Artificial Neural Networks in Evaluation and Optimization of Modified Release Solid Dosage Forms
Abstract
:1. Introduction

2. Artificial Neural Networks
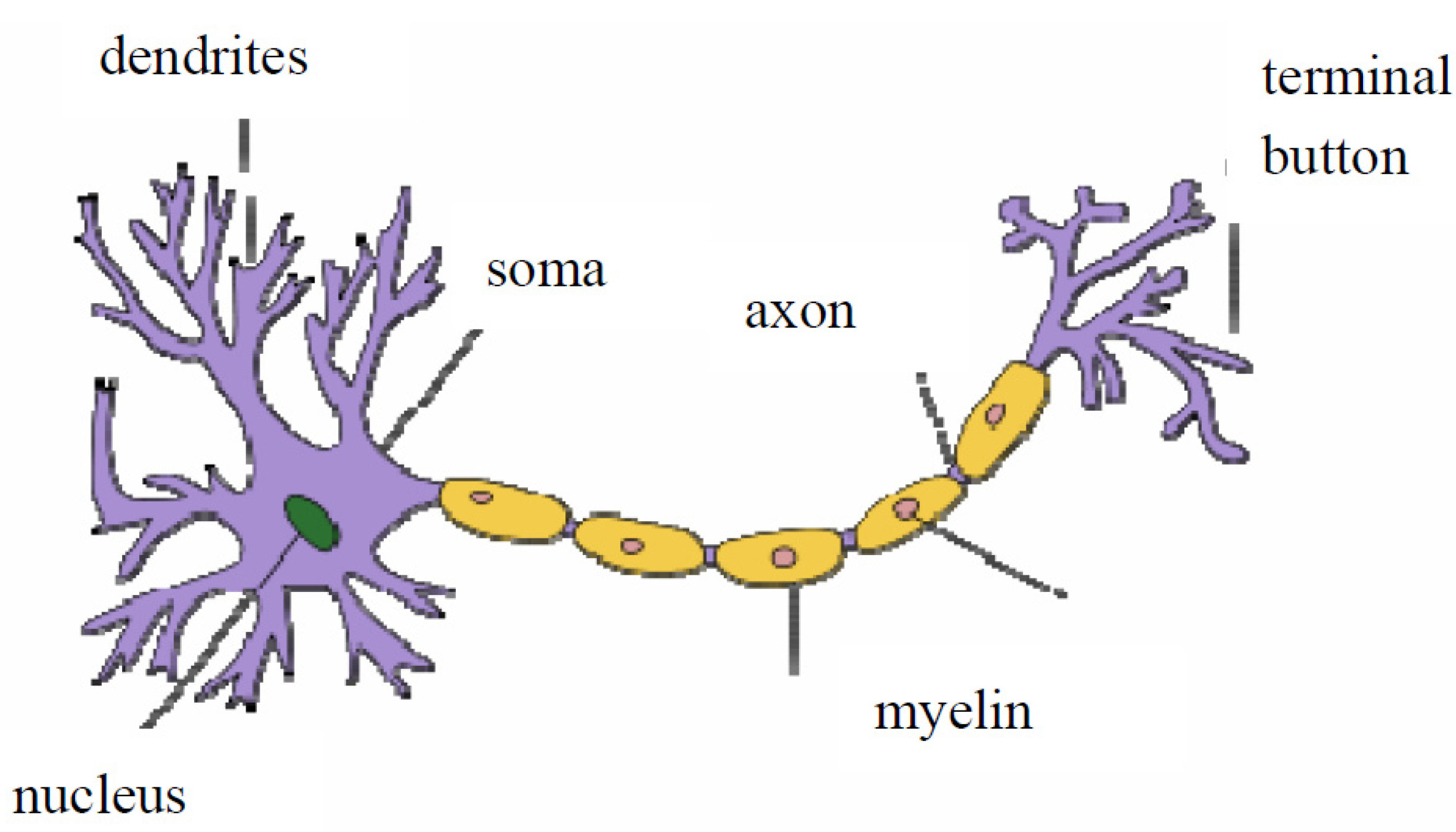
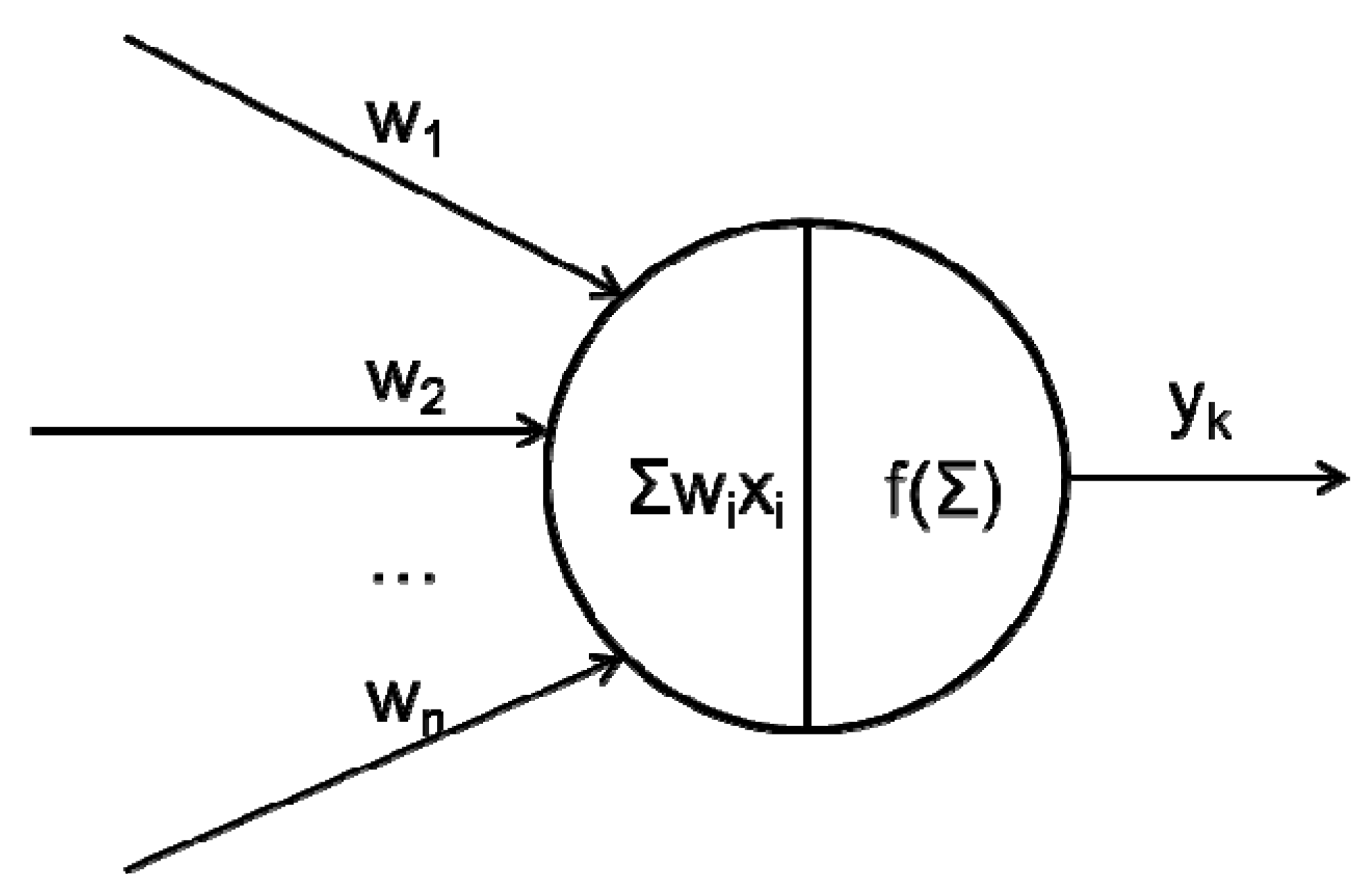
2.1. What Happens between Neurons in a Network?
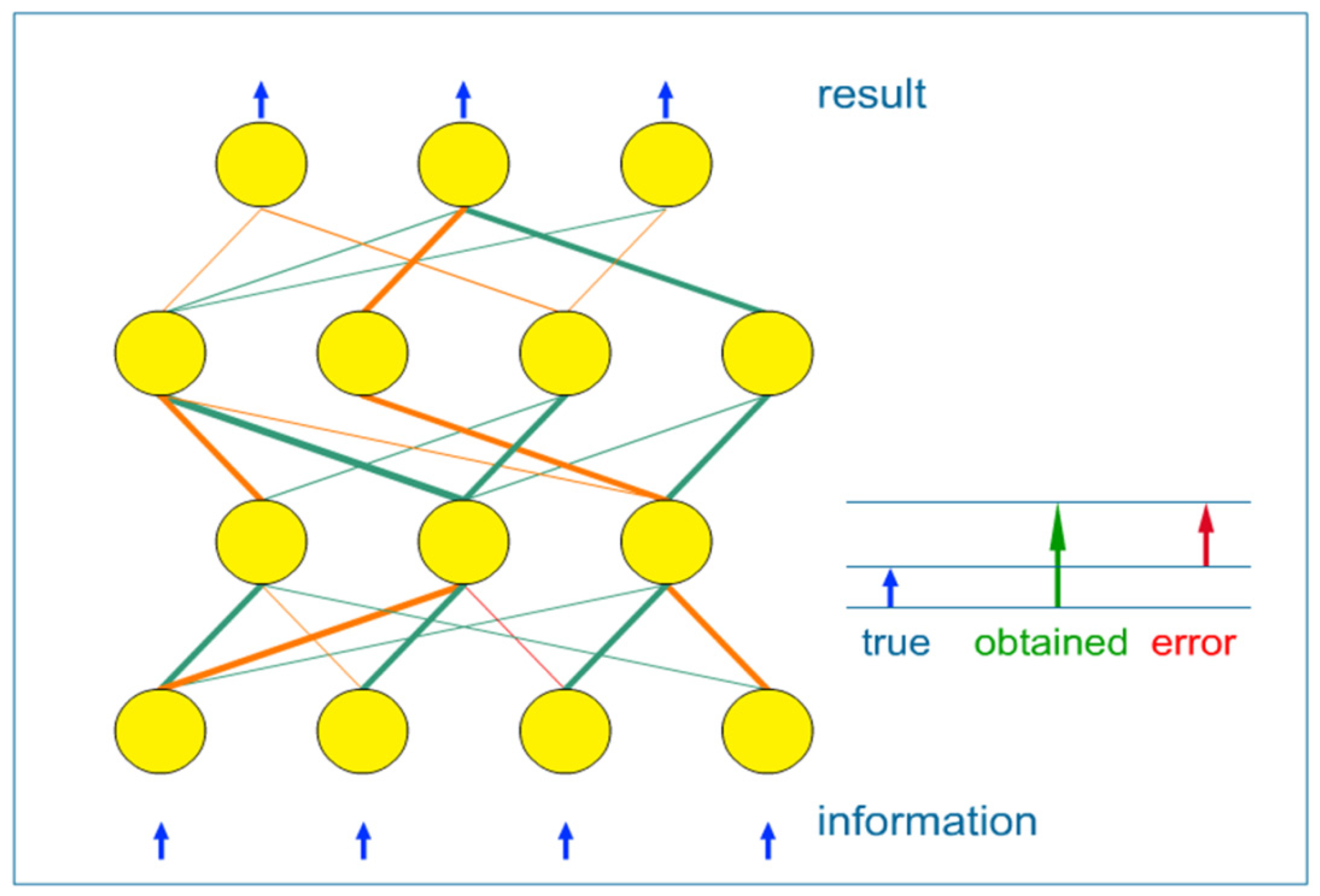
- (1) supervised learning: when network weights are adjusted using the known data, split into input-output pairs. Supervised learning is the most commonly used training method for ANNs and will be further explained in this paper;
- (2) unsupervised learning: it refers to the problem of trying to find hidden structure in unlabeled data. Since the examples given to the learner are unlabeled, there is no error or reward signal to evaluate a potential solution;
- (3) reinforcement learning: which differs from standard supervised learning in that correct input/output pairs are never presented.

| Training of the network |
|
| Usage of the network |
|
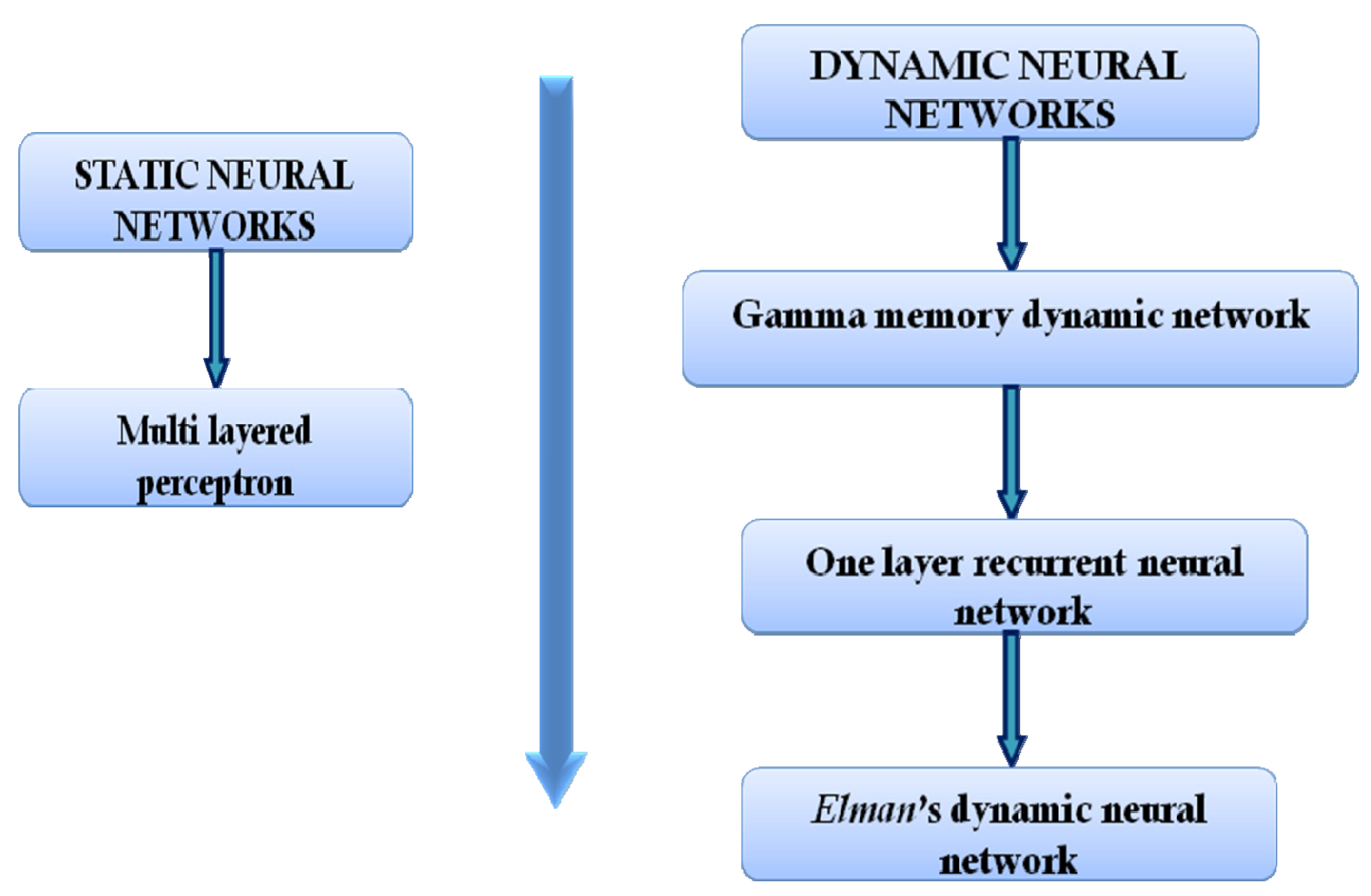
2.2. Basic Topologies (Architectures) of Artificial Neural Networks
2.2.1. Static Networks

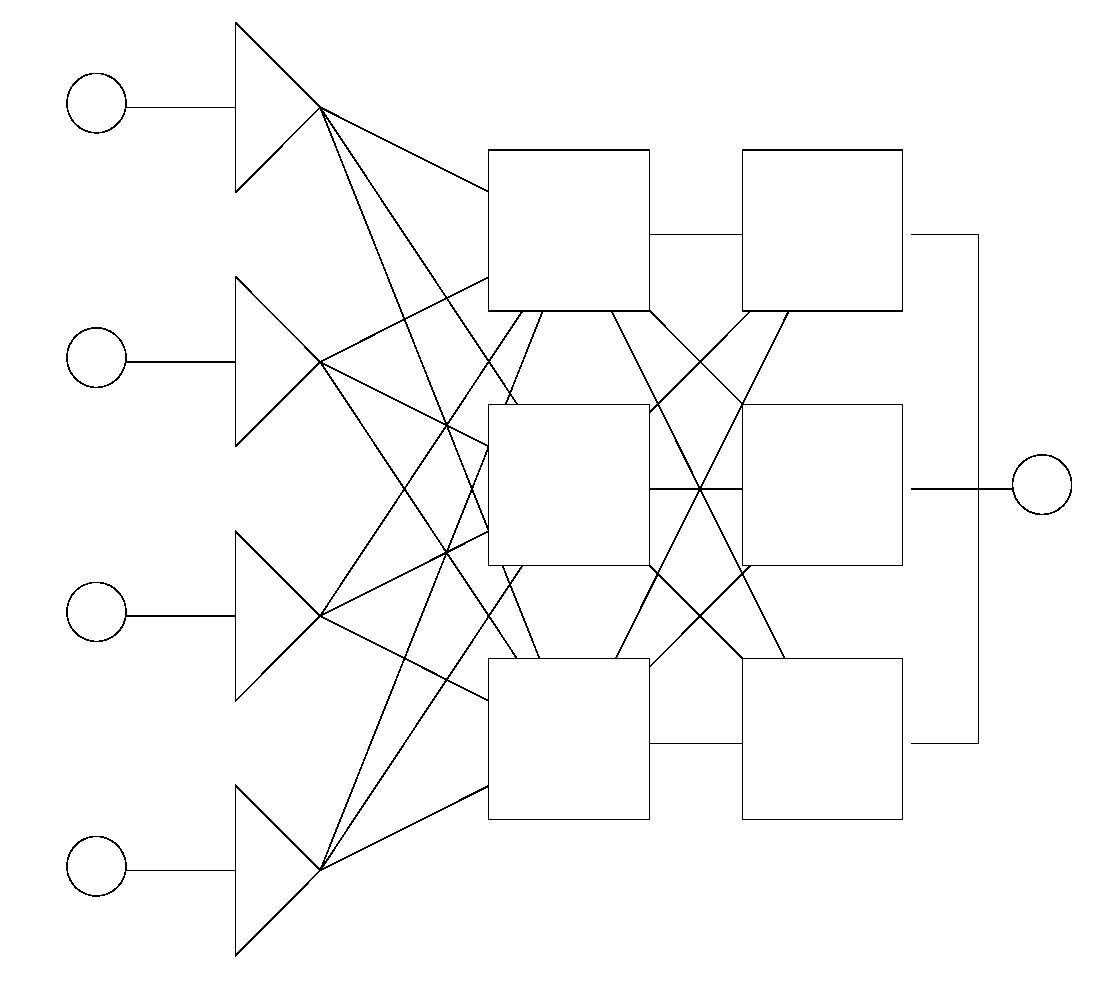
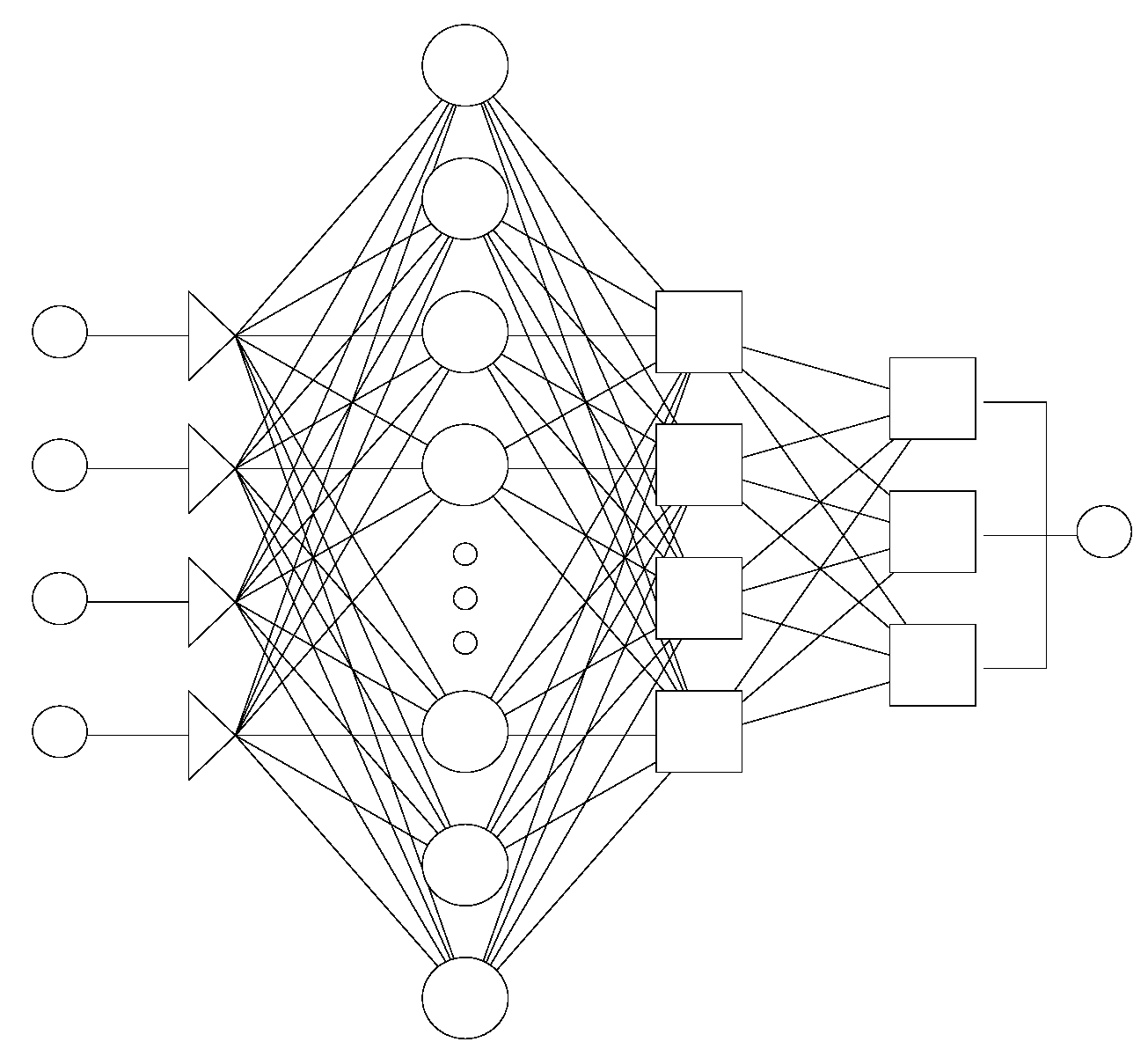


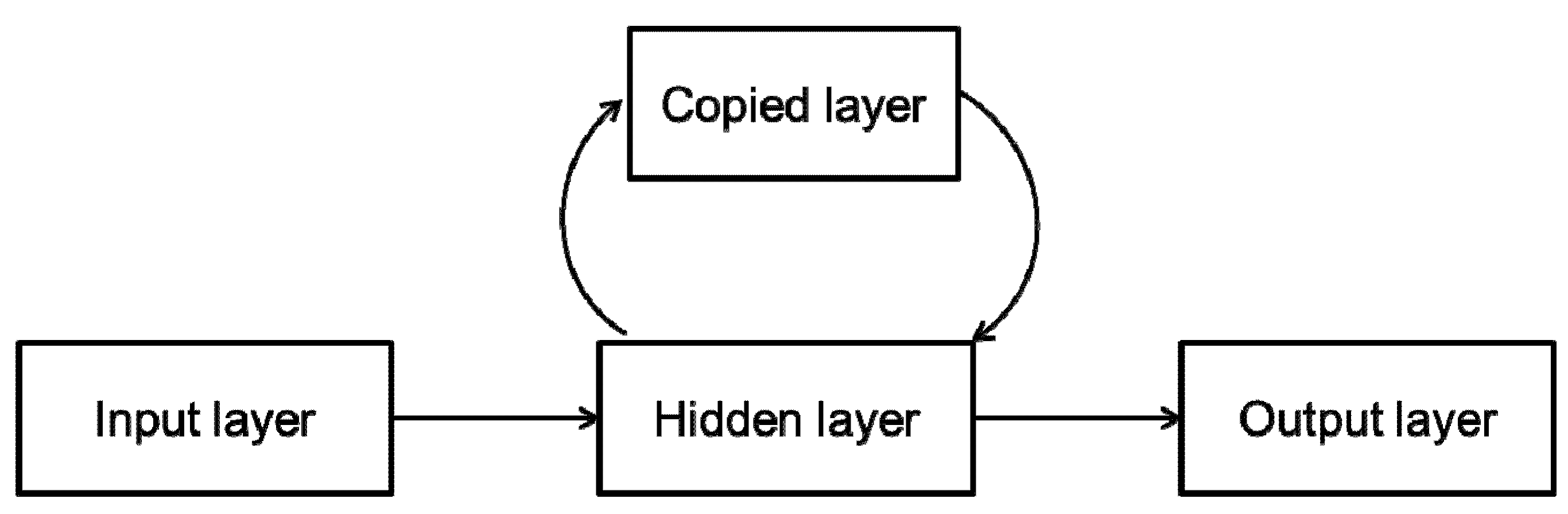
2.2.2. Dynamic Neural Networks

2.3. Application of ANNs in Formulation and Evaluation of Modified Release Dosage Forms
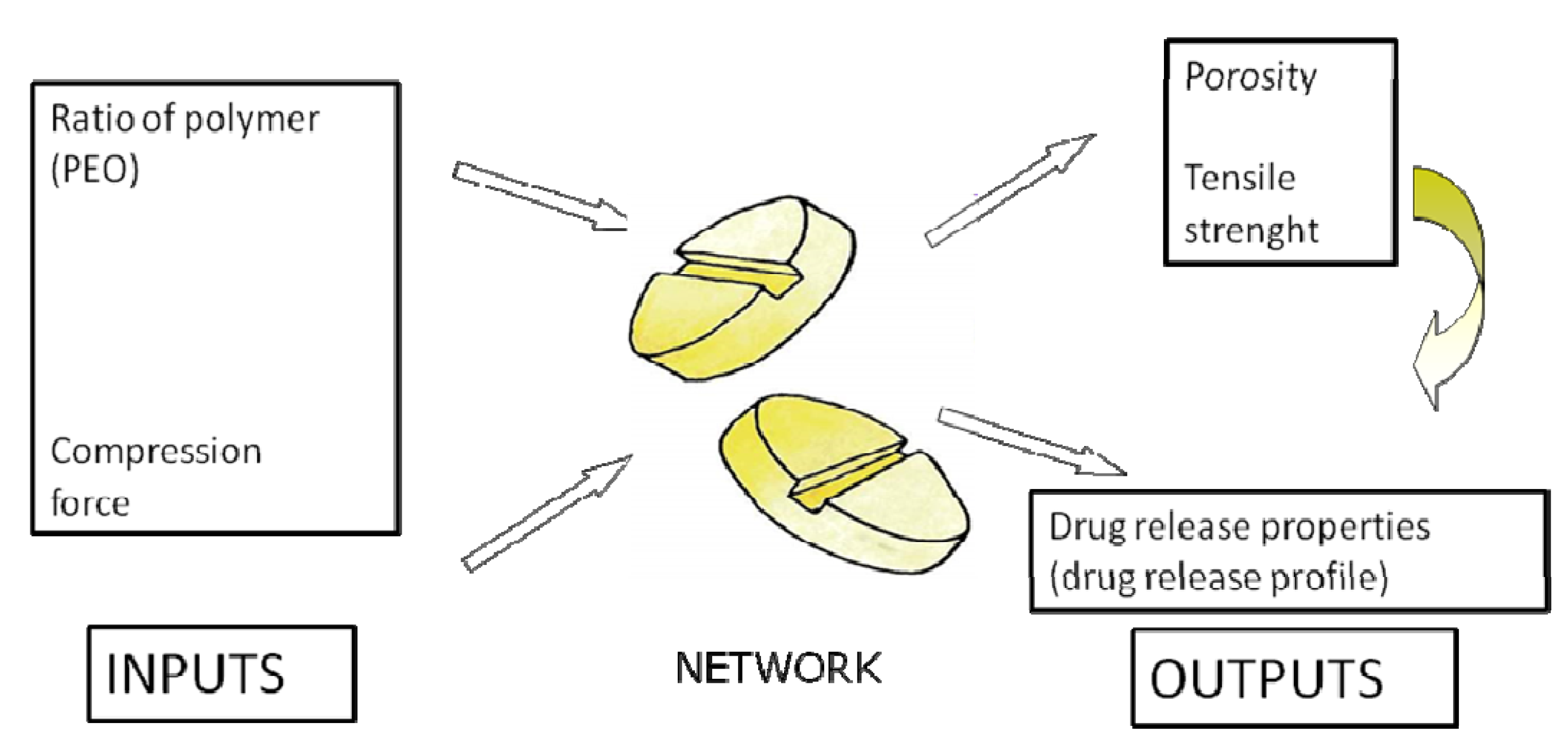
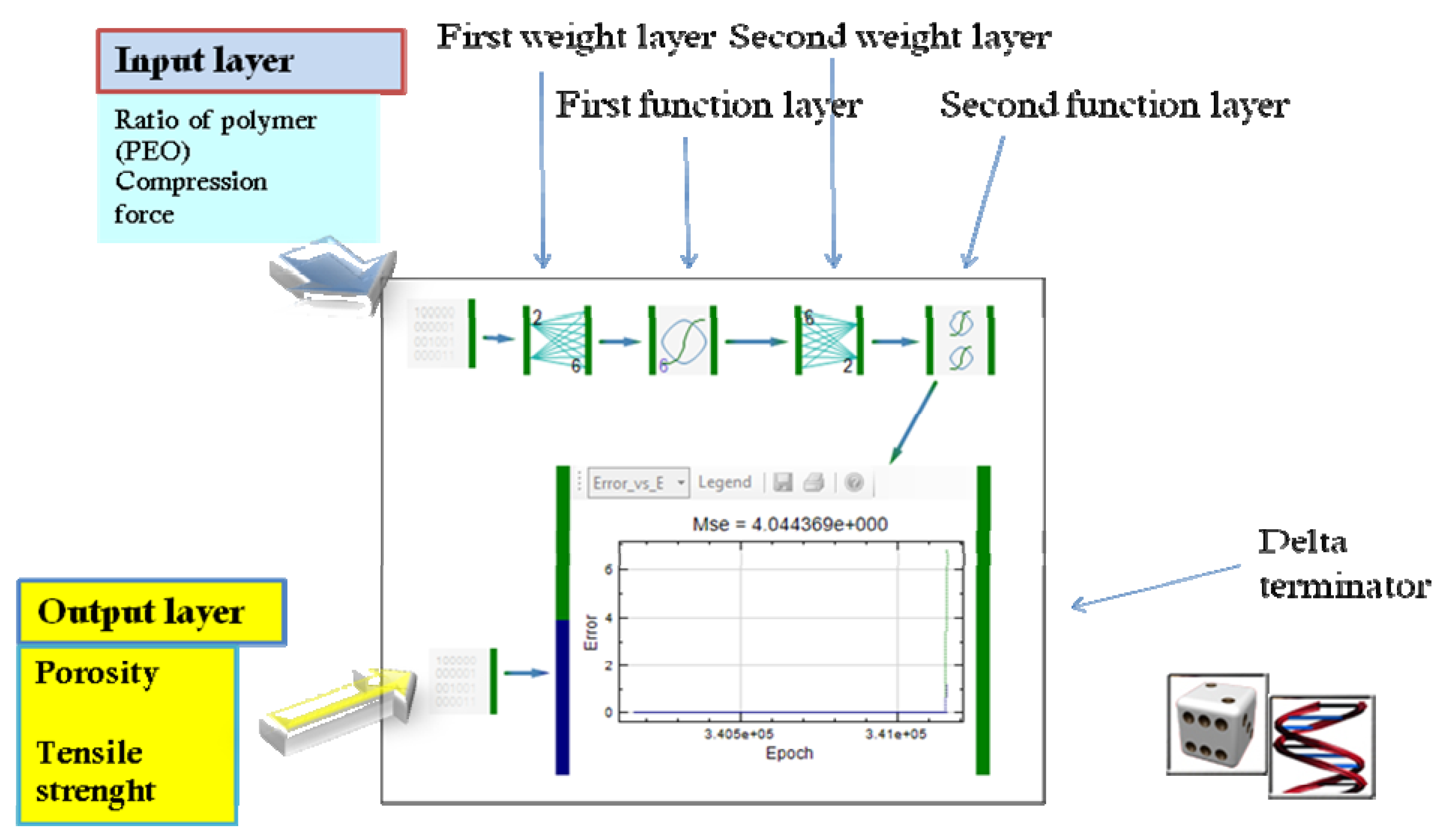
| Formulation | Observed values Porosity (%) and tensile strength (MPa) | Predicted values Porosity (%) and tensile strength (MPa) (r2 = 0.9982) |
|---|---|---|
| Test 1 | 19.55 ± 0.49 1.304 ± 0.042 | 20.34 ± 0.78 1.313 ± 0.155 |
| Test 2 | 17.55 ± 0.55 1.661 ± 0.035 | 17.33 ± 0.78 1.539 ± 0.155 |
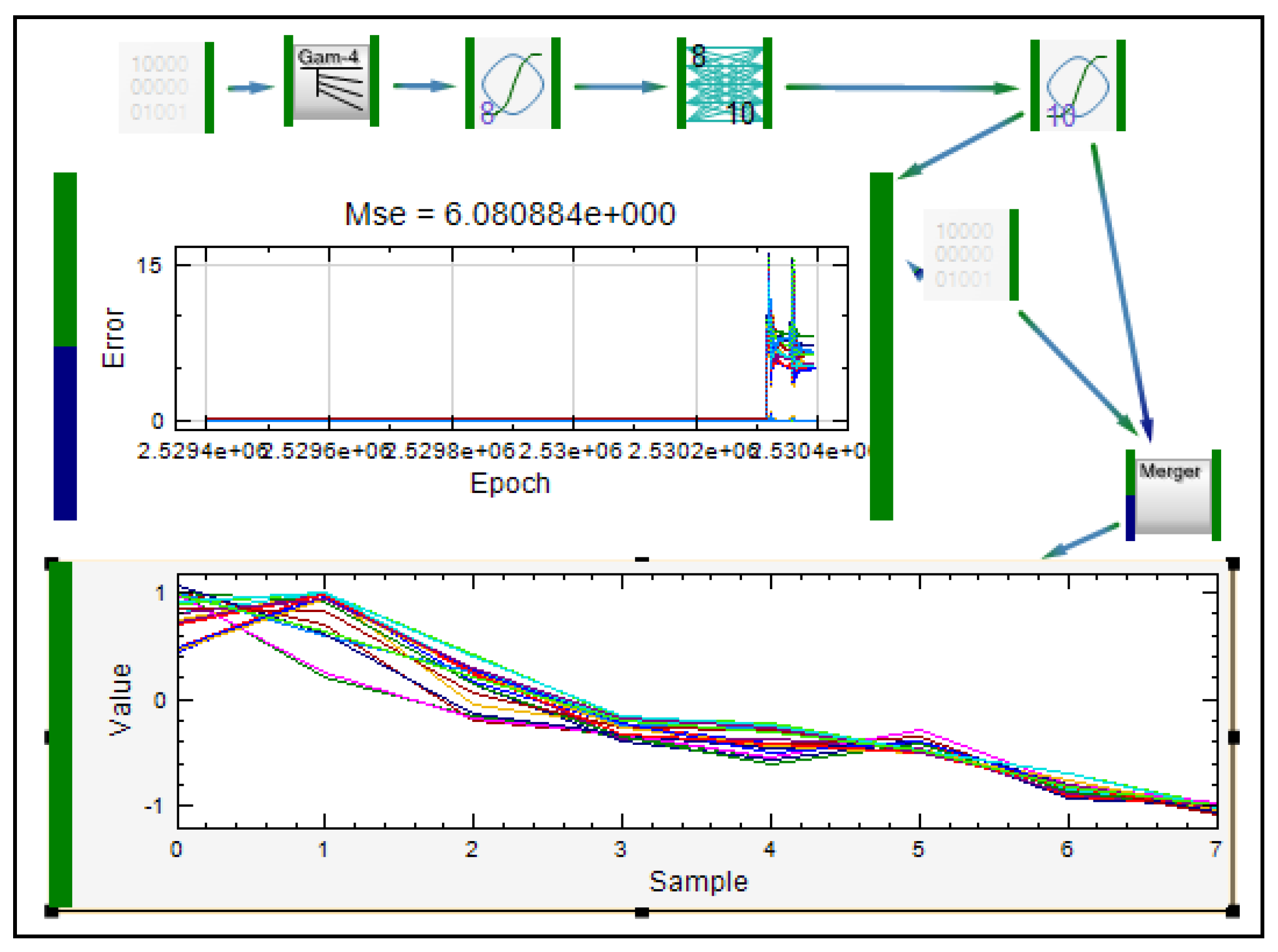
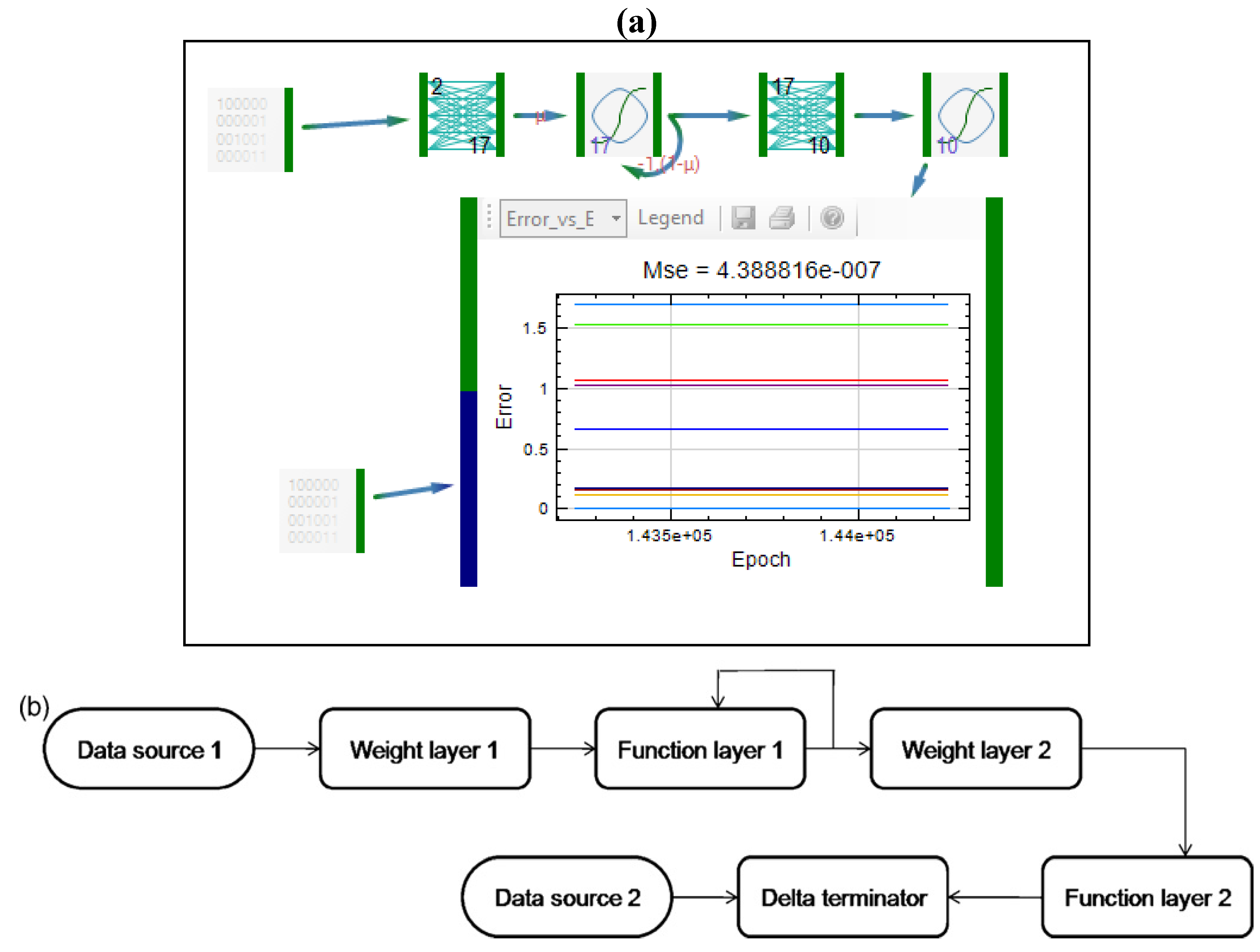
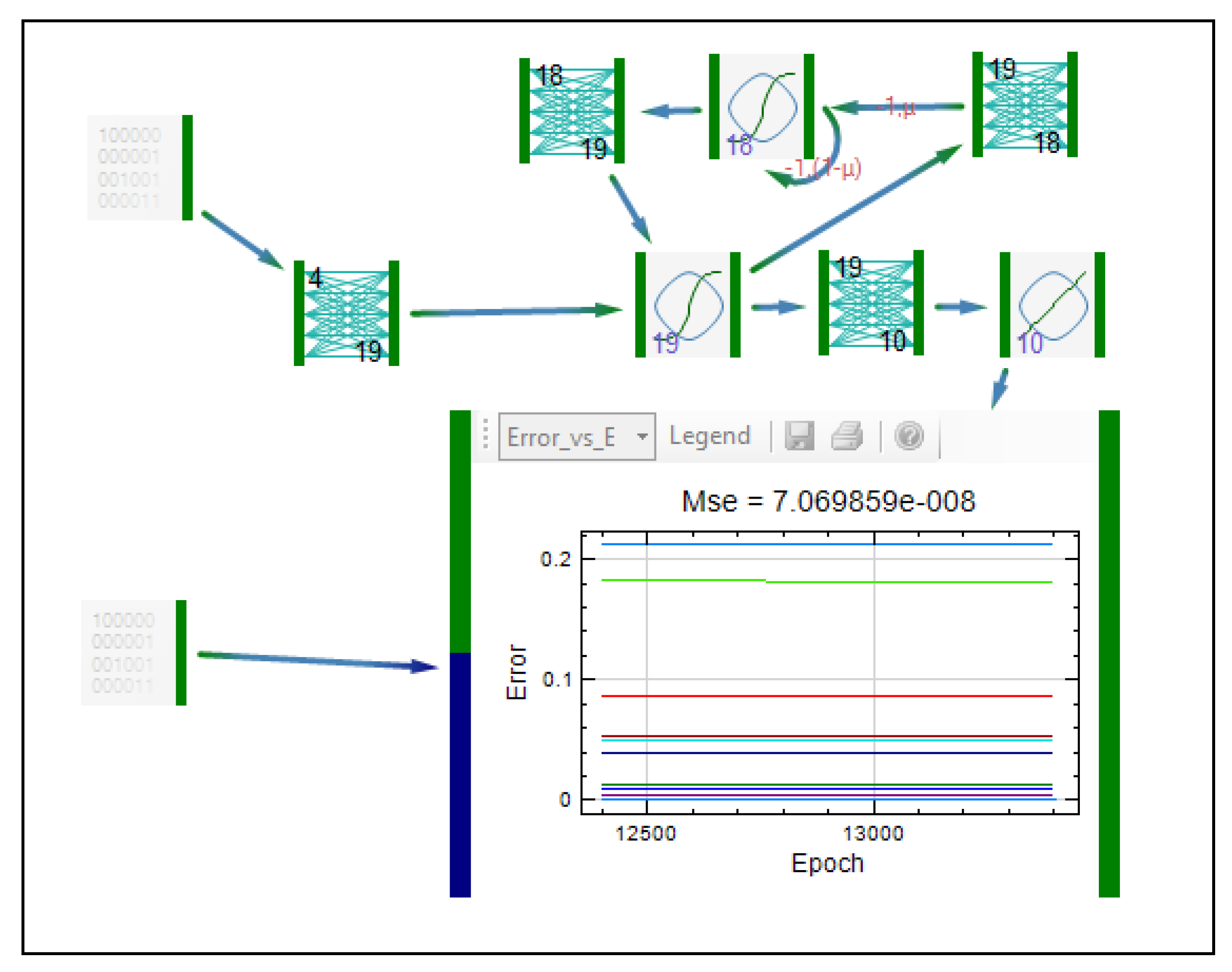
| Formulation, characterization and optimization of modified release formulation | ||
|---|---|---|
| Inputs/outputs/aim | Network type | Authors, year |
