Abstract
Online collaboration and web-based knowledge sharing have gained momentum as major components of the Web 2.0 movement. Consequently, knowledge embedded in such platforms is no longer static and continuously evolves through experts’ micro-contributions. Traditional Information Retrieval and Social Network Analysis techniques take a document-centric approach to expertise modeling by creating a macro-perspective of knowledge embedded in large corpus of static documents. However, as knowledge in collaboration platforms changes dynamically, the traditional macro-perspective is insufficient for tracking the evolution of knowledge and expertise. Hence, Expertise Profiling is presented with major challenges in the context of dynamic and evolving knowledge. In our previous study, we proposed a comprehensive, domain-independent model for expertise profiling in the context of evolving knowledge. In this paper, we incorporate Language Modeling into our methodology to enhance the accuracy of resulting profiles. Evaluation results indicate a significant improvement in the accuracy of profiles generated by this approach. In addition, we present our profile visualization tool, Profile Explorer, which serves as a paradigm for exploring and analyzing time-dependent expertise profiles in knowledge-bases where content evolves overtime. Profile Explorer facilitates comparative analysis of evolving expertise, independent of the domain and the methodology used in creating profiles.
1. Introduction
Organizations are in constant demand of individuals with expertise in specific topics and therefore require extensive profiling systems to enable them to locate experts in a particular knowledge area. However, filling out comprehensive profiling systems and keeping them up to date requires extensive manual effort and has proven to be impractical. A research into expertise profiles at IBM has found that after 10 years of repetitive and consistent pressure from the executives, including periodic emails sent to experts to remind them to update their profiles, only 60% of all IBM profiles are kept up-to-date [1]. This clearly indicates that a manual approach to profiling expertise is not sufficient and an automated solution is required to create and maintain expertise profiles, especially in collaboration platforms where knowledge evolves overtime.
There is growing recognition that access to the intrinsic expertise information is critical to the efficient running of enterprise operations. For example, the employees of geographically dispersed organizations typically have difficulty in determining what others are doing and which resources can best address their problems. Failure to foster exchange within the knowledge community leads to duplication of effort and an overall reduction in productivity levels.
Expertise is not easily identified and is even more difficult to manage on an ongoing basis, which leaves vast resources of tacit knowledge and experience untapped. Online communities and collaboration platforms have emerged as major components of Web 2.0 [2] and the Semantic Web [3] movements, where experts share their knowledge and expertise through contributions to the underlying knowledge base. Collaboration platforms with support for evolving knowledge provide a dynamic environment where content is subject to ongoing evolution through expert contributions. Consequently, knowledge embedded in such platforms is not static as it evolves through incremental refinements to content or micro-contributions, e.g., collaborative authoring/editing of parts of reports, open contributions to domain-specific topics, etc. Examples of such platforms are generic Wikis (e.g., Wikipedia) or specific collaborative knowledge bases predominantly in the biomedical domain, such as AlzSWAN [4]—which captures and manages hypotheses, arguments and counter-arguments in the Alzheimer’s Disease domain or Gene Wiki [5], a sub-project of Wikipedia which supports discussions on genes.
Representing, capturing and finding expertise has been addressed in many different ways by diverse (often complementary) communities. In Information Retrieval the task of “Expert Finding” assumes the existence of a set of documents and of a set of expertise profiles and aims to find the best matches between the profiles and the documents. The field of Social Network Analysis, on the other hand, looks at the graphs connecting individuals in different contexts to infer their expertise from shared domain-specific topics [6]. Finally, in the Semantic Web domain expertise is captured using ontologies and then inferred from axioms and rules defined over instances of these ontologies.
The major issue associated with all these approaches lies in the underlying use and analysis of a large corpus of static documents authored by the experts (typically publications, reports, etc.). However, in the context provided by the current collaborative knowledge bases, documents are neither static (as they are continuously and incrementally refined) nor lead to large corpora authored by individual experts (usually authors edit a fraction of a document, which is closer to their expertise/interest).
In a previous publication [7] we have introduced a model and a methodology—Semantic and Time-dependent Expertise Profiling (STEP)—aimed at capturing micro-contributions in the macro-context of the living documents, as well as the temporality of expertise profiles. The methodology consists of three phases: (i) concept extraction—semantically annotating concepts present within the micro-contributions with ontological entities; (ii) concept consolidation—identifying and grouping concepts representing the same ontological entity; and (iii) profile creation—building short-term expertise profiles (i.e., restricted by a time window—two weeks, one month, etc.) and long-term expertise profiles—as aggregations of short-term profiles. Initially, we applied the methodology in the biomedical domain, mainly because of the existing tool support. In particular, the concept extraction phase relied on the NCBO Annotator [8], hence making the accuracy of the generated profiles dependent on the accuracy of this tool. Nevertheless, experimental results demonstrated that our approach is much better suited for handling evolving knowledge than the existing traditional techniques.
In this paper, we build on our previous work and we investigate the effect of using diverse Language Modeling techniques in the concept extraction phase, in order to minimize the role of any domain-specific annotation tool. More concretely, this paper brings the following contributions:
- we integrate two Statistical Language Modeling mechanisms into the STEP methodology (i.e., Topic models and N-Gram models);
- we evaluate the effect of these models on the resulting expertise profiles by comparing them against the original results;
- we introduce the Profile Explorer, a visualization tool that provides a novel paradigm for browsing and analysing time-dependent expertise and interests which evolve over time.
The remainder of the paper is structured as follows. In Section 2 and Section 3 we discuss the motivation and use cases associated with our research. Section 4 revisits the Semantic and Time-dependent Expertise Profiling (STEP) methodology, while Section 5 introduces the application of language modeling techniques as part of the concept extraction and profile building phases of STEP. Section 6 presents a comparative overview of the experimental results achieved by applying the new language modeling techniques, followed by a discussion of these results and of the limitations of our approach in Section 7. In Section 8 we introduce the “Profile Explorer” visualization paradigm, and before concluding in Section 10, we provide a comprehensive overview of the related work in Section 9.
2. Motivation
Traditional IR techniques analyze a large corpus of static documents; i.e., once written, documents do not change and remain forever in the same form. Consequently, they take a document-centric approach to expertise profiling and create a macro-perspective of the knowledge embedded in static documents—individuals are associated with topics extracted from documents independently of their actual contribution. Social Network Analysis (SNA), on the other hand, considers graphs connecting individuals in different contexts and infers their expertise from the shared domain-specific topics [6], e.g., researchers co-authoring publications with other researchers accumulate part of their co-authors’ expertise, which is extracted from non-coauthored publications.
Both techniques rely on frequency-based statistics, such as Term Frequency (TF) and Inverse Document Frequency (IDF) in the case of IR, or counts of shared in and out links in the case of SNA, to reflect the importance of an item in a document/node in a collection or network. Furthermore, IR methods usually represent documents authored by individuals as bags-of-words and expertise identification is done by associating individual profiles to such bags-of-words—either by ranking candidates based on their similarities to a given topic or by searching for co-occurrences of both the individual and the given topic, in the set of supporting documents. As a remark, such associations can be used to compute semantic similarities between expertise profiles [9].
However, in the context of evolving knowledge, these techniques are insufficient, as micro-contributions generally consist of short fragments of text, which do not provide adequate context. We aim to capture expertise by identifying tacit knowledge in the context of collaboration platforms, and thus focus on a contribution-oriented approach, where expertise topics are associated with an author based on the author’s contributions to the underlying knowledge base, rather than expertise topics that emerge from the knowledge base (documents) as a whole [7]. In addition to clearly focusing on individual contributions, we are able to take advantage of the underlying dynamic environment and create fine-grained time-dependent expertise profiles [10].
Tracking the evolution of micro-contributions enables one to monitor the activity performed by individuals, which in turn, provides a way to show not only the change in personal interests over time, but also the maturation process of an expert’s knowledge (similar to some extent to the maturation process of scientific hypotheses, from simple ideas to scientifically proven facts). From a technical perspective, building expertise profiles from concepts defined in widely adopted ontologies or Linked Data datasets enables individuals who publish their profile on the Web to be intrinsically integrated within the Linked Data Cloud. This provides “expertise seekers” and web crawlers with access to richer, more accurate and more up-to-date profiles and facilitates better consolidation of expertise profiles and a seamless aggregation of communities of experts.
From an academic perspective, a shift in the scientific publishing process seems to gain momentum, from the current document-centric approach to a contribution-oriented approach in which the hypotheses or domain-related innovations (in form of short statements) will replace the current publications. Examples of this new trend can be seen via nano-publications [11] or liquid publications [12]. In this new setting, mapping such micro-contributions to expertise will be essential in order to support the development of reputation metrics. As a note, our current work focuses only on building expertise profiles from micro-contributions, which can then provide the foundations upon which novel trust and reputation models could be applied.
Finally, more fine-grained and time-aware “expertise finders” would help motivate contributing members by rewarding and recognizing experts’ knowledge and expertise. This in turn serves as an incentive for increasing member contributions as such voluntary contributions are vital to the sustainability of these environments. In addition, as knowledge is subject to rapid evolution in highly dynamic scientific research environments, modeling expertise is imperative to the assembly and collaboration of cross-disciplinary teams.
