |
| Wetering 2006 [63] | 402 | 50 (range: 4–71) | NR | NR (Netherlands) | NR (Netherlands) | NR | CSA/TAC + AZA/MMF + CORT |
| |
| Klauke 2008 [35] | 106 | RI (n = 53): 50.2 ± 14.6 noRI (n = 53): 51.3 ± 10.7 | RI (n = 53): 35.5 ± 13.7 noRI (n = 53): 34.2 ± 12.1 | NR | NR | NR | CSA/TAC + AZA/MMF + CORT ± imTOR |
| |
| Benza 2009 [54] | 108 | 52 ± 11 | NR | African 14% Caucasian 86% | African 14% Caucasian 86% | NR | CSA + MMF + CORT |
| |
| Lachance2012 [57] | 158 | 53 (IQR: 43–58) | NR | Caucasian 98% Unknown 2% | NR (Canada) | NR | CSA/TAC + MMF/AZA + CORT |
| |
| Feingold 2012 [27] | 453 | 6.2 ± 6.1 | NR | White 60% Black 13% Hispanic 23% | NR | Yes | CSA or TAC |
| |
| 869 T > C (syn. Leu10> Pro, codon 10) rs1800470 | Baan 2000 [53] | 168 | 44 ± 12 or 48 ± 9 (Cr < or > 250 µmol/L ) | NR | Caucasian 95% Unknown 5% | NR | NR | CSA based schemes |
|
| Lacha 2001 [62] | 298 | NR | NR | NR (Czech Republic) | NR (Czech Republic) | NR | CSA + AZA + CORT |
| |
| Wetering 2006 [63] | 402 | 50 (range: 4–71) | NR | NR (Netherlands) | NR (Netherlands) | NR | CSA/TAC + AZA/MMF + CORT |
| |
| Klauke 2008 [35] | 106 | RI (n = 53): 50.2 ± 14.6 noRI (n = 53): 51.3 ± 10.7 | RI (n = 53): 35.5 ± 13.7 noRI (n = 53): 34.2 ± 12.1 | NR | NR | NR | CSA/TAC + AZA/MMF + CORT ± mTOR |
| |
| Lachance 2012 [57] | 158 | 53 (IQR: 43–58) | NR | Caucasian 98% Unknown 2% | NR (Canada) | NR | CSA/TAC + MMF/AZA + CORT |
| |
| Feingold 2012 [27] | 453 | 6.2 ± 6.1 | NR | White 60% Black 13% Hispanic 23% | NR | Yes | CSA or TAC |
| |
| rs4803455 C > A | Oreschak 2021 [37] | 192 | 49 ± 12 | NR | Caucasian 79% Black 9% Asian 4% Other 8% | NR | Yes | TAC or CSA + MMF or AZA + CORT | Renal function: minor allele C improved renal function in eGFR at 1 year after transplantation (univariable and multivariable analyses). |
| IL-4 | |||||||||
| −590 C > T rs2243250 | Feingold 2012 [27] | 453 | 6.2 ± 6.1 | NR | White 60% Black 13% Hispanic 23% | NR | No | CSA or TAC |
|
| IL-6 | |||||||||
| −174 G/C rs1800795 | Girnita 2008 [52] | 323 | NR (Pediatric) | NR | White 63.5% Black 13.3% Hispanic 23.2% | NR | Yes | TAC or CSA + MMF or AZA + CORT |
|
| Feingold 2012 [27] | 453 | 6.2 ± 6.1 | NR | White 60% Black 13% Hispanic 23% | NR | No | CSA or TAC |
| |
| IL-10 | |||||||||
| 1082 G/A 819 C/T 592 C/A | Girnita 2008 [52] | 323 | NR (Pediatric) | NR | White 63.5% Black 13.3% Hispanic 23.2% | NR | Yes | TAC or CSA + MMF or AZA + CORT |
|
| 1082 G/A | Feingold 2012 [27] | 453 | 6.2 ± 6.1 | NR | White 60% Black 13% Hispanic 23% | NR | Yes | CSA or TAC | Renal function: no significant differences. |
| IL-1RN | |||||||||
| VNTRs in intron 2 rs380092 | Feingold 2012 [27] | 453 | 6.2 ± 6.1 | NR | White 60% Black 13% Hispanic 23% | NR | Yes | CSA or TAC |
|
| IL15RA | |||||||||
| +21G > A | Feingold 2012 [27] | 453 | 6.2 ± 6.1 | NR | White 60% Black 13% Hispanic 23% | NR | Yes | CSA or TAC |
|
| +5165T > A | Feingold 2012 [27] | 453 | 6.2 ± 6.1 | NR | White 60% Black 13% Hispanic 23% | NR | Yes | CSA or TAC |
|
| VEGF | |||||||||
| 2578 C/A 460 C/T 405 C/G | Girnita 2008 [52] | 323 | NR (Pediatric) | NR | White 63.5% Black 13.3% Hispanic 23.2% | NR | Yes | TAC or CSA + MMF or AZA + CORT |
|
| PRKCB | |||||||||
| rs11074606 | Lachance 2012 [57] | 158 | 53 (IQR: 43–58) | NR | Caucasian 98% Unknown 2% | NR (Canada) | NR | CSA/TAC + MMF/AZA + CORT |
|
| NOD2/CARD15 | |||||||||
| rs2066844 C > T | Jordán 2012 [46] | 60 | NR | NR | NR | NR | NR | CSA or TAC |
|
| Sánchez-Lázaro 2015 [15] | 60 | CSA (n = 36): 53 ± 11 TAC (n = 24): 52 ± 10 | CSA (n = 36): 42 ± 12 TAC (n = 24): 43 ± 9 | NR | NR | NR | CSA or TAC + MMF + CORT |
| |
| LINC01121 | |||||||||
| rs17033285 | Asleh 2018 [59] | 251 | 50.3 ± 13 | 32 ± 12 | Caucasian 96.8% American Indian 0.4% Unknown 2.8% | NR | NR | (CSA or TAC) or SIRO + MMF or AZA | Renal function: minor allele 17.3-fold change in eGFR at 1 year after transplantation (improvement in renal function). |
| rs76427116 | Asleh 2018 [59] | 251 | 50.3 ± 13 | 32 ± 12 | Caucasian 96.8% American Indian 0.4% Unknown 2.8% | NR | NR | (CSA or TAC) or SIRO + MMF or AZA | Renal function: minor allele change in eGFR at 1 year after transplantation (improvement in renal function). |
