Artificial Intelligence to Reshape the Healthcare Ecosystem
Abstract
1. Introduction
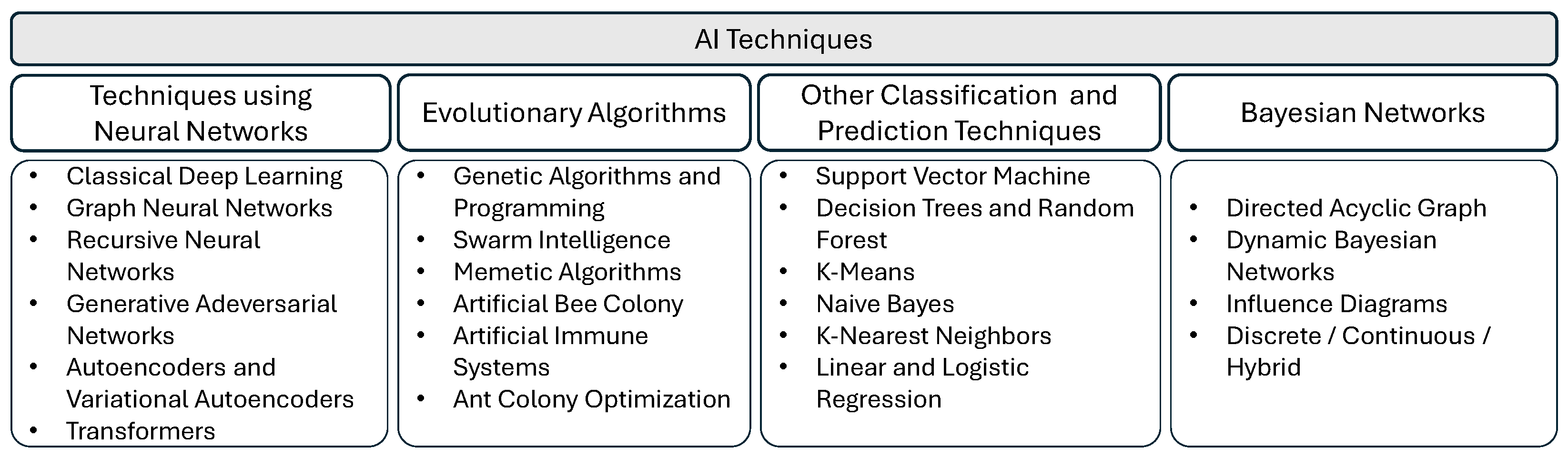
- Classical deep learning, including convolutional neural networks and U-Net architectures;
- Graph neural networks;
- Recurrent neural networks;
- Generative AI;
- Diffusion models;
- Reinforcement learning.
2. AI Techniques for Healthcare
- I—Input nodes: These receive and process external data by adapting them to the internal nodes, to which each node I is connected;
- H—Hidden (internal) nodes: Organized in multiple levels, each of these nodes processes the received signals and transmits the result to the subsequent nodes. They actually perform, independently of each other, the data processing process through the use of algorithms;
- O—Output nodes: Final layer of nodes that collect the processing results of the H-layer and adapt them to the next neural network block.
- ReLU (rectified linear unit):It is widely used due to both its simplicity and its effectiveness in mitigating the vanishing gradient problem.
- Leaky ReLU:where is a small constant that allows a small, non-zero gradient when the unit is inactive. It aims at mitigating the so-called dying ReLU problem.
- Sigmoid:It is an S-shaped curve, that maps input to a range between 0 and 1. It is prone to vanishing gradients.
- Tanh (hyperbolic tangent):It is an S-shaped curve that maps input to a range between −1 and 1. It is also prone to vanishing gradients.
- Swish:An activation function that was shown to outperform ReLU in particular situations, with as a learnable parameter.
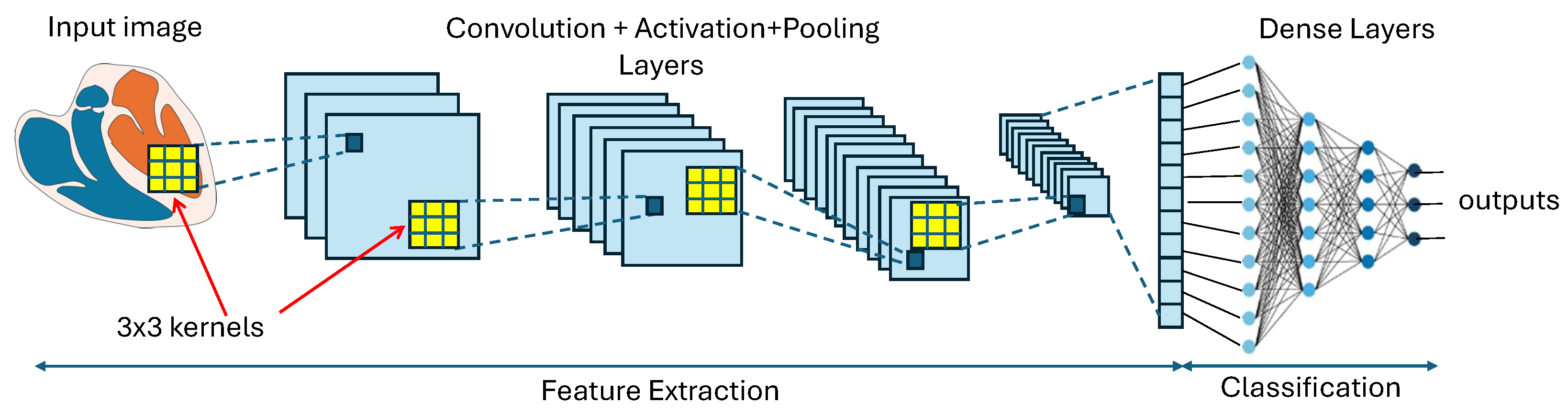
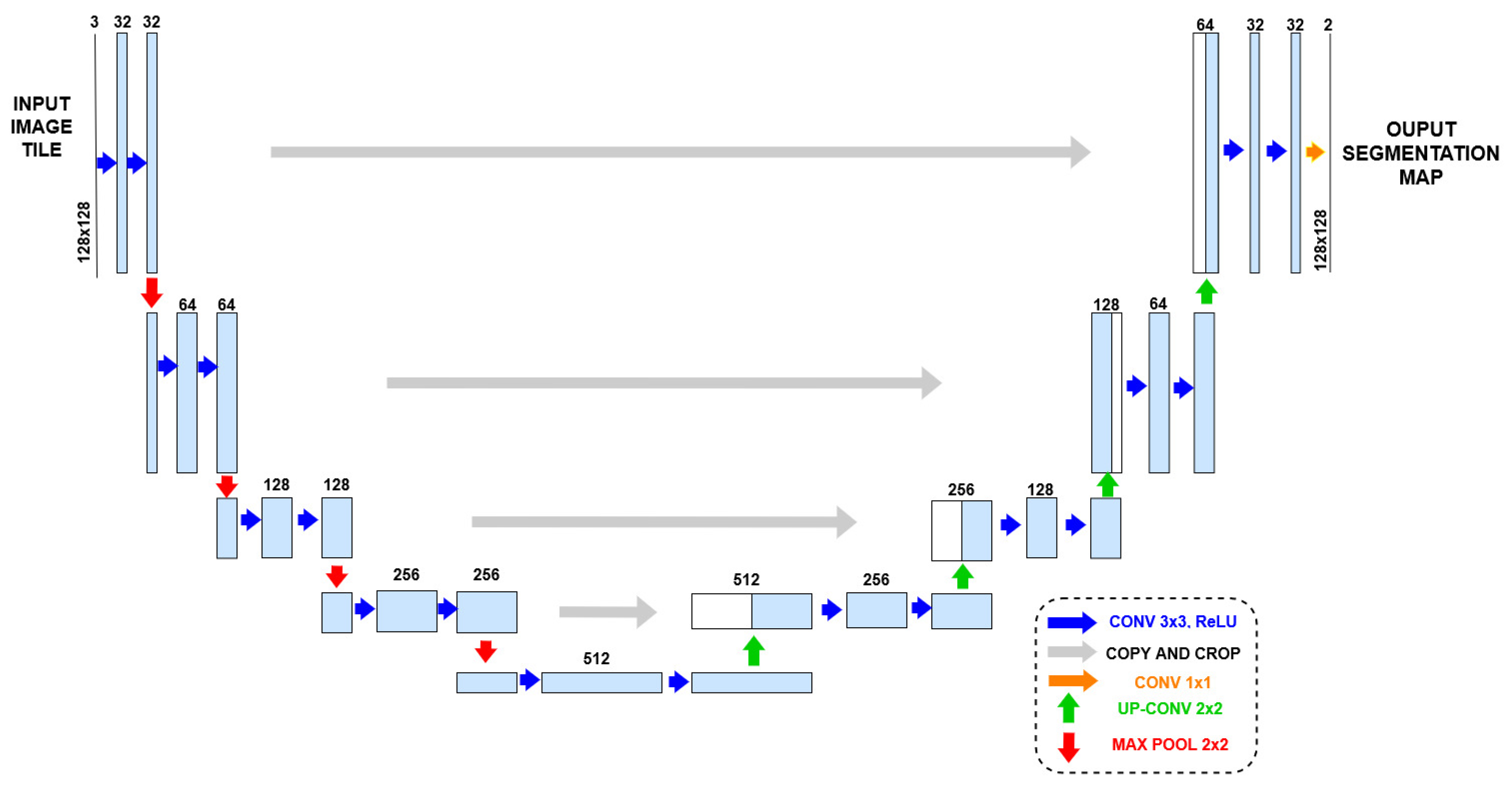
2.1. Deep Learning: Convolutional Neural Networks and U-Net Architectures
- Convolutional layers: These use filters (or kernels) that slide over the input data and multiply by their values to capture local patterns. Each trained filter is expected to focus on a particular region of the input image file. In other words, its role is to offer subsequent layers a local feature, such an edge or a particular shape pattern, to either compose a global feature or an immediate detection.