3. Use Cases
The STEP methodology introduced in [7], and further extended in this paper, has been applied in the context of evolving knowledge in the biomedical domain. More specifically, we have implemented STEP and performed evaluations using contributions extracted from the Molecular and Cellular Biology (MCB) [13] and the Genetics [14] Wiki projects (both sub-projects of Wikipedia). These use cases target completely different knowledge domains and hence provide a different perspective of the fine-grained provenance and enable us to evaluate the abstraction layer that renders our final model into a domain-agnostic form.
The content of the MCB and Genetics projects constantly evolves through incremental changes made by authors to articles in these projects. Below we present a series of examples of micro-contributions, emerged from such incremental changes to existing articles in the MCB project (in this case by author Jpkamil) on different dates:
- 4 February 2008—Lipase article: Lipoprotein lipase functions in the blood to act on triacylglycerides carried on VLDL (very low density lipoprotein) so that cells can take up the freed fatty acids. Lipoprotein lipase deficiency is caused by mutations in the gene encoding lipoprotein lipase.
- 15 February 2008—Lipase article: Pancreatic lipase related protein 1 is very similar to PLRP2 and HPL by amino acid sequence (all three genes probably arose via gene duplication of a single ancestral pancreatic lipase gene). However, PLRP1 is devoid of detectable lipase activity and its function remains unknown, even though it is conserved in other mammals.
The fine-grained structure of the knowledge captured within such micro-contributions enables a completely novel way of modeling and using expertise. Short-term profiles, i.e., expertise profiles created over a restricted time period (e.g., January–June 2008)—allow one to determine bursts of activities related to particular topics, e.g., the level of participation of an individual in a project. In the example provided above, one could infer that Jpkamil has been active within this period in the area of Lipase genes. Similarly, long term profiles, i.e., expertise profiles created over the entire history of an individual—can be used to determine how long were individuals involved in a specific topic, or how recent is their knowledge in this topic.
From an analysis perspective, expertise profiles built from micro-contributions could provide insights into the alignment between the level of involvement of particular individuals in a specific topic and the focus on an underlying project. Moreover, a level of collaboration among experts can be determined (e.g., determining the most active group of experts in a topic over a period of time), which can span within and across multiple projects. Finally, such profiles enable a fine-grained comparison of the expertise of the individuals across topics and time. Consequently, the application of this technology ranges from task and project management to HR and job application processes.
4. STEP Revisited
Micro-contributions represent incremental refinements by authors to an evolving body of knowledge. Examples of such micro-contributions are edits to a Wikipedia article or a Gene page in Gene Wiki [5] or a statement in WikiGenes [15] and OMIM [16]. Regardless of the platform, we are interested in capturing the fine-grained provenance of these micro-contributions including the actions that lead to their creation, as well as the macro-context that hosts these contributions; i.e., paragraph or section of the document in which they appear. Therefore, in the initial phase of our study, we created the Fine-grained Provenance Ontology [17], which combines coarse and fine-grained provenance modeling to capture micro-contributions and their localization in the context of their host living documents. The objective has been to reuse and extend existing, established vocabularies from the Semantic Web that have attracted a considerable user community or are derived from de facto standards. This focus guarantees direct applicability and low entry barriers.
Semantic and Time-dependent Expertise Profiling (STEP) provides a generic methodology for modeling expertise in the context of evolving knowledge. It consists of three main modules, as depicted in Figure 1; (i) Concept Extraction; (ii) Concept Consolidation; and (iii) Profile Creation.
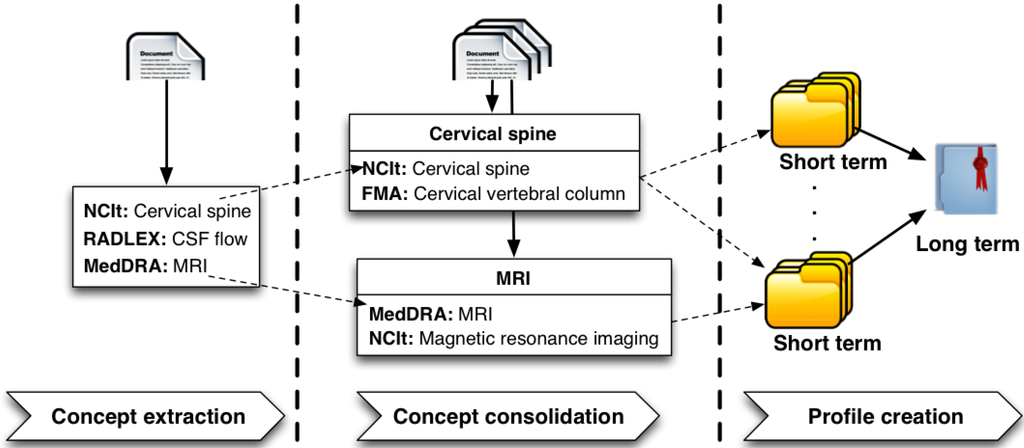
Figure 1.
Semantic and time-dependent expertise profiling methodology.
Below, we provide a brief description of each of these modules, and refer the reader to [7] for the complete and detailed overview.
The Concept Extraction module aims at annotating micro-contributions with concepts from domain-specific ontologies. This can be achieved by utilizing a typical information extraction or semantic annotation process, which is, in principle, domain-dependent. Hence, in order to provide a profile creation framework applicable to any domain, we do not restrict this step to the use of a particular concept extraction tool/technique. As micro-contributions for an expert are annotated with concepts from domain-specific ontologies, clusters containing semantically similar concepts emerge. Every entity identified in a micro-contribution is usually annotated with multiple concepts, as numerous domain-specific ontologies are used in the concept extraction process. Furthermore, concept clusters that represent similar semantics often contain one or more concepts in common. These concepts result from the annotation of terms, which appear more frequently and therefore become members of multiple concept clusters. Other concepts in these clusters convey similar semantics; however, they do not appear as frequently as concepts, which are common to multiple clusters.
The Concept Consolidation module aggregates these less prominent concepts with concepts that are manifestations of the same entities and appear more frequently; in other words, concept consolidation detects the intersection of groups of concepts resulting from annotation of lexically different, but semantically similar entities across micro-contributions and uses their union to create “Virtual Concepts”. Hence it provides a more accurate and coherent view over entities identified within micro-contributions. A “Virtual Concept” represents an abstract entity and contains domain-specific concepts from different ontologies, which are manifestations of the abstract entity. Virtual concepts are the building blocks of expertise profiles generated by STEP and central to short-term and long-term profile creation methods and the visualization and search functionality facilitated by Profile Explorer [18], our expertise profile visualization tool, as described in Section 8.
The Profile Creation module uses the extracted and consolidated concepts to create expertise profiles. The expertise of an individual is dynamic and usually changes with time. In order to capture the temporal aspect of expertise, we differentiate between Short Term and Long Term profiles.
A short-term profile represents a collection of concepts identified and extracted from micro-contributions over a specific period of time. Short-term profiles aim at capturing periodic bursts of expertise in specific topics, over a length of time. In order to compute a short term profile, we use a ranking of all virtual concepts identified within that time span based on an individual weight that takes into account the normalized frequency and the degree of co-occurrence of a virtual concept with other virtual concepts identified within the same period. The equation below lists the mathematical formulation of this weight. The intuition behind this ranking is that the expertise of an individual is more accurately represented by a set of co-occurring concepts forming an expertise context, rather than by individual concepts that occur frequently outside such a context.
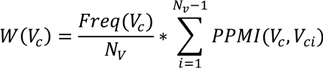
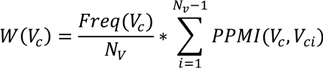
The elements of the equation above are: Vc—the virtual concept for which a weight is calculated, Nv—total number of virtual concepts in the considered time window, and PPMI—the positive pointwise mutual information, as defined below:
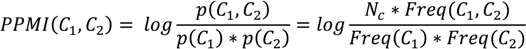 where Nc—the total number of concepts and Freq(C1, C2)—the joint frequency (or co-occurrence) of C1 and C2. PPMI is always positive, i.e., if PPMI(C1, C2) < 0 then PPMI(C1, C2) = 0.
where Nc—the total number of concepts and Freq(C1, C2)—the joint frequency (or co-occurrence) of C1 and C2. PPMI is always positive, i.e., if PPMI(C1, C2) < 0 then PPMI(C1, C2) = 0.
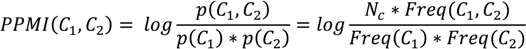
A long-term profile captures the collection of concepts occurring both persistently and uniformly across all short-term profiles for an expert. Unlike other expertise profiling approaches, we consider uniformity as important as persistency; i.e., an individual is considered to be an expert in a topic if this topic is present persistently and its presence is distributed uniformly across all the short-term profiles. Consequently, in computing the ranking of the concepts in the long-term profile, the weight has two components, as listed in the equation below:
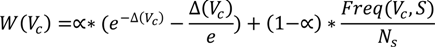 where Ns is the total number of short-term profiles, Freq(Vc, S) is the number of short-term profiles containing Vc, α is a tuning constant and Δ(Vc) is the standard deviation of Vc, computed using the equation below. The standard deviation of Vc shows the extent to which the appearance of the virtual concept in the short-term profiles deviates from a uniform distribution. A standard deviation of 0 represents a perfectly distributed appearance. Consequently, we’ve introduced a decreasing exponential that increases the value of the uniformity factor inversely proportional to the decrease of the standard deviation—i.e., the lower the standard deviation, the higher the uniformity factor.
where Ns is the total number of short-term profiles, Freq(Vc, S) is the number of short-term profiles containing Vc, α is a tuning constant and Δ(Vc) is the standard deviation of Vc, computed using the equation below. The standard deviation of Vc shows the extent to which the appearance of the virtual concept in the short-term profiles deviates from a uniform distribution. A standard deviation of 0 represents a perfectly distributed appearance. Consequently, we’ve introduced a decreasing exponential that increases the value of the uniformity factor inversely proportional to the decrease of the standard deviation—i.e., the lower the standard deviation, the higher the uniformity factor.
