A Systematic Literature Review on Applications of GAN-Synthesized Images for Brain MRI
Abstract
1. Introduction
2. Materials and Methods
2.1. Prior Research
2.2. Motivation
- There is no existing SLR paper that describes the applications of GAN-synthesized brain MRI.
- There is no existing SLR paper that mentions the types of GAN loss functions.
- The paper presents a clear categorical division of brain MRI applications of GAN-synthesized images as well as the details of the technique.
- The paper presents a concise account of distinct loss functions used in GAN training.
- The paper identifies software to preprocess brain MRI.
- The paper compares the various evaluation metrics available for the performance evaluation of synthetic images.
2.3. Research Questions
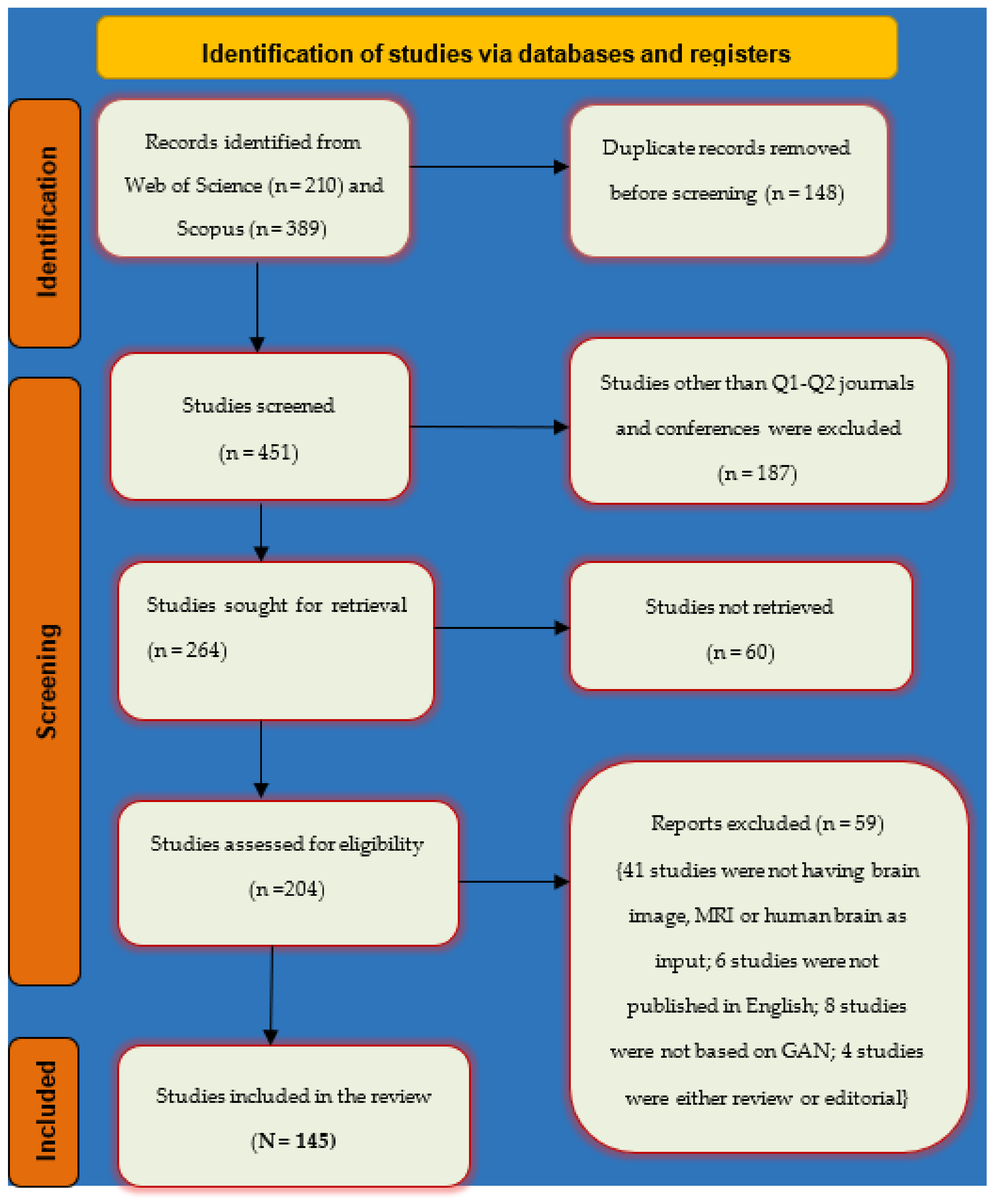
2.4. Search Strategy
2.5. Inclusion and Exclusion Criteria
3. Results
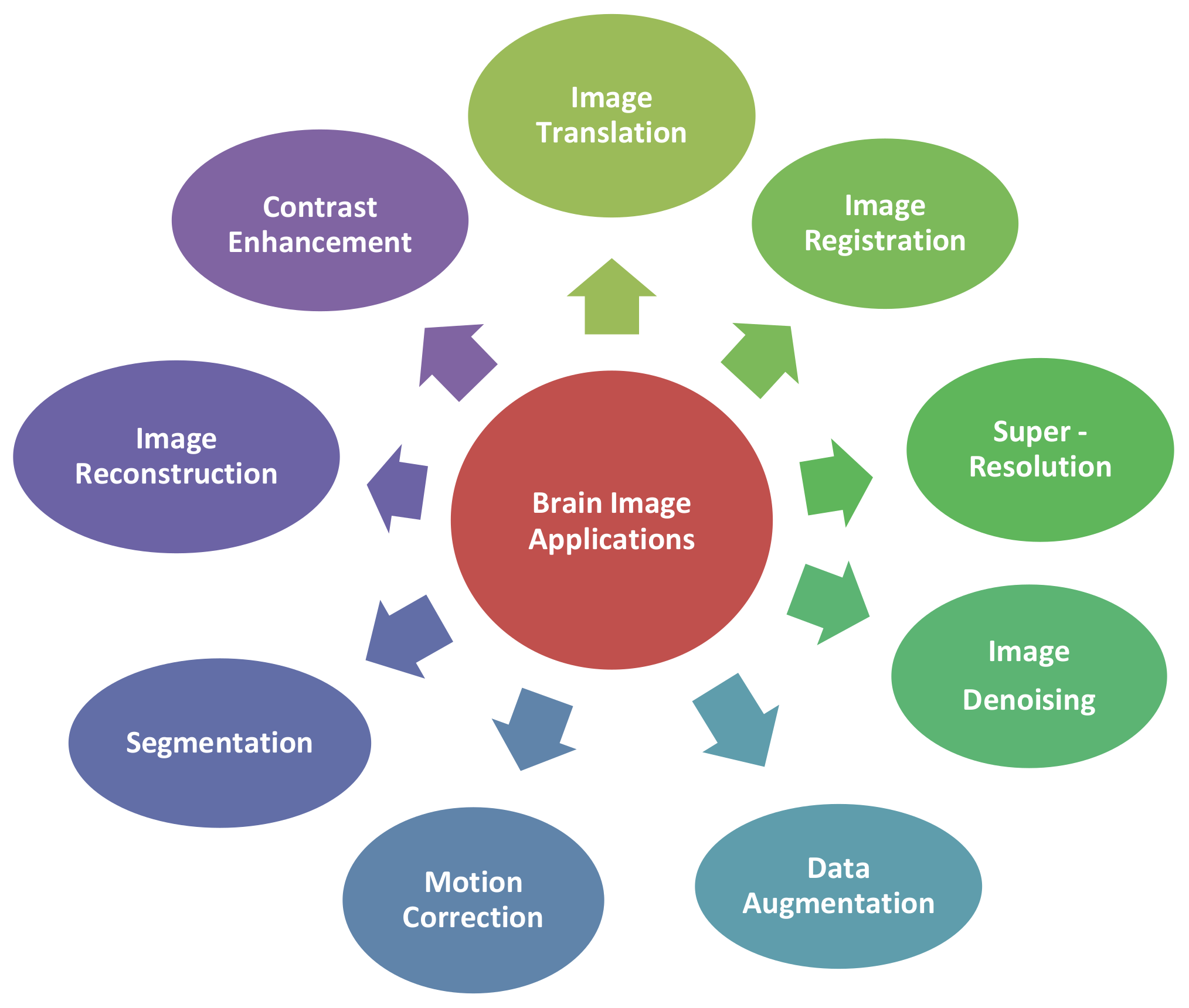
3.1. Applications of GAN-Synthesized Images for Brain MRI (RQ 1)
3.1.1. Image Translation
- A.
- MRI-to-CT Translation:
- B.
- MRI-to-PET Translation:
3.1.2. Image Registration
3.1.3. Image Super-Resolution
3.1.4. Contrast Enhancement
- A.
- Modality Translation:
- B.
- Quality Improvement:
- C.