- Non-linear activation functions in nodes: These functions are applied after each convolutional layer. Non-linearity allows the network to learn and implement complex functions, and is essential for processing images and implement tasks such as recognition and classification.
- Pooling layers: They play a crucial role by performing down-sampling operations along the spatial dimensions of the input images. The benefits of this operation include spatial dimensionality reduction, which reduces the number of parameters in the network and memory requirements to store them, invariance to small translations, rotations, and distortions in the input images, and reduction in the sensitivity of the implemented function to both noise and random variations in input data. Descriptions of some types of pooling functions follow:
- −
- Max pooling: Takes the maximum value within a defined window (e.g., ):where x is the input feature map, y is the output pooled map, and define the window size.
- −
- Average pooling: Computes the average value within a defined window.where k is the window size.
- −
- Global pooling: Applies pooling over the entire spatial dimensions of the feature map. This way, each feature map is reduced to a single value, often used before the following fully connected layers to flatten the feature maps.
- Fully connected layers (also known as dense layers): They take the aspect of the layers of the general ANN shown in Figure 3. They are used to learn complex, non-linear combinations of the features. For example, in a classification network, the fully connected layers map the features to class scores.
- Dropout layer: During training, dropout is often applied to fully connected layers to prevent overfitting. Dropout randomly sets a fraction of the neurons to zero at each training step, forcing the network to learn redundant representations and improving generalization.
2.2. Graph Neural Networks
2.3. Recurrent Neural Networks
2.3.1. Long Short-Term Memory
2.3.2. Gated Recurrent Unit
2.4. Generative AI
2.4.1. Generative Adversarial Networks
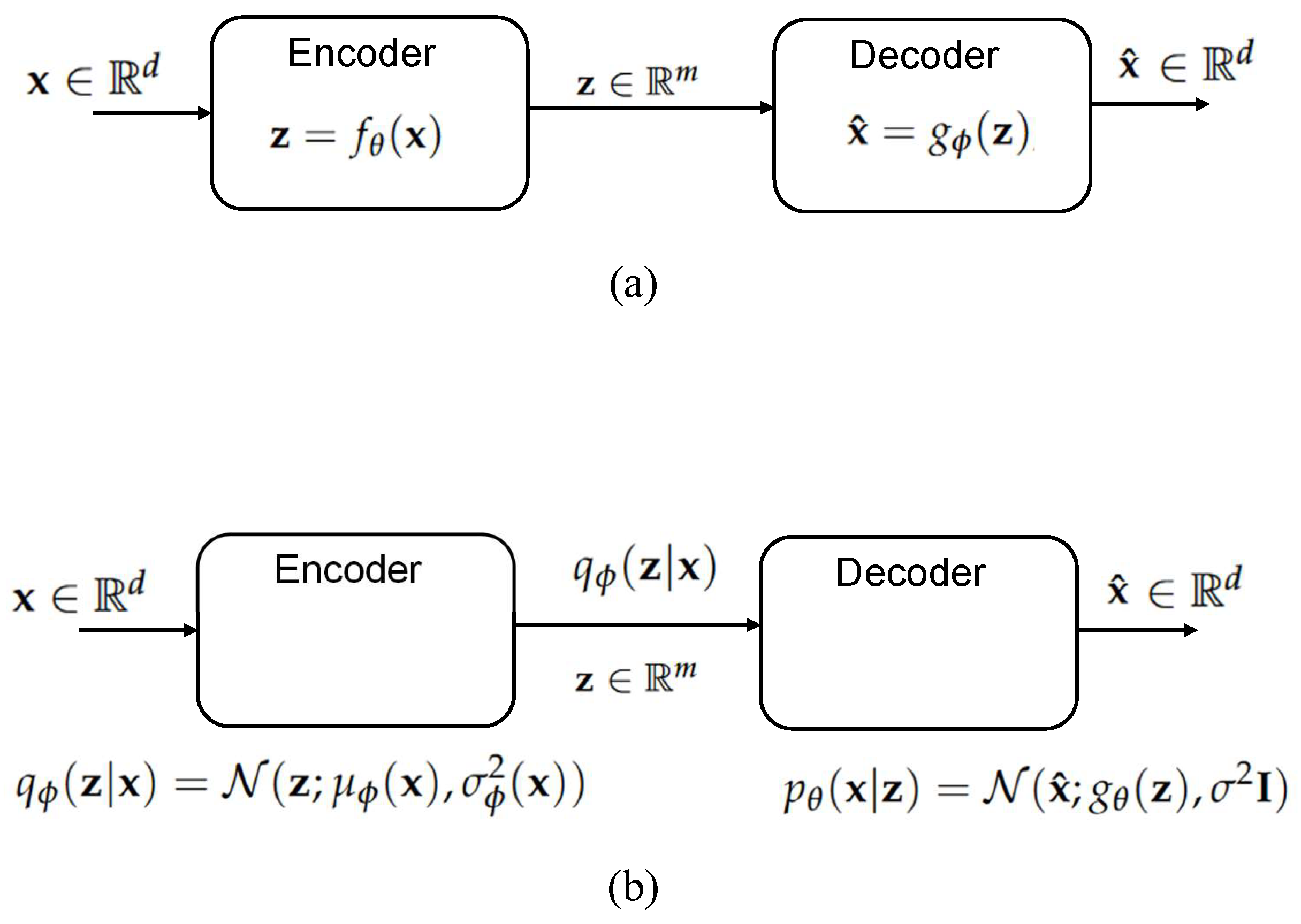
2.4.2. Autoencoders and Variational Autoencoders
Autoencoders
Variational Autoencoders
2.4.3. Recurrent Neural Networks (RNNs)
2.4.4. Transformers
- First it is necessary to linearly project the input embedding X into multiple sets of keys, queries, and values by using the learned weight matrices:where h is the number of attention heads.
- Then, the self-attention mechanism is applied to each set of keys, queries, and values, as follows:
- After this, the outputs of all attention heads are concatenated:
- Finally, the result of the concatenation is linearly projected through the learned weight matrix :
2.5. Diffusion Models
2.6. Reinforcement Learning
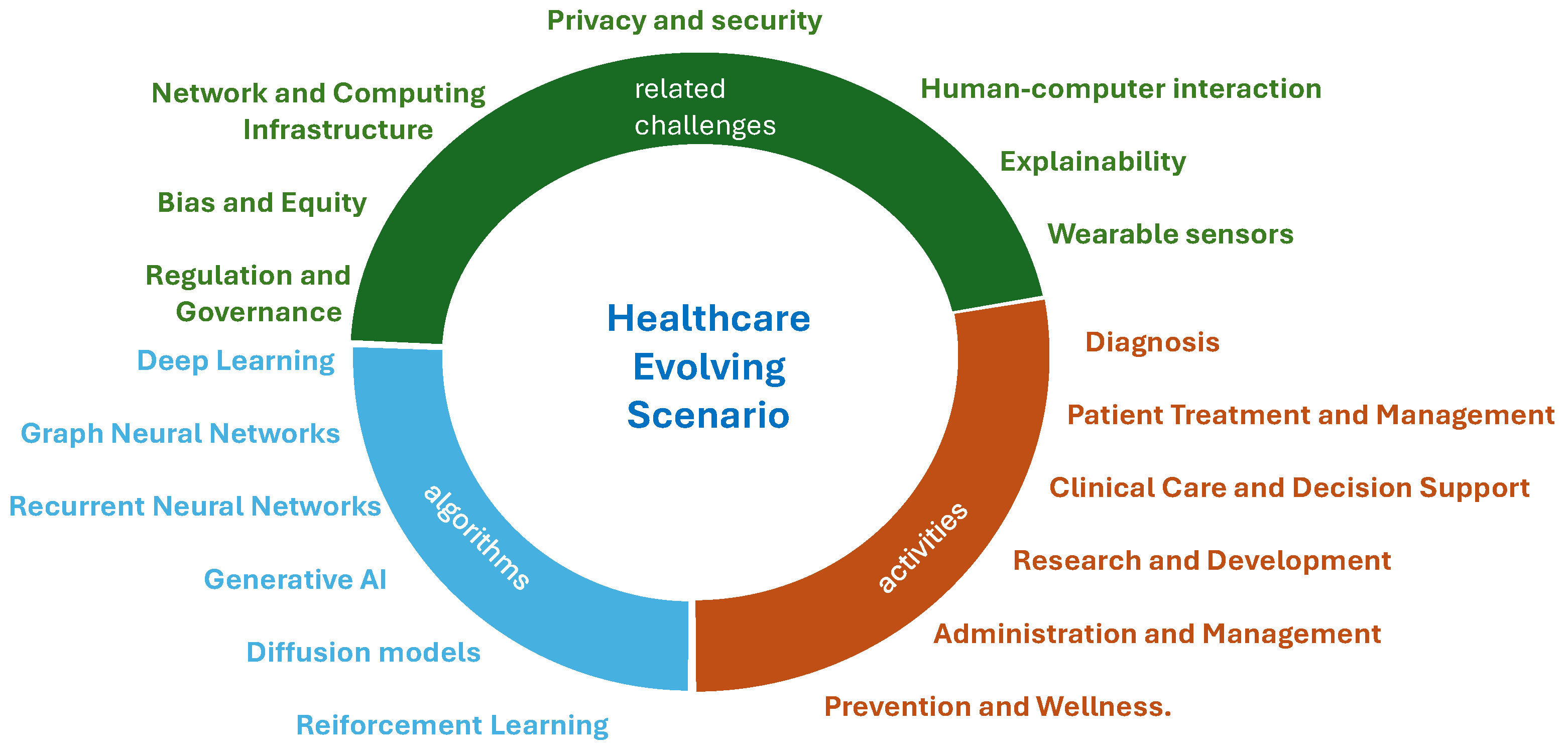
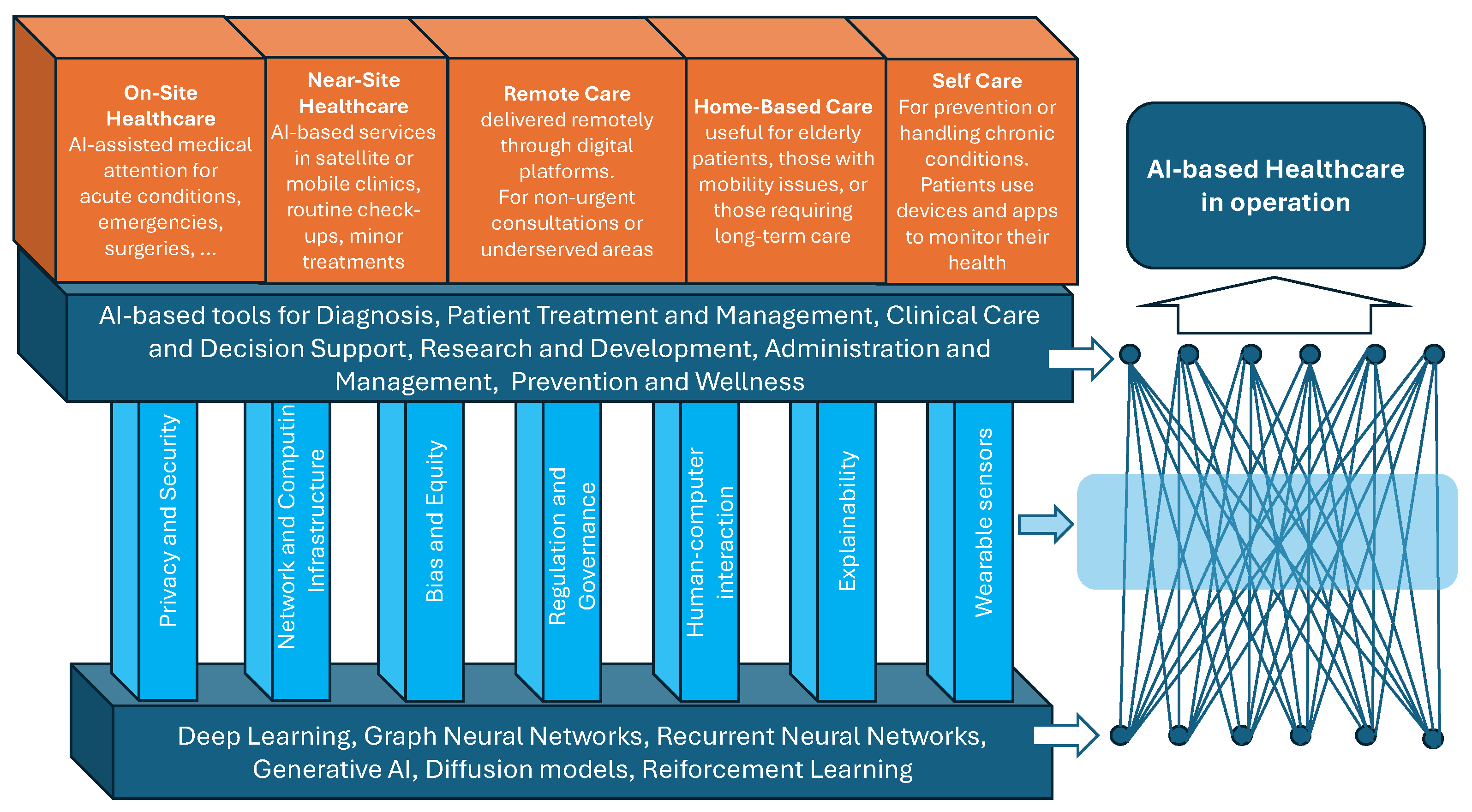
3. AI Penetration in Baseline Healthcare Services
4. Other Related Challenges
4.1. Human–Computer Interaction
4.2. Explainability
4.3. Wearable Sensors
4.4. Privacy and Security
4.5. Network and Computing Infrastructure
4.6. Bias and Equity
4.7. Regulation and Governance
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Maslej, N.; Fattorini, L.; Perrault, R.; Parli, V.; Reuel, A.; Brynjolfsson, E.; Etchemendy, J.; Ligett, K.; Lyons, T.; Manyika, J.; et al. The AI Index 2024 Annual Report; AI Index Steering Committee, Institute for Human-Centered AI, Stanford University: Stanford, CA, USA, 2024. [Google Scholar]
- Artificial Intelligence in Healthcare Market. 2024. Available online: https://www.fortunebusinessinsights.com/industry-reports/artificial-intelligence-in-healthcare-market-100534 (accessed on 22 August 2024).
- Artificial Intelligence in Healthcare: Market Size, Growth, and Trends. 2024. Available online: https://binariks.com/blog/artificial-intelligence-ai-healthcare-market/ (accessed on 22 August 2024).
- Park, H.A. Secure Telemedicine System. In Proceedings of the 2018 International Conference on Computational Science and Computational Intelligence (CSCI), Las Vegas, NV, USA, 12–14 December 2018; pp. 732–737. [Google Scholar] [CrossRef]
- Picozzi, P.; Nocco, U.; Puleo, G.; Labate, C.; Cimolin, V. Telemedicine and Robotic Surgery: A Narrative Review to Analyze Advantages, Limitations and Future Developments. Electronics 2023, 13, 124. [Google Scholar] [CrossRef]