| Design of controlled release formulations. Varying formulation variables were used as inputs and in vitro cumulative percentages of drug released were used as ANN outputs. | MLP | Chen, 1999 [21] |
| Optimization of diclofenac sodium sustained release matrix tablets. Trained model was used to predict release profiles and to optimize the formulation composition. | MLP | Zupancic Bozic, 1997 [31] |
| Design of extended release aspirin tablets. The amount of Eudragit® RS PO/Eudragit® L-polymer and compression pressure were selected as inputs, whereas in vitro dissolution profiles, release order and release constant from the Korsmayer Peppas equation were selected as output parameters. | GRNN | Ibric, 2002, 2003, 2007 [12,27,28] |
| Prediction of drug dissolution profiles. Inputs for the network training were the matrix forming agents’ ratio, the time point of the measurement of percent dissolved, and the difference between the release rate of the preceding two time points of the predicted profile. In vitro dissolution profiles were used as network outputs. | MLP | Peh, 2000 [24] |
| Investigation of controlled drug release. Drug fraction and time were used as network inputs and in vitro dissolution profiles as outputs. | MLP | Reis, 2004 [32] |
| Prediction of dissolution profiles for matrix controlled release theophylline pellet preparation. Inputs for the network training were the matrix forming agents’ ratio, and the time point of the measurement of percent dissolved; in vitro dissolution profiles were used as outputs. | EDNN | Goh, 2002 [18] |
| Modeling of diclofenac sodium release from Carbopol 71G matrix tablets. Polymer and binder content were inputs, while in vitro dissolution profiles were outputs | MLP | Ivic, 2010a [26] |
| Modeling of diclofenac sodium release from polyethylene oxide matrix tablets. Polymer weight ratio and compression force were used as inputs, whereas in vitro dissolution profiles were used as networks outputs. Dissolution profiles were treated as time series using dynamic neural networks. | MLP, GMDNN, OLDNN | Petrović, 2009 [13] |