- Single Network Generation:
3.1.5. Image Denoising
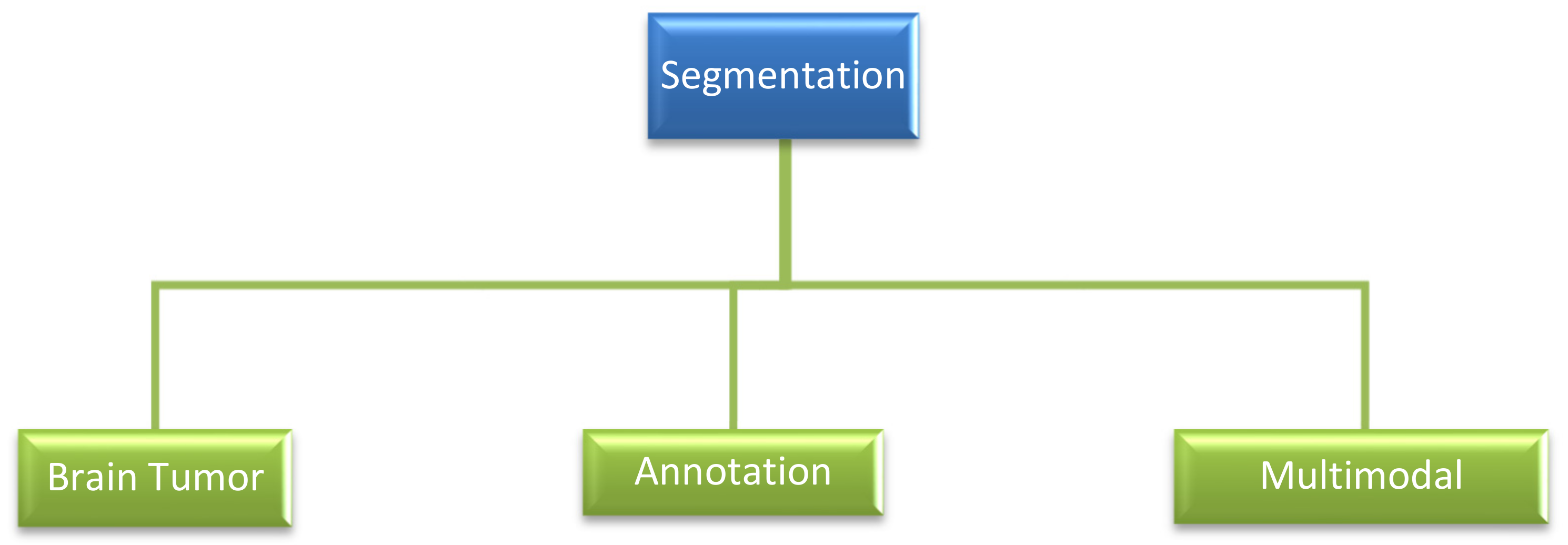
3.1.6. Segmentation
- A.
- Brain Tumor Segmentation:
- B.
- Annotation:
- C.
- Multimodal Segmentation:
3.1.7. Reconstruction
- A.
- MRI Acceleration:
- B.
- MR Slice Reconstruction:
- C.
- Enhancement of Scan Efficiency:
- D.
- Bias-free MRI Scan:
3.1.8. Motion Correction
3.1.9. Data Augmentation
3.2. Loss Functions (RQ 2)
3.3. Preprocessing of Ground Truth Brain MRI (RQ 3)
- Intensity Normalization:
- Skull Stripping:
- Registration:
- Bias Field Correction:
- Center Cropping:
- Data Augmentation:
- Motion Correction:
3.4. Comparative Study of Evaluation Metric (RQ 4)
4. Discussion
4.1. GAN Variants
4.2. Multimodal Image Generation
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Currie, S.; Hoggard, N.; Craven, I.J.; Hadjivassiliou, M.; Wilkinson, I.D. Understanding MRI: Basic MR physics for physicians. Postgrad. Med. J. 2013, 89, 209–223. [Google Scholar] [CrossRef] [PubMed]
- Latif, G.; Kazmi, S.B.; Jaffar, M.A.; Mirza, A.M. Classification and Segmentation of Brain Tumor Using Texture Analysis. In Proceedings of the 9th WSEAS International Conference on Artificial Intelligence, Knowledge Engineering and Data Bases, Stevens Point, WI, USA, 20–22 February 2010; pp. 147–155. [Google Scholar]
- Tiwari, A.; Srivastava, S.; Pant, M. Brain tumor segmentation and classification from magnetic resonance images: Review of selected methods from 2014 to 2019. Pattern Recognit. Lett. 2020, 131, 244–260. [Google Scholar] [CrossRef]
- Lecun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Shen, D.; Liu, T.; Peters, T.M.; Staib, L.H.; Essert, C.; Zhou, S.; Yap, P.-T.; Khan, A. Miccai 2019-Part 4; Springer: Berlin/Heidelberg, Germany, 2019; Volume 1, ISBN 9783030322519. [Google Scholar]
- Gudigar, A.; Raghavendra, U.; Hegde, A.; Kalyani, M.; Ciaccio, E.J.; Rajendra Acharya, U. Brain pathology identification using computer aided diagnostic tool: A systematic review. Comput. Methods Programs Biomed. 2020, 187, 105205. [Google Scholar] [CrossRef] [PubMed]
- Goodfellow, I.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative adversarial networks. Commun. ACM 2020, 63, 139–144. [Google Scholar] [CrossRef]
- Ali, H.; Biswas, R.; Ali, F.; Shah, U.; Alamgir, A.; Mousa, O.; Shah, Z. The role of generative adversarial networks in brain MRI: A scoping review. Insights Imaging 2022, 13, 98. [Google Scholar] [CrossRef]
- Creswell, A.; White, T.; Dumoulin, V.; Arulkumaran, K.; Sengupta, B.; Bharath, A.A. Generative Adversarial Networks: An Overview. IEEE Signal Process. Mag. 2018, 35, 53–65. [Google Scholar] [CrossRef]
- Rashid, M.; Singh, H.; Goyal, V. The use of machine learning and deep learning algorithms in functional magnetic resonance imaging—A systematic review. Expert Syst. 2020, 37, 1–29. [Google Scholar] [CrossRef]
- Vijina, P.; Jayasree, M. A Survey on Recent Approaches in Image Reconstruction. In Proceedings of the 2020 International Conference on Power, Instrumentation, Control and Computing (PICC), Thrissur, India, 17–19 December 2020. [Google Scholar] [CrossRef]
- Kitchenham, B.; Charters, S. Guidelines for Performing Systematic Literature Reviews in Software Engineering; Technical Report EBSE; Keele University: Keele, UK, 2007. [Google Scholar]
- Liu, W.; Hu, G.; Gu, M. The probability of publishing in first-quartile journals. Scientometrics 2016, 106, 1273–1276. [Google Scholar] [CrossRef]
- Isola, P.; Zhu, J.Y.; Zhou, T.; Efros, A.A. Image-to-image translation with conditional adversarial networks. Proc. IEEE Conf. Comput. Vis. Pattern Recognit. 2017, 2017, 5967–5976. [Google Scholar] [CrossRef]
- Yang, Q.; Li, N.; Zhao, Z.; Fan, X.; Chang, E.I.C.; Xu, Y. MRI Cross-Modality Image-to-Image Translation. Sci. Rep. 2020, 10, 3753. [Google Scholar] [CrossRef]
- Kazemifar, S.; McGuire, S.; Timmerman, R.; Wardak, Z.; Nguyen, D.; Park, Y.; Jiang, S.; Owrangi, A. MRI-only brain radiotherapy: Assessing the dosimetric accuracy of synthetic CT images generated using a deep learning approach. Radiother. Oncol. 2019, 136, 56–63. [Google Scholar] [CrossRef]
- Kazemifar, S.; Barragán Montero, A.M.; Souris, K.; Rivas, S.T.; Timmerman, R.; Park, Y.K.; Jiang, S.; Geets, X.; Sterpin, E.; Owrangi, A. Dosimetric evaluation of synthetic CT generated with GANs for MRI-only proton therapy treatment planning of brain tumors. J. Appl. Clin. Med. Phys. 2020, 21, 76–86. [Google Scholar] [CrossRef]
- Bourbonne, V.; Jaouen, V.; Hognon, C.; Boussion, N.; Lucia, F.; Pradier, O.; Bert, J.; Visvikis, D.; Schick, U. Dosimetric validation of a gan-based pseudo-ct generation for mri-only stereotactic brain radiotherapy. Cancers 2021, 13, 1082. [Google Scholar] [CrossRef]
- Tang, B.; Wu, F.; Fu, Y.; Wang, X.; Wang, P.; Orlandini, L.C.; Li, J.; Hou, Q. Dosimetric evaluation of synthetic CT image generated using a neural network for MR-only brain radiotherapy. J. Appl. Clin. Med. Phys. 2021, 22, 55–62. [Google Scholar] [CrossRef]
- Armanious, K.; Jiang, C.; Fischer, M.; Küstner, T.; Nikolaou, K.; Gatidis, S.; Yang, B. MedGAN: Medical image translation using GANs. Comput. Med. Imaging Graph. 2019, 79, 101684. [Google Scholar] [CrossRef]
- Tao, L.; Fisher, J.; Anaya, E.; Li, X.; Levin, C.S. Pseudo CT Image Synthesis and Bone Segmentation from MR Images Using Adversarial Networks with Residual Blocks for MR-Based Attenuation Correction of Brain PET Data. IEEE Trans. Radiat. Plasma Med. Sci. 2020, 5, 193–201. [Google Scholar] [CrossRef]
- Liu, X.; Emami, H.; Nejad-Davarani, S.P.; Morris, E.; Schultz, L.; Dong, M.; Glide-Hurst, C.K. Performance of deep learning synthetic CTs for MR-only brain radiation therapy. J. Appl. Clin. Med. Phys. 2021, 22, 308–317. [Google Scholar] [CrossRef]
- Emami, H.; Dong, M.; Glide-Hurst, C.K. Attention-Guided Generative Adversarial Network to Address Atypical Anatomy in Synthetic CT Generation. In Proceedings of the 2020 IEEE 21st International Conference on Information Reuse and Integration for Data Science (IRI), Las Vegas, NV, USA, 11–13 August 2020; pp. 188–193. [Google Scholar] [CrossRef]
- Abu-Srhan, A.; Almallahi, I.; Abushariah, M.A.M.; Mahafza, W.; Al-Kadi, O.S. Paired-unpaired Unsupervised Attention Guided GAN with transfer learning for bidirectional brain MR-CT synthesis. Comput. Biol. Med. 2021, 136, 104763. [Google Scholar] [CrossRef]
- Lei, Y.; Harms, J.; Wang, T.; Liu, Y.; Shu, H.K.; Jani, A.B.; Curran, W.J.; Mao, H.; Liu, T.; Yang, X. MRI-only based synthetic CT generation using dense cycle consistent generative adversarial networks. Med. Phys. 2019, 46, 3565–3581. [Google Scholar] [CrossRef]
- Uzunova, H.; Ehrhardt, J.; Handels, H. Memory-efficient GAN-based domain translation of high resolution 3D medical images. Comput. Med. Imaging Graph. 2020, 86, 101801. [Google Scholar] [CrossRef] [PubMed]
- Shafai-Erfani, G.; Lei, Y.; Liu, Y.; Wang, Y.; Wang, T.; Zhong, J.; Liu, T.; McDonald, M.; Curran, W.J.; Zhou, J.; et al. MRI-based proton treatment planning for base of skull tumors. Int. J. Part. Ther. 2019, 6, 12–25. [Google Scholar] [CrossRef] [PubMed]
- Gong, K.; Yang, J.; Larson, P.E.Z.; Behr, S.C.; Hope, T.A.; Seo, Y.; Li, Q. MR-Based Attenuation Correction for Brain PET Using 3-D Cycle-Consistent Adversarial Network. IEEE Trans. Radiat. Plasma Med. Sci. 2021, 5, 185–192. [Google Scholar] [CrossRef] [PubMed]
- Matsui, T.; Taki, M.; Pham, T.Q.; Chikazoe, J.; Jimura, K. Counterfactual Explanation of Brain Activity Classifiers Using Image-To-Image Transfer by Generative Adversarial Network. Front. Neuroinform. 2022, 15, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Mehmood, M.; Alshammari, N.; Alanazi, S.A.; Basharat, A.; Ahmad, F.; Sajjad, M.; Junaid, K. Improved colorization and classification of intracranial tumor expanse in MRI images via hybrid scheme of Pix2Pix-cGANs and NASNet-large. J. King Saud Univ.-Comput. Inf. Sci. 2022, 34, 4358–4374. [Google Scholar] [CrossRef]