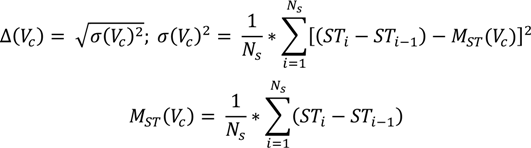 where (STi − STi-1) represents the window difference between short-term profiles in which a virtual concept appears, and MST(Vc) is the mean of all window differences. In practice, we aim to detect uniformity by performing a linear regression over the differences between the short-term profiles that contain the virtual concept.
where (STi − STi-1) represents the window difference between short-term profiles in which a virtual concept appears, and MST(Vc) is the mean of all window differences. In practice, we aim to detect uniformity by performing a linear regression over the differences between the short-term profiles that contain the virtual concept.
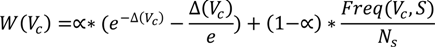
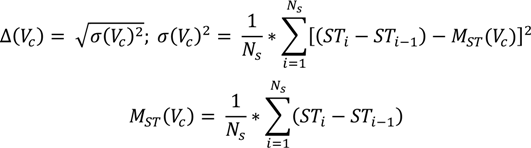
5. Integrating Language Modeling with STEP
So far, STEP has been implemented and tested in the biomedical domain having the NCBO Annotator [8] as an underlying concept extraction technique and the NCBO Recommender [19] as helper in the concept consolidation phase (see [7] for details). The annotator’s underlying technology is domain-agnostic; first, the biomedical free text is provided as input to the concept recognition tool used by the annotator along with a dictionary. The dictionary (or lexicon) is constructed using ontologies configured for use by the annotator. As most concept recognizers take as input a resource and a dictionary to produce annotations, the only customization to the biomedical domain, would be the biomedical ontologies used by the annotator. However, the NCBO Annotator is predominantly used in the biomedical domain and was the only means of annotating micro-contributions in the initial phase of our research [7]. Consequently, it’s had a massive influence on the final results, as effectively, accuracy of the final profiles was heavily dependent on the accuracy of annotations performed by the annotator. Therefore, its versatility has come at the price of extraction efficiency, as an exact match is required between the terms present in text and the labels of ontological concepts, in order for annotations to be detected. For example, a simple usage of the plural of a noun (e.g., Flagella) is enough to miss an ontological concept (such as Flagellum); furthermore, in some cases, only constituents of a phrase are annotated (e.g., “tibial shaft”); aggregating partial annotations does not accurately convey the semantics of the whole term (e.g., consolidating concepts representing “shaft” and “tibial” does not convey the same semantics as concepts representing “tibial shaft”).
We have tried to minimize the impact of this problem by consolidating semantically similar concepts. And while the Concept Consolidation resulted in a significant improvement of the results produced by the annotator, we observed that some instances where either not included in the resulting expertise profiles, or concepts included in the profiles were not ranked accurately.
The exhaustive discovery and resolution of sub-optimal performance by the NCBO Annotator in the context of our use cases, is outside the scope of this research. However, our experiments clearly indicate that the accuracy of resulting profiles is directly influenced by the quality of annotations produced by the annotator. Since the STEP methodology provides a pluggable architecture, in order to reduce the effects of domain-specific annotation tools on the accuracy of the generated profiles, we propose integrating Language Models with the Concept Extraction module of the methodology. The following sections outline our proposed methods, which are domain-agnostic in order to ensure that the overall architecture remains domain-independent.
5.1. Lemmatization
As outlined above, often the NCBO Annotator produces no annotations for terms that vary from their base or dictionary form (lemma). We have therefore performed Lemmatization [20] on micro-contributions prior to extracting concepts using the NCBO Annotator. Lemmatization, which is the algorithmic process of determining the lemma (base or dictionary form) for a given word, improves the accuracy of practical information extraction tasks. Morphological analysis of biomedical text can yield useful results and is more effective when performed by a specialized lemmatization program for biomedicine [21]. In our experiments we have used the BioLemmatizer [22], to conduct morphological processing of micro-contributions prior to concept extraction. It is important to note the distinction between stemming and lemmatization; a stemmer operates on a single word without knowledge of the context, and therefore cannot discriminate between words that have different meanings depending on part of speech. We have therefore opted for pre-processing micro-contributions by lemmatization as it involves understanding context and determining the part of speech of a word in a sentence. This is especially important in the concept consolidation phase, where groups of concepts are consolidated and integrated based on annotations common to these groups; common annotations among multiple concept clusters result from the annotation of lexically different but semantically similar entities, where semantic similarity is detected from the context in which they appear. Lemmatization increases the number of annotated terms in micro-contributions of an expert, which can potentially increase the number of common concepts among groups of concepts, leading to an increase in the number of consolidated concept clusters. This will in turn prevent loss of semantics and inaccurate ranking of concepts in expertise profiles.
5.2. Topic Modeling
Inaccurate annotations during the concept extraction phase may result from mapping parts of a phrase to ontological concepts, where the union of these concepts does not represent the underlying term; as illustrated by the “tibial shaft” example, combining concepts representing “tibial” and “shaft” does not accurately convey the semantics of “tibial shaft”. In order to capture the semantics of phrases more accurately and to capture terms and phrases that may not have been annotated, we propose incorporating two of the Statistical Language Modeling techniques [23], i.e., Topic Modeling [24] and N-gram Modeling [25], with the concept extraction module of the STEP methodology.
Incremental and collaborative refinements to content in collaboration platforms, including micro-contributions in the biomedical domain, usually contain discussion on a variety of topics; in order to discover the abstract topics and the hidden thematic structure of micro-contributions, we have performed topic modeling on all contributions made by an author. Topic models are algorithms for discovering the main themes that pervade a large and otherwise unstructured collection of documents. Topic models can organize the collection according to the discovered themes. Topic modeling algorithms can be applied to massive collections of documents and adapted to many kinds of data. Among other applications, they have been used to find patterns in genetic data, images and social networks [26]. We have opted for the Latent Dirichlet Allocation (LDA) topic model, which allows documents to have a mixture of topics [27]. The intuition behind LDA is that documents exhibit multiple topics; e.g., a discussion regarding Achondroplasia, a disorder of bone growth, in SKELETOME [28], will most likely include information about the possible causes of the disease such as inheritance and genetic mutation, genes and chromosomes, treatment options, medication, etc. LDA is a statistical model of document collections that tries to capture this intuition. A topic is defined to be a distribution over a fixed vocabulary. For example, the “genetics” topic has words about genetics (such as FGFR3 gene, chromosome, etc.) with high probability. The assumption is that these topics are specified before any data has been generated [26]. Each micro-contribution made by an expert is seen as an exhibition of these topics in different proportion. We then annotate the defined topics and words included in those topics to derive domain-specific concepts from domain ontologies.
5.3. N-gram Modeling
Latent Dirichlet Allocation is based on the “bag-of-words” assumption, in that the order of words in a document does not matter. However, word order and phrases are often critical to capturing the meaning of text. N-gram models can be imagined as placing a small window over a sentence or text, in which only n words are visible at the same time. We have therefore performed experiments using the N-gram modeling technique, where every sequence of two adjacent entities (bi-gram model) [29] in micro-contributions for an expert is identified and annotated with concepts from domain ontologies.
It is important to note that while we are still using the NCBO Annotator for annotating terms that result from topic and n-gram modeling, the way in which these terms are identified is independent of the annotation process, differs from the method used by the annotator for identifying terms and is in fact, domain-agnostic. Furthermore, missing or inaccurate annotations are not the result of the “annotation” process, but stem from the method by which terms are identified in the context of the given micro-contributions. Therefore, our proposed mechanisms aim at complementing term/topic extraction given the context of micro-contributions. This process is independent of and a pre-requisite to the annotation process.
We integrated lemmatization followed by topic and n-gram modeling with the concept extraction module of the STEP methodology, in order to improve the accuracy of resulting profiles and facilitate a domain-independent method of identifying entities in micro-contributions. Topic modeling and n-gram modeling techniques were implemented on our datasets as two separate experiments. Concepts resulting from each experiment were then used to create short-term and long-term expertise profiles. The following section outlines our datasets, experiments and evaluations.
6. Experiments
In order to exemplify and get a better understanding of the strengths and limitations of our proposed approach, i.e., integrating Language Modeling with the STEP methodology, we applied it to the biomedical domain. More specifically, we performed the same experiments as in [7], using data from the MCB and Genetics Wiki projects, in order to compare the initial results to the tool agnostic approach discussed in the previous sections. In the following, we detail the characteristics of the datasets, the experimental results and lessons learned.
It is important to note that performing an all-inclusive comparative evaluation of our approach has not been possible, due to the lack of a gold standard. As a baseline, our experiments have used expertise profiles defined and created by experts upon joining the MCB or Genetics Wiki projects; this presents a series of challenges, as discussed later in this section.