- Bhattacharya, S.; Rawat, D. Comparative study of remote surgery techniques. In Proceedings of the 2015 IEEE Global Humanitarian Technology Conference (GHTC), Seattle, WA, USA, 8–11 October 2015; pp. 407–413. [Google Scholar] [CrossRef]
- Femminella, M.; Reali, G.; Valocchi, D.; Nunzi, E. The ARES Project: Network Architecture for Delivering and Processing Genomics Data. In Proceedings of the 2014 IEEE 3rd Symposium on Network Cloud Computing and Applications (ncca 2014), Rome, Italy, 5–7 February 2014; pp. 23–30. [Google Scholar] [CrossRef]
- Reali, G.; Femminella, M.; Nunzi, E.; Valocchi, D. Genomics as a service: A joint computing and networking perspective. Comput. Netw. 2018, 145, 27–51. [Google Scholar] [CrossRef]
- Holmgren, A.J.; Esdar, M.; Hüsers, J.; Coutinho-Almeida, J. Health Information Exchange: Understanding the Policy Landscape and Future of Data Interoperability. Yearb. Med. Inform. 2023, 32, 184–194. [Google Scholar] [CrossRef]
- Health Level Seven. Available online: https://www.hl7.org/ (accessed on 22 August 2024).
- Yetisen, A.K.; Martinez-Hurtado, J.L.; Ünal, B.; Khademhosseini, A.; Butt, H. Wearables in Medicine. Adv. Mater. 2018, 30, 1706910. [Google Scholar] [CrossRef] [PubMed]
- Nunzi, E.; Iorio, A.M.; Camilloni, B. A 21-winter seasons retrospective study of antibody response after influenza vaccination in elderly (60–85 years old) and very elderly (>85 years old) institutionalized subjects. Hum. Vaccines Immunother. 2017, 13, 2659–2668. [Google Scholar] [CrossRef][Green Version]
- Renga, G.; D’Onofrio, F.; Pariano, M.; Galarini, R.; Barola, C.; Stincardini, C.; Bellet, M.M.; Ellemunter, H.; Lass-Flörl, C.; Costantini, C.; et al. Bridging of host-microbiota tryptophan partitioning by the serotonin pathway in fungal pneumonia. Nat. Commun. 2023, 14, 5753. [Google Scholar] [CrossRef]
- Cellina, M.; Cè, M.; Alì, M.; Irmici, G.; Ibba, S.; Caloro, E.; Fazzini, D.; Oliva, G.; Papa, S. Digital Twins: The New Frontier for Personalized Medicine? Appl. Sci. 2023, 13, 7940. [Google Scholar] [CrossRef]
- Chataut, R.; Nankya, M.; Akl, R. 6G Networks and the AI Revolution—Exploring Technologies, Applications, and Emerging Challenges. Sensors 2024, 24, 1888. [Google Scholar] [CrossRef]
- Albawi, S.; Mohammed, T.A.; Al-Zawi, S. Understanding of a convolutional neural network. In Proceedings of the 2017 International Conference on Engineering and Technology (ICET), Antalya, Turkey, 21–23 August 2017; pp. 1–6. [Google Scholar] [CrossRef]
- International Atomic Energy Agency. Diagnostic Radiology Physics—A Handbook for Teachers and Students; Non-Serial Publications; IAEA: Wien, Austria, 2014; Available online: https://www.iaea.org/publications/8841/diagnostic-radiology-physics (accessed on 18 September 2024).
- Horowitz, S.L.; Pavlidis, T. Picture Segmentation by a Tree Traversal Algorithm. J. ACM 1976, 23, 368–388. [Google Scholar] [CrossRef]
- Fu, K.; Mui, J. A survey on image segmentation. Pattern Recognit. 1981, 13, 3–16. [Google Scholar] [CrossRef]
- Ghaheri, A.; Shoar, S.; Naderan, M.; Hoseini, S.S. The Applications of Genetic Algorithms in Medicine. Oman Med. J. 2015, 30, 406–416. [Google Scholar] [CrossRef] [PubMed]
- Ling, S.H.; Lam, H.K. Evolutionary Algorithms in Health Technologies. Algorithms 2019, 12, 202. [Google Scholar] [CrossRef]
- An, Q.; Rahman, S.; Zhou, J.; Kang, J.J. A Comprehensive Review on Machine Learning in Healthcare Industry: Classification, Restrictions, Opportunities and Challenges. Sensors 2023, 23, 4178. [Google Scholar] [CrossRef] [PubMed]
- Langarizadeh, M.; Moghbeli, F. Applying Naive Bayesian Networks to Disease Prediction: A Systematic Review. Acta Inform. Medica 2016, 24, 364. [Google Scholar] [CrossRef]
- McLachlan, S.; Dube, K.; Hitman, G.A.; Fenton, N.E.; Kyrimi, E. Bayesian networks in healthcare: Distribution by medical condition. Artif. Intell. Med. 2020, 107, 101912. [Google Scholar] [CrossRef]
- Kyrimi, E.; Dube, K.; Fenton, N.; Fahmi, A.; Neves, M.R.; Marsh, W.; McLachlan, S. Bayesian networks in healthcare: What is preventing their adoption? Artif. Intell. Med. 2021, 116, 102079. [Google Scholar] [CrossRef]
- QIIME2. Available online: https://qiime2.org/ (accessed on 22 August 2024).