- Nie, D.; Trullo, R.; Lian, J.; Wang, L. Medical Image Synthesis with Deep Convolutional Adversarial Networks. Physiol. Behav. 2016, 176, 100–106. [Google Scholar] [CrossRef]
- Kang, S.K.; Seo, S.; Shin, S.A.; Byun, M.S.; Lee, D.Y.; Kim, Y.K.; Lee, D.S.; Lee, J.S. Adaptive template generation for amyloid PET using a deep learning approach. Hum. Brain Mapp. 2018, 39, 3769–3778. [Google Scholar] [CrossRef]
- Wei, W.; Poirion, E.; Bodini, B.; Durrleman, S.; Ayache, N.; Stankoff, B.; Colliot, O. Predicting PET-derived demyelination from multimodal MRI using sketcher-refiner adversarial training for multiple sclerosis. Med. Image Anal. 2019, 58, 101546. [Google Scholar] [CrossRef]
- Gao, X.; Shi, F.; Shen, D.; Liu, M. Task-Induced Pyramid and Attention GAN for Multimodal Brain Image Imputation and Classification in Alzheimer’s Disease. IEEE J. Biomed. Health Inform. 2022, 26, 36–43. [Google Scholar] [CrossRef]
- Hu, S.; Lei, B.; Member, S.; Wang, S. Networks for Brain MR to PET Synthesis. IEEE Trans. Med. Imaging 2022, 41, 145–157. [Google Scholar] [CrossRef]
- Pan, Y.; Liu, M.; Lian, C.; Xia, Y.; Shen, D. Spatially-Constrained Fisher Representation for Brain Disease Identification with Incomplete Multi-Modal Neuroimages. IEEE Trans. Med. Imaging 2020, 39, 2965–2975. [Google Scholar] [CrossRef]
- Zotova, D.; Jung, J.; Laertizien, C. GAN-Based Synthetic FDG PET Images from T1 Brain MRI Can Serve to Improve Performance of Deep Unsupervised Anomaly Detection Models. In Proceedings of the International Workshop on Simulation and Synthesis in Medical Imaging, Strasbourg, France, 27 September 2021; pp. 142–152. [Google Scholar] [CrossRef]
- Liu, H.; Nai, Y.H.; Saridin, F.; Tanaka, T.; O’ Doherty, J.; Hilal, S.; Gyanwali, B.; Chen, C.P.; Robins, E.G.; Reilhac, A. Improved amyloid burden quantification with nonspecific estimates using deep learning. Eur. J. Nucl. Med. Mol. Imaging 2021, 48, 1842–1853. [Google Scholar] [CrossRef]
- DL, H. Medical image registration. Phys. Med. Biol. 2001, 46, R1–R45. [Google Scholar] [CrossRef]
- Salehi, S.S.; Khan, S.; Erdogmus, D.; Gholipour, A. Real-time Deep Pose Estimation with Geodesic Loss for Image-to-Template Rigid Registration. Physiol. Behav. 2019, 173, 665–676. [Google Scholar] [CrossRef]
- Zheng, Y.; Sui, X.; Jiang, Y.; Che, T.; Zhang, S.; Yang, J.; Li, H. SymReg-GAN: Symmetric Image Registration with Generative Adversarial Networks. IEEE Trans. Pattern Anal. Mach. Intell. 2021, 44, 5631–5646. [Google Scholar] [CrossRef]
- Liu, X.; Zhao, H.; Zhang, S.; Tang, Z. Brain Image Parcellation Using Multi-Atlas Guided Adversarial Fully Convolutional Network. In Proceedings of the 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), Venice, Italy, 8–11 April 2019; pp. 723–726. [Google Scholar]
- Tang, Z.; Liu, X.; Li, Y.; Yap, P.T.; Shen, D. Multi-Atlas Brain Parcellation Using Squeeze-and-Excitation Fully Convolutional Networks. IEEE Trans. Image Process. 2020, 29, 6864–6872. [Google Scholar] [CrossRef]
- Fan, J.; Cao, X.; Wang, Q.; Yap, P.-T.; Shen, D. Adversarial Learning for Mono- or Multi-Modal Registration. Med. Image Anal. 2019, 58, 101545. [Google Scholar] [CrossRef]
- Mahapatra, D.; Ge, Z. Training Data Independent Image Registration with Gans Using Transfer Learning and Segmentation Information. In Proceedings of the International Symposium on Biomedical Imaging (ISBI 2019), Venice, Italy, 8–11 April 2019; pp. 709–713. [Google Scholar]
- Yang, C.Y.; Huang, J.B.; Yang, M.H. Exploiting self-similarities for single frame super-resolution. Lect. Notes Comput. Sci. 2011, 6494, 497–510. [Google Scholar] [CrossRef]
- Greenspan, H.; Peled, S.; Oz, G.; Kiryati, N. MRI inter-slice reconstruction using super-resolution. Lect. Notes Comput. Sci. 2001, 2208, 1204–1206. [Google Scholar] [CrossRef]
- Zhu, J.; Yang, G.; Lio, P. How can we make gan perform better in single medical image super-resolution? A lesion focused multi-scale approach. In Proceedings of the 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), Venice, Italy, 8–11 April 2019; pp. 1669–1673. [Google Scholar] [CrossRef]
- Chong, C.K.; Ho, E.T.W. Synthesis of 3D MRI Brain Images with Shape and Texture Generative Adversarial Deep Neural Networks. IEEE Access 2021, 9, 64747–64760. [Google Scholar] [CrossRef]
- Ahmad, W.; Ali, H.; Shah, Z.; Azmat, S. A new generative adversarial network for medical images super resolution. Sci. Rep. 2022, 12, 9533. [Google Scholar] [CrossRef] [PubMed]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. In Proceedings of the 3rd International Conference on Learning Representations, ICLR 2015, San Diego, CA, USA, 7–9 May 2015; pp. 1–14. [Google Scholar]
- Hongtao, Z.; Shinomiya, Y.; Yoshida, S. 3D Brain MRI Reconstruction based on 2D Super-Resolution Technology. IEEE Trans. Syst. Man. Cybern. Syst. 2020, 2020, 18–23. [Google Scholar] [CrossRef]
- Zhang, H.; Shinomiya, Y.; Yoshida, S. 3D MRI Reconstruction Based on 2D Generative Adversarial Network Super-Resolution. Sensors 2021, 21, 2978. [Google Scholar] [CrossRef] [PubMed]
- Delannoy, Q.; Pham, C.H.; Cazorla, C.; Tor-Díez, C.; Dollé, G.; Meunier, H.; Bednarek, N.; Fablet, R.; Passat, N.; Rousseau, F. SegSRGAN: Super-resolution and segmentation using generative adversarial networks—Application to neonatal brain MRI. Comput. Biol. Med. 2020, 120, 103755. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Tan, C.; Yang, J.; Yang, G.; Lio’, P. Arbitrary Scale Super-Resolution for Medical Images. Int. J. Neural Syst. 2021, 31. [Google Scholar] [CrossRef]
- Pham, C.; Meunier, H.; Bednarek, N.; Fablet, R.; Passat, N.; Rousseau, F.; De Reims, C.H.U.; Champagne-ardenne, D.R. Simultaneous Super-Resolution and Segmentation Using A Generative Adversarial Network: Application To Neonatal Brain MRI. In Proceedings of the 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), Venice, Italy, 8–11 April 2019; pp. 991–994. [Google Scholar]
- Han, S.; Carass, A.; Schar, M.; Calabresi, P.A.; Prince, J.L. Slice profile estimation from 2D MRI acquisition using generative adversarial networks. Proc. Int. Symp. Biomed. Imaging 2021, 2021, 145–149. [Google Scholar] [CrossRef]
- Zhou, X.; Qiu, S.; Joshi, P.S.; Xue, C.; Killiany, R.J.; Mian, A.Z.; Chin, S.P.; Au, R.; Kolachalama, V.B. Enhancing magnetic resonance imaging-driven Alzheimer’s disease classification performance using generative adversarial learning. Alzheimer’s Res. Ther. 2021, 13, 60. [Google Scholar] [CrossRef]
- You, S.; Lei, B.; Wang, S.; Chui, C.K.; Cheung, A.C.; Liu, Y.; Gan, M.; Wu, G.; Shen, Y. Fine Perceptive GANs for Brain MR Image Super-Resolution in Wavelet Domain. IEEE Trans. Neural Networks Learn. Syst. 2022, 1–13. [Google Scholar] [CrossRef]
- Sun, L.; Chen, J.; Xu, Y.; Gong, M.; Yu, K.; Batmanghelich, K. Hierarchical Amortized GAN for 3D High Resolution Medical Image Synthesis. IEEE J. Biomed. Health Inform. 2022, 26, 3966–3975. [Google Scholar] [CrossRef]
- Sui, Y.; Afacan, O.; Jaimes, C.; Gholipour, A.W.S. Scan-Specific Generative Neural Network for MRI Super-Resolution Reconstruction. IEEE Trans. Med. Imaging 2022, 41, 1383–1399. [Google Scholar] [CrossRef]
- Katti, G.; Ara, S.A. A shireen Magnetic resonance imaging (MRI)–A review. Int. J. Dent. Clin. 2011, 3, 65–70. [Google Scholar]
- Revett, K. An Introduction to Magnetic Resonance Imaging: From Image Acquisition to Clinical Diagnosis. In Innovations in Intelligent Image Analysis. Studies in Computational Intelligence; Springer: Berlin/Heidelberg, Germany, 2011; pp. 127–161. ISBN 978-3-642-17933-4. [Google Scholar]
- Hofer, S.; Frahm, J. Topography of the human corpus callosum revisited-Comprehensive fiber tractography using diffusion tensor magnetic resonance imaging. Neuroimage 2006, 32, 989–994. [Google Scholar] [CrossRef]
- Mzoughi, H.; Njeh, I.; Wali, A.; Slima, M.B.; BenHamida, A.; Mhiri, C.; Mahfoudhe, K. Ben Deep Multi-Scale 3D Convolutional Neural Network (CNN) for MRI Gliomas Brain Tumor Classification. J. Digit. Imaging 2020, 33, 903–915. [Google Scholar] [CrossRef]
