TriagE-NLU: A Natural Language Understanding System for Clinical Triage and Intervention in Multilingual Emergency Dialogues
Abstract
1. Introduction
- We develop TriagE-NLU, a multilingual, patient-aware natural language understanding system that combines semantic parsing and intervention classification for emergency telemedicine.
- We introduce EUREKA (Emergency Understanding Representation for Evidence-based Knowledge and Action). This unified semantic schema encodes symptoms, onset patterns, risk factors, and inferred urgency to map patient dialogue to structured intent.
- We construct TriageX, the first synthetic, clinically annotated dataset for multilingual emergency dialogue understanding, and train our model using federated averaging (FedAvg) to support privacy-preserving, cross-institutional deployment.
2. Related Work
3. Proposed TriagE-NLU System
- Multimodal Input Processing: The system accepts heterogeneous input streams, including transcribed input speech or dialogue between patients and clinicians, structured patient metadata (e.g., age, pre-existing conditions, vital signs), and temporal and contextual markers (e.g., symptom onset duration, location metadata).
- Semantic Parsing Engine: This module leverages multilingual transformer models fine-tuned for low-resource clinical contexts. It maps utterances and contextual signals to a structured representation based on a new proposed EUREKA schema (Emergency Understanding Representation for Evidence-based Knowledge and Action), which will be described in Section 4, that encodes symptom mentions, temporal qualifiers, patient risk factors, and urgency indicators.
- Intervention Grounding Module: Parsed semantic frames are fed into a decision engine that aligns recognized medical states with appropriate interventions using a rules-and-learned-policy hybrid model informed by clinical guidelines (e.g., ACLS, WHO triage protocols). For example, a combination of “slurred speech”, “unilateral weakness”, and “<10 min onset” is mapped to “stroke protocol activation”.
- Federated Learning Update Loop: Each deployment instance of TriagE-NLU (e.g., in a regional hospital network) trains on local usage patterns while sharing model updates through secure aggregation. This ensures continuous adaptation without exposing sensitive clinical dialogue, maintaining both data locality and generalization capabilities.
Novelty Positioning
- Patient-Aware Semantic Understanding: Unlike prior work focused on retrospective clinical texts, our system models spontaneous speech or dialogue between patients and clinicians. It fuses natural language with structured patient data (e.g., age, onset time, and vital signs), enabling patient-specific semantic parsing in time-sensitive emergency settings.
- Grounding Semantic Representations in Interventions: To our knowledge, TriagE-NLU is the first to bridge high-level semantic parsing with clinically actionable output. While prior systems have modeled symptom classification or medical Q&A, none have mapped parsed utterances to structured intervention suggestions in multilingual telemedicine contexts.
- Multilingual and Low-Resource Triage Generalization: Most existing systems are monolingual or assume idealized input. We propose a multilingual, low-resource capable system that performs fast semantic parsing across languages, using weak supervision and cross-lingual transfer. This enables deployment in under-resourced regions where emergency response support is critically needed.
- Federated and Privacy-Preserving Learning: TriagE-NLU integrates federated learning for continual improvement across decentralized clinical environments. This addresses key privacy and legal barriers to training on sensitive conversations while allowing regional adaptation to medical protocols, linguistic variance, and cultural expression of symptoms.
- Absence of Existing Benchmarks: We introduce TriageX, a benchmark dataset of simulated multilingual emergency teleconsultation inputs. While dialogue datasets exist in general-purpose or open-domain contexts, there is no publicly available resource that combines medical urgency, multiple languages, and structured intervention annotation, establishing a new benchmark task for understanding semantic medical triage.
4. Semantic Representation: EUREKA Schema Proposal
4.1. Design Goals
4.2. Schema Structure
4.3. Parsing Workflow
5. Training Methodology
5.1. Data Preparation
5.2. Semantic Parsing Model
5.3. Intervention Classification Model
5.4. Federated Learning Protocol
5.5. Cross-Lingual Alignment and Adaptation
5.6. Evaluation Strategy
6. Benchmark Construction: TriageX Dataset
6.1. Motivation and Design Principles
6.2. Data Sources and Simulation Framework
6.3. Schema and Annotation Protocol
6.4. Languages and Demographics
6.5. Benchmark Tasks
7. Results and Performance Analysis
7.1. Use Case: Rural Telemedicine Stroke Suspect Case
7.2. TriagE-NLU Evaluation
- mT5-Base: Fine-tuned on TriageX using standard seq2seq loss for EUREKA parsing only.