| BTBD7P | |||||||||
| rs4917601 | Asleh 2018 [59] | 251 | 50.3 ± 13 | 32 ± 12 | Caucasian 96.8% American Indian 0.4% Unknown 2.8% | NR | NR | (CSA or TAC) or SIRO + MMF or AZA | Renal function: minor allele 11.6-fold change in eGFR at 1 year after transplantation (improvement in renal function). Similar effect with rs4617520, rs7095911, rs11195513, rs4465313, rs7923594, and rs4918638. |
| MARCH | |||||||||
| rs9762450 | Asleh 2018 [59] | 251 | 50.3 ± 13 | 32 ± 12 | Caucasian 96.8% American Indian 0.4% Unknown 2.8% | NR | NR | (CSA or TAC) or SIRO + MMF or AZA | Renal function: minor allele 7.8–8.7-fold change in eGFR at 1 year after transplantation (improvement in renal function). |
| ELAVL2 | |||||||||
| rs12057071 rs13294337 rs1431304 rs2891188 rs7024224 rs10966079 rs10966081 | Asleh 2018 [59] | 251 | 50.3 ± 13 | 32 ± 12 | Caucasian 96.8% American Indian 0.4% Unknown 2.8% | NR | NR | (CSA or TAC) or SIRO + MMF or AZA | Renal function: minor allele 12.1–13.2-fold change in eGFR at 1 year after transplantation (improvement in renal function). |
| HMHB1 | |||||||||
| rs918378 rs10463361 rs72795604 rs11167832 | Asleh 2018 [59] | 251 | 50.3 ± 13 | 32 ± 12 | Caucasian 96.8% American Indian 0.4% Unknown 2.8% | NR | NR | (CSA or TAC) or SIRO + MMF or AZA | Renal function: minor allele 13.6–14.2-fold change in eGFR at 1 year after transplantation (improvement in renal function). |
| rs918378 | Snipelisky 2017 [60] | 287 | NR | NR | NR (USA) | NR | NR | CNI | Renal function: minor allele 14.18-fold change in eGFR at 1 year after transplantation (improvement in renal function). |
| LOC339894 | |||||||||
| Snipelisky 2017 [60] | 287 | NR | NR | NR (USA) | NR | NR | CNI | Renal function: minor allele 12.61-fold change in eGFR at 1 year after transplantation (improvement in renal function). | |
| LOC10042392 | |||||||||
| rs4617520 | Snipelisky 2017 [60] | 287 | NR | NR | NR (USA) | NR | NR | CNI | Renal function: minor allele 11.95-fold change in eGFR at 1 year after transplantation (improvement in renal function). |
| MMP12 | |||||||||
| rs652438 | Snipelisky 2017 [60] | 287 | NR | NR | NR (USA) | NR | NR | CNI | Renal function: minor allele 16.7-fold change in eGFR at 1 year after transplantation (improvement in renal function). |
| C12orf75 | |||||||||
| rs1230081 | Snipelisky 2017 [60] | 287 | NR | NR | NR (USA) | NR | NR | CNI | Renal function: minor allele 14.55-fold change in eGFR at 1 year after transplantation (improvement in renal function). |
| PLCB1 | |||||||||
| rs170549 A > G | Oreschak 2021 [37] | 192 | 49 ± 12 | NR | Caucasian 79% Black 9% Asian 4% Other 8% | NR | Yes | TAC or CSA + MMF or AZA + CORT | Renal function: minor allele A was associated with renal dysfunction in eGFR at 1 year after transplantation (univariable and multivariable analyses). |
| ACE | |||||||||
| Intron 16; Insertion (I); Deletion (D) rs4646994 | Pethig 2000 [56] | 146 | 46 ± 11 | NR | NR (Germany) | NR (Germany) | NR | CSA + AZA + CORT | CAV: DD genotype was associated with higher CAV than II and DI genotypes at 6 years post-transplant. |
| Feingold 2012 [27] | 453 | 6.2 ± 6.1 | NR | White 60% Black 13% Hispanic 23% | NR | Yes | CSA or TAC | Renal function: no significant differences. | |
| CTGF/CCN2 | |||||||||
| 945 C > G rs6918698 | Pantou 2012 [55] | 72 | 38 ± 14 (14–63) | NR | NR (Greece) | NR (Greece) | Yes | EVE + CNI + CORT |
|
| G > A (rs13218743) | Oreschak 2021 [37] | 148 | 49 ± 13 | NR | European American 77.7% African American 10.8% Asian 4.1% Other 7.4% | NR | NR | TAC/CSA ± MMF/AZA + CORT |
|
| HNF1A | |||||||||
| A > C (p.I27L) (rs1169288) | Oreschak 2021 [37] | 148 | 49 ± 13 | NR | European American 77.7% African American 10.8% Asian 4.1% Other 7.4% | NR | NR | TAC/CSA ± MMF/AZA + CORT |
|
| T > C (rs2393791) | Oreschak 2021 [37] | 148 | 49 ± 13 | NR | European American 77.7% African American 10.8% Asian 4.1% Other 7.4% | NR | NR | TAC/CSA ± MMF/AZA + CORT |
|
| HO-1 | |||||||||
| −489 A > T rs2071746 | Feingold 2012 [27] | 453 | 6.2 ± 6.1 | NR | White 60% Black 13% Hispanic 23% | NR | Yes | CSA or TAC |
|
| 326 A > G rs17879828 | Feingold 2012 [27] | 453 | 6.2 ± 6.1 | NR | White 60% Black 13% Hispanic 23% | NR | No | CSA or TAC |
|
