1. Introduction
Variational autoencoders (VAEs), graph variational autoencoders (GVAEs) and their variants [
1,
2,
3], the deep generative models that have emerged in recent years, integrate the auto-encoder and variational inference, not only keep the excellent expression of the neural network but solve the randomness of probabilistic latent variables in the generation process. At present, the development of VAEs in the probability models has two primary directions. The first is to design a more expressive variational posterior, the other one is to replace the standard normal distribution prior of VAEs with learning more intricate prior [
4,
5,
6,
7,
8]. Although works in the first direction have made some signs of progress in various kinds of learning tasks, delving into learning more about whether the variational posterior is valid is still interesting. Reference [
9] proved that even with a very strong posterior, VAEs always can not match the learned posterior with N(0, I) prior. So we focus on learning a prior with abundant semantics for GVAEs.
The performance of GVAEs is mainly dependent on the validity of their data representation. Learning the representation of graph data, also known as network representation learning (NRL) [
10,
11,
12], encodes nodes in the network and denotes them in a unified low-dimensional latent space. Hence, one way to improve GVAEs prior is to use more flexible probability distribution modeling latent variables. Gaussian distribution [
13,
14,
15,
16] is the most widely used probability distribution of latent variables in VAE models [
17,
18,
19]. This kind of phenomenon is because of the well-known “central-limit theorem” [
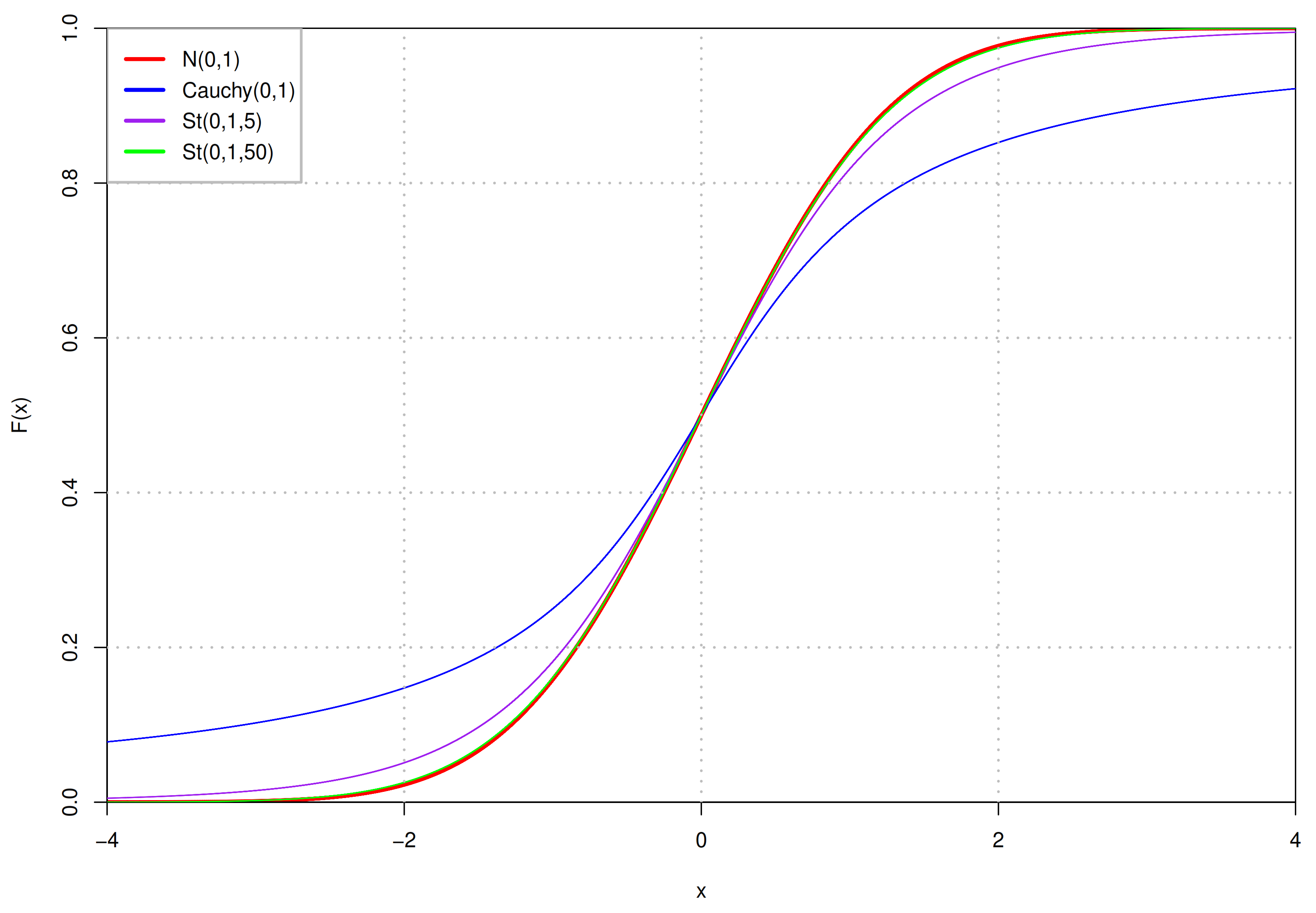
20]. If the sample size is large enough, the distribution of the samples’ mean is approximately Gaussian, irrelevant to the distribution of samples in the population. The mean value of the samples’ mean distribution does not have to be zero, it depends on the value range of samples. More specifically, if there are a set of non-negative samples, their mean value is also non-negative. In addition, Gaussian distribution is the natural selection in the case of known mean and variance owing to its information entropy is the largest under this condition. However, it also has some disadvantages, especially its “empirical rule”, which makes up about
of the numerical values distributed within three standard deviations centered on the mean. When the latent space dimension increases, its ability to capture more information will decline. Nevertheless, the choice of heavy-tailed Student’s t (St) distribution [
21,
22,
23,
24] can significantly remedy this defect. For the reason that it can adaptively model the networks according to the degree of freedom (dof) parameter. The dof parameter controls the curve shape changes of the St distribution. The smaller the dof, the flatter the St distribution curve. Conversely, the larger the dof, the curve is closer to the Gaussian distribution with the same mean and scale parameters. Because most of the real-world networks are not random, the degree distribution in different networks more or less has the power-law property [