| Drug release control and system understanding of sucrose esters matrix tablets. Networks inputs were HLB values of sucrose esters (SEs), SEs concentration, tablet volume, tablet porosity and tablet tensile strength. In vitro dissolution profiles and parameters indicative of burst release mean dissolution time and release exponent were used as outputs. | MLP | Chansanroj, 2011 [33] |
| A number of unique ANN configurations are presented, that have been evaluated for their ability to determine an IVIVC from different formulations of the same product. In vitro dissolution data were used as inputs and associated outputs were pharmacokinetic time points from nine patients enrolled in a crossover study. | MLP, GRNN, RNN | Dowell, 1999 [34] |
| Development of level A in vitro–in vivo correlation. Inputs for the network training were in vitro dissolution samples whereas in vivo dissolution profiles calculated by numerical deconvolution for each volunteer individually were used as outputs. | GRNN | Parojčić, 2007 [35] |
| Prediction of relative lung bioavailability and clinical effect of salbutamol when delivered to healthy volunteers and asthmatic patients from dry powder inhalers (DPIs). Training of the ANN network was performed using in vitro aerodynamic characteristics of the formulation and demographic data of volunteers/patients as input parameters, whilst in vivo data (urinary excretion of the drug and its metabolite) were networks outputs. | MLP | De Matas, 2008 [36] |
| Prediction of kinetics of doxorubicin release from sulfopropyl dextran ion-exchange microspheres. Three independent variables, drug loading level, concentration of NaCl and CaCl2 in the release medium were used as the ANN inputs and the fractional release of doxorubicin at four different time points as the outputs. | MLP, HNN | Li, 2005 [37] |
| Prediction of drug release profiles in transdermal iontophoresis. Neural networks inputs were the process conditions of pH, ionic strength and current, as well as the time point. The output was the predicted permeation rate of the drug (diclofenac sodium). | RBFNN | Lim, 2003 [38] |