- Wang, G.; Gong, E.; Banerjee, S.; Martin, D.; Tong, E.; Choi, J.; Chen, H.; Wintermark, M.; Pauly, J.M.; Zaharchuk, G. Synthesize High-Quality Multi-Contrast Magnetic Resonance Imaging from Multi-Echo Acquisition Using Multi-Task Deep Generative Model. IEEE Trans. Med. Imaging 2020, 39, 3089–3099. [Google Scholar] [CrossRef]
- Dar, S.U.H.; Yurt, M.; Karacan, L.; Erdem, A.; Erdem, E.; Cukur, T. Image Synthesis in Multi-Contrast MRI with Conditional Generative Adversarial Networks. IEEE Trans. Med. Imaging 2019, 38, 2375–2388. [Google Scholar] [CrossRef]
- Sharma, A.; Hamarneh, G. Missing MRI Pulse Sequence Synthesis Using Multi-Modal Generative Adversarial Network. IEEE Trans. Med. Imaging 2020, 39, 1170–1183. [Google Scholar] [CrossRef]
- Alogna, E.; Giacomello, E.; Loiacono, D. Brain Magnetic Resonance Imaging Generation using Generative Adversarial Networks. In Proceedings of the 2020 IEEE Symposium Series on Computational Intelligence (SSCI), Canberra, Australia, 1–4 December 2020; pp. 2528–2535. [Google Scholar] [CrossRef]
- Liu, X.; Xing, F.; El Fakhri, G.; Woo, J. A unified conditional disentanglement framework for multimodal brain mr image translation. Proc. Int. Symp. Biomed. Imaging 2021, 2021, 10–14. [Google Scholar] [CrossRef]
- Qu, Y.; Deng, C.; Su, W.; Wang, Y.; Lu, Y.; Chen, Z. Multimodal Brain MRI Translation Focused on Lesions. ACM Int. Conf. Proc. Ser. 2020, 352–359. [Google Scholar] [CrossRef]
- Liu, X.; Yu, A.; Wei, X.; Pan, Z.; Tang, J. Multimodal MR Image Synthesis Using Gradient Prior and Adversarial Learning. IEEE J. Sel. Top. Signal Process. 2020, 14, 1176–1188. [Google Scholar] [CrossRef]
- Yu, B.; Zhou, L.; Wang, L.; Shi, Y.; Fripp, J.; Bourgeat, P. Ea-GANs: Edge-Aware Generative Adversarial Networks for Cross-Modality MR Image Synthesis. IEEE Trans. Med. Imaging 2019, 38, 1750–1762. [Google Scholar] [CrossRef]
- Gao, Y.; Liu, Y.; Wang, Y.; Shi, Z.; Yu, J. A Universal Intensity Standardization Method Based on a Many-to-One Weak-Paired Cycle Generative Adversarial Network for Magnetic Resonance Images. IEEE Trans. Med. Imaging 2019, 38, 2059–2069. [Google Scholar] [CrossRef]
- Han, C.; Hayashi, H.; Rundo, L.; Araki, R.; Shimoda, W.; Muramatsu, S.; Furukawa, Y.; Mauri, G.; Nakayama, H. GAN-based synthetic brain MR image generation. Proc. Int. Symp. Biomed. Imaging 2018, 2018, 734–738. [Google Scholar] [CrossRef]
- Yu, B.; Zhou, L.; Wang, L.; Shi, Y.; Fripp, J.; Bourgeat, P. Sample-Adaptive GANs: Linking Global and Local Mappings for Cross-Modality MR Image Synthesis. IEEE Trans. Med. Imaging 2020, 39, 2339–2350. [Google Scholar] [CrossRef]
- Tomar, D.; Lortkipanidze, M.; Vray, G.; Bozorgtabar, B.; Thiran, J.P. Self-Attentive Spatial Adaptive Normalization for Cross-Modality Domain Adaptation. IEEE Trans. Med. Imaging 2021, 40, 2926–2938. [Google Scholar] [CrossRef]
- Shen, L.; Zhu, W.; Wang, X.; Xing, L.; Pauly, J.M.; Turkbey, B.; Harmon, S.A.; Sanford, T.H.; Mehralivand, S.; Choyke, P.L.; et al. Multi-Domain Image Completion for Random Missing Input Data. IEEE Trans. Med. Imaging 2021, 40, 1113–1122. [Google Scholar] [CrossRef] [PubMed]
- Rachmadi, M.F.; Valdés-Hernández, M.D.C.; Makin, S.; Wardlaw, J.; Komura, T. Automatic spatial estimation of white matter hyperintensities evolution in brain MRI using disease evolution predictor deep neural networks. Med. Image Anal. 2020, 63, 101712. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.H.; Do, W.J.; Park, S.H. Improving resolution of MR images with an adversarial network incorporating images with different contrast. Med. Phys. 2018, 45, 3120–3131. [Google Scholar] [CrossRef] [PubMed]
- Hamghalam, M.; Wang, T.; Lei, B. High tissue contrast image synthesis via multistage attention-GAN: Application to segmenting brain MR scans. Neural Netw. 2020, 132, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Yang, G.; Papanastasiou, G.; Tsaftaris, S.A.; Newby, D.E.; Gray, C.; Macnaught, G.; MacGillivray, T.J. DiCyc: GAN-based deformation invariant cross-domain information fusion for medical image synthesis. Inf. Fusion 2021, 67, 147–160. [Google Scholar] [CrossRef]
- Ma, B.; Zhao, Y.; Yang, Y.; Zhang, X.; Dong, X.; Zeng, D.; Ma, S.; Li, S. MRI image synthesis with dual discriminator adversarial learning and difficulty-aware attention mechanism for hippocampal subfields segmentation. Comput. Med. Imaging Graph. 2020, 86, 101800. [Google Scholar] [CrossRef]
- Yang, X.; Lin, Y.; Wang, Z.; Li, X.; Cheng, K.T. Bi-Modality Medical Image Synthesis Using Semi-Supervised Sequential Generative Adversarial Networks. IEEE J. Biomed. Health Inform. 2020, 24, 855–865. [Google Scholar] [CrossRef]
- Hagiwara, A.; Otsuka, Y.; Hori, M.; Tachibana, Y.; Yokoyama, K.; Fujita, S.; Andica, C.; Kamagata, K.; Irie, R.; Koshino, S.; et al. Improving the quality of synthetic FLAIR images with deep learning using a conditional generative adversarial network for pixel-by-pixel image translation. Am. J. Neuroradiol. 2019, 40, 224–230. [Google Scholar] [CrossRef]
- Naseem, R.; Islam, A.J.; Cheikh, F.A.; Beghdadi, A. Contrast Enhancement: Cross-modal Learning Approach for Medical Images. Proc. IST Int’l. Symp. Electron. Imaging: Image Process. Algorithms Syst. 2022, 34, IPAS-344. [Google Scholar] [CrossRef]
- Choi, Y.; Choi, M.; Kim, M.; Ha, J.W.; Kim, S.; Choo, J. StarGAN: Unified Generative Adversarial Networks for Multi-domain Image-to-Image Translation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 8789–8797. [Google Scholar] [CrossRef]
- Dai, X.; Lei, Y.; Fu, Y.; Curran, W.J.; Liu, T.; Mao, H.; Yang, X. Multimodal MRI synthesis using unified generative adversarial networks. Med. Phys. 2020, 47, 6343–6354. [Google Scholar] [CrossRef]
- Xin, B.; Hu, Y.; Zheng, Y.; Liao, H. Multi-Modality Generative Adversarial Networks with Tumor Consistency Loss for Brain MR Image Synthesis. Proc. Int. Symp. Biomed. Imaging 2020, 2020, 1803–1807. [Google Scholar] [CrossRef]
- Mohan, J.; Krishnaveni, V.; Guo, Y. A survey on the magnetic resonance image denoising methods. Biomed. Signal Process. Control 2014, 9, 56–69. [Google Scholar] [CrossRef]
- Bermudez, C.; Plassard, A.; Davis, T.; Newton, A.; Resnick, S.; Landmana, B. Learning Implicit Brain MRI Manifolds with Deep Learning. Physiol. Behav. 2017, 176, 139–148. [Google Scholar] [CrossRef]
- Ran, M.; Hu, J.; Chen, Y.; Chen, H.; Sun, H.; Zhou, J.; Zhang, Y. Denoising of 3D magnetic resonance images using a residual encoder–decoder Wasserstein generative adversarial network. Med. Image Anal. 2019, 55, 165–180. [Google Scholar] [CrossRef]
- Christilin, D.M.A.B.; Mary, D.M.S. Residual encoder-decoder up-sampling for structural preservation in noise removal. Multimed. Tools Appl. 2021, 80, 19441–19457. [Google Scholar] [CrossRef]
- Li, Z.; Tian, Q.; Ngamsombat, C.; Cartmell, S.; Conklin, J.; Filho, A.L.M.G.; Lo, W.C.; Wang, G.; Ying, K.; Setsompop, K.; et al. High-fidelity fast volumetric brain MRI using synergistic wave-controlled aliasing in parallel imaging and a hybrid denoising generative adversarial network (HDnGAN). Med. Phys. 2022, 49, 1000–1014. [Google Scholar] [CrossRef]
- Akkus, Z.; Galimzianova, A.; Hoogi, A.; Rubin, D.L.; Erickson, B.J. Deep Learning for Brain MRI Segmentation: State of the Art and Future Directions. J. Digit. Imaging 2017, 30, 449–459. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Qin, Z.; Ding, Y.; Lan, T. Brain Tumor Segmentation with Generative Adversarial Nets. In Proceedings of the 2019 2nd International Conference on Artificial Intelligence and Big Data (ICAIBD), Chengdu, China, 25–28 May 2019; pp. 301–305. [Google Scholar] [CrossRef]
- Cheng, G.; Ji, H.; He, L. Correcting and reweighting false label masks in brain tumor segmentation. Med. Phys. 2021, 48, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Elazab, A.; Wang, C.; Safdar Gardezi, S.J.; Bai, H.; Wang, T.; Lei, B.; Chang, C. Glioma Growth Prediction via Generative Adversarial Learning from Multi-Time Points Magnetic Resonance Images. Proc. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. EMBS 2020, 2020, 1750–1753. [Google Scholar] [CrossRef]