- mBART: A multilingual encoder/decoder model.
- MBART + MLP: A multilingual encoder/decoder model with an MLP classification head for intervention prediction.
- XLM-RoBERTa: A multilingual language model.
- XLM-RoBERTa + BiLSTM: Token-level slot tagging using a BiLSTM-CRF on top of frozen XLM-R embeddings.
7.3. Multilingual Transfer and Zero-Shot Generalization
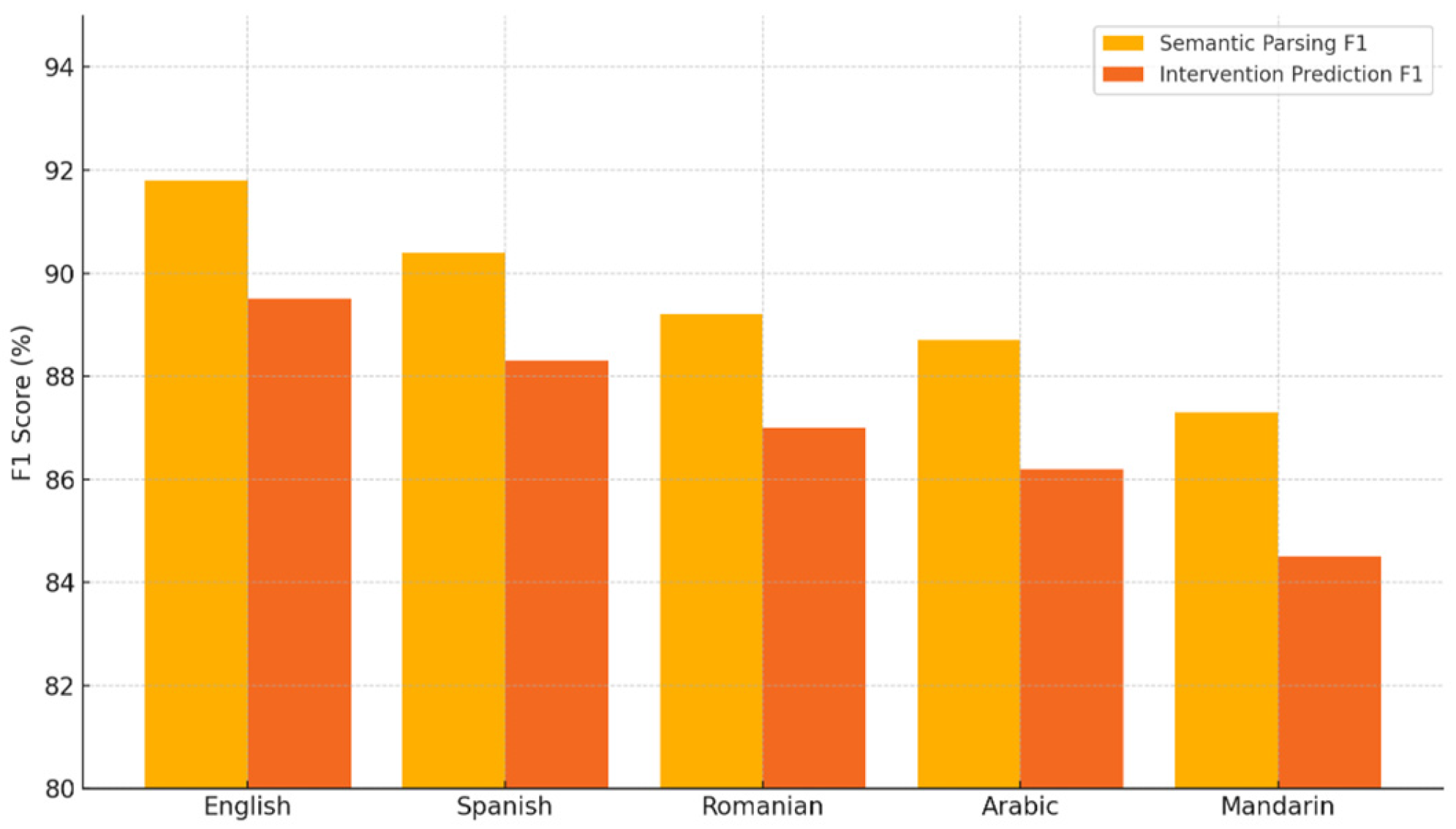
- English achieves the highest performance, with 0.92 F1 for semantic parsing and 0.89 for intervention prediction, indicating strong performance in high-resource language settings.