25,
26]. For instance, in the American airline network, nodes are airports, and the links between them are airlines. The nodes such as John F. Kennedy International Airport and Los Angeles International Airport are hubs with the power-law property. However, there are just several hubs in a network, and most nodes have a handful of links [
27]. In addition, each latent vector is a node representation, so that we can fit the structure of real-world networks by applying the more general St distribution to model node representations. Concretely, ensuring the variational posterior of latent representation is close to a St distribution with zero mean and one scale, while dof remains unchanged [
4]. They solve the problem that Gaussian prior never rejects outliers in the modeling. Furthermore, they deal with data with image characteristics, which is different from this paper.
As we probe into the data with St distribution, it is a pivotal point that how to statistically model data at an applicable complexity. If finite models used for analysis, we must specify the number of latent features in advance, but it is unknown beforehand and needs to learn from the data. In the paper citation network, latent features are represented by the keywords of a paper. If the latent dimension is small, the latent features are general keywords, such as data mining and the uncertainty of artificial intelligence. If the latent dimension is large, the latent features may be more specific keywords, such as anomaly detection, Bayesian network. By adding latent dimensions, the topic related to the paper can be dug out; then we can find neglected correlation relationship of some papers. However, if the latent dimension is too large, some useless latent features will appear, such as Chebyshev’s inequality. Papers that apply the same inequality are regarded as relevant, but this decision is inaccurate in most cases.
The Bayesian nonparametric approach [
28,
29,
30,
31] gives us a way to solve this problem, which assumes that latent structures grow with data so that an infinite number of latent features can exist in the model. Under this condition, the determination of model complexity is incorporated into the data analysis process without setting manually based on expert knowledge or supervision information. The Dirichlet Process (DP) [
32,
33,
34] is a typical Bayesian nonparametric method, which defines a binary matrix and each row of the matrix represents a node representation, each dimension captures a specific aspect of nodes. DP, as a prior of St distribution, can find possible features of all nodes in networks and also help discover important missing information or links.