- Elazab, A.; Wang, C.; Gardezi, S.J.S.; Bai, H.; Hu, Q.; Wang, T.; Chang, C.; Lei, B. GP-GAN: Brain tumor growth prediction using stacked 3D generative adversarial networks from longitudinal MR Images. Neural Netw. 2020, 132, 321–332. [Google Scholar] [CrossRef] [PubMed]
- Sandhiya, B.; Priyatharshini, R.; Ramya, B.; Monish, S.; Sai Raja, G.R. Reconstruction, identification and classification of brain tumor using gan and faster regional-CNN. In Proceedings of the 2021 3rd International Conference on Signal Processing and Communication (ICPSC), Coimbatore, India, 13–14 May 2021; pp. 238–242. [Google Scholar] [CrossRef]
- Alex, V.; Safwan, K.P.M.; Chennamsetty, S.S.; Krishnamurthi, G. Generative adversarial networks for brain lesion detection. Med. Imaging 2017 Image Process. 2017, 10133, 101330G. [Google Scholar] [CrossRef]
- City, I. Transforming Intensity Distribution of Brain Lesions via Conditional Gans for Segmentation. In Proceedings of the 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI), Iowa City, IA, USA, 3–7 April 2020; pp. 1499–1502. [Google Scholar]
- Thirumagal, E.; Saruladha, K. Design of FCSE-GAN for dissection of brain tumour in MRI. In Proceedings of the 2020 International Conference on Smart Technologies in Computing, Electrical and Electronics (ICSTCEE), Bengaluru, India, 9–10 October 2020; pp. 61–65. [Google Scholar] [CrossRef]
- Tokuoka, Y.; Suzuki, S.; Sugawara, Y. An inductive transfer learning approach using cycleconsistent adversarial domain adaptation with application to brain tumor segmentation. In Proceedings of the 2019 6th International Conference on Biomedical and Bioinformatics Engineering, Shanghai, China, 13–15 November 2019; pp. 44–48. [Google Scholar] [CrossRef]
- Huo, Y.; Xu, Z.; Moon, H.; Bao, S.; Assad, A.; Moyo, T.K.; Savona, M.R.; Abramson, R.G.; Landman, B.A. SynSeg-Net: Synthetic Segmentation without Target Modality Ground Truth. IEEE Trans. Med. Imaging 2019, 38, 1016–1025. [Google Scholar] [CrossRef]
- Kossen, T.; Subramaniam, P.; Madai, V.I.; Hennemuth, A.; Hildebrand, K.; Hilbert, A.; Sobesky, J.; Livne, M.; Galinovic, I.; Khalil, A.A.; et al. Synthesizing anonymized and labeled TOF-MRA patches for brain vessel segmentation using generative adversarial networks. Comput. Biol. Med. 2021, 131, 104254. [Google Scholar] [CrossRef]
- Yu, W.; Lei, B.; Ng, M.K.; Cheung, A.C.; Shen, Y.; Wang, S. Tensorizing GAN with High-Order Pooling for Alzheimer’s Disease Assessment. IEEE Trans. Neural Netw. Learn. Syst. 2021, 33, 4945–4959. [Google Scholar] [CrossRef]
- Wu, X.; Bi, L.; Fulham, M.; Feng, D.D.; Zhou, L.; Kim, J. Unsupervised brain tumor segmentation using a symmetric-driven adversarial network. Neurocomputing 2021, 455, 242–254. [Google Scholar] [CrossRef]
- Asma-Ull, H.; Yun, I.D.; Han, D. Data Efficient Segmentation of Various 3D Medical Images Using Guided Generative Adversarial Networks. IEEE Access 2020, 8, 102022–102031. [Google Scholar] [CrossRef]
- Tong, N.; Gou, S.; Yang, S. Shape constrained fully convolutional DenseNet with adversarial training for multiorgan segmentation on head and neck CT and low-field MR images. Med. Phys. 2019, 46, 2669–2682. [Google Scholar] [CrossRef]
- Yuan, W.; Wei, J.; Wang, J.; Ma, Q.; Tasdizen, T. Unified generative adversarial networks for multimodal segmentation from unpaired 3D medical images. Med. Image Anal. 2020, 64, 101731. [Google Scholar] [CrossRef]
- Chen, Y.; Yang, X.; Cheng, K.; Li, Y.; Liu, Z.; Shi, Y. Efficient 3D Neural Networks with Support Vector Machine for Hippocampus Segmentation. In Proceedings of the 2020 International Conference on Artificial Intelligence and Computer Engineering (ICAICE), Beijing, China, 23–25 October 2020; pp. 337–341. [Google Scholar] [CrossRef]
- Fu, X.; Chen, C.; Li, D. Survival prediction of patients suffering from glioblastoma based on two-branch DenseNet using multi-channel features. Int. J. Comput. Assist. Radiol. Surg. 2021, 16, 207–217. [Google Scholar] [CrossRef]
- Zhang, C.; Song, Y.; Liu, S.; Lill, S.; Wang, C.; Tang, Z.; You, Y.; Gao, Y.; Klistorner, A.; Barnett, M.; et al. MS-GAN: GAN-Based Semantic Segmentation of Multiple Sclerosis Lesions in Brain Magnetic Resonance Imaging. In Proceedings of the 2018 Digital Image Computing: Techniques and Applications (DICTA), Canberra, Australia, 10–13 December 2018; pp. 1–8. [Google Scholar] [CrossRef]
- Lee, D.; Yoo, J.; Tak, S.; Ye, J.C. Deep residual learning for accelerated MRI using magnitude and phase networks. IEEE Trans. Biomed. Eng. 2018, 65, 1985–1995. [Google Scholar] [CrossRef]
- Shaul, R.; David, I.; Shitrit, O.; Riklin Raviv, T. Subsampled brain MRI reconstruction by generative adversarial neural networks. Med. Image Anal. 2020, 65, 101747. [Google Scholar] [CrossRef]
- Quan, T.M.; Nguyen-Duc, T.; Jeong, W.K. Compressed Sensing MRI Reconstruction Using a Generative Adversarial Network with a Cyclic Loss. IEEE Trans. Med. Imaging 2018, 37, 1488–1497. [Google Scholar] [CrossRef]
- Li, G.; Lv, J.; Wang, C. A Modified Generative Adversarial Network Using Spatial and Channel-Wise Attention for CS-MRI Reconstruction. IEEE Access 2021, 9, 83185–83198. [Google Scholar] [CrossRef]
- Lv, J.; Li, G.; Tong, X.; Chen, W.; Huang, J.; Wang, C.; Yang, G. Transfer learning enhanced generative adversarial networks for multi-channel MRI reconstruction. Comput. Biol. Med. 2021, 134, 104504. [Google Scholar] [CrossRef]
- Do, W.-J.; Seo, S.; Han, Y.; Chul Ye, J.; Hong Choi, S.; Park, S.-H. Reconstruction of multicontrast MR images through deep learning. Med. Phys. 2019, 47, 983–997. [Google Scholar] [CrossRef]
- Gu, J.; Li, Z.; Wang, Y.; Yang, H.; Qiao, Z.; Yu, J. Deep Generative Adversarial Networks for Thin-Section Infant MR Image Reconstruction. IEEE Access 2019, 7, 68290–68304. [Google Scholar] [CrossRef]
- Han, C.; Rundo, L.; Murao, K.; Noguchi, T.; Shimahara, Y.; Milacski, Z.Á.; Koshino, S.; Sala, E.; Nakayama, H.; Satoh, S. MADGAN: Unsupervised medical anomaly detection GAN using multiple adjacent brain MRI slice reconstruction. BMC Bioinform. 2021, 22, 31. [Google Scholar] [CrossRef] [PubMed]
- Chai, Y.; Xu, B.; Zhang, K.; Lepore, N.; Wood, J.C. MRI restoration using edge-guided adversarial learning. IEEE Access 2020, 8, 83858–83870. [Google Scholar] [CrossRef] [PubMed]
- Wegmayr, V.; Horold, M.; Buhmann, J.M. Generative aging of brain MRI for early prediction of MCI-AD conversion. Proc. Int. Symp. Biomed. Imaging 2019, 2019, 1042–1046. [Google Scholar] [CrossRef]
- Guo, X.; Wu, L.; Zhao, L. Deep Graph Translation. IEEE Trans. Neural Netw. Learn. Syst. 2022, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Nebli, A.; Rekik, I. Adversarial brain multiplex prediction from a single brain network with application to gender fingerprinting. Med. Image Anal. 2021, 67, 101843. [Google Scholar] [CrossRef]
- Wang, L. 3D Cgan Based Cross-Modality Mr Image Synthesis for Brain Tumor Segmentation. In Proceedings of the 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), Washington, DC, USA, 4–7 April 2018; pp. 626–630. [Google Scholar]
- Dar, S.U.H.; Yurt, M.; Shahdloo, M.; Ildiz, M.E.; Tinaz, B.; Cukur, T. Prior-guided image reconstruction for accelerated multi-contrast mri via generative adversarial networks. IEEE J. Sel. Top. Signal Process. 2020, 14, 1072–1087. [Google Scholar] [CrossRef]
- Chen, Y.; Jakary, A.; Avadiappan, S.; Hess, C.P.; Lupo, J.M. QSMGAN: Improved Quantitative Susceptibility Mapping using 3D Generative Adversarial Networks with increased receptive field. Neuroimage 2020, 207, 116389. [Google Scholar] [CrossRef]
- Ji, J.; Liu, J.; Han, L.; Wang, F. Estimating Effective Connectivity by Recurrent Generative Adversarial Networks. IEEE Trans. Med. Imaging 2021, 40, 3326–3336. [Google Scholar] [CrossRef]
- Finck, T.; Li, H.; Grundl, L.; Eichinger, P.; Bussas, M.; Mühlau, M.; Menze, B.; Wiestler, B. Deep-Learning Generated Synthetic Double Inversion Recovery Images Improve Multiple Sclerosis Lesion Detection. Investig. Radiol. 2020, 55, 318–323. [Google Scholar] [CrossRef]