- Nextflow. Available online: https://www.nextflow.org/ (accessed on 22 August 2024).
- Tian, Y.; Gou, W.; Ma, Y.; Shuai, M.; Liang, X.; Fu, Y.; Zheng, J.S. The Short-Term Variation of Human Gut Mycobiome in Response to Dietary Intervention of Different Macronutrient Distributions. Nutrients 2023, 15, 2152. [Google Scholar] [CrossRef] [PubMed]
- Costantini, C.; Nunzi, E.; Spolzino, A.; Merli, F.; Facchini, L.; Spadea, A.; Melillo, L.; Codeluppi, K.; Marchesi, F.; Marchesini, G.; et al. A High-Risk Profile for Invasive Fungal Infections Is Associated with Altered Nasal Microbiota and Niche Determinants. Infect. Immun. 2022, 90, e00048-22. [Google Scholar] [CrossRef]
- Nunzi, E.; Mezzasoma, L.; Bellezza, I.; Zelante, T.; Orvietani, P.; Coata, G.; Giardina, I.; Sagini, K.; Manni, G.; Di Michele, A.; et al. Microbiota-Associated HAF-EVs Regulate Monocytes by Triggering or Inhibiting Inflammasome Activation. Int. J. Mol. Sci. 2023, 24, 2527. [Google Scholar] [CrossRef]
- Di Tommaso, P.; Chatzou, M.; Floden, E.W.; Barja, P.P.; Palumbo, E.; Notredame, C. Nextflow enables reproducible computational workflows. Nat. Biotechnol. 2017, 35, 316–319. [Google Scholar] [CrossRef]
- Langer, B.E.; Amaral, A.; Baudement, M.O.; Bonath, F.; Charles, M.; Chitneedi, P.K.; Clark, E.L.; Di Tommaso, P.; Djebali, S.; Ewels, P.A.; et al. Empowering bioinformatics communities with Nextflow and nf-core. bioRxiv 2024. [Google Scholar] [CrossRef]
- Dubey, S.R.; Singh, S.K.; Chaudhuri, B.B. Activation Functions in Deep Learning: A Comprehensive Survey and Benchmark. arXiv 2021, arXiv:2109.14545. [Google Scholar] [CrossRef]
- Benedetti, P.; Femminella, M.; Reali, G. Mixed-Sized Biomedical Image Segmentation Based on U-Net Architectures. Appl. Sci. 2022, 13, 329. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. arXiv 2015, arXiv:1505.04597. [Google Scholar]
- Wang, G.; Li, W.; Zuluaga, M.; Aughwane, R.; Patel, P.; Aertsen, M.; Doel, T.; David, A.; Deprest, J.; Ourselin, S.; et al. Interactive Medical Image Segmentation Using Deep Learning With Image-Specific Fine Tuning. IEEE Trans. Med. Imaging 2018, 37, 1562–1573. [Google Scholar] [CrossRef]
- Wang, G.; Zuluaga, M.A.; Li, W.; Pratt, R.; Patel, P.A.; Aertsen, M.; Doel, T.; David, A.L.; Deprest, J.A.; Ourselin, S.; et al. DeepIGeoS: A Deep Interactive Geodesic Framework for Medical Image Segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2019, 41, 1559–1572. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. In Proceedings of the 3rd International Conference on Learning Representations, ICLR 2015, San Diego, CA, USA, 7–9 May 2015. [Google Scholar]
- Çiçek, O.; Abdulkadir, A.; Lienkamp, S.S.; Brox, T.; Ronneberger, O. 3D U-Net: Learning Dense Volumetric Segmentation from Sparse Annotation. arXiv 2016, arXiv:1606.06650. [Google Scholar] [CrossRef]
- An End-to-End Platform for Machine Learning. 2024. Available online: https://www.tensorflow.org/?hl=en (accessed on 6 September 2024).
- Pytorch. 2024. Available online: https://pytorch.org/ (accessed on 6 September 2024).
- Cuda Toolkit. 2024. Available online: https://developer.nvidia.com/cuda-toolkit (accessed on 6 September 2024).
- Scarselli, F.; Gori, M.; Tsoi, A.C.; Hagenbuchner, M.; Monfardini, G. The Graph Neural Network Model. IEEE Trans. Neural Netw. 2009, 20, 61–80. [Google Scholar] [CrossRef]
- Yan, Y.; He, S.; Yu, Z.; Yuan, J.; Liu, Z.; Chen, Y. Investigation of Customized Medical Decision Algorithms Utilizing Graph Neural Networks. arXiv 2024, arXiv:2405.17460. [Google Scholar] [CrossRef]
- Ahmedt-Aristizabal, D.; Armin, M.A.; Denman, S.; Fookes, C.; Petersson, L. Graph-Based Deep Learning for Medical Diagnosis and Analysis: Past, Present and Future. Sensors 2021, 21, 4758. [Google Scholar] [CrossRef]
- Meng, X.; Zou, T. Clinical applications of graph neural networks in computational histopathology: A review. Comput. Biol. Med. 2023, 164, 107201. [Google Scholar] [CrossRef] [PubMed]
- Saxena, R.R.; Saxena, R. Applying Graph Neural Networks in Pharmacology. TechRxiv 2024. [Google Scholar] [CrossRef]
- Paul, S.G.; Saha, A.; Hasan, M.Z.; Noori, S.R.H.; Moustafa, A. A Systematic Review of Graph Neural Network in Healthcare-Based Applications: Recent Advances, Trends, and Future Directions. IEEE Access 2024, 12, 15145–15170. [Google Scholar] [CrossRef]
- Zhang, S.; Tong, H.; Xu, J.; Maciejewski, R. Graph convolutional networks: A comprehensive review. Comput. Soc. Netw. 2019, 6, 11. [Google Scholar] [CrossRef]
- Hamilton, W.L.; Ying, R.; Leskovec, J. Inductive Representation Learning on Large Graphs. arXiv 2017, arXiv:1706.02216. [Google Scholar] [CrossRef]
- Cai, C.; Wang, D.; Wang, Y. Graph Coarsening with Neural Networks. arXiv 2021, arXiv:2102.01350. [Google Scholar] [CrossRef]
- Veličković, P.; Cucurull, G.; Casanova, A.; Romero, A.; Liò, P.; Bengio, Y. Graph Attention Networks. arXiv 2017, arXiv:1710.10903. [Google Scholar] [CrossRef]
- PyTorch Geometric. Available online: https://pytorch-geometric.readthedocs.io/en/latest/ (accessed on 22 August 2024).
- Deep Graph Library. Available online: https://www.dgl.ai/ (accessed on 22 August 2024).
- Graph Nets. Available online: https://github.com/google-deepmind/graph_nets (accessed on 22 August 2024).
- Spektral. Available online: https://graphneural.network/ (accessed on 22 August 2024).
- StellarGraph. Available online: https://stellargraph.readthedocs.io/en/stable/ (accessed on 22 August 2024).
- Lipton, Z.C.; Berkowitz, J.; Elkan, C. A Critical Review of Recurrent Neural Networks for Sequence Learning. arXiv 2015, arXiv:1506.00019. [Google Scholar] [CrossRef]
- Al-Askar, H.; Radi, N.; MacDermott, A. Recurrent Neural Networks in Medical Data Analysis and Classifications. In Applied Computing in Medicine and Health; Elsevier: Amsterdam, The Netherlands, 2016; pp. 147–165. [Google Scholar] [CrossRef]
- Al Olaimat, M.; Bozdag, S.; for the Alzheimer’s Disease Neuroimaging Initiative. TA-RNN: An attention-based time-aware recurrent neural network architecture for electronic health records. Bioinformatics 2024, 40, i169–i179. [Google Scholar] [CrossRef]
- Choi, E.; Schuetz, A.; Stewart, W.F.; Sun, J. Using recurrent neural network models for early detection of heart failure onset. J. Am. Med. Inform. Assoc. 2017, 24, 361–370. [Google Scholar] [CrossRef]
- Riasi, A.; Delrobaei, M.; Salari, M. A decision support system based on recurrent neural networks to predict medication dosage for patients with Parkinson’s disease. Sci. Rep. 2024, 14, 8424. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.J.; Ortiz-Soriano, V.; Neyra, J.A.; Chen, J. KIT-LSTM: Knowledge-guided Time-aware LSTM for Continuous Clinical Risk Prediction. medRxiv 2022. [Google Scholar] [CrossRef]
- Julie, G.; Jaisakthi, S.M.; Robinson, Y.H. (Eds.) Handbook of Deep Learning in Biomedical Engineering and Health Informatics, 1st ed.; Apple Academic Press Inc.: Palm Bay, FL, USA; CRC Press: Boca Raton, FL, USA, 2022; OCLC: 1237707833. [Google Scholar]
- Saha, A.; Samaan, M.; Peng, B.; Ning, X. A Multi-Layered GRU Model for COVID-19 Patient Representation and Phenotyping from Large-Scale EHR Data. In Proceedings of the 14th ACM International Conference on Bioinformatics, Computational Biology, and Health Informatics, Houston, TX, USA, 3–6 September 2023; pp. 1–6. [Google Scholar] [CrossRef]
- Kiser, A.C.; Eilbeck, K.; Bucher, B.T. Developing an LSTM Model to Identify Surgical Site Infections using Electronic Healthcare Records. AMIA Summits Transl. Sci. Proc. 2023, 2023, 330–339. [Google Scholar]
- Hochreiter, S.; Schmidhuber, J. Long Short-Term Memory. Neural Comput. 1997, 9, 1735–1780. [Google Scholar] [CrossRef]
- Cho, K.; van Merrienboer, B.; Gulcehre, C.; Bahdanau, D.; Bougares, F.; Schwenk, H.; Bengio, Y. Learning Phrase Representations Using RNN Encoder-Decoder for Statistical Machine Translation. arXiv 2014, arXiv:1406.1078. [Google Scholar] [CrossRef]
- Apache MxNet. Available online: https://mxnet.apache.org/versions/1.9.1/ (accessed on 22 August 2024).