| Optimization of drug release from compressed multi unit particle system (MUPS) using generalized regression neural network (GRNN) | GRNN | Ivic, 2010b [39] |
3. Future Prospects
Acknowledgments
Conflict of Interest
References
- Snee, R.D. Quality by design-four years and three myths later. Pharm. Proc. 2009, 2, 14–16. [Google Scholar]
- Lewis, G.A.; Mathieu, D.; Luu, P.T. Pharmaceutical Experimental Design; Marcel Dekker: New York, NY, USA, 1999. [Google Scholar]
- Bishop, C.M. Pattern Recognition and Machine Learning; Springer: Berlin, Germany, 2006. [Google Scholar]
- Agatonovic-Kustrin, S.; Beresford, R. Basic concepts of artificial neural network (ANN) modeling and its application in pharmaceutical research. J. Pharm. Biomed. Anal. 2000, 22, 717–727. [Google Scholar] [CrossRef]
- Sun, Y.; Peng, Y.; Chen, Y.; Shukla, A. Application of artificial neural networks in the design of controlled release drug delivery systems. Adv. Drug Deliv. Rev. 2003, 55, 1201–1215. [Google Scholar] [CrossRef]
- Basheer, I.A.; Hajmeer, M. Artificial neural networks: Fundamentals, computing, design and application. J. Microbiol. Methods 2000, 43, 3–31. [Google Scholar] [CrossRef]
- Rosenblatt, F. Principles of Neurodynamics: Perceptrons and the Theory of Brain Mechanisms; Spartan Books: Washington DC, USA, 1961. [Google Scholar]
- Rumelhart, D.E.; Geoffrey, E.H.; Williams, R.J. Parallel Distributed Processing: Explorations in the Microstructure of Cognition, Volume 1: Foundations; MIT Press: Massachusetts, UK, 1986. [Google Scholar]
- Zhang, J.; Man, K.F. Time series prediction using recurrent neural network in multi-dimension embedding phase space. IEEE Syst. Man Cybern. Soc. 1998, 2, 11–14. [Google Scholar]
- Specht, D.F. A general regression neural network. IEEE Trans. Neural Netw. 1991, 2, 568–576. [Google Scholar] [CrossRef]
- Patterson, D. Artificial Neural Networks; Prentice Hall: Singapore, 1996. [Google Scholar]