6.1. Datasets
The Molecular and Cellular Biology Wiki project aims at organizing information in articles related to molecular and cell biology in Wikipedia. Similarly, the Genetics Wiki project aims at organizing improvement and maintenance of genetics articles in Wikipedia. The underlying articles in both projects are constantly updated through expert contributions. Wikipedia allows authors to state opinions and raise issues in discussion pages. These incremental additions to content, or micro-contributions, give the knowledge captured within the environment a dynamic character.
We have collected micro-contributions for 22 authors from the MCB project and 7 authors from the Genetics project over the course of the last 5 years. These contributions resulted in a total of approximately 4,000 updates, with an average of 270 tokens per micro-contribution and an average of 137 micro-contributions per author.
The 29 designated authors, for whom we’ve collected micro-contributions, were the only ones that had provided a personal perspective on their expertise when joining the corresponding project. Although a much larger number of participants are available, not all of them provide a sufficiently detailed description of their expertise. We were interested in expertise profiles that contain areas of expertise, rather than the position of the participant (e.g., “post doc” or “graduate student”) or their interest in the project (e.g., “improving Wikipedia entries”, “expanding stub articles”). Each of the 29 authors selected from all the participants provided an average of 4.5 expertise topics in their profiles. We used these topics to create long-term profiles for each author, which we have used as our baseline. An example of such a profile is the one for author “AaronM” that specifies: “cytoskeleton”, “cilia”, “flagella” and “motor proteins” as his expertise.
6.2. Experimental Results
Previously [7], we conducted experiments with the STEP methodology using the NCBO Annotator as a concept extraction technique. In this study, we have incorporated topic modeling and n-gram modeling into STEP, in order to enhance the accuracy of generated profiles by minimizing the effects of domain-specific annotation tools on the final results. This section presents our experiments and the comparative analysis performed among expertise profiles generated by the following approaches: (i) expertise profiles created by the generic STEP methodology, where concepts are extracted from micro-contributions using the NCBO Annotator tool; i.e., the original approach [7]; (ii) expertise profiles created by integrating Topic Modeling with the Concept Extraction module in STEP; i.e., the topic modeling approach and (iii) expertise profiles created by integrating N-gram Modeling with the Concept Extraction module in STEP; i.e., the n-gram modeling approach. All methods were performed on micro-contributions for the 29 authors selected from the Molecular and Cellular Biology and Genetics Wiki projects. In addition, we have included comparisons among profiles generated by the above-mentioned approaches and profiles created by two traditional IR-inspired expertise-finding systems. As previously mentioned, the baseline consists of expertise profiles created by the designated authors upon joining the corresponding projects.
In terms of efficiency measures, we have considered F-score, Precision and Recall as defined in the context of Information Retrieval. In the context of our experiments, the value of F-score provides a measure of the accuracy of profiles generated by each approach through considering both Precision and Recall. F-score can be interpreted as a weighted average of the precision and recall, with its best value at 1 and worst score at 0. For a given expertise profile, Precision is the number of correct concepts (concepts matching the baseline) divided by the number of all returned concepts (total number of concepts in the generated profile) and Recall is the number of correct concepts (concepts matching the baseline) divided by the number of concepts that should have been returned (total number of concepts in the baseline).
As described in Section 4, virtual concepts included in expertise profiles are associated with a weight. Figure 2 tracks the values of F-Score for different concept weight thresholds. If we do not set a concept weight threshold (all virtual concepts are included in generated profiles) or set thresholds less than 0.5 (virtual concepts with weight < 0.5 are included in generated profiles), the original approach achieves the highest F-score. This is due to the fact that profiles generated by topic modeling and n-gram modeling approaches contain more noise since they include additional concepts representing the topics and n-grams derived from experts’ micro-contributions.
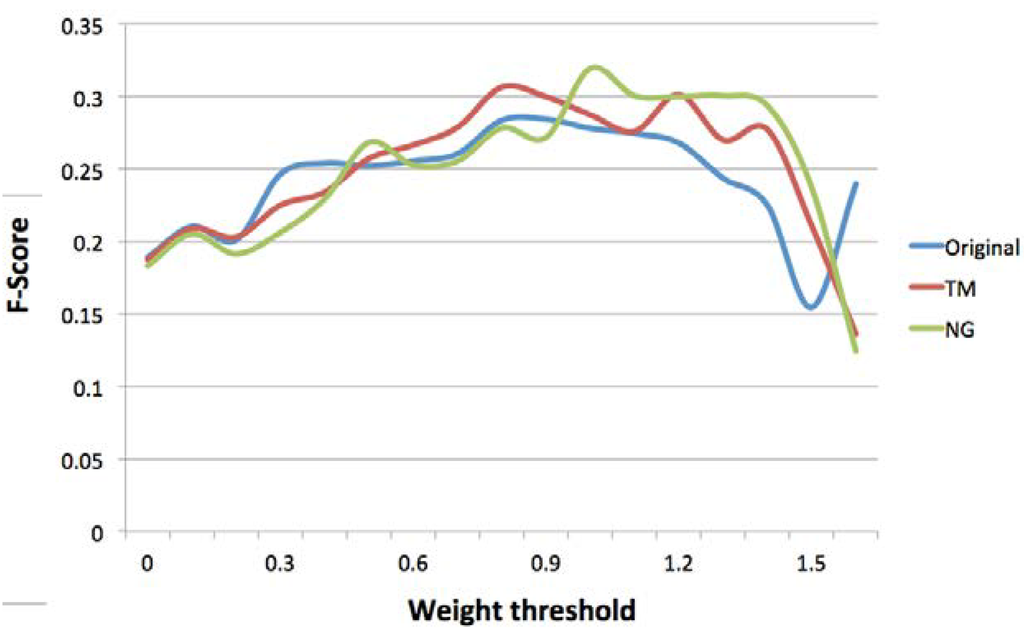
Figure 2.
F-score at different concept weight thresholds.
Subsequently increasing the concept weight threshold from 0.5 to 1.2 results in consistently higher F-score values achieved by topic modeling and n-gram modeling approaches, with the highest F-score achieved by n-gram modeling at concept weight threshold of 1 (31.94%). The enhanced accuracy of profiles generated by topic and n-gram modeling approaches, is partly due to the presence of concepts representing the topics and n-grams derived by these approaches, as well as a reduction of noise in the profiles as a result of increasing the concept weight threshold.
Increasing the concept weight threshold from 1.2 to 1.5, results in the decline of the value of F-score for all approaches, with n-gram modeling achieving the highest accuracy at all thresholds in this range. The decline in the value of F-score is due to the exclusion of a large number of concepts from generated profiles as a result of higher thresholds, achieving a mining value for the topic modeling and n-gram modeling approaches at a weight threshold of 1.6.
Figure 3 depicts the relationship between precision and recall for different concept weight thresholds. If we do not set any threshold on the weight of the concepts in the long-term profiles, the original approach achieves the best precision; i.e., precision is 10.86% for a recall of 72.94%, followed by topic modeling (precision: 10.79% and recall: 72.94%) and n-gram modeling (precision: 10.42% and recall: 76.82%). Overall, at concept weight thresholds of less than 0.5, topic modeling and n-gram modeling approaches have resulted in lower precision, as additional concepts included in the profiles (i.e., concepts representing topics and n-grams derived from experts’ micro-contributions) have contributed to more noise in the profiles (demonstrated by a higher recall).
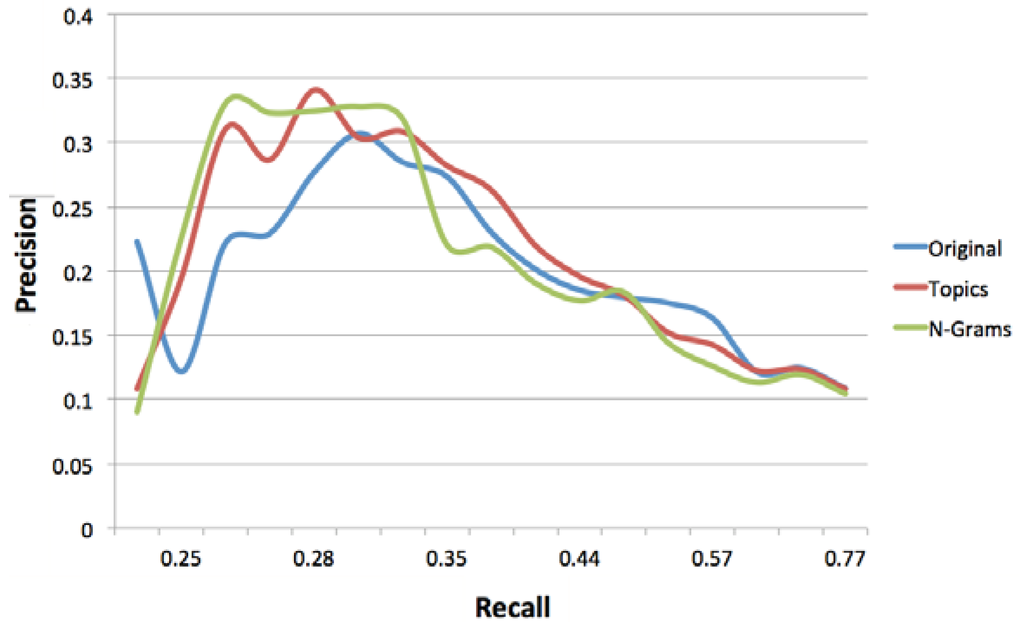
Figure 3.