| NOS2 | |||||||||
| Ser608Leu rs2297518 | Feingold 2012 [27] | 453 | 6.2 ± 6.1 | NR | White 60% Black 13% Hispanic 23% | NR | Yes | CSA or TAC |
|
| +38 C > G rs10459953 | Feingold 2012 [27] | 453 | 6.2 ± 6.1 | NR | White 60% Black 13% Hispanic 23% | NR | Yes | CSA or TAC |
|
| FAS | |||||||||
| −670 A > G rs71800682 | Feingold 2012 [27] | 453 | 6.2 ± 6.1 | NR | White 60% Black 13% Hispanic 23% | NR | Yes | CSA or TAC |
|
| FASL | |||||||||
| −844 C > T rs763110 | Feingold 2012 [27] | 453 | 6.2 ± 6.1 | NR | White 60% Black 13% Hispanic 23% | NR | Yes | CSA or TAC |
|
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
Abbreviations
| CNI | Calcineurin inhibitor |
| MMF | Mycophenolate mofetil |
| mTOR | Mammalian target of rapamycin |
| DARE | Database of abstracts or reviews of effects |
| CYP | Cytochrome P450 |
| eGFR | Estimated glomerular filtration rate |
| POR | Cytochrome P450 oxidoreductase |
| NR1I2 | Nuclear Receptor Subfamily 1 Group I Member 2 |
| PXR | Pregnane X receptor |
| UGT | Uridine glycosyltransferase |
| MPA | Mycophenolic acid |
| IMPDH | Inosine monophosphate dehydrogenase |
| TPMT | Thiopurine methyltransferase |
| ABCB1 | ATP-binding cassette transporter B family member 1 |
| P-gp | P-glycoprotein |
| ABCB2 | ATP-binding cassette transporter B family member 2 |
| MRP2 | Multidrug resistance protein 2 |
| SLCO1B1 | Solute carrier organic anion transporter family member 1B1 |
| SLC13A1 | Solute carrier family 13 member 1 |
| HLA-G | Human leukocyte antigen G |
| TNF-α | Tumor necrosis factor alfa |
| TGF-ß1 | Transforming growth factor- ß1 |
| CTGF | Connective tissue growth factor |
| NOD2 | Nucleotide-binding oligomerization domain containing 2 |
| CARD15 | Caspase-activating recruitment domain 15 |
| ACE | Angiostensin-converting enzyme |
| RAAS | Renin–angiotensin–aldosterone system |
| GWAS | Genome-wide association study |
| SNP | Single nucleotide polymorphisms |
| HWE | Hardy–Weinberg equilibrium |
| A | Adenine |
| G | Guanine |
| C | Citosine |
| T | Timine |
| U | Uracil |
| NR | Not reported |
| TAC | Tacrolimus |
| CORT | Corticosteroids |
| CL/F | Apparent plasma clearance of drug after extravascular administration |
| POD | Postoperative day |
| CL | Clearance |
| PRED | Prednisone |
| CSA | Cyclosporine |
| AZA | Azathioprine |
| SIR | Sirolimus |
| BSX | Basiliximab |
| ATG | Anti-thymocyte globulin |
| AUC | Area under the curve |
| Cmax | Concentration maximum |
| t max | Time maximum |
| Css | Concentration stationary state |
| USA | United States of America |
| C0/D | Initial concentration–dose relationship |
| TDD | Total daily dose |
| CI | Confidence interval |
| RI | Renal insufficiency |
| ng | Nanograms |
| ml | Milliliters |
| IQR | Interquartile range |
| min | Minutes |
| m2 | Square meters |
| C0 | Initial concentration |
| m | Months |
| y | Years |
| mg | Milligrams |
| kg | Kilograms |
| d | Days |
| PK | Pharmacokinetics |
| EVE | Everolimus |
| V/F | Apparent volume of distribution after extravascular administration |
| L | Liters |
| h | Hours |
| vs. | Versus |
| AUC0–12 | Area under the curve from 0 to 12 h |
| R2 | Coefficient of determination |
| Cmin | Concentration minimum |
| HTx | Heart transplantation |
| POR | Cytochrome P450 oxidoreductase |
| NR1I2 | Nuclear receptor subfamily 1 group I member 2 |
| PXR | Pregnane X receptor |
| UGT | Uridine glycosyltransferase |
| CMV | Citomegalovirus |
| AcMPAG | Acyl glucuronide of mycophenolic acid |
| IMPDH | Inosine monophosphate dehydrogenase |
| TPMT | Thiopurine methyltransferase |
| HZ | Heterozygous |
| WT | Wild type |
| ABCB1 | ATP-binding cassette transporter B family member 1 |
| AUC0–4 | Area under the curve from 0 to 4 h |
| OR | Odds ratio |
| V/F | Apparent volume of distribution after extravascular administration |
| EBPR | Endomyocardial biopsy-proven rejection |
| AE | Adverse events |
| MDRD | Modification of diet in renal disease |
| RHC | Rejection with hemodynamic compromise |
| bp | Base pairs |
| CMR | Cell-mediated rejection |
| CAV | Cardiac allograft vasculopathy |
| TNF-B | Tumor necrosis factor beta |
| Cr | Creatinine |
| µmol | Micromol |
| IL | Interleukin |
| VEGF | Vascular endothelial growth factor |