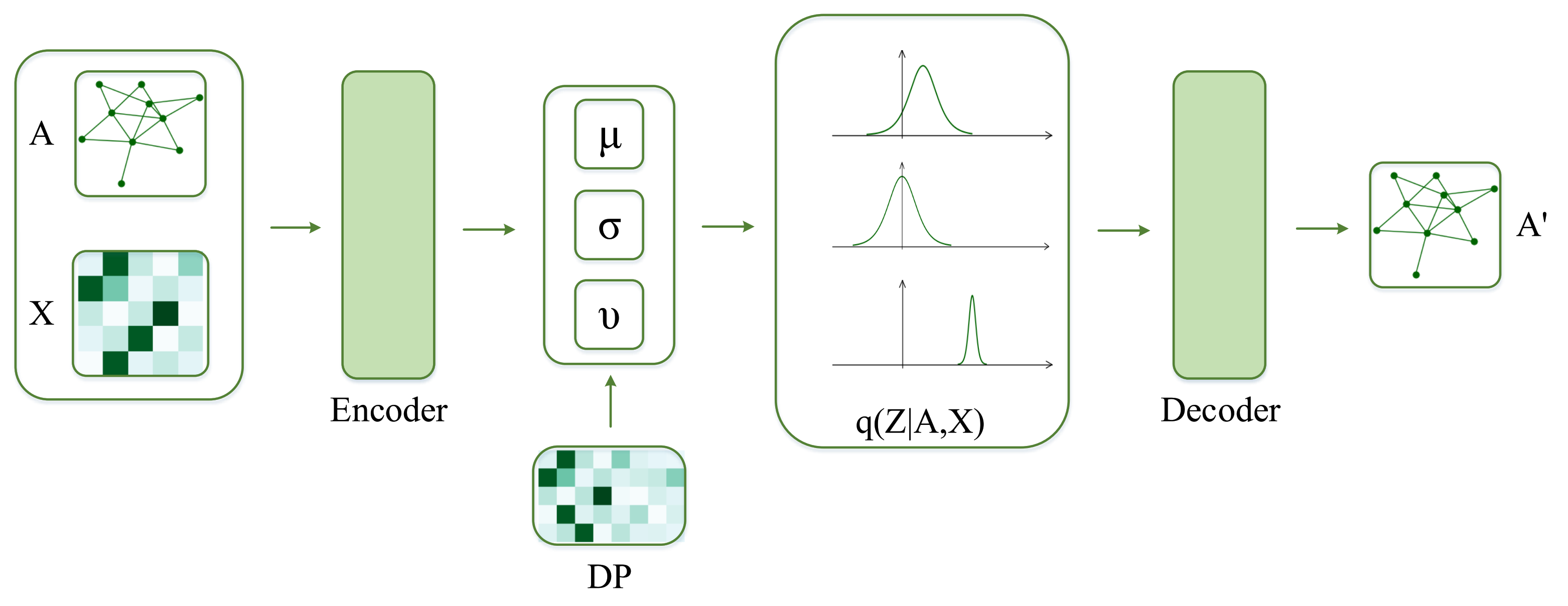
In this paper, we propose DP-St prior for GVAE. In detail, we use St distribution as the probability distribution of a latent variable and solve the problem of limited detection ability of the traditional Gaussian distribution. At the same time, we apply the DP to explore more prior knowledge of nodes in a network, solve the problem that a fixed capacity can not precisely capture the complexity of the available networks well. We integrate DP and St distribution to construct an unbounded latent representation with sufficient expression ability, longer tails, and a deeper dimension to provide tremendous flexibility for latent variables. Our contributions are summarized as follows:
We proposed DP-St prior, DP is the prior of St distribution and combined it with GVAE for network representation learning. The DP-St prior is a heavy-tailed nonparametric process, which can adapt to the dimension of latent variables, generate manifold prior knowledge for GVAE;
We illustrate that the adjustment of the heavy-tailed level of St distribution is conducive to filter out useless latent features and retain the most representative ones. It can help detect missing links in the network and accurately model the real-world networks.