Precision-recall curve at different concept weight thresholds.
Increasing the concept weight threshold to 0.5 will result in an increase in the precision achieved by all methods, with n-gram modeling achieving the highest precision (18.35%), albeit at the expense of the recall (49.96%). Subsequently increasing the threshold will result in further improvements to the precision achieved by all approaches, however at the expense of lower recall values. The best precision is achieved by the topic modeling approach at the concept weight threshold of 1.2 (34.08%) followed by the n-gram modeling approach (32.47%) and the original approach (27.68%). The results indicate that a higher accuracy is achieved by topic modeling and n-gram modeling approaches by setting an appropriate concept weight threshold, where the noise is reduced. Increasing the concept weight threshold over and above 1.2, results in a significant decrease in both the precision and recall values achieved by all methods; this is due to the exclusion of a large number of concepts with weights below such high thresholds.
Experimental results indicate that at concept weight thresholds greater than 0.4, topic modeling and n-gram modeling approaches have consistently achieved higher accuracy in comparison to the original approach. Topic modeling has demonstrated the highest precision at the threshold of 1.2, although at the expense of recall. Overall, the N-gram modeling approach has achieved the highest accuracy (F-score: 31.94%) at the concept weight threshold of 1. The enhanced accuracy is due to the fact that the n-gram modeling approach derives n-grams by taking into account word order and context (higher precision) and multiple words and phrases (higher recall).
7. Discussion
In the initial phase of our research, we demonstrated the effectiveness of the generic STEP methodology for profiling expertise using micro-contributions in the context of online communities where content evolves overtime [7]. In other words, we demonstrated that implementing our proposed methodology in the context of evolving knowledge (via a typical concept extraction technique), produces profiles with higher accuracy than traditional IR approaches, which perform expertise profiling using large corpus of static documents such as publications and reports. Furthermore, we captured the temporal aspect of expertise by creating short-term and long-term profiles.
In this paper, we have demonstrated that incorporating Language Modeling into the generic STEP methodology provides a domain-independent method for deriving the semantics of micro-contributions from short fragments of text while reducing the influence of domain-specific tools; i.e., the NCBO Annotator, on concept extraction and resulting profiles. This is achieved by using topic and n-gram modeling, statistical language modeling techniques that can be applied to any domain, for extracting topics and terms from micro-contributions. The resulting topics and terms are subsequently annotated and mapped to ontological concepts and used in building short-term and long-term profiles.
The experimental results shed a positive light onto the performance of our proposed method. By setting an appropriate threshold, i.e., concept weight threshold of 1, n-gram modeling incorporated with STEP delivers a significantly improved accuracy (F-score: 31.94%). While these results can be further improved, they are encouraging as they illustrate that incorporating domain-independent methods significantly enhances the accuracy of profiles generated using micro-contributions in the context of collaboration platforms where knowledge is subject to ongoing changes.
The difference in abstraction between the content of micro-contributions and expertise profiles created by the authors and used as our baseline plays a crucial role in evaluation. Micro-contributions are generally very specific; i.e., the terminology describes specific domain aspects, while expertise profiles defined by experts consist of mostly general terms (e.g., genetics, bioinformatics, microbiology, etc.). This makes direct comparison very challenging. The use of ontologies enables us to take into account more than just the actual concepts extracted from micro-contributions, by looking at their ontological parents or children. Consequently, we would be able to realize a comparison at a similar abstraction level, which could improve the evaluation results.
8. Visualization of Expertise Profiles
As described previously, STEP aims at capturing the temporality of expertise by differentiating between short term and long-term profiles. A short-term profile represents a collection of concepts extracted from micro-contributions over a period of time. The goal of the long-term profile, on the other hand, is to capture the collection of concepts occurring both persistently and uniformly across all short-term profiles for an expert.
Capturing the temporal aspect of expertise offers significant value as it facilitates tracking and analyzing changes in interests and expertise overtime. In order to provide a user friendly and intuitive framework for the visualization and analysis of evolving interests and expertise overtime, we have created the “Profile Explorer” [18] visualization tool. Profile Explorer facilitates visualization of short term and long term profiles and provides a framework for conducting comparative analysis of experts and expertise by linking an expert’s long term profile with short term profiles and underlying contributions. Profile Explorer creates a paradigm that facilitates visualization, search and comparative analysis of expertise profiles. We have specifically de-coupled Profile Explorer from our methodology and the biomedical domain, i.e., the domain within which our experiments are conducted, in order to provide a visualization paradigm for analyzing expertise that is independent of methodology or domain.
Furthermore, visualizing the temporal aspect captured by our expertise model facilitates detailed analysis of interests and expertise by allowing us to determine: (i) the level of activity in particular topics over time; (ii) the burst of activity in particular topics (short term profiles); (iii) the amount of time an expert has been contributing to specific topics; (iv) how recently an expert has made contributions to a specific topic; (v) the most/least active experts in particular topic/s; (vi) if participants’ activities are in line with the focus of the project or underlying articles (as concepts in short-term profiles can be linked to terms in micro-contributions through the Fine-grained provenance ontology); (vii) the level of collaboration among authors; e.g., determine the group of authors who have had the highest level of activity in a particular topic over a period of time; (viii) the ontology/s which have best described the contributions to a particular project; i.e., viewing a project from the point of view of a particular domain; (ix) the expertise of authors contributing to multiple projects; (x) comparison of expertise captured from contributions to multiple projects.
Profile Explorer utilizes TimelineJS [30] to demonstrate the temporal aspect of expertise profiles. TimelineJS is a storytelling, user friendly and intuitive timeline built in JavaScript. It can pull in media from different sources and has built in support for Twitter, Flickr, Google Maps, YouTube, Vimeo, Dailymotion, Wikipedia, SoundCloud and more media types in the future. Profile Explorer also utilizes Data-driven Documents (D3) [31] to illustrate the content of expertise profiles (i.e., concepts from domain-specific ontologies) as word clouds. Data-driven Documents (D3) is a JavaScript library for binding data to graphics using HTML, SVG and CSS, animation and interaction.
The most important features that clearly distinguish Profile Explorer from other expertise visualization tools and networks are (i) capturing and visualizing the time-dependent aspect of expertise; (ii) conducting comparative analysis based on semantics represented by ontological concepts and (iii) the ability to visualize and analyze expertise profiles independent of domain or methodology. While tools such as SciVal Experts [32] and BiomedExperts [33] provide a visual interface to experts’ profiles, they only provide an overall view of an expert’s expertise and are therefore unable to facilitate comparative analysis of the evolution of expertise and interests overtime.
The following illustrates the Profile Explorer tool using snapshots from the live system [18] and a use case (username: Jpkamil) from the Molecular and Cellular Biology (MCB) Wiki Project [13]. For this user, the goal is to find all short term profiles where the concept “Lipase” or semantically similar concepts have been identified as a topic of expertise; i.e., periods of activity where this user has made contributions to this topic or semantically similar topics. Then, from among the short term profiles identified, we aim to find contributions that contain the concept “Hymenoptera” or semantically similar concepts.
Figure 4 depicts the profile timeline for this user. The system has been configured to create short-term profiles expanding over two week intervals. As depicted in Figure 4, the timeline displays all short term profiles and the long term profile for the user; each short term profile is labeled with a date corresponding to the start of the two week period to which the short term profile belongs; e.g., 17 march 2006 represents the expertise topics derived from contributions made by this author over the two week interval starting from 17 March 2006.
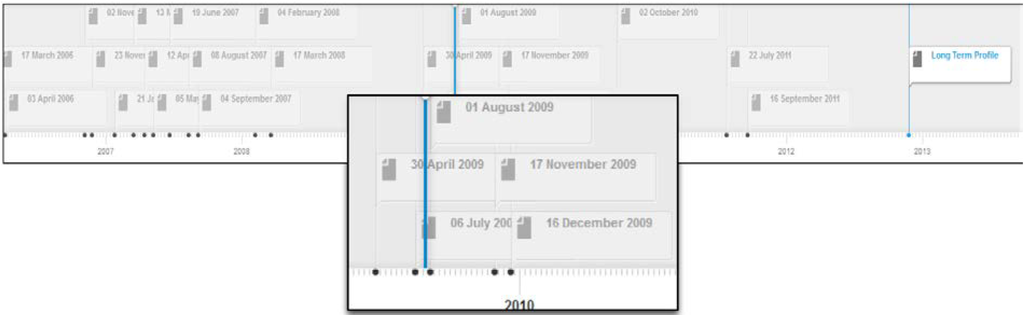
Figure 4.
Profile timeline for user Jpkamil.
Selecting the Long Term Profile label in the timeline will display the long-term profile cloud for this author, as depicted in Figure 5. Each label in the cloud represents a domain concept and its size is proportional to the weight of the concept in the profile.