| PRKCB | Protein kinase C beta |
| LINC01121 | Long intergenic non-protein coding RNA 1121 |
| BTBD7P | Bric-a-brac/tramtrack/broad complex domain containing 7 pseudogene 2 |
| MARCH1 | Membrane-associated ring-CH-type finger 1 |
| ELAVL2 | Embryonic lethal abnormal vision-like RNA binding protein 2 |
| HMHB1 | Histocompatibility minor HB-1 |
| MMP12 | Matrix metalloproteinase-12 |
| PLCB1 | Phospholipase C beta 1 |
| CTGF | Connective tissue growth factor |
| HNF1A | Hepatocyte nuclear factor 1 homeobox A |
| HO-1 | Heme oxygenase 1 |
| NOS2 | Nitric oxide synthase 2 |
| FAS | Fas cell surface death receptor |
| FASL | Fas cell surface death receptor ligand |
References
- Sutaria, N.; Sylvia, L.; DeNofrio, D. Immunosuppression and Heart Transplantation. Handb. Exp. Pharmacol. 2022, 272, 117–137. [Google Scholar] [PubMed]
- Herrero, M.J.; Almenar, L.; Jordán, C.; Sánchez, I.; Poveda, J.L.; Aliño, S.F. Clinical Interest of Pharmacogenetic Polymorphisms in the Immunosuppressive Treatment After Heart Transplantation. Transplant. Proc. 2010, 42, 3181–3182. [Google Scholar] [CrossRef] [PubMed]
- Gijsen, V.M.; van Schaik, R.H.; Elens, L.; Soldin, O.P.; Soldin, S.J.; Koren, G.; de Wildt, S.N. CYP3A4*22 and CYP3A combined genotypes both correlate with tacrolimus disposition in pediatric heart transplant recipients. Pharmacogenomics 2013, 14, 1027–1036. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Chen, J.; Lin, X.; Qiu, H.; Zhang, J. Population Pharmacokinetic Analysis for Model-Based Therapeutic Drug Monitoring of Tacrolimus in Chinese Han Heart Transplant Patients. Eur. J. Drug Metab. Pharmacokinet. 2023, 48, 89–100. [Google Scholar] [CrossRef]
- Liu, M.; Hernandez, S.; Aquilante, C.L.; Deininger, K.M.; Lindenfeld, J.; Schlendorf, K.H.; Van Driest, S.L. Composite CYP3A (CYP3A4 and CYP3A5) phenotypes and influence on tacrolimus dose adjusted concentrations in adult heart transplant recipients. Pharmacogenomics J. 2024, 24, 4. [Google Scholar] [CrossRef]
- Zheng, H.; Webber, S.; Zeevi, A.; Schuetz, E.; Zhang, J.; Bowman, P.; Boyle, G.; Law, Y.; Miller, S.; Lamba, J.; et al. Tacrolimus Dosing in Pediatric Heart Transplant Patients Is Related to CYP3A5 and MDR1 Gene Polymorphisms. Am. J. Transplant. 2003, 3, 477–483. [Google Scholar] [CrossRef]
- Liu, L.; Huang, X.; Zhou, Y.; Han, Y.; Zhang, J.; Zeng, F.; Huang, Y.; Zhou, H.; Zhang, Y. CYP3A4/5 genotypes and age codetermine tacrolimus concentration and dosage in pediatric heart transplant recipients. Int. Immunopharmacol. 2022, 111, 109164. [Google Scholar] [CrossRef]
- Uno, T.; Wada, K.; Matsuda, S.; Terada, Y.; Oita, A.; Kawase, A.; Takada, M. Impact of the CYP3A5*1 Allele on the Pharmacokinetics of Tacrolimus in Japanese Heart Transplant Patients. Eur. J. Drug Metab. Pharmacokinet. 2018, 43, 665–673. [Google Scholar] [CrossRef]
- Deininger, K.M.; Vu, A.; Page, R.L., 2nd; Ambardekar, A.V.; Lindenfeld, J.; Aquilante, C.L. CYP3A pharmacogenetics and tacrolimus disposition in adult heart transplant recipients. Clin. Transplant. 2016, 30, 1074–1081. [Google Scholar] [CrossRef]
- Kniepeiss, D.; Renner, W.; Trummer, O.; Wagner, D.; Wasler, A.; Khoschsorur, G.A.; Truschnig-Wilders, M.; Tscheliessnigg, K.-H. The role of CYP3A5 genotypes in dose requirements of tacrolimus and everolimus after heart transplantation. Clin. Transplant. 2011, 25, 146–150. [Google Scholar] [CrossRef]
- de Denus, S.; Zakrzewski, M.; Barhdadi, A.; Leblanc, M.H.; Racine, N.; Bélanger, F.; Carrier, M.; Ducharme, A.; Dubé, M.-P.; Turgeon, J.; et al. Association Between Renal Function and CYP3A5 Genotype in Heart Transplant Recipients Treated with Calcineurin Inhibitors. J. Heart Lung Transplant. 2011, 30, 326–331. [Google Scholar] [CrossRef]
- Kniepeiss, D.; Wagner, D.; Wasler, A.; Tscheliessnigg, K.H.; Renner, W. The role of CYP2C8 genotypes in dose requirement and levels of everolimus after heart transplantation. Wien. Klin. Wochenschr. 2013, 125, 393–395. [Google Scholar] [CrossRef] [PubMed]
- Lesche, D.; Sigurdardottir, V.; Setoud, R.; Oberhänsli, M.; Carrel, T.; Fiedler, G.M.; Largiadèr, C.R.; Mohacsi, P.; Sistonen, J. CYP3A53 and POR28 Genetic Variants Influence the Required Dose of Tacrolimus in Heart Transplant Recipients. Ther. Drug Monit. 2014, 36, 710–715. [Google Scholar] [CrossRef]