- Spanish follows closely with 0.90 parsing F1 and 0.88 intervention F1, maintaining robust performance.
- Romanian and Arabic, both lower-resource languages, show a modest drop: 0.89 and 0.88% parsing F1, and 0.87 and 0.86 intervention F1, respectively.
- Mandarin exhibits the lowest scores in both categories: 0.87 parsing F1 and 0.84 intervention F1, suggesting room for improvement in very low-resource or complex linguistic settings.
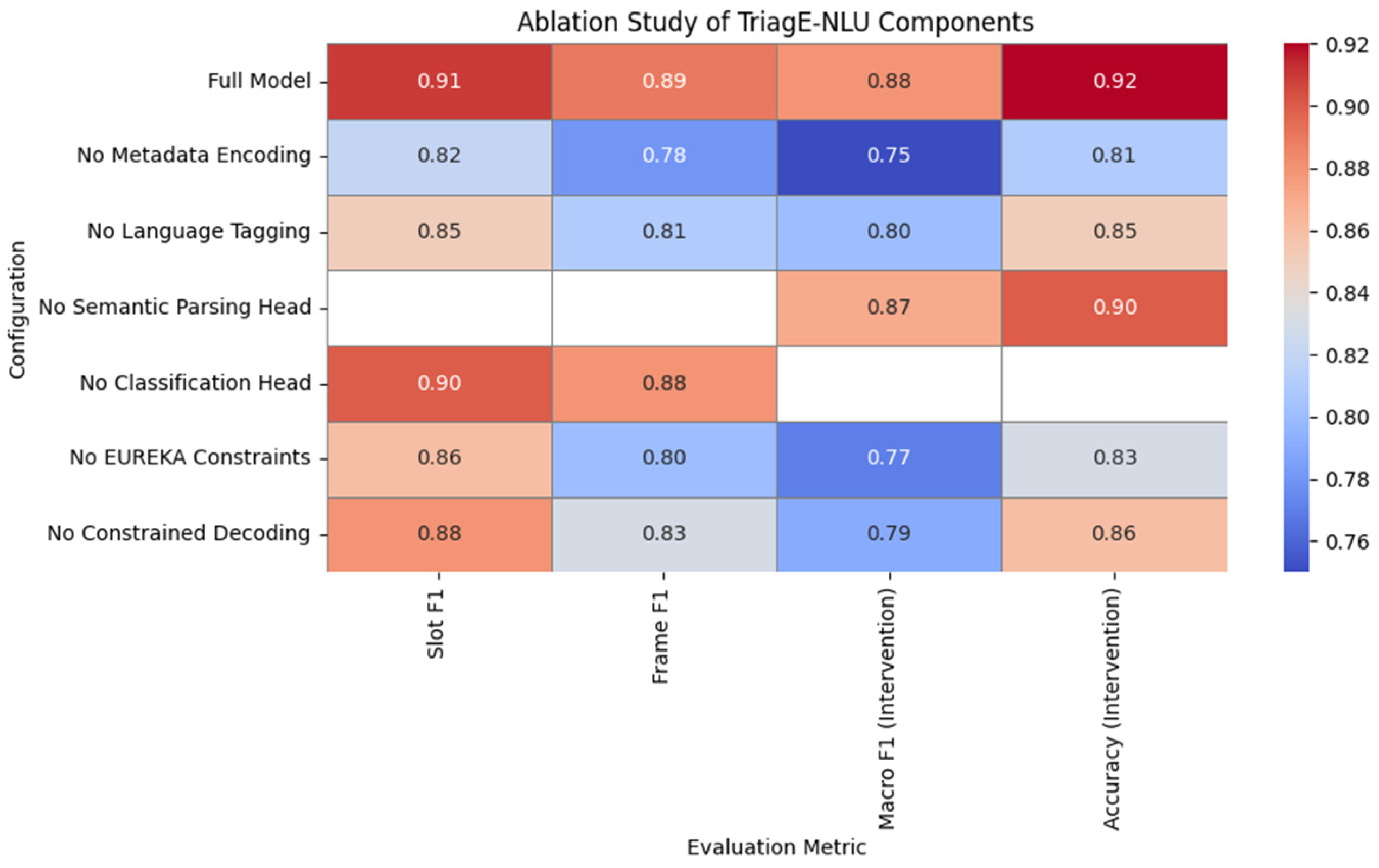
7.4. Ablation Studies
8. Conclusions and Future Work
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Appendix A. TriagE-NLU Implementation
- Preprocessing and Data Handling involve several steps. First, multilingual inputs from the TriageX dataset are loaded and tokenized. Patient metadata is then normalized and merged with the corresponding input text to provide contextual grounding. Finally, the target EUREKA frames are converted into linearized string formats suitable for sequence generation tasks.
- Semantic Parsing (mT5) is performed by fine-tuning the google/mT5-Base model in a sequence-to-sequence setup. The input follows the format: “<lang>: <dialogue> [SEP] <age> [SEP] <conditions>”, and the output corresponds to a serialized EUREKA frame represented as a JSON-like string. To ensure structural validity, constrained decoding rules are applied during generation to enforce correct EUREKA slot formatting.
- Intervention Classification Head is implemented by attaching an MLP classifier either on top of the encoder output or the parsed EUREKA frame. This classifier is trained using a multitask learning approach, either jointly with the semantic parser or in a sequential setup, depending on the configuration.
- Federated Learning Simulation uses Flower to simulate clients.
- The Evaluation process includes computing frame-level and slot-level F1 scores for the EUREKA semantic parsing task, as well as measuring accuracy and Macro F1 for intervention classification. Additionally, performance is analyzed separately for each language to assess cross-lingual generalization.
References
- Jerfy, A.; Selden, O.; Balkrishnan, R. The Growing Impact of Natural Language Processing in Healthcare and Public Health. INQUIRY 2024, 61, 00469580241290095. [Google Scholar] [CrossRef]
- Biswas, P.; Arockiam, D. Review on Multi-lingual Sentiment Analysis in Health Care. J. Electr. Syst. 2024, 20, 3394–3405. [Google Scholar] [CrossRef]