- Zhao, Y.; Ma, B.; Jiang, P.; Zeng, D.; Wang, X.; Li, S. Prediction of Alzheimer’s Disease Progression with Multi-Information Generative Adversarial Network. IEEE J. Biomed. Health Inform. 2021, 25, 711–719. [Google Scholar] [CrossRef]
- Ren, Z.; Li, J.; Xue, X.; Li, X.; Yang, F.; Jiao, Z.; Gao, X. Reconstructing seen image from brain activity by visually-guided cognitive representation and adversarial learning. Neuroimage 2021, 228, 117602. [Google Scholar] [CrossRef]
- Goldfryd, T.; Gordon, S.; Raviv, T.R. Deep Semi-Supervised Bias Field Correction of Mr Images. In Proceedings of the 2021 IEEE 18th International Symposium on Biomedical Imaging (ISBI), Nice, France, 13–16 April 2021; pp. 1836–1840. [Google Scholar]
- Meliadò, E.F.; Raaijmakers, A.J.E.; Sbrizzi, A.; Steensma, B.R.; Maspero, M.; Savenije, M.H.F.; Luijten, P.R.; van den Berg, C.A.T. A deep learning method for image-based subject-specific local SAR assessment. Magn. Reson. Med. 2020, 83, 695–711. [Google Scholar] [CrossRef]
- Parkes, L.; Fulcher, B.; Yücel, M.; Fornito, A. An evaluation of the efficacy, reliability, and sensitivity of motion correction strategies for resting-state functional MRI. Neuroimage 2018, 171, 415–436. [Google Scholar] [CrossRef]
- Yendiki, A.; Koldewyn, K.; Kakunoori, S.; Kanwisher, N.; Fischl, B. Spurious group differences due to head motion in a diffusion MRI study. Neuroimage 2014, 88, 79–90. [Google Scholar] [CrossRef]
- Johnson, P.M.; Drangova, M. Conditional generative adversarial network for 3D rigid-body motion correction in MRI. Magn. Reson. Med. 2019, 82, 901–910. [Google Scholar] [CrossRef]
- Armanious, K.; Gatidis, S.; Nikolaou, K.; Yang, B.; Thomas, K. Retrospective Correction of Rigid and Non-Rigid Mr Motion Artifacts Using Gans. In Proceedings of the 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), Venice, Italy, 8–11 April 2019; pp. 1550–1554. [Google Scholar]
- Küstner, T.; Armanious, K.; Yang, J.; Yang, B.; Schick, F.; Gatidis, S. Retrospective correction of motion-affected MR images using deep learning frameworks. Magn. Reson. Med. 2019, 82, 1527–1540. [Google Scholar] [CrossRef]
- Wolterink, J.M.; Dinkla, A.M.; Savenije, M.H.F.; Seevinck, P.R.; van den Berg, C.A.T.; Išgum, I. Deep MR to CT synthesis using unpaired data. Lect. Notes Comput. Sci. 2017, 10557, 14–23. [Google Scholar] [CrossRef]
- Armanious, K.; Jiang, C.; Abdulatif, S.; Küstner, T.; Gatidis, S.; Yang, B. Unsupervised medical image translation using Cycle-MeDGAN. In Proceedings of the 2019 27th European Signal Processing Conference (EUSIPCO), A Coruna, Spain, 2–6 September 2019. [Google Scholar] [CrossRef]
- Sajjad, M.; Khan, S.; Muhammad, K.; Wu, W.; Ullah, A.; Baik, S.W. Multi-grade brain tumor classification using deep CNN with extensive data augmentation. J. Comput. Sci. 2019, 30, 174–182. [Google Scholar] [CrossRef]
- Rejusha, R.R.T.; Vipin Kumar, S.V.K. Artificial MRI Image Generation using Deep Convolutional GAN and its Comparison with other Augmentation Methods. In Proceedings of the 2021 International Conference on Communication, Control and Information Sciences (ICCISc), Idukki, India, 16–18 June 2021. [Google Scholar] [CrossRef]
- Zhang, X.; Yang, Y.; Wang, H.; Ning, S.; Wang, H. Deep Neural Networks with Broad Views for Parkinson’s Disease Screening. In Proceedings of the 2019 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), San Diego, CA, USA, 18–21 November 2019; pp. 1018–1022. [Google Scholar] [CrossRef]
- Ge, C.; Gu, I.Y.H.; Store Jakola, A.; Yang, J. Cross-Modality Augmentation of Brain Mr Images Using a Novel Pairwise Generative Adversarial Network for Enhanced Glioma Classification. In Proceedings of the 2019 IEEE International Conference on Image Processing (ICIP), Taipei, Taiwan, 22–25 September 2019; pp. 559–563. [Google Scholar] [CrossRef]
- Deepak, S.; Ameer, P.M. MSG-GAN Based Synthesis of Brain MRI with Meningioma for Data Augmentation. In Proceedings of the 2020 IEEE International Conference on Electronics, Computing and Communication Technologies (CONECCT), Bangalore, India, 2–4 July 2020. [Google Scholar] [CrossRef]
- Han, C.; Rundo, L.; Araki, R.; Nagano, Y.; Furukawa, Y.; Mauri, G.; Nakayama, H.; Hayashi, H. Combining noise-to-image and image-to-image GANs: Brain MR image augmentation for tumor detection. IEEE Access 2019, 7, 156966–156977. [Google Scholar] [CrossRef]
- Ge, C.; Gu, I.Y.H.; Jakola, A.S.; Yang, J. Enlarged Training Dataset by Pairwise GANs for Molecular-Based Brain Tumor Classification. IEEE Access 2020, 8, 22560–22570. [Google Scholar] [CrossRef]
- Sanders, J.W.; Chen, H.S.M.; Johnson, J.M.; Schomer, D.F.; Jimenez, J.E.; Ma, J.; Liu, H.L. Synthetic generation of DSC-MRI-derived relative CBV maps from DCE MRI of brain tumors. Magn. Reson. Med. 2021, 85, 469–479. [Google Scholar] [CrossRef] [PubMed]
- Mukherkjee, D.; Saha, P.; Kaplun, D.; Sinitca, A.; Sarkar, R. Brain tumor image generation using an aggregation of GAN models with style transfer. Sci. Rep. 2022, 12, 9141. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Lu, Y.; Mane, R.; Guan, C. Deep Learning for Neuroimaging Segmentation with a Novel Data Augmentation Strategy. Proc. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. EMBS 2020, 2020, 1516–1519. [Google Scholar] [CrossRef]
- Biswas, A.; Bhattacharya, P.; Maity, S.P.; Banik, R. Data Augmentation for Improved Brain Tumor Segmentation. IETE J. Res. 2021, 1–11. [Google Scholar] [CrossRef]
- Geng, X.; Yao, Q.; Jiang, K.; Zhu, Y.Q. Deep Neural Generative Adversarial Model based on VAE + GAN for Disorder Diagnosis. In Proceedings of the 2020 International Conference on Internet of Things and Intelligent Applications (ITIA), Zhenjiang, China, 27–29 November 2020. [Google Scholar] [CrossRef]
- Barile, B.; Marzullo, A.; Stamile, C.; Durand-Dubief, F.; Sappey-Marinier, D. Data augmentation using generative adversarial neural networks on brain structural connectivity in multiple sclerosis. Comput. Methods Programs Biomed. 2021, 206, 106113. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Du, C.; Wang, S.; Wang, H.; He, H. Multi-subject data augmentation for target subject semantic decoding with deep multi-view adversarial learning. Inf. Sci. 2021, 547, 1025–1044. [Google Scholar] [CrossRef]
- Budianto, T.; Nakai, T.; Imoto, K.; Takimoto, T.; Haruki, K. Dual-encoder Bidirectional Generative Adversarial Networks for Anomaly Detection. In Proceedings of the 2020 19th IEEE International Conference on Machine Learning and Applications (ICMLA), Miami, FL, USA, 14–17 December 2020; pp. 693–700. [Google Scholar] [CrossRef]
- Platscher, M.; Zopes, J.; Federau, C. Image translation for medical image generation: Ischemic stroke lesion segmentation. Biomed. Signal Process. Control 2022, 72, 103283. [Google Scholar] [CrossRef]
- Gu, Y.; Peng, Y.; Li, H. AIDS Brain MRIs Synthesis via Generative Adversarial Networks Based on Attention-Encoder. In Proceedings of the 2020 IEEE 6th International Conference on Computer and Communications (ICCC), Chengdu, China, 11–14 December 2020; pp. 629–633. [Google Scholar] [CrossRef]
- Kneoaurek, K.; Ivanovic2, M.; Weber, D.A. Medical image registration. Eur. News 2000, 31, 5–8. [Google Scholar] [CrossRef]
- Sun, Y.; Gao, K.; Wu, Z.; Li, G.; Zong, X.; Lei, Z.; Wei, Y.; Ma, J.; Yang, X.; Feng, X.; et al. Multi-Site Infant Brain Segmentation Algorithms: The iSeg-2019 Challenge. IEEE Trans. Med. Imaging 2021, 40, 1363–1376. [Google Scholar] [CrossRef]
- Song, X.W.; Dong, Z.Y.; Long, X.Y.; Li, S.F.; Zuo, X.N.; Zhu, C.Z.; He, Y.; Yan, C.G.; Zang, Y.F. REST: A Toolkit for resting-state functional magnetic resonance imaging data processing. PLoS ONE 2011, 6, e25031. [Google Scholar] [CrossRef]
- Gu, Y.; Zeng, Z.; Chen, H.; Wei, J.; Zhang, Y.; Chen, B.; Li, Y.; Qin, Y.; Xie, Q.; Jiang, Z.; et al. MedSRGAN: Medical images super-resolution using generative adversarial networks. Multimed. Tools Appl. 2020, 79, 21815–21840. [Google Scholar] [CrossRef]