- Ibrić, S.; Jovanović, M.; Djurić, Z.; Parojčić, J.; Solomun, L. The application of generalized regression neural network in the modeling and optimization of aspirin extended release tablets with Eudragit RS PO as matrix substance. J. Control. Release 2002, 82, 213–222. [Google Scholar] [CrossRef]
- Petrović, J.; Ibrić, S.; Betz, G.; Parojčić, J.; Đurić, Z. Application of dynamic neural networks in the modeling of drug release from polyethylene oxide matrix tablets. Eur. J. Pharm. Sci. 2009, 38, 172–180. [Google Scholar] [CrossRef]
- Waibel, A.; Hanazawa, T.; Hinton, G.; Shikano, K.; Land, K. Phoneme recognition using time-delay neural networks. IEEE Trans. ASSP 1989, 37, 328–339. [Google Scholar] [CrossRef]
- Samarasinghe, S. Neural Networks for Applied Sciences and Engineering; Auerbach Publications: New York, NY, USA, 2006. [Google Scholar]
- Köker, R. Design and performance of an intelligent predictive controller for a six-degree-of-freedom robot using the Elman network. Inf. Sci. 2006, 12, 1781–1799. [Google Scholar]
- Elman, J.L. Finding structure in time. Cogn. Sci. 1990, 2, 179–211. [Google Scholar]
- Goh, W.Y.; Lim, C.P.; Peh, K.K.; Subari, K. Application of a recurrent neural network to prediction of drug dissolution profiles. Neural Comput. Appl. 2002, 10, 311–317. [Google Scholar] [CrossRef]
- Hussain, A.S.; Yu, X.; Johnson, R.D. Application of neural computing in pharmaceutical product development. Pharm. Res. 1991, 8, 1248–1252. [Google Scholar] [CrossRef]
- Hussain, A.S.; Shivanand, P.; Johnson, R.D. Application of neural computing in pharmaceutical product development: Computer aided formulation design. Drug Dev. Ind. Pharm. 1994, 20, 1739–1752. [Google Scholar] [CrossRef]
- Chen, Y.; McCall, T.W.; Baichwal, A.R.; Meyer, M.C. The application of an artificial neural network and pharmacokinetic simulations in the design of controlled-release dosage forms. J. Control. Release 1999, 59, 33–41. [Google Scholar] [CrossRef]
- Turkoglu, M.; Aydin, I.; Murray, M.; Sakr, A. Modeling of a roller-compaction process using neural networks and genetic algorithms. Eur. J. Pharm. Biopharm. 1999, 48, 239–245. [Google Scholar] [CrossRef]