- Supriyono, A.P.W.; Suyono, F.K. Advancements in Natural Language Processing: Implications, Challenges, and Future Directions. Telemat. Inform. Rep. 2024, 16, 100173. [Google Scholar] [CrossRef]
- Basra, J.; Saravanan, R.; Rahul, A. A survey on Named Entity Recognition—datasets, tools, and methodologies. Nat. Lang. Process. J. 2023, 3, 100017. [Google Scholar] [CrossRef]
- Mandale-Jadhav, A. Text Summarization Using Natural Language Processing. J. Electr. Syst. 2025, 20, 3410–3417. [Google Scholar] [CrossRef]
- Upadhyay, P.; Agarwal, R.; Dhiman, S.; Sarkar, A.; Chaturvedi, S. A comprehensive survey on answer generation methods using NLP. Nat. Lang. Process. J. 2024, 8, 100088. [Google Scholar] [CrossRef]
- Sun, H.; Peng, J.; Yang, W.; He, L.; Du, B.; Yan, R. Enhancing Medical Dialogue Generation through Knowledge Refinement and Dynamic Prompt Adjustment. arXiv 2025, arXiv:2506.10877. [Google Scholar] [CrossRef]
- Li, X.; Peng, L.; Wang, Y.P.; Zhang, W. Open challenges and opportunities in federated foundation models towards biomedical healthcare. BioData Min. 2025, 18, 2. [Google Scholar] [CrossRef]
- Li, I.; Rao, S.; Solares, J.R.A.; Hassaine, A.; Ramakrishnan, R.; Canoy, D.; Zhu, Y.; Rahimi, K.; Salimi-Khorshidi, G. BEHRT: Transformer for Electronic Health Records. Sci. Rep. 2020, 10, 7155. [Google Scholar] [CrossRef]
- Alsentzer, E.; Murphy, J.; Boag, W.; Weng, W.-H.; Jindi, D.; Naumann, T.; McDermott, M. Publicly available clinical BERT embeddings. In Proceedings of the 2nd Clinical Natural Language Processing Workshop, Minneapolis, MN, USA, 7 June 2019; pp. 72–78. [Google Scholar] [CrossRef]
- Kocaballi, A.B.; Berkovsky, S.; Quiroz, J.C.; Laranjo, L.; Tong, H.L.; Rezazadegan, D.; Briatore, A.; Coiera, E. The Personalization of Conversational Agents in Health Care: Systematic Review. J. Med. Internet Res. 2019, 21, e15360. [Google Scholar] [CrossRef]
- Milne-Ives, M.; de Cock, C.; Lim, E.; Shehadeh, M.H.; de Pennington, N.; Mole, G.; Normando, E.; Meinert, E. The effectiveness of artificial intelligence conversational agents in healthcare. J. Med. Internet Res. 2020, 22, e20346. [Google Scholar] [CrossRef] [PubMed]
- Zeng, G.; Yang, W.; Ju, Z.; Yang, Y.; Wang, S.; Zhang, R.; Zhou, M.; Zeng, J.; Dong, X.; Zhang, R.; et al. MedDialog: Large-scale Medical Dialogue Dataset. In Proceedings of the 2020 Conference on Empirical Methods in Natural Language Processing (EMNLP), Online, 16–20 November 2020. [Google Scholar] [CrossRef]