- Microsoft Cognitive Toolkit. Available online: https://cntk.ai (accessed on 22 August 2024).
- Chainer. Available online: https://chainer.org/ (accessed on 22 August 2024).
- Rayan, A.; Holyl Alruwaili, S.; Alaerjan, A.S.; Alanazi, S.; Taloba, A.I.; Shahin, O.R.; Salem, M. Utilizing CNN-LSTM techniques for the enhancement of medical systems. Alex. Eng. J. 2023, 72, 323–338. [Google Scholar] [CrossRef]
- Liu, Y.; Song, Z.; Xu, X.; Rafique, W.; Zhang, X.; Shen, J.; Khosravi, M.R.; Qi, L. Bidirectional GRU networks-based next POI category prediction for healthcare. Int. J. Intell. Syst. 2022, 37, 4020–4040. [Google Scholar] [CrossRef]
- Chen, Y.; Esmaeilzadeh, P. Generative AI in Medical Practice: In-Depth Exploration of Privacy and Security Challenges. J. Med. Internet Res. 2024, 26, e53008. [Google Scholar] [CrossRef]
- Blease, C.; Torous, J.; McMillan, B.; Hägglund, M.; Mandl, K.D. Generative Language Models and Open Notes: Exploring the Promise and Limitations. JMIR Med. Educ. 2024, 10, e51183. [Google Scholar] [CrossRef]
- Biswas, A.; Talukdar, W. Intelligent Clinical Documentation: Harnessing Generative AI for Patient-Centric Clinical Note Generation. Int. J. Innov. Sci. Res. Technol. (IJISRT) 2024, 9, 994–1008. [Google Scholar] [CrossRef]
- Jabbar, A.; Li, X.; Omar, B. A Survey on Generative Adversarial Networks: Variants, Applications, and Training. arXiv 2020, arXiv:2006.05132. [Google Scholar] [CrossRef]
- Gonzalez-Abril, L.; Angulo, C.; Ortega, J.A.; Lopez-Guerra, J.L. Generative Adversarial Networks for Anonymized Healthcare of Lung Cancer Patients. Electronics 2021, 10, 2220. [Google Scholar] [CrossRef]
- Vaccari, I.; Orani, V.; Paglialonga, A.; Cambiaso, E.; Mongelli, M. A Generative Adversarial Network (GAN) Technique for Internet of Medical Things Data. Sensors 2021, 21, 3726. [Google Scholar] [CrossRef]
- Abedi, M.; Hempel, L.; Sadeghi, S.; Kirsten, T. GAN-Based Approaches for Generating Structured Data in the Medical Domain. Appl. Sci. 2022, 12, 7075. [Google Scholar] [CrossRef]
- Karras, T.; Laine, S.; Aila, T. A Style-Based Generator Architecture for Generative Adversarial Networks. arXiv 2018, arXiv:1812.04948. [Google Scholar] [CrossRef]
- Singh, A.; Ogunfunmi, T. An Overview of Variational Autoencoders for Source Separation, Finance, and Bio-Signal Applications. Entropy 2021, 24, 55. [Google Scholar] [CrossRef]
- Zemouri, R.; Levesque, M.; Boucher, E.; Kirouac, M.; Lafleur, F.; Bernier, S.; Merkhouf, A. Recent Research and Applications in Variational Autoencoders for Industrial Prognosis and Health Management: A Survey. In Proceedings of the 2022 Prognostics and Health Management Conference (PHM-2022 London), London, UK, 7–29 May 2022; pp. 193–203. [Google Scholar] [CrossRef]
- Chen, S.; Guo, W. Auto-Encoders in Deep Learning—A Review with New Perspectives. Mathematics 2023, 11, 1777. [Google Scholar] [CrossRef]
- Bu, Y.; Zou, S.; Liang, Y.; Veeravalli, V.V. Estimation of KL Divergence: Optimal Minimax Rate. IEEE Trans. Inf. Theory 2018, 64, 2648–2674. [Google Scholar] [CrossRef]
- Zhu, F.; Ye, F.; Fu, Y.; Liu, Q.; Shen, B. Electrocardiogram generation with a bidirectional LSTM-CNN generative adversarial network. Sci. Rep. 2019, 9, 6734. [Google Scholar] [CrossRef] [PubMed]
- Morid, M.A.; Sheng, O.R.L.; Dunbar, J. Time Series Prediction Using Deep Learning Methods in Healthcare. ACM Trans. Manag. Inf. Syst. 2023, 14, 1–29. [Google Scholar] [CrossRef]
- Liao, Y.; Liu, H.; Spasić, I. Deep learning approaches to automatic radiology report generation: A systematic review. Inform. Med. Unlocked 2023, 39, 101273. [Google Scholar] [CrossRef]
- Hripcsak, G.; Austin, J.H.M.; Alderson, P.O.; Friedman, C. Use of Natural Language Processing to Translate Clinical Information from a Database of 889,921 Chest Radiographic Reports. Radiology 2002, 224, 157–163. [Google Scholar] [CrossRef]
- Hossain, E.; Rana, R.; Higgins, N.; Soar, J.; Barua, P.D.; Pisani, A.R.; Turner, K. Natural Language Processing in Electronic Health Records in relation to healthcare decision-making: A systematic review. Comput. Biol. Med. 2023, 155, 106649. [Google Scholar] [CrossRef]
- Luo, R.; Sun, L.; Xia, Y.; Qin, T.; Zhang, S.; Poon, H.; Liu, T.Y. BioGPT: Generative Pre-trained Transformer for Biomedical Text Generation and Mining. arXiv 2022, arXiv:2210.10341. [Google Scholar] [CrossRef]
- Yenduri, G.; Ramalingam, M.; Selvi, G.C.; Supriya, Y.; Srivastava, G.; Maddikunta, P.K.R.; Raj, G.D.; Jhaveri, R.H.; Prabadevi, B.; Wang, W.; et al. GPT (Generative Pre-Trained Transformer)— A Comprehensive Review on Enabling Technologies, Potential Applications, Emerging Challenges, and Future Directions. IEEE Access 2024, 12, 54608–54649. [Google Scholar] [CrossRef]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, L.; Polosukhin, I. Attention Is All You Need. arXiv 2017, arXiv:1706.03762. [Google Scholar] [CrossRef]
- Devlin, J.; Chang, M.W.; Lee, K.; Toutanova, K. BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding. arXiv 2018, arXiv:1810.04805. [Google Scholar] [CrossRef]
- Wang, Y.; Pan, Y.; Yan, M.; Su, Z.; Luan, T.H. A Survey on ChatGPT: AI–Generated Contents, Challenges, and Solutions. IEEE Open J. Comput. Soc. 2023, 4, 280–302. [Google Scholar] [CrossRef]
- Mastropaolo, A.; Scalabrino, S.; Cooper, N.; Nader Palacio, D.; Poshyvanyk, D.; Oliveto, R.; Bavota, G. Studying the Usage of Text-To-Text Transfer Transformer to Support Code-Related Tasks. In Proceedings of the 2021 IEEE/ACM 43rd International Conference on Software Engineering (ICSE), Madrid, Spain, 22–30 May 2021; pp. 336–347. [Google Scholar] [CrossRef]
- Chen, X.; Pun, C.M.; Wang, S. MedPrompt: Cross-Modal Prompting for Multi-Task Medical Image Translation. arXiv 2023, arXiv:2310.02663. [Google Scholar] [CrossRef]
- Chen, Z.; Cano, A.H.; Romanou, A.; Bonnet, A.; Matoba, K.; Salvi, F.; Pagliardini, M.; Fan, S.; Köpf, A.; Mohtashami, A.; et al. MEDITRON-70B: Scaling Medical Pretraining for Large Language Models. arXiv 2023, arXiv:2311.16079. [Google Scholar] [CrossRef]
- Cao, H.; Tan, C.; Gao, Z.; Xu, Y.; Chen, G.; Heng, P.A.; Li, S.Z. A Survey on Generative Diffusion Models. IEEE Trans. Knowl. Data Eng. 2024, 36, 2814–2830. [Google Scholar] [CrossRef]
- Rombach, R.; Blattmann, A.; Lorenz, D.; Esser, P.; Ommer, B. High-Resolution Image Synthesis with Latent Diffusion Models. In Proceedings of the 2022 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), New Orleans, LA, USA, 18–24 June 2022; pp. 10674–10685. [Google Scholar] [CrossRef]
- Croitoru, F.A.; Hondru, V.; Ionescu, R.T.; Shah, M. Diffusion Models in Vision: A Survey. IEEE Trans. Pattern Anal. Mach. Intell. 2023, 45, 10850–10869. [Google Scholar] [CrossRef]
- Alakhdar, A.; Poczos, B.; Washburn, N. Diffusion Models in De Novo Drug Design. arXiv 2024, arXiv:2406.08511. [Google Scholar] [CrossRef]
- Diffusers. Available online: https://huggingface.co/docs/diffusers/index (accessed on 25 August 2024).
- NVIDIA Clara. Available online: https://docs.nvidia.com/clara/index.html (accessed on 25 August 2024).