- Lesche, D.; Sigurdardottir, V.; Setoud, R.; Englberger, L.; Fiedler, G.M.; Largiadèr, C.R.; Mohacsi, P.; Sistonen, J. Influence of CYP3A5 genetic variation on everolimus maintenance dosing after cardiac transplantation. Clin. Transplant. 2015, 29, 1213–1220. [Google Scholar] [CrossRef]
- Sánchez-Lázaro, I.; Herrero, M.J.; Jordán-De Luna, C.; Bosó, V.; Almenar, L.; Rojas, L.; Martínez-Dolz, L.; E Megías-Vericat, J.; Sendra, L.; Miguel, A.; et al. Association of SNPs with the Efficacy and Safety of Immunosuppressant Therapy After Heart Transplantation. Pharmacogenomics 2015, 16, 971–979. [Google Scholar] [CrossRef] [PubMed]
- Sigurdardottir, V.; Lesch, D.; Setoud, R.; Oberhaensli, M.; Carrel, T.; Largiader, C.R.; Sistonen, J.; Mohacsi, P. Influence of Genetic Variation in Pharmacokinetic (PK) and -Dynamic (PD) Pathways of Immunosuppressants on Drug Response in Heart Transplant Patients. J. Heart Lung Transplant. 2013, 32, S135. [Google Scholar] [CrossRef]
- Ting, L.S.L.; Benoit-Biancamano, M.O.; Bernard, O.; Riggs, K.W.; Guillemette, C.; Ensom, M.H.H. Pharmacogenetic impact of UDP-glucuronosyltransferase metabolic pathway and multidrug resistance-associated protein 2 transport pathway on mycophenolic acid in thoracic transplant recipients: An exploratory study. Pharmacotherapy 2010, 30, 1097–1108. [Google Scholar] [CrossRef]
- Sombogaard, F.; van Schaik, R.H.; Mathot, R.A.; Budde, K.; van der Werf, M.; Vulto, A.G.; Weimar, W.; Glander, P.; Essioux, L.; van Gelder, T. Interpatient variability in IMPDH activity in MMF-treated renal transplant patients is correlated with IMPDH type II 3757T > C polymorphism. Pharmacogenet. Genom. 2009, 19, 626–634. [Google Scholar] [CrossRef] [PubMed]
- Ohmann, E.L.; Burckart, G.J.; Chen, Y.; Pravica, V.; Brooks, M.M.; Zeevi, A.; Webber, S.A. Inosine 5’-monophosphate dehydrogenase 1 haplotypes and association with mycophenolate mofetil gastrointestinal intolerance in pediatric heart transplant patients. Pediatr. Transplant. 2010, 14, 891–895. [Google Scholar] [CrossRef]
- Ohmann, E.L.; Burckart, G.J.; Brooks, M.M.; Chen, Y.; Pravica, V.; Girnita, D.M.; Zeevi, A.; Webber, S.A. Genetic polymorphisms influence mycophenolate mofetil-related adverse events in pediatric heart transplant patients. J. Heart Lung Transplant. 2010, 29, 509–516. [Google Scholar] [CrossRef]
- Relling, M.V.; Gardner, E.E.; Sandborn, W.J.; Schmiegelow, K.; Pui, C.H.; Yee, S.W.; Stein, C.M.; Carrillo, M.; E Evans, W.; Hicks, J.K.; et al. Clinical pharmacogenetics implementation consortium guidelines for thiopurine methyltransferase genotype and thiopurine dosing: 2013 update. Clin. Pharmacol. Ther. 2013, 93, 324–325. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.J.; Geske, J.R.; Boilson, B.A.; Frantz, R.P.; Edwards, B.S.; Kushwaha, S.S.; Kremers, W.K.; Weinshilboum, R.M.; Pereira, N.L. TPMT genetic variants are associated with increased rejection with azathioprine use in heart transplantation. Pharmacogenet. Genom. 2013, 23, 658–665. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Molina, B.; Tavira, B.; Lambert, J.L.; Bernardo, M.J.; Alvarez, V.; Coto, E. Effect of CYP3A5, CYP3A4, and ABCB1 Genotypes as Determinants of Tacrolimus Dose and Clinical Outcomes After Heart Transplantation. Transplant. Proc. 2012, 44, 2635–2638. [Google Scholar] [CrossRef]
- Isla Tejera, B.; Aumente Rubio, M.D.; Martínez-Moreno, J.; Reyes Malia, M.; Arizón, J.M.; Suárez García, A. Pharmacogenetic Analysis of the Absorption Kinetics of Cyclosporine in a Population of Spanish Cardiac Transplant Patients. Farm. Hosp. 2009, 33, 324–329. [Google Scholar] [CrossRef]
- Antignac, M.; Bezian, E.; Lemaitre, F.; Fernandez, C.; Varnous, S.; Urien, S.; Becquemont, L.; Farinotti, R. Population pharmacokinetics of everolimus in cardiac adult recipients: Impact of genetic polymorphism and drug interactions. Fundam. Clin. Pharmacol. 2010, 24, 53–54. [Google Scholar]
- Lemaitre, F.; Bezian, E.; Goldwirt, L.; Fernandez, C.; Farinotti, R.; Varnous, S.; Urien, S.; Antignac, M. Population pharmacokinetics of everolimus in cardiac recipients: Comedications, ABCB1, and CYP3A5 polymorphisms. Ther. Drug Monit. 2012, 34, 686–694. [Google Scholar] [CrossRef] [PubMed]