- Takayama, K.; Morva, A.; Fujikawa, M.; Hattori, Y.; Obata, Y.; Nagai, T. Formula optimization of theophylline controlled-release tablet based on artificial neural networks. J. Control. Release 2000, 68, 175–186. [Google Scholar] [CrossRef]
- Peh, K.K.; Lim, C.P.; Quek, S.S.; Khoh, K.H. Use of artificial neural networks to predict drug dissolution profiles and evaluation of network performance using similarity factor. Pharm. Res. 2000, 17, 1384–1389. [Google Scholar] [CrossRef]
- Vaithiyalingam, S.; Khan, M.A. Optimization and characterization of controlled release multi-particulate beads formulated with a customized cellulose acetate butyrate dispersion. Int. J. Pharm. 2002, 234, 179–193. [Google Scholar] [CrossRef]
- Ivić, B.; Ibrić, S.; Cvetković, N.; Petrović, A.; Trajković, S.; Djurić, Z. Application of design of experiments and multilayer perceptrons neural network in the optimization of diclofenac sodium extended release tablets with Carbopol® 71G. Chem. Pharm. Bull. 2010, 58, 947–949. [Google Scholar] [CrossRef]
- Ibrić, S.; Jovanović, M.; Djurić, Z.; Parojčić, J.; Petrović, S.D.; Solomun, L.J.; Stupar, B. Artificial neural networks in the modeling and optimization of aspirin extended release tablets with Eudragit L 100 as matrix substance. AAPS Pharm. Sci. Tech. 2003, 4, 62–70. [Google Scholar]
- Ibrić, S.; Jovanović, M.; Djurić, Z.; Parojčić, J.; Solomun, L.; Lučić, B. Generalized regression neural networks in prediction of drug stability. J. Pharm. Pharmacol. 2007, 59, 745–750. [Google Scholar]
- Petrović, J.; Ibrić, S.; Betz, G.; Djurić, Z. Optimization of matrix tablets controlled drug release using Elman dynamic neural networks and decision trees. Int. J. Pharm. 2012, 428, 57–67. [Google Scholar] [CrossRef]
- Djuriš, J. Characterization of controlled release hydrophilic and lipid matrix tablets using in silico methods.
- Zupancic Bozic, D.; Vrecer, F.; Kozjek, F. Optimization of diclofenac sodium dissolution from sustained release formulations using an artificial neural network. Eur. J. Pharm. Sci. 1997, 5, 163–169. [Google Scholar] [CrossRef]