- Zang, T.; Cai, Z.; Wang, C.; Qiu, M.; Yang, B.; He, X. SMedBERT: A Knowledge-Enhanced Dialogue Generation Model for Medical Consultation. In Proceedings of the 59th Annual Meeting of the Association for Computational Linguistics and the 11th International Joint Conference on Natural Language Processing (Volume 1: Long Papers), Online, 1–6 August 2021. [Google Scholar] [CrossRef]
- Sutton, R.T.; Pincock, D.; Baumgart, D.C.; Sadowski, D.C.; Fedorak, R.N.; Kroeker, K.I. An overview of clinical decision support systems: Benefits, risks, and strategies for success. NPJ Digit. Med. 2020, 3, 17. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Dadu, A.; Nalls, M.; Faghri, F.; Sun, J. Instruction Tuning Large Language Models to Understand Electronic Health Records. In Proceedings of the Advances in Neural Information Processing Systems (NeurIPS 2024), Vancouver, BC, Canada, 10–15 December 2024; Available online: https://proceedings.neurips.cc/paper_files/paper/2024/file/62986e0a78780fe5f17b495aeded5bab-Paper-Datasets_and_Benchmarks_Track.pdf (accessed on 24 June 2025).
- Rieke, N.; Hancox, J.; Li, W.; Milletari, F.; Roth, H.R.; Albarqouni, S.; Bakas, S.; Galtier, M.N.; Landman, B.A.; Maier-Hein, K.; et al. The future of digital health with federated learning. NPJ Digit. Med. 2020, 3, 119. [Google Scholar] [CrossRef] [PubMed]
- Sheller, M.J.; Edwards, B.; Reina, G.A.; Martin, J.; Pati, S.; Kotrotsou, A.; Milchenko, M.; Xu, W.; Marcus, D.; Colen, R.R.; et al. Federated learning in medicine: Facilitating multi-institutional collaborations without sharing patient data. Sci. Rep. 2020, 10, 12598. [Google Scholar] [CrossRef]
- Xue, L.; Constant, N.; Roberts, A.; Kale, M.; Al-Rfou, R.; Siddhant, A.; Barua, A.; Raffel, C. mT5: A massively multilingual pre-trained text-to-text transformer. In Proceedings of the 2021 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Online, 6–11 June 2021. [Google Scholar] [CrossRef]
- Tang, Y.; Tran, C.; Li, X.; Chen, P.-J.; Goyal, N.; Chaudhary, V.; Gu, J.; Fan, A. Multilingual Translation from Denoising Pre-Training. In Proceedings of the Findings of the Association for Computational Linguistics: ACL-IJCNLP 2021, Online, 1–6 August 2021; pp. 3450–3466. Available online: https://aclanthology.org/2021.findings-acl.304/ (accessed on 24 June 2025).