- Feingold, B.; Brooks, M.M.; Zeevi, A.; Ohmann, E.L.; Burckart, G.J.; Ferrell, R.E.; Chinnock, R.; Canter, C.; Addonizio, L.; Bernstein, D.; et al. Renal Function and Genetic Polymorphisms in Pediatric Heart Transplant Recipients. J. Heart Lung Transplant. 2012, 31, 1003–1008. [Google Scholar] [CrossRef]
- Mirza, S.; Wilson, N.; Van Zyl, J.; Nguyen, P.; Sam, P.; Hall, S.; Askar, M.; Patel, R. CYP3A5 extensive metabolizer phenotype may be associated with an increase in class 2 donor specific antibodies and antibody-mediated rejection. Am. J. Transplant. 2021, 21, 365. [Google Scholar]
- Uno, T.; Wada, K.; Matsuda, S.; Terada, Y.; Terakawa, N.; Oita, A.; Yokoyama, S.; Kawase, A.; Hosomi, K.; Takada, M. Effects of clotrimazole on tacrolimus pharmacokinetics in patients with heart transplants with different CYP3A5 genotypes. Eur. J. Clin. Pharmacol. 2019, 75, 67–75. [Google Scholar] [CrossRef]
- Déri, M.; Szakál-Tóth, Z.; Fekete, F.; Mangó, K.; Incze, E.; Minus, A.; Merkely, B.; Sax, B.; Monostory, K. CYP3A-status is associated with blood concentration and dose-requirement of tacrolimus in heart transplant recipients. Sci. Rep. 2021, 11, 21389. [Google Scholar] [CrossRef]
- Gijsen, V.; Mital, S.; van Schaik, R.H.; Soldin, O.P.; Soldin, S.J.; van der Heiden, I.P.; Nulman, I.; Koren, G.; de Wildt, S.N. Age and CYP3A5 Genotype Affect Tacrolimus Dosing Requirements After Transplant in Pediatric Heart Recipients. J. Heart Lung Transplant. 2011, 30, 1352–1359. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.Y.; Chen, W.Q.; Chen, Z.G.; Huang, J.; Liao, Z.K.; Liu, Q.; Zheng, Z.; Song, Y.-H.; Wang, W.; Hu, S.-S. The Effects of CYP3A5 Genetic Polymorphisms on Serum Tacrolimus Dose-Adjusted Concentrations and Long-Term Prognosis in Chinese Heart Transplantation Recipients. Eur. J. Drug Metab. Pharmacokinet. 2019, 44, 771–776. [Google Scholar] [CrossRef]
- Pasternak, A.L.; Marshall, V.D.; Gersch, C.L.; Rae, J.M.; Englesbe, M.; Park, J.M. Evaluating the Impact of CYP3A5 Genotype on Post-Transplant Healthcare Resource Utilization in Pediatric Renal and Heart Transplant Recipients Receiving Tacrolimus. Pharmgenomics Pers. Med. 2021, 12, 319–326. [Google Scholar] [CrossRef] [PubMed]
- Pasternak, A.L.; Park, J.M.; Pai, M.P. Predictive Capacity of Population Pharmacokinetic Models for the Tacrolimus Dose Requirements of Pediatric Solid Organ Transplant Recipients. Ther. Drug Monit. 2023, 45, 95–101. [Google Scholar] [CrossRef]
- Klauke, B.; Wirth, A.; Zittermann, A.; Bohms, B.; Tenderich, G.; Körfer, R.; Milting, H. No Association Between Single Nucleotide Polymorphisms and the Development of Nephrotoxicity After Orthotopic Heart Transplantation. J. Heart Lung Transplant. 2008, 27, 741–745. [Google Scholar] [CrossRef] [PubMed]
- Jordán de Luna, C.; Herrero Cervera, M.J.; Sánchez Lázaro, I.; Almenar Bonet, L.; Poveda Andrés, J.L.; Aliño Pellicer, S.F. Pharmacogenetic Study of ABCB1 and CYP3A5 Genes During the First Year Following Heart Transplantation Regarding Tacrolimus or Cyclosporine Levels. Transplant. Proc. 2011, 43, 2241–2243. [Google Scholar] [CrossRef]
- Oreschak, K.; Saba, L.M.; Rafaels, N.; Ambardekar, A.V.; Deininger, K.M.; Page II, R.L.; Lindenfeld, J.; Aquilante, C.L. Variants in mycophenolate and CMV antiviral drug pharmacokinetic and pharmacodynamic genes and leukopenia in heart transplant recipients. J. Heart Lung Transplant. 2021, 40, 917–925. [Google Scholar] [CrossRef]
- Barnard, J.B.; Richardson, S.; Sheldon, S.; Fildes, J.; Pravica, V.; Hutchinson, I.V.; Leonard, C.T.; Yonan, N. The MDR1/ABCB1 gene, a high-impact risk factor for cardiac transplant rejection. Transplantation 2006, 82, 1677–1682. [Google Scholar] [CrossRef]
- De Iudicibus, S.; Castronovo, G.; Gigante, A.; Stocco, G.; Decorti, G.; Di Lenarda, R. Role of MDR1 gene polymorphisms in gingival overgrowth induced by cyclosporine in transplant patients. J. Periodontal Res. 2008, 43, 665–672. [Google Scholar] [CrossRef]