- Reis, M.A.A.; Sinisterra, R.D.; Belchior, J.C. An alternative approach based on artificial neural networks to study controlled drug release. J. Pharm. Sci. 2004, 93, 418–430. [Google Scholar] [CrossRef]
- Chansanroj, K.; Petrović, J.; Ibrić, S.; Betz, G. Drug release control and system understanding of sucrose esters matrix tablets by artificial neural networks. Eur. J. Pharm. Sci. 2011, 44, 321–331. [Google Scholar] [CrossRef]
- Dowell, J.A.; Hussain, A.; Devane, J.; Young, D. Artificial neural networks applied to the in vitro–in vivo correlation of an extended-release formulations: Initial trials and experience. J. Pharm. Sci. 1999, 88, 154–160. [Google Scholar] [CrossRef]
- Parojčić, J.; Ibrić, S.; Đurić, Z.; Jovanović, M.; Corrigan, O.I. An investigation into the usefulness of generalized regression neural network analysis in the development of level A in vitro–in vivo correlation. Eur. J. Pharm. Sci. 2007, 30, 264–272. [Google Scholar] [CrossRef]
- De Matas, M.; Shao, Q.; Richardson, C.H.; Chrystyn, H. Evaluation of in vitro in vivo correlations for dry powder inhaler delivery using artificial neural networks. Eur. J. Pharm. Sci. 2008, 33, 80–90. [Google Scholar] [CrossRef]
- Li, Y.; Rauth, A.M.; Wu, X.Y. Prediction of kinetics of doxorubicin release from sulfopropyl dextran ion-exchange microspheres using artificial neural networks. Eur. J. Pharm. Sci. 2005, 24, 401–410. [Google Scholar] [CrossRef]
- Lim, C.P.; Quek, S.S.; Peh, K.K. Prediction of drug release profiles using an intelligent learning system: An experimental study in transdermal iontophoresis. J. Pharm. Biomed. Anal. 2003, 31, 159–168. [Google Scholar] [CrossRef]
- Ivic, B.; Ibric, S.; Betz, G.; Djuric, Z. Optimization of drug release from compressed multi unit particle system (MUPS) using generalized regression neural network (GRNN). Arch. Pharm. Res. 2010, 33, 103–113. [Google Scholar] [CrossRef]
- Chen, Y.; Jiao, T.; McCall, T.W.; Baichwal, A.R.; Meyer, M.C. Comparison of four artificial neural network software programs used to predict the in vitro dissolution of controlled-release tablets. Pharm. Dev. Tech. 2002, 7, 373–379. [Google Scholar] [CrossRef]