- Conneau, A.; Khandelwal, K.; Goyal, N.; Chaudhary, V.; Wenzek, G.; Guzman, F.; Grave, E.; Ott, M.; Zettlemoyer, L.; Stoyanov, V. Unsupervised Cross-lingual Representation Learning at Scale. In Proceedings of the 58th Annual Meeting of the Association for Computational Linguistics, Online, 5–10 July 2020. [Google Scholar] [CrossRef]
- Yang, X.; Chen, A.; PourNejatian, N.; Shin, H.C.; Smith, K.E.; Parisien, C.; Compas, C.; Martin, C.; Flores, M.G.; Zhang, Y.; et al. GatorTron: A Large Clinical Language Model to Unlock Patient Information from Unstructured Electronic Health Records. arXiv 2022, arXiv:2203.03540. [Google Scholar] [CrossRef]
- Liu, W.; Tang, J.; Cheng, Y.; Li, W.; Zheng, Y.; Liang, X. MedDG: An Entity-Centric Medical Consultation Dataset for Entity-Aware Medical Dialogue Generation. arXiv 2022, arXiv:2010.07497. [Google Scholar] [CrossRef]
- Liu, T.; Gu, J.; Goyal, N.; Li, X.; Edunov, S.; Ghazvininejad, M.; Lewis, M.; Zettlemoyer, L. Multilingual Denoising Pre-training for Neural Machine Translation. arXiv 2020, arXiv:2001.08210. [Google Scholar] [CrossRef]
- Kusumawardani, R.P.; Kusumawati, K.N. Named entity recognition in the medical domain for Indonesian language health consultation services using bidirectional-lstm-crf algorithm. Procedia Comput. Sci. 2024, 245, 1146–1156. [Google Scholar] [CrossRef]
- Rojas-Carabali, W.; Agrawal, R.; Gutierrez-Sinisterra, L.; Baxter, S.L.; Cifuentes-González, C.; Wei, Y.C.; Abisheganaden, J.; Kannapiran, P.; Wong, S.; Lee, B.; et al. Natural Language Processing in medicine and ophthalmology: A review for the 21st-century clinician. Asia Pac. J. Ophthalmol. 2024, 13, 100084. [Google Scholar] [CrossRef]
- Benjamin, C.; Henry, I.; Bergman, S.S.; Gabriele, P.; Joseph, S.; Alun, H.D. Natural language processing techniques applied to the electronic health record in clinical research and practice-an introduction to methodologies. Comput. Biol. Med. 2025, 188, 09808. [Google Scholar] [CrossRef]
- Thatoi, P.; Choudhary, R.; Shiwlani, A.; Qureshi, H.A.; Kuma, S. Natural Language Processing (NLP) in the Extraction of Clinical Information from Electronic Health Records (EHRs) for Cancer Prognosis. Int. J. Membr. Sci. Technol. 2023, 10, 2676–2694. [Google Scholar]
- Sezgin, E.; Hussain, S.A.; Rust, S.; Huang, Y. Extracting Medical Information from Free-Text and Unstructured Patient-Generated Health Data Using Natural Language Processing Methods: Feasibility Study with Real-world Data. JMIR Form. Res. 2023, 7, e43014. [Google Scholar] [CrossRef]
- Yang, C.; Deng, J.; Chen, X.; An, Y. SPBERE: Boosting span-based pipeline biomedical entity and relation extraction via entity information. J. Biomed. Inform. 2023, 145, 104456. [Google Scholar] [CrossRef]
- Haleem, A.; Javaid, M.; Singh, R.P.; Suman, R. Telemedicine for healthcare: Capabilities, features, barriers, and applications. Sens. Int. 2021, 2, 100117. [Google Scholar] [CrossRef]