- Taegtmeyer, A.B.; Breen, J.B.; Smith, J.; Burke, M.; Leaver, N.; Pantelidis, P.; Lyster, H.; Yacoub, M.H.; Barton, P.J.R.; Banner, N.R. ATP-Binding Cassette Subfamily B Member 1 Polymorphisms Do Not Determine Cyclosporin Exposure, Acute Rejection or Nephrotoxicity After Heart Transplantation. Transplantation 2010, 89, 75–82. [Google Scholar] [CrossRef]
- Kunicki, P.K.; Kowalska, E.; Waś, J.; Wróbel, A.; Hurkacz, M.; Machowska, M.; Sobieszczánska-Malek, M.; Komuda, K.; Karczmarz, M.; Jerzak-Wodzyńska, G.; et al. Everolimus steady-state concentration may be related to ABCB1 genotype: Possible impact on therapeutic drug monitoring. Clin. Chem. Lab. Med. 2016, 54, eA260. [Google Scholar]
- Oreschak, K.; Deininger, K.M.; Ambardekar, A.V.; Page II, R.L.; Lindenfeld, J.; Aquilante, C.L. Effect of ABCC2 polymorphisms on tacrolimus disposition in heart transplant recipients. J. Heart Lung Transplant. 2018, 37, S115. [Google Scholar] [CrossRef]
- Burckart, G.J.; Figg, W.D., 2nd; Brooks, M.M.; Green, D.J.; Troutman, S.M.; Ferrell, R.; Chinnock, R.; Canter, C.; Addonizio, L.; Bernstein, D.; et al. Multi-institutional study of outcomes after pediatric heart transplantation: Candidate gene polymorphism analysis of ABCC2. J. Pediatr. Pharmacol. Ther. 2014, 19, 16–24. [Google Scholar] [CrossRef]
- Han, N.; Yun, H.Y.; Kim, I.W.; Oh, Y.J.; Kim, Y.S.; Oh, J.M. Population pharmacogenetic pharmacokinetic modeling for flip-flop phenomenon of enteric-coated mycophenolate sodium in kidney transplant recipients. Eur. J. Clin. Pharmacol. 2014, 70, 1211–1219. [Google Scholar] [CrossRef]
- Chowbay, B.; Cumaraswamy, S.; Cheung, Y.B.; Zhou, Q.; Lee, E.J. Genetic polymorphisms in MDR1 and CYP3A4 genes in Asians and the influence of MDR1 haplotypes on cyclosporin disposition in heart transplant recipients. Pharmacogenetics 2003, 13, 89–95. [Google Scholar] [CrossRef]
- Jordán, C.; Bosó, V.; Sánchez, I.; Herrero, M.J.; Ruiz, J.; Almenar, L.; Poveda, J.L.; Aliño, S.F. Single Nucleotide Polymorphisms, SNPs, Associated with the Efficacy and Security of Immunosuppressive Treatment in Heart Transplantation. Transplantation 2012, 94, 154. [Google Scholar] [CrossRef]
- Oreschak, K.; Deininger, K.M.; Ambardekar, A.V.; Page II, R.L.; Lindenfeld, J.; Aquilante, C.L. Effect of ABCB1 haplotypes on tacrolimus disposition in heart transplant recipients. J. Heart Lung Transplant. 2017, 36, S64. [Google Scholar] [CrossRef]
- Torres, M.I.; Luque, J.; Lorite, P.; Isla-Tejera, B.; Palomeque, T.; Aumente, M.D.; Arizon, J.; Peña, J. 14-Base pair polymorphism of human leukocyte antigen-G as genetic determinant in heart transplantation and cyclosporine therapy monitoring. Hum. Immunol. 2009, 70, 830–835. [Google Scholar] [CrossRef]
- Adamson, M.B.; Di Giovanni, B.; Ribeiro, R.V.P.; Yu, F.; Lazarte, J.; Rao, V.; Delgado, D.H. HLA-G +3196 polymorphism as a risk factor for cell mediated rejection following heart transplant. Hum. Immunol. 2020, 81, 134–140. [Google Scholar] [CrossRef]
- Azzawi, M.; Hasleton, P.S.; Turner, D.M.; Yonan, N.; Deiraniya, A.K.; Sinnott, P.J.; Hutchinson, I.V. Tumor necrosis factor-alpha gene polymorphism and death due to acute cellular rejection in a subgroup of heart transplant recipients. Hum. Immunol. 2001, 62, 140–142. [Google Scholar] [CrossRef]
- Ternstrom, L.; Jeppsson, A.; Ricksten, A.; Nilsson, F. Tumor necrosis factor gene polymorphism and cardiac allograft vasculopathy. J. Heart Lung Transplant. 2005, 24, 433–438. [Google Scholar] [CrossRef] [PubMed]
- Girnita, D.M.; Brooks, M.M.; Webber, S.A.; Burckart, G.J.; Ferrell, R.; Zdanowicz, G.; DeCroo, S.; Smith, L.; Chinnock, R.; Canter, C.; et al. Genetic polymorphisms impact the risk of acute rejection in pediatric heart transplantation: A multi-institutional study. Transplantation 2008, 85, 1632–1639. [Google Scholar] [CrossRef] [PubMed]
- Baan, C.C.; Balk, A.H.; Holweg, C.T.; van Riemsdijk, I.C.; Maat, L.P.; Vantrimpont, P.J.; Niesters, H.G.; Weimar, W. Renal failure after clinical heart transplantation is associated with the TGF-beta 1 codon 10 gene polymorphism. J. Heart Lung Transplant. 2000, 19, 866–872. [Google Scholar] [CrossRef] [PubMed]
- Benza, R.L.; Coffey, C.S.; Pekarek, D.M.; Barchue, J.P.; Tallaj, J.A.; Passineau, M.J.; Grenett, H.E. Transforming growth factor-beta polymorphisms and cardiac allograft rejection. J. Heart Lung Transplant. 2009, 28, 1057–1062. [Google Scholar] [CrossRef]