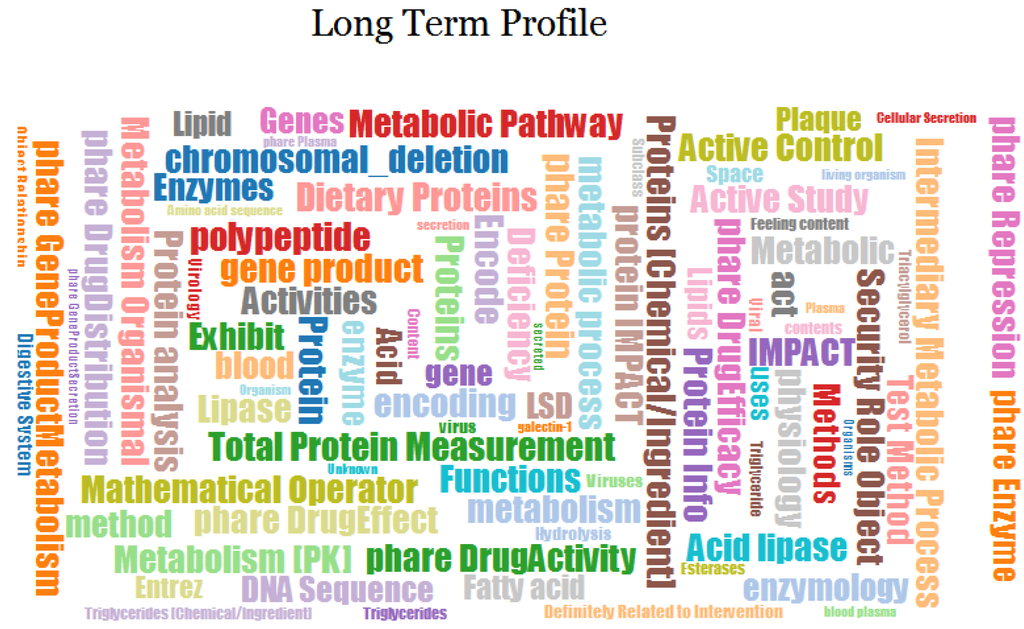
Figure 5.
Long Term Profile for user Jpkamil.
In addition to visualization, Profile Explorer provides profile search functionality. Search terms can be selected from the word cloud representing the long-term profile of an expert. For example, in order to find short-term profiles, which represent contributions/expertise in the topic “Lipase”, we’ll select this term in the word cloud of the long-term profile. Figure 6 depicts the screen when the search term has been selected in the word cloud. Selecting a search term and invoking the search functionality will display the profile timeline, highlighting all short term profiles which contain concepts that represent the search term; i.e., “Lipase” or other concepts which represent terms that are semantically similar to the selected term. This is achieved through virtual concepts, the building blocks of all profiles, as described in Section 4.
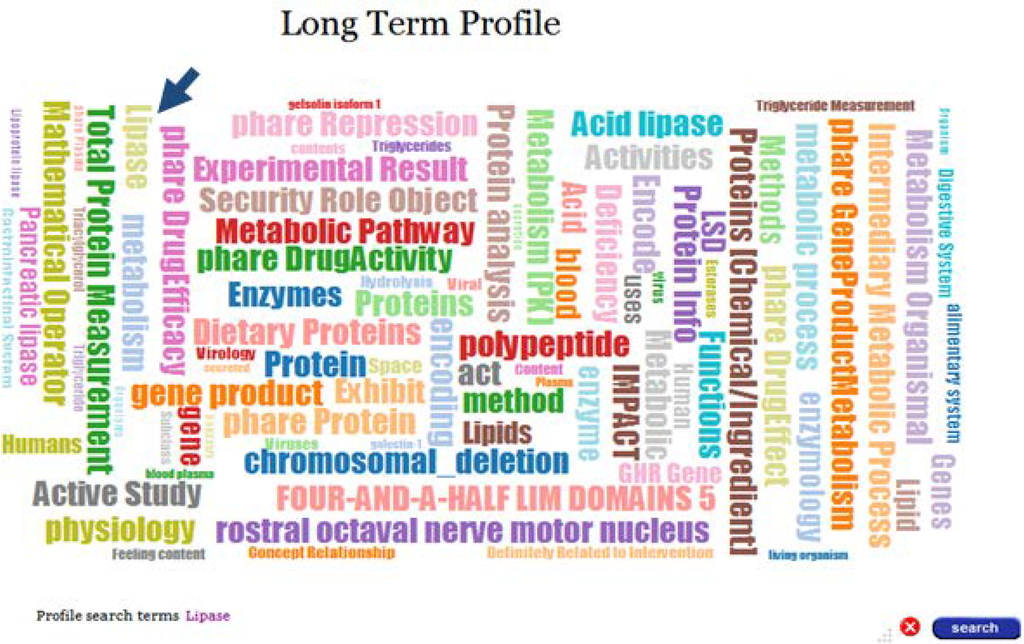
Figure 6.
Selected search term in the Long Term Profile.
Figure 7 depicts a portion of the profile timeline for this expert, after the search functionality has been invoked. This also provides a birds-eye view over the bursts of activities of the user under scrutiny in the context of the “Lipase” expertise.
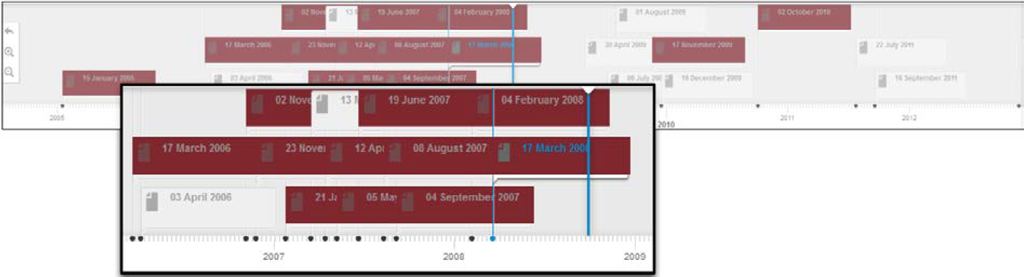
Figure 7.
Profile timeline—Search.
Selecting a highlighted short term profile in the timeline displays its corresponding word cloud, where the label of ontological concepts representing the search term/s are highlighted with a dark border. Figure 8 depicts the word cloud for the short-term profile, 17 March 2008. As mentioned above, concepts that belong to the family of concepts (Virtual Concept) representing “Lipase”, have been highlighted. As depicted in Figure 8, three concepts have been highlighted; “Lipase” (the selected term), “Triacylglycerol lipase” and “Lipase Adverse Event” representing semantically similar concepts. Therefore, the search function selects profiles which contain concepts representing the semantics of search term/s, rather than containing concepts with labels that match the search term/s. Furthermore, as also depicted in Figure 8, the Profile search terms are displayed on the screen and extended to include semantically similar terms.

Figure 8.
Short Term Profile cloud—Search.
The Profile Explorer also facilitates search in the context of short-term profiles, where search term/s are selected in the word cloud of a short-term profile for the expert. This will display a timeline of all the micro-contributions made by the author within the time period corresponding to the short-term profile. Micro-contributions containing the search term/s are highlighted in the timeline. As with the search performed in the context of the long-term profile, this search is also based on the semantics of search terms; i.e., micro-contributions are selected and highlighted based on the presence of concepts, which are semantically similar to the concepts selected for search. In this example, if we select the term “Hymenoptera” in the word cloud of this short-term profile, the timeline of micro-contributions for this period will appear where each micro-contribution is identified and labeled by the topic that it represents. Micro-contributions containing the search terms are highlighted (Figure 9).

Figure 9.
Micro-contribution timeline.
Selecting a micro-contribution displays its content with terms matching the search terms highlighted and underlined. As depicted in Figure 10, concepts that represent the selected term (Hymenoptera) and semantically similar concepts (wasp) are highlighted. The search terms are displayed on the screen and extended to include semantically similar terms.
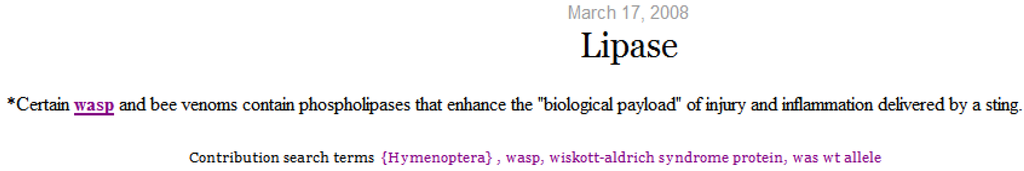
Figure 10.
Micro-contribution content.
9. Related Work
Expertise profiling is an active research topic in a wide variety of applications and domains, including biomedical, scientific and education. In this section, we present a brief overview of the related efforts, with particular emphasis on the Information Retrieval (IR) and the Semantic Web domains. The two most popular and well performing approaches in the TREC (Text Retrieval Conference) [34] expert search task are profile-centric and document-centric approaches. These studies use the co-occurrence model and techniques such as Bag-of-Words or Bag-of-Concepts on documents that are typically large and rich in content. Often a weighted, multiple-sized, window-based approach in an IR model is used for association discovery [35]. Alternatively, the effectiveness of exploiting the dependencies between query terms for expert finding is demonstrated [36]. Other studies present solutions through effective use of ontologies and techniques such as spreading to link additional related terms to a user profile by referring to background knowledge [9].