In the experiments, the improvement of link prediction and semi-supervised node classification tasks are demonstrated. In particular, our model can effectively improve the prediction accuracy of link prediction in low-dimensional latent variables caused by St distribution modeling. It can also obtain the optimal prediction results in the semi-supervised node classification task when the labeled training data is small.
2. Related Work
Exploring the proper form of latent representation is an essential task for many representation learning applications. Moreover, we have seen that learning a prior of latent variables is indispensable to the VAE models. From the theoretical perspective, some recent studies demonstrate the necessity of learning VAE prior. Reference [
35] highlighted the significance of the probability distribution of latent space by analyzing some variants of variational evidence lower bound objective (ELBO). Since it is necessary to complete the VAE optimization process by maximizing the ELBO, and this process is equivalent to minimizing the KL divergence between the variational posterior and prior of latent variables. Furthermore, Reference [
36] illustrates the negative impact of applying a simple prior for VAE models. That is to say, it will result in few active latent variables, and finally produce over-regularized models with unpromising latent representation, which is not conducive to the tasks of presentation learning.
Considering the importance of latent representation for carrying out various learning tasks, some models improved VAE from the perspective of the Gaussian distribution of latent variables, which is commonly used in most VAE models and their variants. Reference [
37] elaborate that applying the nonparametric process to a neural network can automatically ascertain some critical parameters in the model, such as model capacity. They use the stick-breaking process to generate latent variables, and the Stick-Breaking VAE model is constructed by various parameterization methods. This model shows more discriminative ability of latent representation than Gaussian VAEs. Reference [
4] replace the Gaussian distribution of latent variables in the traditional VAE models with a higher error-tolerant St distribution and apply the standard St distribution as a prior, and demonstrate that heavy-tailed latent variables can aggrandize their flexibility when the data contains outliers. Nonetheless, only aggrandizing the tail thickness is far from enough to make the latent variables adapt to the dimension with sufficient flexibility and obtain ample prior knowledge to represent the data. Nonetheless, only enlarging the tail thickness is far from enough to make the latent variables adapt to the dimension with sufficient flexibility and obtain ample prior knowledge to represent the data. All the works mentioned above are used to deal with tasks related to image data, while we intend to do the presentation learning tasks on network data.
There are also a few works that employ the DP prior. In [
38], they focus on the task of various classification through learning logistic-regression methods, and they illustrate that the DP is helpful to learn the similarity between different classification tasks. The DP-GP-LVM (Dirichlet Process-Gaussian Process-Latent Variable Models) [
39] algorithm presents a Bayesian nonparametric latent variable model, apply the Gaussian process prior and DP prior at the same time. More precisely, they are used for producing latent variables and learning structure, respectively. However, neither of these works has considered the application of DP prior to latent variables. Our model takes full advantage of DP as a nonparametric method in the adaptive dimension and facilitates the learning of appropriate representation for latent variables.
5. Experiments
We report the performance of our DP-St GVAE on two tasks—unsupervised link prediction and semi-supervised node classification. Our model has been put into effect on three benchmark citation datasets (Cora, Citeseer, and PubMed) [
44], and the datasets are described in
Table 1 in detail. Each node in these datasets represents literature, and has a unique attribute vector, the connection between literature indicates that there is a reference relationship between them.
In all experiments, the models are realized by TensorFlow, and ADAM is used for training. The encoder is composed of GCN [
45] with parameter
, and we let function
use activation function tanh for the transformation
. In our DP-St GVAE model, the sticking-breaking hyperparameter
is set to 10, and the Uniform distribution hyperparameter
is set to 10.
5.1. Link Prediction
To assess our model’s ability to perform link prediction tasks, we compared it with two algorithms: GVAE [
3] and standard tGVAE [
4]. Our model improves the problem that the standard St distribution as a prior can not fully discover the latent information for a large portion of nodes in networks, and in our experiments, it is applied to the network data, whose encoder and decoder are changed to use the same settings as our method, then we denoted it as St GVAE.