- Cherstvy, A.G.; Thapa, S.; Wagner, C.E.; Metzler, R. Non-Gaussian, non-ergodic, and non-Fickian diffusion of tracers in mucin hydrogels. Soft Matter 2019, 15, 2526–2551. [Google Scholar] [CrossRef]
- Al-Hamadani, M.; Fadhel, M.; Alzubaidi, L.; Harangi, B. Reinforcement Learning Algorithms and Applications in Healthcare and Robotics: A Comprehensive and Systematic Review. Sensors 2024, 24, 2461. [Google Scholar] [CrossRef]
- Kaelbling, L.P.; Littman, M.L.; Moore, A.W. Reinforcement learning: A survey. J. Artif. Int. Res. 1996, 4, 237–285. [Google Scholar] [CrossRef]
- Shakya, A.K.; Pillai, G.; Chakrabarty, S. Reinforcement learning algorithms: A brief survey. Expert Syst. Appl. 2023, 231, 120495. [Google Scholar] [CrossRef]
- Mnih, V.; Kavukcuoglu, K.; Silver, D.; Graves, A.; Antonoglou, I.; Wierstra, D.; Riedmiller, M. Playing Atari with Deep Reinforcement Learning. arXiv 2013, arXiv:1312.5602. [Google Scholar] [CrossRef]
- Mnih, V.; Kavukcuoglu, K.; Silver, D.; Rusu, A.A.; Veness, J.; Bellemare, M.G.; Graves, A.; Riedmiller, M.; Fidjeland, A.K.; Ostrovski, G.; et al. Human-level control through deep reinforcement learning. Nature 2015, 518, 529–533. [Google Scholar] [CrossRef]
- Schulman, J.; Wolski, F.; Dhariwal, P.; Radford, A.; Klimov, O. Proximal Policy Optimization Algorithms. arXiv 2017, arXiv:1707.06347. [Google Scholar] [CrossRef]
- Mnih, V.; Badia, A.P.; Mirza, M.; Graves, A.; Lillicrap, T.; Harley, T.; Silver, D.; Kavukcuoglu, K. Asynchronous Methods for Deep Reinforcement Learning. In Proceedings of the 33rd International Conference on Machine Learning, New York, NY, USA, 20–22 June 2016; Volume 48, pp. 1928–1937. [Google Scholar]
- Coronato, A.; Naeem, M.; De Pietro, G.; Paragliola, G. Reinforcement learning for intelligent healthcare applications: A survey. Artif. Intell. Med. 2020, 109, 101964. [Google Scholar] [CrossRef]
- Yu, C.; Liu, J.; Nemati, S. Reinforcement Learning in Healthcare: A Survey. arXiv 2019, arXiv:1908.08796. [Google Scholar] [CrossRef]
- Eckardt, J.N.; Wendt, K.; Bornhäuser, M.; Middeke, J.M. Reinforcement Learning for Precision Oncology. Cancers 2021, 13, 4624. [Google Scholar] [CrossRef] [PubMed]
- OpenAI. Gymnasium—An API Standard for Reinforcement Learning with a Diverse Collection of Reference Environments. 2024. Available online: https://gymnasium.farama.org/index.html (accessed on 6 September 2024).
- Stable Baselines3. Stable-Baselines3 Docs - Reliable Reinforcement Learning Implementations. 2017. Available online: https://stable-baselines3.readthedocs.io/en/master/ (accessed on 14 July 2020).
- Li, X.; Qian, W.; Xu, D.; Liu, C. Image Segmentation Based on Improved Unet. J. Phys. Conf. Ser. 2021, 1815, 012018. [Google Scholar] [CrossRef]
- Lu, H.; She, Y.; Tie, J.; Xu, S. Half-UNet: A Simplified U-Net Architecture for Medical Image Segmentation. Front. Neuroinform. 2022, 16, 911679. [Google Scholar] [CrossRef]
- Siddique, N.; Paheding, S.; Elkin, C.P.; Devabhaktuni, V. U-Net and Its Variants for Medical Image Segmentation: A Review of Theory and Applications. IEEE Access 2021, 9, 82031–82057. [Google Scholar] [CrossRef]
- Gao, Y.; Huang, R.; Chen, M.; Wang, Z.; Deng, J.; Chen, Y.; Yang, Y.; Zhang, J.; Tao, C.; Li, H. FocusNet: Imbalanced Large and Small Organ Segmentation with an End-to-End Deep Neural Network for Head and Neck CT Images. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2019, Proceedings of the 22nd International Conference, Shenzhen, China, 13–17 October 2019; Proceedings, Part III; Springer: Berlin/Heidelberg, Germany, 2019; pp. 829–838. [Google Scholar] [CrossRef]
- Valindria, V.V.; Lavdas, I.; Cerrolaza, J.J.; Aboagye, E.O.; Rockall, A.G.; Rueckert, D.; Glocker, B. Small Organ Segmentation in Whole-body MRI using a Two-stage FCN and Weighting Schemes. In Proceedings of the MLMI@MICCAI, Granada, Spain, 16 September 2018. [Google Scholar]
- Saood, A.; Hatem, I. COVID-19 lung CT image segmentation using deep learning methods: U-Net versus SegNet. BMC Med. Imaging 2021, 21, 19. [Google Scholar] [CrossRef]
- Han, G.; Zhang, M.; Wu, W.; He, M.; Liu, K.; Qin, L.; Liu, X. Improved U-Net based insulator image segmentation method based on attention mechanism. Energy Rep. 2021, 7, 210–217. [Google Scholar] [CrossRef]
- Fujima, N.; Sakashita, T.; Homma, A.; Harada, T.; Shimizu, Y.; Tha, K.K.; Kudo, K.; Shirato, H. Non-invasive prediction of the tumor growth rate using advanced diffusion models in head and neck squamous cell carcinoma patients. Oncotarget 2017, 8, 33631–33643. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, K.; Ren, Y.; Ma, L.; Fan, Y.; Yang, Z.; Yang, Q.; Shi, J.; Sun, Y. Deep learning-based prediction of treatment prognosis from nasal polyp histology slides. Int. Forum Allergy Rhinol. 2023, 13, 886–898. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Chen, L.; Cai, Y.; Wu, D. Hierarchical Graph Neural Network for Patient Treatment Preference Prediction with External Knowledge. In Advances in Knowledge Discovery and Data Mining; Kashima, H., Ide, T., Peng, W.C., Eds.; Series Title: Lecture Notes in Computer Science; Springer Nature: Cham, Switzerland, 2023; Volume 13937, pp. 204–215. [Google Scholar] [CrossRef]
- He, H.; Zhao, S.; Xi, Y.; Ho, J.C. MedDiff: Generating Electronic Health Records using Accelerated Denoising Diffusion Model. arXiv 2023, arXiv:2302.04355. [Google Scholar] [CrossRef]
- Ashton, J.J.; Young, A.; Johnson, M.J.; Beattie, R.M. Using machine learning to impact on long-term clinical care: Principles, challenges, and practicalities. Pediatr. Res. 2023, 93, 324–333. [Google Scholar] [CrossRef] [PubMed]
- Proios, D.; Yazdani, A.; Bornet, A.; Ehrsam, J.; Rekik, I.; Teodoro, D. Leveraging patient similarities via graph neural networks to predict phenotypes from temporal data. In Proceedings of the 2023 IEEE 10th International Conference on Data Science and Advanced Analytics (DSAA), Thessaloniki, Greece, 9–13 October 2023; pp. 1–10. [Google Scholar] [CrossRef]
- Whiles, B.B.; Bird, V.G.; Canales, B.K.; DiBianco, J.M.; Terry, R.S. Caution! AI Bot Has Entered the Patient Chat: ChatGPT Has Limitations in Providing Accurate Urologic Healthcare Advice. Urology 2023, 180, 278–284. [Google Scholar] [CrossRef] [PubMed]
- Ceritli, T.; Ghosheh, G.O.; Chauhan, V.K.; Zhu, T.; Creagh, A.P.; Clifton, D.A. Synthesizing Mixed-type Electronic Health Records using Diffusion Models. arXiv 2023, arXiv:2302.14679. [Google Scholar] [CrossRef]
- Liu, S.; See, K.C.; Ngiam, K.Y.; Celi, L.A.; Sun, X.; Feng, M. Reinforcement Learning for Clinical Decision Support in Critical Care: Comprehensive Review. J. Med. Internet Res. 2020, 22, e18477. [Google Scholar] [CrossRef]
- Wu, W.; Zhang, J.; Xie, H.; Zhao, Y.; Zhang, S.; Gu, L. Automatic detection of coronary artery stenosis by convolutional neural network with temporal constraint. Comput. Biol. Med. 2020, 118, 103657. [Google Scholar] [CrossRef]
- Jun, T.J.; Kweon, J.; Kim, Y.H.; Kim, D. T-Net: Nested encoder–decoder architecture for the main vessel segmentation in coronary angiography. Neural Netw. 2020, 128, 216–233. [Google Scholar] [CrossRef]
- Moon, S.; Zhung, W.; Yang, S.; Lim, J.; Kim, W.Y. PIGNet: A physics-informed deep learning model toward generalized drug–target interaction predictions. Chem. Sci. 2022, 13, 3661–3673. [Google Scholar] [CrossRef]
- Kazerouni, A.; Aghdam, E.K.; Heidari, M.; Azad, R.; Fayyaz, M.; Hacihaliloglu, I.; Merhof, D. Diffusion models in medical imaging: A comprehensive survey. Med. Image Anal. 2023, 88, 102846. [Google Scholar] [CrossRef]
- Hung, A.L.Y.; Zhao, K.; Zheng, H.; Yan, R.; Raman, S.S.; Terzopoulos, D.; Sung, K. Med-cDiff: Conditional Medical Image Generation with Diffusion Models. Bioengineering 2023, 10, 1258. [Google Scholar] [CrossRef]
- Malik, A.; Solaiman, B. AI in hospital administration and management: Ethical and legal implications. In Research Handbook on Health, AI and the Law; Solaiman, B., Cohen, I.G., Eds.; Edward Elgar Publishing: Cheltenham, UK, 2024; pp. 21–40. [Google Scholar] [CrossRef]
- Pham, T.; Tao, X.; Zhang, J.; Yong, J.; Zhou, X.; Gururajan, R. MeKG: Building a Medical Knowledge Graph by Data Mining from MEDLINE. In Brain Informatics; Liang, P., Goel, V., Shan, C., Eds.; Series Title: Lecture Notes in Computer Science; Springer International Publishing: Cham, Switzerland, 2019; Volume 11976, pp. 159–168. [Google Scholar] [CrossRef]
- Chen, Z.; Dou, Y.; De Causmaecker, P. Neural networked-assisted method for the nurse rostering problem. Comput. Ind. Eng. 2022, 171, 108430. [Google Scholar] [CrossRef]