- Tan, C.; Degim, I.T. Development of sustained release formulation of an antithrombotic drug and application of fuzzy logic. Pharm. Dev. Tech. 2012, 17, 242–250. [Google Scholar] [CrossRef]
- Barmpalexis, P.; Kachrimanis, K.; Tsakonas, A.; Georgarakis, E. Symbolic regression via genetic programming in the optimization of a controlled release pharmaceutical formulation. Chemom. Intell. Lab. Syst. 2011, 107, 75–82. [Google Scholar] [CrossRef]
- Barmpalexis, P.; Kachrimanis, K.; Georgarakis, E. Solid dispersions in the development of a nimodipine floating tablet formulation and optimization by artificial neural networks and genetic programming. Eur. J. Pharm. Biopharm. 2011, 77, 122–131. [Google Scholar] [CrossRef]
- Colbourn, E.A.; Roskilly, S.J.; Rowe, R.C.; York, P. Modelling formulations using gene expression programming-A comparative analysis with artificial neural networks. Eur. J. Pharm. Sci. 2011, 44, 366–374. [Google Scholar] [CrossRef]
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Ibrić, S.; Djuriš, J.; Parojčić, J.; Djurić, Z. Artificial Neural Networks in Evaluation and Optimization of Modified Release Solid Dosage Forms. Pharmaceutics 2012, 4, 531-550. https://doi.org/10.3390/pharmaceutics4040531
Ibrić S, Djuriš J, Parojčić J, Djurić Z. Artificial Neural Networks in Evaluation and Optimization of Modified Release Solid Dosage Forms. Pharmaceutics. 2012; 4(4):531-550. https://doi.org/10.3390/pharmaceutics4040531
Chicago/Turabian StyleIbrić, Svetlana, Jelena Djuriš, Jelena Parojčić, and Zorica Djurić. 2012. "Artificial Neural Networks in Evaluation and Optimization of Modified Release Solid Dosage Forms" Pharmaceutics 4, no. 4: 531-550. https://doi.org/10.3390/pharmaceutics4040531
APA StyleIbrić, S., Djuriš, J., Parojčić, J., & Djurić, Z. (2012). Artificial Neural Networks in Evaluation and Optimization of Modified Release Solid Dosage Forms. Pharmaceutics, 4(4), 531-550. https://doi.org/10.3390/pharmaceutics4040531