Algorithms have been proposed to find experts in Wikipedia. One such study attempts to find experts in Wikipedia content or among Wikipedia users [37]. It uses semantics from Wordnet and Yago in order to disambiguate expertise topics and to improve the retrieval effectiveness. However, this study is unable to use the standards proposed for the evaluation of retrieval systems, as relevance assessments are required for representing the ground truth for a list of queries. Furthermore, none of the IR evaluation metrics can be used, since relevance judgments are not available on the Wikipedia collection or the list of queries to run. A relevant initiative to this task is the Web People Search task, which was organized as part of the SemEval-2007 [38] evaluation exercise. This task consists of clustering a set of documents that mention an ambiguous person name according to the actual entities referred to using that name. However, the problem here is that the evaluated task is people name disambiguation and not expert finding. The INEX initiative [39], which provides an infrastructure for the evaluation of content-oriented retrieval of XML documents based on a set of topics, is also relevant but does not consider the expert finding task. To accomplish this task, the study aims to build a gold standard via manually and voluntary defined expertise profiles by Wikipedia users.
Such studies contribute to the task of expert finding and in the majority of cases, propose methods for finding experts, given a query or knowledge area in which experts are sought. Not only is expert finding a different task to expert profiling, but the methods applied in such studies rely on a large corpus of static documents (e.g., publications) and therefore are not suitable in the context of shorter text, such as micro-contributions in the context of living and evolving documents. Another study, which introduces the task of expert profiling, also relies on queries for extracting expert profiles [40]; the first model uses traditional IR techniques to obtain a set of relevant documents for a given knowledge area (query) and aggregates the relevance of those documents that are associated with the given person. The second model represents both candidates and knowledge areas (queries) as a set of keywords and the skills of an individual are estimated based on the overlap between these sets.
The Entity and Association Retrieval System (EARS) [41] is an open source toolkit for entity-oriented search and discovery in large test collections. EARS, implements a generative probabilistic modeling framework for capturing associations between entities and topics. Currently, EARS supports two main tasks: finding entities (“which entities are associated with topic X?”) and profiling entities (“what topics is an entity associated with?”). EARS employs two main families of models, both based on generative language modeling techniques, for calculating the probability of a query topic (q) being associated with an entity (e), P(q|e). According to one family of models (Model 1) it builds a textual representation (i.e., language model) for each entity, according to the documents associated with that entity. From this representation, it then estimates the probability of the query topic given the entity’s language model. In the second group of models (Model 2), it first identifies important documents for a given topic, and then determines which entities are most closely associated with these documents. We have conducted experiments with EARS using our biomedical use cases (see [7]); however, this system also relies on a given set of queries. Furthermore, as with other studies that target expert finding, EARS relies on a large corpus of static publications, while we aim at building expert profiles from micro-contributions, without relying on any queries.
Finally, in the same category of expertise finding, we find SubSift (short for submission sifting), which is a family of RESTful Web services for profiling and matching text [42]. It was originally designed to match submitted conference or journal papers to potential peer reviewers, based on the similarity between the papers’ abstracts and the reviewers’ publications as found in online bibliographic databases. In this context, the software has already been used to support several major data mining conferences. SubSift, similar to the approaches discussed above, relies on significant amounts of data and uses traditional IR techniques such as TF-IDF, Bag-of-Words and Vector based modeling to profile and compare collections of documents.
The ExpertFinder framework uses and extends existing vocabularies that have attracted a considerable user community already such as FOAF, SIOC, SKOS and DublinCore [43]. Algorithms are also proposed for building expertise profiles using Wikipedia by searching for experts via the content of Wikipedia and its users, as well as techniques that use semantics for disambiguation and search extension [40]. We have leveraged these prior efforts to enable the integration of expertise profiles via a shared understanding based on widely adopted vocabularies and ontologies. This approach will also lead to a seamless aggregation of communities of experts.
WikiGenes combines a dynamic collaborative knowledgebase for the life sciences with explicit authorship. Authorship tracking technology enables users to directly identify the source of every word. The rationale behind WikiGenes is to provide a platform for the scientific community to collect, communicate and evaluate knowledge about genes, chemicals, diseases and other biomedical concepts in a bottom-up approach. WikiGenes links every contribution to its author, as this link is essential to assess origin, authority and reliability of information. This is especially important in the Wiki model, with its dynamic content and large number of authors [44]. Although WikiGenes links every contribution to its author, it does not associate authors with profiles. More importantly, it does not perform semantic analysis on the content of contributions to extract expertise.
As more and more Web users participate in online discussions and micro-blogging, a number of studies have emerged, which focus on aspects such as content recommendation and discovery of users’ topics of interest, especially in Twitter. Early results in discovering Twitter users’ topics of interest are proposed by examining, disambiguating and categorizing entities mentioned in their tweets using a knowledgebase. A topic profile is then developed, by discerning the categories that appear most frequently and that cover all of the entities [45]. The feasibility of linking individual tweets with news articles has also been analyzed for enriching and contextualizing the semantics of user activities on Twitter in order to generate valuable user profiles for the Social Web [46]. This analysis has revealed that the exploitation of tweet-news relations has significant impact on user modeling and allows for the construction of more meaningful representations of Twitter activities. As with other traditional IR methods, this study applies Bag-of-Words and TF-IDF methods for establishing similarity between tweets and news articles and requires a large corpus. In addition, there are fundamental differences between micro-contributions in the context of evolving knowledge bases, contributions to forum discussions and Twitter messages; namely, online knowledge bases do not have to be tailored towards various characteristics of tweets such as presence of @, shortening of words, usage of slang, noisy postings, etc. Also, forum participations are a much richer medium for textual analysis as they are generally much longer than tweets and therefore provide a more meaningful context and usually conform better to the grammatical rules of written English. More importantly, twitter messages do not evolve, whilst we specifically aim at capturing expertise in the context of evolving knowledge.
The Saffron system provides users with a personalized view of the most important expertise topics, researchers and publications, by combining structured data from various sources on the Web with information extracted from unstructured documents using Natural Language Processing techniques [47]. It uses the Semantic Web Dog Food (SWDF) [48] corpus to rank expertise and makes a distinction between the frequency of an expertise topic occurring in the context of a skill type and the overall occurrence of an expertise topic. Saffron also extends information about people by crawling Linked Open Data (LOD) [49] from seed URLs in SWDF. The semantics of the SWDF and crawled data represented using Semantic Web technologies is consolidated to build a holistic view represented via the social graph of an expert.
Existing social networks such as BiomedExperts (BME) [33] provide a source for inferring implicit relationships between concepts of the expertise profiles by analyzing relationships between researchers; i.e., co-authorship. BME is the world’s first pre-populated scientific social network for life science researchers. It gathers data from PubMed [50] on authors’ names and affiliations and uses that data to create publication and research profiles for each author. It builds conceptual profiles of text, called Fingerprints, from documents, Websites, emails and other digitized content and matches them with a comprehensive list of pre-defined fingerprinted concepts to make research results more relevant and efficient.
10. Conclusions and Future Work
In this paper, we have proposed the integration of two Statistical Language Modeling techniques, with the Semantic and Time-dependent Expertise Profiling (STEP) methodology; i.e., our fundamental methodology for expertise profiling using micro-contributions in the context of evolving knowledge. We took advantage of the pluggable architecture of the STEP methodology and integrated the Concept Extraction module with lemmatization as a pre-processing step, followed by Topic Modeling and N-gram Modeling. Our evaluation results demonstrate a significant improvement in the accuracy of profiles generated by incorporating Language Modeling into STEP, as this approach facilitates a domain-independent method for the identification of entities in micro-contributions and minimizes the effects of domain-specific tools on the generated profiles.
In addition, we presented Profile Explorer, a framework that serves as a paradigm for visualization, search and comparative analysis of expertise profiles independent of the methodology or domain. Profile Explorer facilitates tracking and analyzing changes in interests and expertise overtime by capturing the temporality of expertise. Future work will focus on providing additional functionality such as comparative analysis of expertise profiles; e.g., determining time periods when two experts were focused on a common set of expertise topics, clustering micro-contributions based on concepts and clustering experts based on expertise.
Furthermore, direct comparison of expertise profiles generated by STEP and expertise profiles created by authors and used as our baseline is very challenging as the two groups of profiles are defined at different levels of abstraction. Expertise profiles created by STEP using micro-contributions are generally very specific; i.e., the terminology describes specific domain aspects, while expertise profiles defined by experts upon joining projects consist of mostly general terms (e.g., genetics, bioinformatics, microbiology, etc.). Therefore, future work will focus on devising a method for defining both sets of profiles with the same level of abstraction by utilizing the hierarchy of concepts and the structure of ontologies. In doing so, we will create domain-specific views over the expertise of an individual; i.e., we will implement ontological lenses over long-term profiles and profiles described by authors. Using ontological lenses created over profiles and the structure of corresponding ontologies, we will define both sets of profiles at a common level of granularity, which will in turn facilitate direct and more accurate comparisons among profiles generated by STEP and profiles defined by authors.
Acknowledgments
This research is funded by the Australian Research Council (ARC) under the Linkage grant SKELETOME-LP100100156 and the Discovery Early Career Researcher Award (DECRA)-DE120100508.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Sampson, M. Expertise Profiles—How Links to Contributions Changed the Dynamics at IBM. Available online: http://currents.michaelsampson.net/2011/07/expertise-profiles.html (accessed on 30 September 2013).