- Sai, S.; Gaur, A.; Sai, R.; Chamola, V.; Guizani, M.; Rodrigues, J.J.P.C. Generative AI for Transformative Healthcare: A Comprehensive Study of Emerging Models, Applications, Case Studies, and Limitations. IEEE Access 2024, 12, 31078–31106. [Google Scholar] [CrossRef]
- Wang, C.; Li, X.; Ma, W.; Wang, X. Diffusion models over the life cycle of an innovation: A bottom-up and top-down synthesis approach. Public Adm. Dev. 2020, 40, 105–118. [Google Scholar] [CrossRef]
- Yu, C.; Liu, J.; Nemati, S.; Yin, G. Reinforcement Learning in Healthcare: A Survey. ACM Comput. Surv. 2023, 55, 1–36. [Google Scholar] [CrossRef]
- Kim, D.; Lee, J.; Woo, Y.; Jeong, J.; Kim, C.; Kim, D.K. Deep Learning Application to Clinical Decision Support System in Sleep Stage Classification. J. Pers. Med. 2022, 12, 136. [Google Scholar] [CrossRef]
- Diaz Ochoa, J.G.; Mustafa, F.E. Graph neural network modelling as a potentially effective method for predicting and analyzing procedures based on patients’ diagnoses. Artif. Intell. Med. 2022, 131, 102359. [Google Scholar] [CrossRef] [PubMed]
- Das Swain, V.; Saha, K. Teacher, Trainer, Counsel, Spy: How Generative AI can Bridge or Widen the Gaps in Worker-Centric Digital Phenotyping of Wellbeing. In Proceedings of the 3rd Annual Meeting of the Symposium on Human-Computer Interaction for Work, Newcastle upon Tyne, UK, 25–27 June 2024; pp. 1–13. [Google Scholar] [CrossRef]
- Liu, L.; Zhao, K. Report on Methods and Applications for Crafting 3D Humans. arXiv 2024, arXiv:2406.01223. [Google Scholar] [CrossRef]
- Pike, A.C.; Robinson, O.J. Reinforcement Learning in Patients With Mood and Anxiety Disorders vs Control Individuals: A Systematic Review and Meta-analysis. JAMA Psychiatry 2022, 79, 313. [Google Scholar] [CrossRef]
- Wang, X.; Abubaker, S.M.; Babalola, G.T.; Tulk Jesso, S. Co-Designing an AI Chatbot to Improve Patient Experience in the Hospital: A human-centered design case study of a collaboration between a hospital, a university, and ChatGPT. In Proceedings of the Extended Abstracts of the CHI Conference on Human Factors in Computing Systems, Honolulu, HI, USA, 11–16 May 2024; pp. 1–10. [Google Scholar] [CrossRef]
- Qu, J.; Zhang, Y.; Tang, W.; Cheng, W.; Zhang, Y.; Bu, L. Developing a virtual reality healthcare product based on data-driven concepts: A case study. Adv. Eng. Inform. 2023, 57, 102118. [Google Scholar] [CrossRef]
- Balcombe, L.; De Leo, D. Human-Computer Interaction in Digital Mental Health. Informatics 2022, 9, 14. [Google Scholar] [CrossRef]
- Priya, K.V.; Dinesh Peter, J. Enhanced Defensive Model Using CNN against Adversarial Attacks for Medical Education through Human Computer Interaction. Int. J. Hum.—Comput. Interact. 2023, 1–13. [Google Scholar] [CrossRef]
- Slegers, K. Human–Computer Interaction. In The International Encyclopedia of Health Communication, 1st ed.; Ho, E.Y., Bylund, C.L., Van Weert, J.C.M., Eds.; Wiley: Hoboken, NJ, USA, 2022; pp. 1–7. [Google Scholar] [CrossRef]
- Moini, J.; Akinso, O.; Ferdowsi, K.; Moini, M. The role of computers in health care. In Health Care Today in the United States; Elsevier: Amsterdam, The Netherlands, 2023; pp. 485–498. [Google Scholar] [CrossRef]
- Reddy, S. Explainability and artificial intelligence in medicine. Lancet Digit. Health 2022, 4, e214–e215. [Google Scholar] [CrossRef] [PubMed]
- Beger, J. The crucial role of explainability in healthcare AI. Eur. J. Radiol. 2024, 176, 111507. [Google Scholar] [CrossRef]
- The Precise4Q consortium; Amann, J.; Blasimme, A.; Vayena, E.; Frey, D.; Madai, V.I. Explainability for artificial intelligence in healthcare: A multidisciplinary perspective. BMC Med. Inform. Decis. Mak. 2020, 20, 310. [Google Scholar] [CrossRef]
- Chaddad, A.; Peng, J.; Xu, J.; Bouridane, A. Survey of Explainable AI Techniques in Healthcare. Sensors 2023, 23, 634. [Google Scholar] [CrossRef] [PubMed]
- Sadeghi, Z.; Alizadehsani, R.; Cifci, M.A.; Kausar, S.; Rehman, R.; Mahanta, P.; Bora, P.K.; Almasri, A.; Alkhawaldeh, R.S.; Hussain, S.; et al. A review of Explainable Artificial Intelligence in healthcare. Comput. Electr. Eng. 2024, 118, 109370. [Google Scholar] [CrossRef]
- Frasca, M.; La Torre, D.; Pravettoni, G.; Cutica, I. Explainable and interpretable artificial intelligence in medicine: A systematic bibliometric review. Discov. Artif. Intell. 2024, 4, 15. [Google Scholar] [CrossRef]
- Shajari, S.; Kuruvinashetti, K.; Komeili, A.; Sundararaj, U. The Emergence of AI-Based Wearable Sensors for Digital Health Technology: A Review. Sensors 2023, 23, 9498. [Google Scholar] [CrossRef]
- Wang, C.; He, T.; Zhou, H.; Zhang, Z.; Lee, C. Artificial intelligence enhanced sensors - enabling technologies to next-generation healthcare and biomedical platform. Bioelectron. Med. 2023, 9, 17. [Google Scholar] [CrossRef]
- Katsoulakis, E.; Wang, Q.; Wu, H.; Shahriyari, L.; Fletcher, R.; Liu, J.; Achenie, L.; Liu, H.; Jackson, P.; Xiao, Y.; et al. Digital twins for health: A scoping review. NPJ Digit. Med. 2024, 7, 77. [Google Scholar] [CrossRef]
- Johnson, Z.; Saikia, M.J. Digital Twins for Healthcare Using Wearables. Bioengineering 2024, 11, 606. [Google Scholar] [CrossRef] [PubMed]
- Keshta, I. AI-driven IoT for smart health care: Security and privacy issues. Inform. Med. Unlocked 2022, 30, 100903. [Google Scholar] [CrossRef]
- Wang, C.; Zhang, J.; Lassi, N.; Zhang, X. Privacy Protection in Using Artificial Intelligence for Healthcare: Chinese Regulation in Comparative Perspective. Healthcare 2022, 10, 1878. [Google Scholar] [CrossRef]
- Selvanambi, R.; Bhutani, S.; Veauli, K. Security and Privacy for Electronic Healthcare Records Using AI in Blockchain. In Research Anthology on Convergence of Blockchain, Internet of Things, and Security; IGI Global: Hershey, PA, USA, 2022; pp. 768–777. [Google Scholar] [CrossRef]
- Taherdoost, H. Privacy and Security of Blockchain in Healthcare: Applications, Challenges, and Future Perspectives. Sci 2023, 5, 41. [Google Scholar] [CrossRef]
- Ahad, A.; Ali, Z.; Mateen, A.; Tahir, M.; Hannan, A.; Garcia, N.M.; Pires, I.M. A Comprehensive review on 5G-based Smart Healthcare Network Security: Taxonomy, Issues, Solutions and Future research directions. Array 2023, 18, 100290. [Google Scholar] [CrossRef]
- Abir, S.M.A.A.; Abuibaid, M.; Huang, J.S.; Hong, Y. Harnessing 5G Networks for Health Care: Challenges and Potential Applications. In Proceedings of the 2023 International Conference on Smart Applications, Communications and Networking (SmartNets), Istanbul, Turkiye, 25–27 July 2023; pp. 1–6. [Google Scholar] [CrossRef]
- Pradhan, B.; Das, S.; Roy, D.S.; Routray, S.; Benedetto, F.; Jhaveri, R.H. An AI-Assisted Smart Healthcare System Using 5G Communication. IEEE Access 2023, 11, 108339–108355. [Google Scholar] [CrossRef]
- Punugoti, R.; Dutt, V.; Anand, A.; Bhati, N. Exploring the Impact of Edge Intelligence and IoT on Healthcare: A Comprehensive Survey. In Proceedings of the 2023 International Conference on Sustainable Computing and Smart Systems (ICSCSS), Coimbatore, India, 14–16 June 2023; pp. 1108–1114. [Google Scholar] [CrossRef]
- Izhar, M.; Naqvi, S.A.A.; Ahmed, A.; Abdullah, S.; Alturki, N.; Jamel, L. Enhancing Healthcare Efficacy Through IoT-Edge Fusion: A Novel Approach for Smart Health Monitoring and Diagnosis. IEEE Access 2023, 11, 136456–136467. [Google Scholar] [CrossRef]
- Alekseeva, D.; Ometov, A.; Lohan, E.S. Towards the Advanced Data Processing for Medical Applications Using Task Offloading Strategy. In Proceedings of the 2022 18th International Conference on Wireless and Mobile Computing, Networking and Communications (WiMob), Thessaloniki, Greece, 10–12 October 2022; pp. 51–56. [Google Scholar] [CrossRef]
- Ur Rasool, R.; Ahmad, H.F.; Rafique, W.; Qayyum, A.; Qadir, J.; Anwar, Z. Quantum Computing for Healthcare: A Review. Future Internet 2023, 15, 94. [Google Scholar] [CrossRef]
- Shuford, J. Exploring Ethical Dimensions in AI: Navigating Bias and Fairness in the Field. J. Artif. Intell. Gen. Sci. (JAIGS) 2024, 3, 103–124. [Google Scholar] [CrossRef]
- Goh, E.; Bunning, B.; Khoong, E.; Gallo, R.; Milstein, A.; Centola, D.; Chen, J.H. ChatGPT Influence on Medical Decision-Making, Bias, and Equity: A Randomized Study of Clinicians Evaluating Clinical Vignettes. medRxiv 2023. [Google Scholar] [CrossRef]
- Capraro, V.; Lentsch, A.; Acemoglu, D.; Akgun, S.; Akhmedova, A.; Bilancini, E.; Bonnefon, J.F.; Brañas-Garza, P.; Butera, L.; Douglas, K.M.; et al. The impact of generative artificial intelligence on socioeconomic inequalities and policy making. arXiv 2024, arXiv:2401.05377. [Google Scholar] [CrossRef]
- Alzubaidi, L.; Al-Sabaawi, A.; Bai, J.; Dukhan, A.; Alkenani, A.H.; Al-Asadi, A.; Alwzwazy, H.A.; Manoufali, M.; Fadhel, M.A.; Albahri, A.S.; et al. Towards Risk-Free Trustworthy Artificial Intelligence: Significance and Requirements. Int. J. Intell. Syst. 2023, 2023, 1–41. [Google Scholar] [CrossRef]