- Roychowdhury, S.; Roychowdhury, S. A Modular Framework to Predict Alzheimer’s Disease Progression Using Conditional Generative Adversarial Networks. In Proceedings of the 2020 International Joint Conference on Neural Networks (IJCNN), Glasgow, UK, 19–24 July 2020; pp. 12–19. [Google Scholar] [CrossRef]
- Rezaei, M.; Yang, H.; Meinel, C. Generative Adversarial Framework for Learning Multiple Clinical Tasks. In Proceedings of the 2018 Digital Image Computing: Techniques and Applications (DICTA), Canberra, Australia, 10–13 December 2018; pp. 1–8. [Google Scholar] [CrossRef]
- Gulrajani, I.; Ahmed, F.; Arjovsky, M.; Dumoulin, V.; Courville, A. Improved training of wasserstein GANs. Adv. Neural Inf. Process. Syst. 2017, 2017, 5768–5778. [Google Scholar]
- Zhu, J.-Y.; Park, T.; Isola, P.; Efros, A.A. Unpaired image-to-image translation using cycle-consistent adversarial networks. In Proceedings of the IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 22–29 October 2017; pp. 2223–2232. [Google Scholar]
- Mathieu, M.; Couprie, C.; LeCun, Y. Deep multi-scale video prediction beyond mean square error. In Proceedings of the 4th International Conference on Learning Representations, ICLR 2016, Conference Track Proceedings, San Juan, Puerto Rico, 2–4 May 2016; pp. 1–14. [Google Scholar]






| Ref. No. | Year | Objective | Imaging Modality | DL Methods | Type |
|---|---|---|---|---|---|
| [8] | 2022 | It summarizes GAN’s role in brain MRI. | MRI | GAN | Scoping Review |
| [9] | 2018 | The paper gives GAN training, architecture, and a few application details. | All type | GAN | Overview |
| [10] | 2020 | It summarizes machine learning and DL classification methods. | MRI | CNN 1, RNN 2, GAN, DBM 3 | Review |
| [11] | 2020 | It discusses GAN’s application in radiology. They have quantitatively compared the performance metrics for synthetic images. | CT, MRI, PET, and X-ray | GAN, CNN | SLR |
| Number | Research Questions | Motivation |
|---|---|---|
| RQ 1 | What are the applications of GAN-synthesized images for brain MRI? | The question divides the available literature into more clear categories. |
| RQ 2 | What are the most commonly used loss functions in GAN-synthesized image applications for brain MRI? | Loss function affects the training of GAN. |
| RQ 3 | What are the preprocessing steps performed on ground truth brain MRI? | Preprocessing steps performed on ground truth brain MRIs are crucial for the fidelity of the successive GAN operations. |
| RQ 4 | How to compare the existing evaluation metrics for GAN-synthesized brain MRI? | This question encourages a comparative study of available evaluation metrics. |
| Database | Query | Initial Result |
|---|---|---|
| Web of Science | ((MR Imaging) OR (MRI) OR (magnetic resonance imaging)) AND ((Brain Imaging) OR (Brain Images)) AND (GAN OR Generative Adversarial Network) | 210 |
| Scopus | ((MR Imaging) OR (MRI) OR (magnetic resonance imaging)) AND ((Brain Imaging) OR (Brain Images)) AND (GAN OR Generative Adversarial Network) | 389 |
| Inclusion Criteria | Exclusion Criteria |
|---|---|
|
|
|
|
|
|
|
|
|
|
|
| Ref. No. | GAN Model | Technique |
|---|---|---|
| MRI-to-CT | ||
| [16] | GAN | MI to avoid the issue of unregistered data |
| [17] | CGAN | MI and binary cross entropy are the discriminator loss functions to achieve the task-specific goal |
| [18] | CGAN | Pixel loss penalizes pixel-wise differences between the real and SCT scan |
| [19] | CGAN | The method calculates dosimetric accuracy by SCT generation |
| [20] | MedGAN | Non-adversarial losses (combination of style loss and content loss) to obtain high- and low-frequency details of image |
| [21] | CGAN | Residual blocks are inserted into the CGAN network |
| [22] | GAN | Image-guided radiation therapy |
| [23] | AttentionGAN | The attention network helps to predict the regions of interest |
| [24] | UAGGAN | The model identifies the area of the image and puts in a suitable translation to that location |
| [25] | CycleGAN | A dense block allows better one-to-one mapping |
| [26] | CGAN | Constant image patch size. Memory requirement is independent of the image size |
| MRI-to-PET | ||
| [31] | LAGAN | Locality adaptive convolution where the same kernel for every input modality |
| [32] | GAN | Generate adaptive PET templates |
| [33] | Sketcher-Refiner GAN | Generates PET-derived myelin content map from four MRI modalities |
| [34] | TPA-GAN | Integrates pyramid convolution and attention module |
| [35] | BMGAN | Use of image contexts and latent vectors for a generation |
| Ref. No. | GAN Model | Technique |
|---|---|---|
| [40] | CGAN | Slice-to-volume registration |
| [41] | CycleGAN | Symmetric registration |
| [42] | GAN | Multi-atlas-based brain image parcellation |
| [43] | GAN | Multi-atlas-guided deep learning parcellation |
| [44] | GAN | 3D image registration |
| [45] | GAN | Transfer learning for registration |
| Ref. No. | GAN Model | Technique |
|---|---|---|
| [48] | MSGAN | Lesion-Focused SR method |
| [49] | SRGAN | Use of shaping network |
| [50] | SRGAN | Progressive upscaling method to generate true colors |
| [52] | ESRGAN | Slices from 3 latitudes are used for SR |
| [53] | NESRGAN | Noise and interpolated sampling |
| [54] | MedSRGAN | Residual whole map attention to interpolate |
| [55] | GAN | Medical image arbitrary-scale super-resolution method |
| [57] | GAN | Improving resolution of through-plane slices |
| [58] | GAN | The image resolution of 1.5-T scanner is made equivalent to 3-T scanner. |
| [59] | FPGAN | Use a divide-and-conquer manner with multiple subbands in the wavelet domain |
| [60] | End-to-end GAN | Uses a hierarchical structure |
| Loss Function | Description | Probability— Based (Yes/No) | Ref. No. |
|---|---|---|---|
| Commonly used loss functions | |||
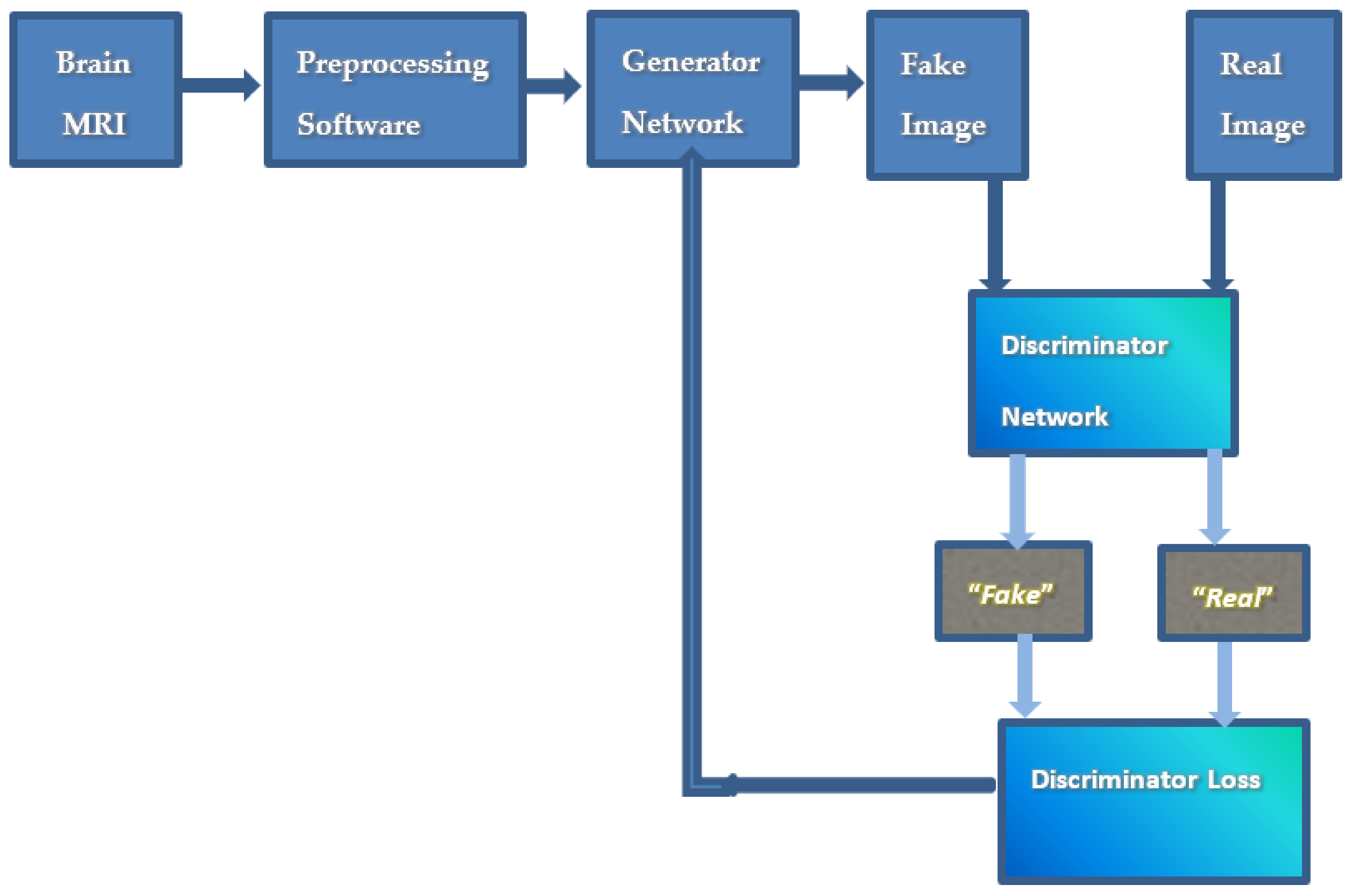
| Adversarial loss | The adversarial loss function is created in the repeated production and classification cycle. The generator minimizes the loss function, and the discriminator maximizes it. (G,D) = Ex,y [log (D(x,y)] + Ex,y [log(1 − D(x,G(x,z))] where y is the ground truth image, G is the generator network, G (x,z) is generated image, D is the discriminator network. | Yes | [20,25,35,41,45,53,54,55,57,59,67,72,74,77,81,83] |
| Cycle consistency loss | A cycle consistency loss allows the generator to learn a one-to-one mapping from the input image field to the target image field. | Yes | [24,28,45,67,71,74,81,82,88] |
| L1 loss | The L1 loss also called mean absolute error (MAE), is a pixel-wise error that shows over-smoothing in resultant images. where n is the number of voxels in an image, ||.||1 is the sum of voxel-wise residuals | No | [19,20,21,23,25,26,33,34,35,37,52,55] |
| L2 loss | The L2 loss also called Mean Squared Distance (MSD) indicates the error between generated and original images and gives faint images. where ||.||2 is the sum of squared voxel-wise residuals of intensity value. | No | [50,51,68,69,80] |
| Perceptual loss | Pixel-reconstruction losses give blurry effects in the final outputs and cannot express the image’s perceptual quality. The perceptual loss is the Euclidean distance in feature space to extract semantic features from target images. where ∅ is a feature extractor, and w, h, and d represent the dimensions of feature maps. | No | [20,35,37,52,53,55,56,66,67,92] |
| Wasserstein loss | WGAN evaluates the Earth Mover’s distance by training the discriminator network and is bounded by a Lipschitz constraint. where x is sampled from real image ‘r’ and noise ‘n’ is the hyperparameter. | Yes | [29,48,49,56,86,92,93,103] |
| Other loss functions | |||
| Attention regularization loss | It ensures learning orthogonal attention maps. | No | [77] |
| Binary cross entropy (BCE) loss | The negative of the logarithm function is used for predicting the probability during binary classification. | Yes | [34,42,43,50,58,65,69] |