For this task, we will follow the settings of GVAE and make the encoder use GCN of two layers to conduct four groups of comparative experiments, the dimensions of latent space are 8, 16, 32, and 64, and the corresponding GCN hidden layer sizes are 16, 32, 64, and 128 respectively. All experiments use a learning rate of 0.01. When the learning rate is 0.1, it is difficult for the model to converge in training. When the learning rate is 0.001, the model converges very slowly, and its performance in learning tasks is lower than that in the learning rate of 0.01. The validation set and the test set contain 5% and 10% of the network edges. AUC (area under ROC curve) and AP (average accuracy) are used as the assessment criteria for the correct classification of edge types in the networks.
We show link prediction performance on three citation datasets in
Table 2,
Table 3 and
Table 4, and our model is almost better than the contrastive approaches. As we can see from the experimental results, with the augment of the latent space dimension, the predictive performances of GVAE and St GVAE models are increasing, and our model presents a trend like a parabola. This is because our model can automatically select the most appropriate latent dimension, and the latent features under this dimension are conducive to the reconstruction of network structure. Moreover, there are St GVAE with a large predicted improvement span in the low-dimension interval, and GVAE with a uniform rising trend about the dimension. When the dimension of latent space is low, such as 8, the performance of St GVAE is the weakest among the three models, because, in the case of fewer latent features, the use of heavy-tailed St distribution can, to some extent, obscure the differences between various features. This indicates the importance of constructing the St distribution with DP because DP can help find the most critical features for each node among these few latent features. This ranking of importance can digest the disadvantage of heavy-tailed distribution in low-dimensional latent space.
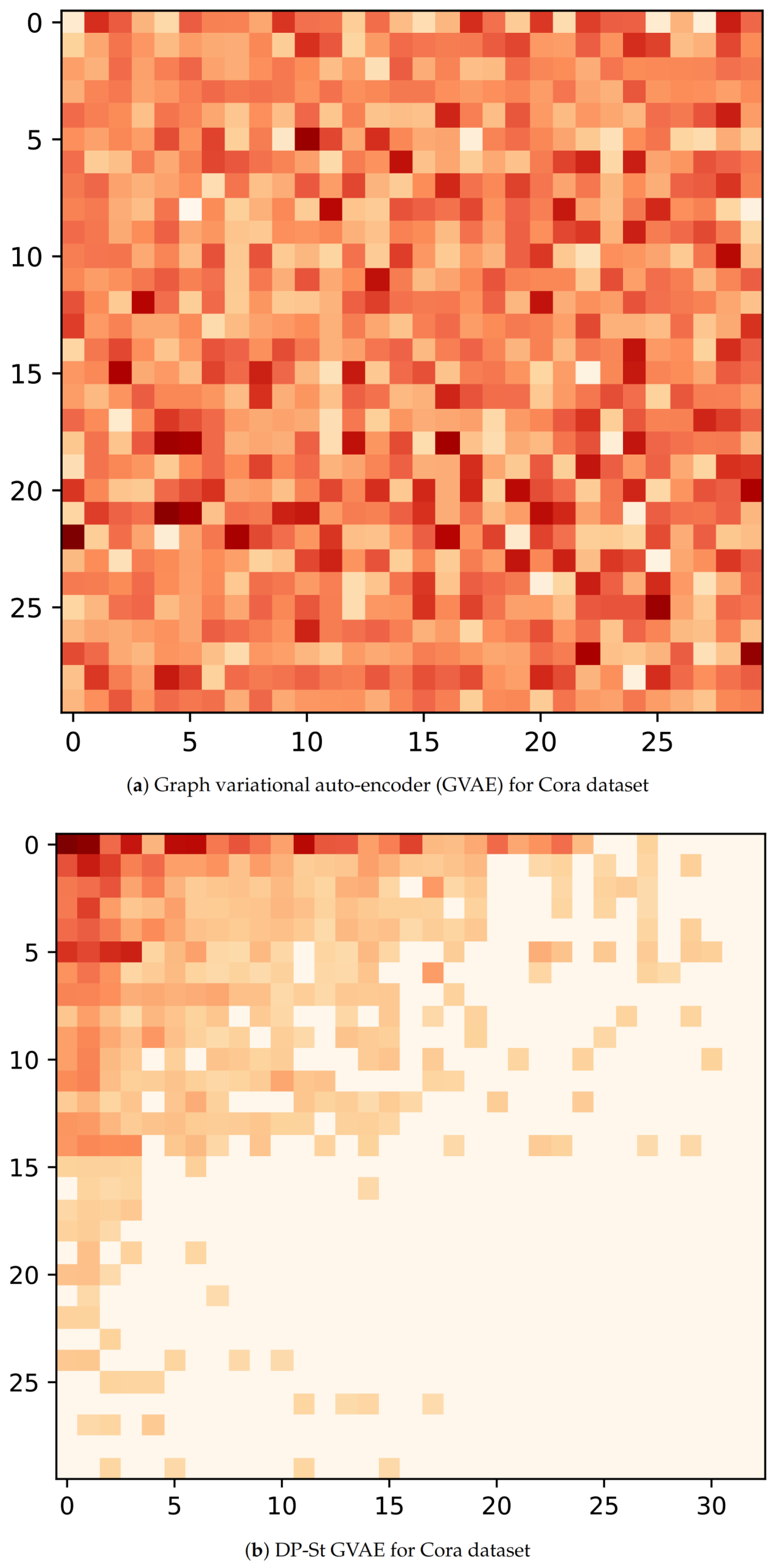
Now, to understand the latent representation
Z, we visualize its posterior probability,
, for GVAE and DP-St GVAE models on Cora and Citeseer datasets. With regard to the node selection for visualization, we first randomly select ten nodes with the most and the least number of latent features with high probability respectively, and then randomly choose 10 nodes in the remaining node-set. In addition, the dimensions of latent variables in the DP-St GVAE model are automatically determined, and for the convenience of comparison, we select similar dimensions for the GVAE model. From
Figure 4a and