- O’Reilly, T.; Musser, J. Web 2.0: Principles and Best Practices; O’Reilly Media: Sebastopol, CA, USA, 2006. [Google Scholar]
- Berners-Lee, T.; Hendler, J.; Lassila, O. The semantic web. Sci. Am. 2001, 284, 34–43. [Google Scholar] [CrossRef]
- Clark, T.; Kinoshita, J. AlzForum and SWAN: The present and future of scientific Web communities. Brief. Bioinforma. 2007, 8, 163–171. [Google Scholar] [CrossRef]
- Gene Wiki. Available online: http://en.wikipedia.org/wiki/Gene_Wiki (accessed on 30 September 2013).
- Zhang, J.; Tang, J.; Li, J. Expert finding in a social network. Adv. Databases 2007, 4443, 1066–1069. [Google Scholar]
- Ziaimatin, H.; Groza, T.; Bordea, G.; Buitelaar, P.; Hunter, J. Expertise profiling in evolving knowledge-curation platforms. Glob. Sci. Technol. Forum J. Comput. 2012, 2, 118–127. [Google Scholar]
- Jonquet, C.; Shah, N.; Musen, M. The Open Biomedical Annotator. In Proceedings of the Summit of Translational Bioinformatics, San Francisco, CA, USA, 15–17 March 2009; pp. 56–60.
- Thiagarajan, R.; Manjunath, G.; Stumptner, M. Finding Experts by Semantic Matching of User Profiles; Technical Report HPL-2008-172; HP Laboratories: Karlruhe, Germany, 2008. [Google Scholar]
- Ziaimatin, H. DC Proposal: Capturing Knowledge Evolution and Expertise in Community-Driven Knowledge Curation Platforms. In Proceedings of the International Semantic Web Conference, Bonn, Germany, 23–27 October 2011.
- Mons, B.; Velterop, J. Nano-Publication in the E-Science Era. In Proceedings of the Workshop on Semantic Web Applications in Scientific Discourse, Washington, DC, USA, 25–29 October 2009.
- Casati, F.; Giunchiglia, F.; Marchese, M. Liquid Publications, Scientific Publications Meet the Web; Technical Rep. DIT-07-073, Informatica e Telecomunicazioni; University of Trento: Trento, Italy, 2007. [Google Scholar]
- Wikipedia:WikiProject Molecular and Cellular Biology. Available online: http://en.wikipedia.org/wiki/Wikipedia:MCB (accessed on 30 September 2013).
- Wikipedia:WikiProject Genetics. Available online: http://en.wikipedia.org/wiki/Wikipedia:WikiProject_Genetics (accessed on 30 September 2013).
- Hoffmann, R. A Wiki for the Life Sciences where Authorship Matters. Available online: http://www.nature.com/ng/journal/v40/n9/full/ng.f.217.html (accessed on 30 September 2013).
- OMIM Online Mendelian Inheritance in Man. Available online: http://omim.org (accessed on 30 September 2013).
- Ziaimatin, H.; Groza, T.; Hunter, J. Expertise Modelling in Community-driven Knowledge Curation Platforms. In Proceedings of the 7th Australasian Ontology Workshop, Co-Located with AI 2011, Perth, Australia, 4 December 2011.
- Ziaimatin, H. Profile Explorer (tested only on Firefox). Available online: http://skeletome.metadata.net/dpro/handler/profile/explorer (accessed on 30 September 2013).
- Jonquet, C.; Musen, M.; Shah, N. Building a biomedical ontology recommender web service. J. Biomed. Semant. 2010, 1 (Suppl 1), S1:1–S1:18. [Google Scholar]
- Stemming and Lemmatization. Available online: http://nlp.stanford.edu/IR-book/html/htmledition/stemming-and-lemmatization-1.html (accessed on 30 September 2013).
- Lemmatisation. Available online: http://en.wikipedia.org/wiki/Lemmatisation (accessed on 30 September 2013).
- Liu, H.; Christiansen, T.; Baumgartner, W.A.; Verspoor, K. BioLemmatizer: A lemmatization tool for morphological processing of biomedical text. J. Biomed. Semant. 2012, 3, 3:1–3:29. [Google Scholar]
- Language Model. Available online: http://en.wikipedia.org/wiki/Language_model (accessed on 30 September 2013).
- Blei, D.M. Topic Modeling. Available online: http://www.cs.princeton.edu/~blei/topicmodeling.html (accessed on 30 September 2013).
- De Kok, D.; Brouwer, H. Natural Language Processing for the Working Programmer. Available online: http://nlpwp.org/book/index.xhtml (accessed on 30 September 2013).
- Blei, D.M. Probabilistic topic models. Commun. ACM 2011, 55, 77–84. [Google Scholar] [CrossRef]
- Blei, D.M.; Ng, A.; Jordan, M. Latent Dirichlet allocation. J. Mach. Learn. Res. 2003, 3, 993–1022. [Google Scholar]
- Groza, T.; Zankl, A.; Li, Y.-F.; Hunter, J. Using Semantic Web Technologies to Build a Community-Driven Knowledge Curation Platform for the Skeletal Dysplasia Domain. In Proceedings of the 10th International Semantic Web Conference, Bonn, Germany, 23–27 October 2011; pp. 81–96.
- N-gram. Available online: http://en.wikipedia.org/wiki/N-gram (accessed on 1 October 2013).
- Timeline JS. Available online: http://timeline.verite.co (accessed on 1 October 2013).
- Data-Driven Documents. Available online: http://d3js.org (accessed on 1 October 2013).
- SciVal Experts. Available online: http://info.scival.com/experts (accessed on 1 October 2013).
- BiomedExperts. Available online: http://www.biomedexperts.com/ (accessed on 1 October 2013).
- Text REtrieval Conference (TREC). Available online: http://trec.nist.gov/ (accessed on 1 October 2013).
- Zhu, J.; Song, D.; Rueger, S. Integrating multiple windows and document features for expert finding. J. Am. Soc. Inf. Sci. Technol. 2009, 60, 694–715. [Google Scholar]
- Yang, L.; Zhang, W. A Study of the Dependencies in Expert Finding. In Proceedings of the 2010 Third International Conference on Knowledge Discovery and Data Mining, Phuket, Thailand, 9–10 January 2010.
- Demartini, G. Finding Experts Using Wikipedia. In Proceedings of the ExpertFinder Workshop, Co-Located with ISWC 2007, Busan, Korea, 11–15 November 2007.
- SemEval-2007. Available online: http://nlp.cs.swarthmore.edu/semeval/ (accessed on 1 October 2013).
- Fuhr, N.; Govert, N.; Kazai, G.; Lalmas, M. INEX: INitiative for the Evaluation of XML Retrieval. In Proceedings of the SIGIR 2002 Workshop on XML and Information Retrieval, Tampere, Finland, 11–15 August 2002.
- Balog, K.; de Rijke, M. Determining Expert Profiles (with an Application to Expert Finding). In Proceedings of the 20th International Joint Conference on Artificial Intelligence, Hyderabad, India, 6–12 January 2007; pp. 2657–2662.
- Balog, K. EARS. Available online: http://code.google.com/p/ears/ (accessed on 1 October 2013).
- Price, S.; Flach, P.A.; Spiegler, S.; Bailey, C.; Rogers, N. SubSift Web Services and Workflows for Profiling and Comparing Scientists and Their Published Works. In Proceedings of the 2010 IEEE 6th International Conference on e-Science, Brisbane, Australia, 7–10 December 2010.
- Aleman-Meza, B.; Bojars, U.; Boley, H.; Breslin, J.; Mochol, M.; Nixon, L.; Polleres, A.; Zhdanova, A. Combining RDF Vocabularies for Expert Finding. In Proceedings of the 4th European Semantic Web Conference, Innsbruck, Austria, 3–7 June 2007; pp. 235–250.
- Hoffmann, R. A wiki for the life sciences where authorship matters. Nat. Genet. 2008, 40, 1047–1051. [Google Scholar] [CrossRef]
- Michelson, M.; Macskassy, S. Discovering Users’ Topics of Interest on Twitter: A First Look. In Proceedings of the 4th Workshop on Analytics for Noisy Unstructuredco-located with the 19th ACM CIKM Conference, Toronto, Canada, 26–30 October 2010; pp. 73–80.
- Abel, F.; Gao, Q.; Houben, G.; Tao, K. Semantic Enrichment of Twitter Posts for User Profile Construction on the Social Web. In Proceedings of the 8th Extended Semantic Web Conference, Heraklion, Greece, 29 May–2 June 2011; pp. 375–389.
- Monaghan, F.; Bordea, G.; Samp, K.; Buitelaar, P. Exploring Your Research: Sprinkling some Saffron on Semantic Web Dog Food. In Proceedings of the Semantic Web Challenge at the International Semantic Web Conference, Shanghai, China, 7–11 November 2010.
- Moeller, K.; Heath, T.; Handschuh, S.; Domingue, J. Recipes for Semantic Web Dog Food—The ESWC and ISWC Metadata Projects. In Proceedings of the 6th International Semantic Web Conference, Busan, Korea, 11–15 November 2007; pp. 802–815.
- Bizer, C.; Heath, T.; Berners-Lee, T. Linked data—The story so far. Int. J. Semant. Web Inf. Syst. 2009, 5, 1–22. [Google Scholar]
- PubMed. Available online: http://www.ncbi.nlm.nih.gov/pubmed/ (accessed on 1 October 2013).
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).