| Classification loss | It is the average cross-entropy value and the discriminator’s logistic sigmoid result. | Yes | [58,102] |
| Cycle-perceptual loss | This loss captures the high-level perceptual errors between original and dummy images. | No | [142] |
| Fidelity loss | The fidelity loss factor indicates the dissimilarity between the fake and the spatial normalized image and is generally added to the discriminator loss function. | No | [115,116] |
| Gradient difference (GD) loss | The GD loss is the gradient difference between the original and dummy images that retain the sharpness in the synthetic images. | No | [86,88,89] |
| Identity Loss | This loss is responsible for colors and intensities conservation. | Yes | [77] |
| Image alignment loss | It is based on normalized mutual information (NMI) and used for information fusion. | Yes | [82] |
| Mean p distance (MPD) | The lp-norm or mean p distance (MPD) measures the distance between synthetic and original images. | No | [25,27] |
| Mutual information loss | Mutual Information (MI) finds the “information content” in one variable when another variable is fully observed and used as the loss function. | Yes | [16,17,103] |
| Multi-scale L1 loss | Multi-scale features variance between the predicted multi-channel probability map and the actual image. | No | [42,43] |
| Registration loss | This loss penalizes the variance between the translated & transformed image and stimulates local smoothness. | No | [41] |
| Self-adaptive Charbonnier loss | It is the pixel-wise differences between real and fake images. | No | [140] |
| Style-transfer loss | Style-transfer loss enhances the texture and fine structure of the desired target images. | Yes | [140,142] |
| Supervision loss | This loss, denoted by cumulative squared error, measures pixel shifts between original and synthetic images. | No | [41] |
| Symmetry loss | It stresses inverse consistency in the predicted transformations. | No | [41] |
| Synthetic consistency loss | This loss balances the mean absolute error (MAE) and gradient difference (GD), indicating how the generated image lags behind the target image. | No | [72] |
| Voxel-wise loss | This loss can be imposed as a pixel-level penalty between the translated and the original image applicable to only paired datasets. | Yes | [66,77,83] |
| Preprocessing Software | URL | Use | Ref. No. |
|---|---|---|---|
| Freesurfer | http://surfer.nmr.mgh.harvard.edu (accessed on 26 August 2022) | Skull-stripping, Registration, fMRI Analysis | [114,162] |
| Functional magnetic resonance imaging of the Brain Software Library (FSL) | http://fsl.fmrib.ox.ac.uk/ (accessed on 26 August 2022) | Registration, alignment, Skull-stripping | [115,162] |
| Advanced Normalization Tool (ANT) | http://stnava.github.io/ANTs/ (accessed on 26 August 2022) | Registration | [5,49,136,163,164] |
| Statistical Parameter Mapping (SPM) | http://www.fil.ion.ucl.ac.uk/spm (accessed on 26 August 2022) | Skull-stripping | [74,121] |
| Velocity (Varian) | https://www.varian.com/ (accessed on 26 August 2022) | Registration | [25,27,165] |
| Data Processing Assistant for Resting-State fMRI (DPARSF) | http://www.restfmri.net (accessed on 26 August 2022) | Data processing of fMRI | [166] |
| Elastix | https://elastix.lumc.nl/ (accessed on 26 August 2022) | Registration | [68,167] |
| BrainSuite | http://brainsuite.org/ (accessed on 26 August 2022) | Skull-stripping | [162] |
| Evaluation Metric | FR/NR | Description | Assessment Method | Ref. No. |
|---|---|---|---|---|
| Average symmetric surface distance (ASSD) | FR | ASSD measures the average of all Euclidean distances between two image volumes. | Segmented image | [53,77,168] |
| Blind/ Reference-less Image Spatial Quality Evaluator (BRISQUE) | NR | BRISQUE focuses natural scene statistics (NSS) such as ringing, blur, and blocking. It quantifies the reduction of naturalness by locally normalizing the luminance coefficients. | Whole image | [33,58,86] |
| Dice Similarity Coefficient (DSC) | FR | DSC measures the spatial overlap and provides a reproducibility validation score for image segmentation. | Segmented image | [70,77,79,83,96,99,144] |
| Frechet Inception Distance (FID) | FR | The distance between Gaussian distributions of synthetic and real images is FID or the Wasserstein-2 distance. | Whole image | [35,59,84,99,120] |
| Hausdorff Distance (HD)95 | FR | HD measures the maximum Euclidean distance between all surface points of two image volumes. | Segmented image | [115] |
| Jaccard similarity coefficient (JSC) | FR | It is a value used to compare the similarity and diversity of images recognized as Intersection over Union. | Segmented image | [98,99,159] |
| Maximum Mean Discrepancy (MMD) | FR | MMD measures the dissimilarity between the probability distribution of real images over the space of natural images and parameterized distribution of the generated images. | Whole image | [32] |
| Mutual Information Distance (MID) | FR | MID measures the association between corresponding synthetic images in different modalities. It first evaluates the mutual information of synthetic image pairs and real image pairs and then computes their absolute difference. | Whole image | [84,99] |
| Normalized Mean Absolute Error (NMAE) | FR | NMAE measures the estimation errors of a specific color component between the original and synthetic images. | Whole image | [33,74,88] |
| Normalized Mutual Information (NMI) | NR | NMI expresses the amount of information synthetic images carry regarding the original image. | Whole image | [140,148] |
| Normalized Cross-Correlation (NCC) | FR | NCC evaluates the degree to which the synthetic and original image signals are similar. It is an elementary approach to match two image patch positions. | Segmented image | [25] |
| Naturalness Image Quality Evaluator (NIQE) | NR | NIQE is a distance-based measure of natural images’ divergence from statistical consistency. The metric quantifies image quality according to the level of distortions. | Whole image | [33,41,58,88] |
| Peak Signal-to-Noise Ratio (PSNR) | FR | It is an expression for the ratio between the maximum possible power of the original image and the power of the generated image. | Whole image | [48,56,59,73,74,75,85,88] |
| Structural Similarity Index Measure (SSIM) | FR | SSIM score indicates the perceptual difference between original and synthetic images. It compares the visible structures in the image such as Luminance, Contrast, and Structure. | Whole image | [38,59,73,74,75,88] |
| Root-Mean-Square Error (RMSE) | FR | It measures the differences between the predicted value by an estimator and the actual value of a definite variable. | Whole image | [68,72,75,99,140] |
| Universal Quality Index (UQI) | FR | Image distortion is the product of loss of correlation, luminance, and contrast distortion. | Whole image | [14] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tavse, S.; Varadarajan, V.; Bachute, M.; Gite, S.; Kotecha, K. A Systematic Literature Review on Applications of GAN-Synthesized Images for Brain MRI. Future Internet 2022, 14, 351. https://doi.org/10.3390/fi14120351
Tavse S, Varadarajan V, Bachute M, Gite S, Kotecha K. A Systematic Literature Review on Applications of GAN-Synthesized Images for Brain MRI. Future Internet. 2022; 14(12):351. https://doi.org/10.3390/fi14120351
Chicago/Turabian StyleTavse, Sampada, Vijayakumar Varadarajan, Mrinal Bachute, Shilpa Gite, and Ketan Kotecha. 2022. "A Systematic Literature Review on Applications of GAN-Synthesized Images for Brain MRI" Future Internet 14, no. 12: 351. https://doi.org/10.3390/fi14120351
APA StyleTavse, S., Varadarajan, V., Bachute, M., Gite, S., & Kotecha, K. (2022). A Systematic Literature Review on Applications of GAN-Synthesized Images for Brain MRI. Future Internet, 14(12), 351. https://doi.org/10.3390/fi14120351