Figure 5a, we can see that many nodes have multiple latent features with high probability, and the latent representation contains rich information and complex correlation. However, in
Figure 4b and
Figure 5b, the latent representation has higher sparsity, and the latent features of each node are more explicit. In the same dimension of latent space, the latent representation generated by GVAE will bring more disturbing information and severely restrict the nodes to find the most representative latent features. After adding a DP-St prior, the various information can be screened, and those useless features that have a negative impact on the target task can be discarded to produce appropriate latent representation.
5.2. Semi-Supervised Node Classification
For the semi-supervised node classification task, the goal is to make the label prediction for nodes without labels. So, in this task, we add the cross-entropy term of labels to the objective function (12). We make use of the random split partition method [
46], randomly dividing all nodes in each dataset into three subsets, training set, validation set, and test set. The number of known labels has been separated into three groups, with 5, 10, and 20 labels per class. For this experiment, we compare our model with the GVAE and St GVAE models used in the link prediction task and added several related works in this task—Graph attention network (GAT) [
47], and Graph convolutional neural network (GCN) [
45].
Table 5,
Table 6 and
Table 7, respectively, show the results on the Cora, Citeseer, and PubMed datasets. The experimental results illustrate that our model achieves the optimal classification result on the PubMed dataset, because there is heavy-tailed degree distribution, and our model can precisely grasp the hidden information and obtain a more obvious accuracy improvement. On the Cora and Citesser datasets, our model is able to achieve more competitive results when the number of labels is known to be smaller. When the number of known labels is tiny, we can see that the node classification performance of GVAE is less than
on the Citeseer dataset, suggesting a high error rate in the generated node representation. Furthermore, it is obvious that the St GVAE model improves the prediction accuracy by replacing the Gaussian distribution with the St distribution, indicating that the traditional Gaussian distribution leaves out some essential information for nodes, and the St distribution can avoid this problem. Finally, when we construct a DP prior for the St distribution, each node can capture the most important hidden information, which provides convenience for discovering the correlation between nodes, and thus the affiliation of nodes.