- Winter, J.S.; Davidson, E. Governance of artificial intelligence and personal health information. Digit. Policy Regul. Gov. 2019, 21, 280–290. [Google Scholar] [CrossRef]
- World Health Organization. Ethics and Governance of Artificial INTELLIGENCE for Health; World Health Organization: Geneva, Switzerland, 2024. [Google Scholar]
- Zhou, K.; Gattinger, G. The Evolving Regulatory Paradigm of AI in MedTech: A Review of Perspectives and Where We Are Today. Ther. Innov. Regul. Sci. 2024, 58, 456–464. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, A.; Karhade, M. Global AI Governance in Healthcare: A Cross-Jurisdictional Regulatory Analysis. arXiv 2024, arXiv:2406.08695. [Google Scholar] [CrossRef]
- Bouderhem, R. Shaping the future of AI in healthcare through ethics and governance. Humanit. Soc. Sci. Commun. 2024, 11, 416. [Google Scholar] [CrossRef]




| AI Method | Best-Suited Healthcare Tasks | Critical Limitations | Essential Requirements | Input Data | Available Technologies | Readiness Level |
|---|---|---|---|---|---|---|
| DL | Segmentation activity of internal organs, early and advanced diagnosis for different pathologies and syndromes | High computational cost for training, high training data volumes | Annotated data, high-performance computing infrastructure, domain expertise for training | Medical images (X-rays, CTs), EHRs, omics data | TensorFlow, PyTorch, Keras, CUDA Toolkit | Medium to High |
| GNN | Drug discovery, protein affinity prediction, modeling complex relationships in EHRs | Scalability issues, difficult interpretability of results, complex data preprocessing | Need for graph-structured data, high-performance computing infrastructure, graph processing tools | Molecular structures, biomedical pathways, EHR graph data | TensorFlow, PyTorch, Deep Graph Library, Spektral, StellarGraph | Low to medium with rapid growth rate |
| RNN | Time-series analysis of health data (e.g., EHR), continuous patient monitoring, telemedicine, protein affinity prediction | Vanishing gradient, long training times, highly variable health status | Data preprocessing and alignment, high-quality data, significant computational resources | Time-series data, EHR sequences, wearable sensor data | TensorFlow, PyTorch, Keras | Medium |
| Generative AI | Clinical documentation, conversational agents, synthetic medical data generation | Possible misleading data, high computational requirements, ethical concerns | Reliable validation of results, high-quality training data, ethical oversights | Clinical images, EHRs, omics data | BERT, GPT, StyleGAN, BioGPT, GPT-4 Medprompt, MediTron-70B | Medium to high, with growing usage for synthetic data for training purposes |
| Diffusion Models | Image reconstruction, denoising of medical images, generation of high-resolution medical images | High computational cost, training complexity, privacy leaks for federated learning | High computational resources, suitable process initialization, high-quality training data | Medical images (MRI, CT scans), noisy or incomplete training images | PyTorch, TensorFlow, Diffusers, NVIDIA Clara | Low, with rapid growth rate |
| RL | Personalized treatments, robotic surgery, support for clinical decision making | Slow training, complex experimental setup, ethical concerns, safety concerns | Emulated environments, real-world feedback data, ethical compliance and safety assessment | Patient data for state model, wearable sensor data, treatment outcomes | TensorFlow, PyTorch, Gymnasium, Stable-Baselines3 | Medium to High |
| Area | Deep Learning | GNN | RNN | Generative AI | Diffusion Models | Reinforcement Learning |
|---|---|---|---|---|---|---|
| Diagnosis | Analysis of radiological images, such as X-rays or CT scans [35,118,119,120,121,122]. Interpretation of laboratory test results, such as blood tests and genetic tests. In [123], the performance of U-Net for segmenting COVID-19 lesions on lung CT-scans is analyzed. | Symptom-based diagnosis: Use of algorithms to diagnose diseases based on symptoms reported by patients [45]. | Aid in medical diagnosis by the analysis of sequential medical data [61,62,63]. | Symptom-based diagnosis: Use of algorithms to diagnose diseases based on symptoms reported by patients. Ref. [124] adds the attention mechanism into the original U-Net architecture for improving the ability to segment small items. | Non-invasive prediction of tumor growth rate by using diffusion models is shown in [125]. | Ref. [113] shows an application of RL techniques for discovering new treatments and personalizing existing ones. |
| Patient Treatment and Management | Processing of histopathology of nasal polyps prognostic information by deep learning for patient treatment is shown in [126]. | Hierarchical GNN for patient treatment preference prediction, integrating doctors’ information and their viewing activities as external knowledge with EMRs to construct the graph [127]. | Virtual assistant that monitors patients’ vitals over time to detect anomalies [64]. | Management of EHR system when incomplete [74]. | Diffusion model application in EHR. Solution to perform class-conditional sampling for preserving label information [128]. | Application to dynamic treatment regimes in chronic diseases [114]. |
| Clinical Care and Decision Support | A study about the utility of machine learning in pediatrics is presented in [129]. | GNN model that learns patient representation using different network configurations and feature modes [130]. | Solutions based on the analysis of sequential medical data [61,62,63]. | Analysis of the accuracy of ChatGPT-derived patient counseling responses based on clinical care guidelines in urology [131]. | Diffusion models generating high-quality realistic mixed-type tabular EHRs, preserving privacy, used for data augmentation [132]. | Survey including recommendation systems to physicians based on clinical guidelines and patient data analysis [133]. |
| Research and Development | Blood vessel segmentation is investigated in [134,135]. In [134,135] a U-Net is used for coronary artery stenosis detection on X-ray coronary angiograms. | Drug discovery: Using AI to identify new drug compounds and predict their efficacy and safety [136]. | Analysis of epidemiological data and for tracking infections, and much more [65,66]. | Research for improving images in healthcare for challenging situations [137]. | Generation of high-quality data for training AI algorithms [138]. | Precision robotics application to healthcare [106]. Ref. [115] focuses on precision oncology and identifies current challenges and pitfalls. |
| Administration and Management | The potentials of deep learning in hospital administration and management, including ethical and legal issues are presented in [139]. | Implementation of a knowledge graph for discovering insights in medical subject headings [140]. | Management of resources, scheduling activities, and improving the operational efficiency of hospitals. For example, see [141]. | Transformative healthcare for automating clinical documentation and processing of patient information [142]. | Diffusion models have also been proposed within the life cycle of innovation [143]. | Many examples in the great survey in [144]. |
| Prevention and Wellness | A deep learning algorithm for sleep stage scoring, making use of a single EEG channel, is presented in [145]. | Use of a patient graph structure with basic information like age, gender, and diagnosis, and the trained GNN models for identifying therapies [146]. | Analysis of sequential medical data [61,62,63]. | Empowering workers and anticipating harms in integrating large language models with workplace technologies [147]. | Use of 3D avatars in applications for fitness and wellness [148]. | RL in patients with mood and anxiety disorders [149]. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reali, G.; Femminella, M. Artificial Intelligence to Reshape the Healthcare Ecosystem. Future Internet 2024, 16, 343. https://doi.org/10.3390/fi16090343
Reali G, Femminella M. Artificial Intelligence to Reshape the Healthcare Ecosystem. Future Internet. 2024; 16(9):343. https://doi.org/10.3390/fi16090343
Chicago/Turabian StyleReali, Gianluca, and Mauro Femminella. 2024. "Artificial Intelligence to Reshape the Healthcare Ecosystem" Future Internet 16, no. 9: 343. https://doi.org/10.3390/fi16090343
APA StyleReali, G., & Femminella, M. (2024). Artificial Intelligence to Reshape the Healthcare Ecosystem. Future Internet, 16(9), 343. https://doi.org/10.3390/fi16090343