- Pantou, M.P.; Manginas, A.; Alivizatos, P.A.; Degiannis, D. Connective tissue growth factor (CTGF/CCN2): A protagonist in cardiac allograft vasculopathy development? J. Heart Lung Transplant. 2012, 31, 881–887. [Google Scholar] [CrossRef]
- Pethig, K.; Heublein, B.; Hoffmann, A.; Borlak, J.; Wahlers, T.; Haverich, A. ACE-gene polymorphism is associated with the development of allograft vascular disease in heart transplant recipients. J. Heart Lung Transplant. 2000, 19, 1175–1182. [Google Scholar] [CrossRef]
- Lachance, K.; Barhdadi, A.; Mongrain, I.; Normand, V.; Zakrzewski, M.; Leblanc, M.H.; Racine, N.; Carrier, M.; Ducharme, A.; Turgeon, J.; et al. PRKCB is associated with calcineurin inhibitor-induced renal dysfunction in heart transplant recipients. Pharmacogenet. Genom. 2012, 22, 336–343. [Google Scholar] [CrossRef]
- Oetjens, M.; Bush, W.S.; Birdwell, K.A.; Dilks, H.H.; Bowton, E.A.; Denny, J.C.; Wilke, R.A.; Roden, D.M.; Crawford, D.C. Utilization of an EMR-Biorepository to Identify the Genetic Predictors of Calcineurin-Inhibitor Toxicity in Heart Transplant Recipients. Pac. Symp. Biocomput. 2014, 253–264. [Google Scholar] [PubMed] [PubMed Central]
- Asleh, R.; Snipelisky, D.; Hathcock, M.; Kremers, W.; Liu, D.; Batzler, A.; Jenkins, G.; Kushwaha, S.; Pereira, N.L. Genomewide association study reveals novel genetic loci associated with change in renal function in heart transplant recipients. Clin. Transplant. 2018, 32, e13395. [Google Scholar] [CrossRef]
- Snipelisky, D.; Hathcock, M.; Kremers, W.; Batzler, A.; Jenkins, G.; Kushwaha, S.; Pereira, N. Genome wide association study reveals novel genetic loci associated with renal function in heart transplant recipients receiving calcineurin inhibitor therapy. Eur. J. Heart Fail. 2017, 19, 157–158. [Google Scholar]
- Densem, C.G.; Hutchinson, I.V.; Yonan, N.; Brooks, N.H. TGF-beta gene polymorphism does not influence the rise in creatinine following cardiac transplantation. Transpl. Immunol. 1999, 7, 247–249. [Google Scholar] [CrossRef] [PubMed]
- Lácha, J.; Hubácek, J.A.; Viklický, O.; Málek, I.; Hutchinson, I.; Vítko, S. TGF-beta1 gene polymorphism is a risk factor for renal dysfunction in heart transplant recipients. Transplant. Proc. 2001, 33, 1567–1569. [Google Scholar] [CrossRef] [PubMed]
- van de Wetering, J.; Weimar, C.H.E.; Balk, A.H.M.M.; Roodnat, J.I.; Holweg, C.T.; Baan, C.C.; van Domburg, R.T.; Weimar, W. The impact of transforming growth factorbeta1 gene polymorphism on end-stage renal failure after heart transplantation. Transplantation 2006, 82, 1744–1748. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Megías-Vericat, J.E.; Palanques-Pastor, T.; Fernández-Sánchez, M.; Guerrero-Hurtado, E.; Gil-Candel, M.; Solana-Altabella, A.; Ballesta-López, O.; Centelles-Oria, M.; García-Pellicer, J.; Poveda-Andrés, J.L. Systematic Review of Pharmacogenetics of Immunosuppressants in Heart Transplantation. Cardiogenetics 2025, 15, 18. https://doi.org/10.3390/cardiogenetics15020018
Megías-Vericat JE, Palanques-Pastor T, Fernández-Sánchez M, Guerrero-Hurtado E, Gil-Candel M, Solana-Altabella A, Ballesta-López O, Centelles-Oria M, García-Pellicer J, Poveda-Andrés JL. Systematic Review of Pharmacogenetics of Immunosuppressants in Heart Transplantation. Cardiogenetics. 2025; 15(2):18. https://doi.org/10.3390/cardiogenetics15020018
Chicago/Turabian StyleMegías-Vericat, Juan Eduardo, Tomás Palanques-Pastor, Mireya Fernández-Sánchez, Eduardo Guerrero-Hurtado, Mayte Gil-Candel, Antonio Solana-Altabella, Octavio Ballesta-López, María Centelles-Oria, Javier García-Pellicer, and José Luis Poveda-Andrés. 2025. "Systematic Review of Pharmacogenetics of Immunosuppressants in Heart Transplantation" Cardiogenetics 15, no. 2: 18. https://doi.org/10.3390/cardiogenetics15020018
APA StyleMegías-Vericat, J. E., Palanques-Pastor, T., Fernández-Sánchez, M., Guerrero-Hurtado, E., Gil-Candel, M., Solana-Altabella, A., Ballesta-López, O., Centelles-Oria, M., García-Pellicer, J., & Poveda-Andrés, J. L. (2025). Systematic Review of Pharmacogenetics of Immunosuppressants in Heart Transplantation. Cardiogenetics, 15(2), 18. https://doi.org/10.3390/cardiogenetics15020018