- Mermin-Bunnell, K.; Zhu, Y.; Hornback, A.; Damhorst, G.; Walker, T.; Robichaux, C.; Mathew, L.; Jaquemet, N.; Peters, K.; Johnson, T.M.; et al. Use of Natural Language Processing of Patient-Initiated Electronic Health Record Messages to Identify Patients With COVID-19 Infection. JAMA Netw. Open. 2023, 6, e2322299. [Google Scholar] [CrossRef]
- Guevara, M.; Chen, S.; Thomas, S.; Chaunzwa, T.L.; Franco, I.; Kann, B.H.; Moningi, S.; Qian, J.M.; Goldstein, M.; Harper, S.; et al. Large language models to identify social determinants of health in electronic health records. NPJ Digit. Med. 2024, 7, 6. [Google Scholar] [CrossRef] [PubMed]







| Feature/Capability | Prior Work in Clinical NLP | TriagE-NLU |
|---|---|---|
| Dialogue Understanding | Mostly retrospective text [26,27] | Supports multi-turn emergency dialogue |
| Patient-Aware Semantic Parsing | Typically text-only models [9,10] | Integrates age, vitals, onset metadata |
| Multilingual and Low-Resource Generalization | Monolingual focus [13,21] | Designed for multilingual settings via transfer learning |
| Grounding in Medical Interventions | Most focus on diagnosis or symptom extraction, not intervention mapping [28,29] | Maps parsed intent to clinical actions |
| Federated Learning for Privacy Adaptation | Centralized learning [30] | Federated training with privacy-preserving aggregation |
| Emergency Telemedicine Focus | General clinical use or outpatient consults [31,32] | Tailored to high-stakes, urgent scenarios |
| Triage Benchmark with Interventions | No available datasets with multilingual, intervention-labeled emergency conversations | Introduces TriageX dataset |
| Model | Slot F1 | Frame F1 | Macro F1 (Intervention) | Accuracy |
|---|---|---|---|---|
| TriagE-NLU | 0.91 | 0.89 | 0.88 | 0.92 |
| mT5-Base (Seq2Seq) | 0.84 | — | — | 0.85 |
| mBART | — | — | 0.81 | 0.83 |
| mBART + MLP | — | — | 0.83 | 0.81 |
| XLM-ROBERTa | — | — | 0.80 | 0.82 |
| XLM-ROBERTa + BiLSTM | 0.79 | — | — | 0.78 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Balaban, B.-M.; Sacală, I.; Petrescu-Niţă, A.-C. TriagE-NLU: A Natural Language Understanding System for Clinical Triage and Intervention in Multilingual Emergency Dialogues. Future Internet 2025, 17, 314. https://doi.org/10.3390/fi17070314
Balaban B-M, Sacală I, Petrescu-Niţă A-C. TriagE-NLU: A Natural Language Understanding System for Clinical Triage and Intervention in Multilingual Emergency Dialogues. Future Internet. 2025; 17(7):314. https://doi.org/10.3390/fi17070314
Chicago/Turabian StyleBalaban, Béatrix-May, Ioan Sacală, and Alina-Claudia Petrescu-Niţă. 2025. "TriagE-NLU: A Natural Language Understanding System for Clinical Triage and Intervention in Multilingual Emergency Dialogues" Future Internet 17, no. 7: 314. https://doi.org/10.3390/fi17070314
APA StyleBalaban, B.-M., Sacală, I., & Petrescu-Niţă, A.-C. (2025). TriagE-NLU: A Natural Language Understanding System for Clinical Triage and Intervention in Multilingual Emergency Dialogues. Future Internet, 17(7), 314. https://doi.org/10.3390/fi17070314







