In Vitro Efficacy of PEI-Derived Lipopolymers in Silencing of Toxic Proteins in a Neuronal Model of Huntington’s Disease
Abstract
1. Introduction
2. Materials and Methods
2.1. Lipid-Substituted PEI-Derived Lipopolimers and Trans-Booster Additive
2.2. Materials
2.3. Cell Culture
2.4. Transfection of siRNAs Using Leu-Fect Lipopolymers
2.5. MTT Assay for Cytotoxicity
2.6. Flow Cytometry Analysis
2.7. Image Analysis of muHTT Aggregates
2.8. RT-qPCR
2.9. Western Blotting
2.10. Statistical Analysis
3. Results
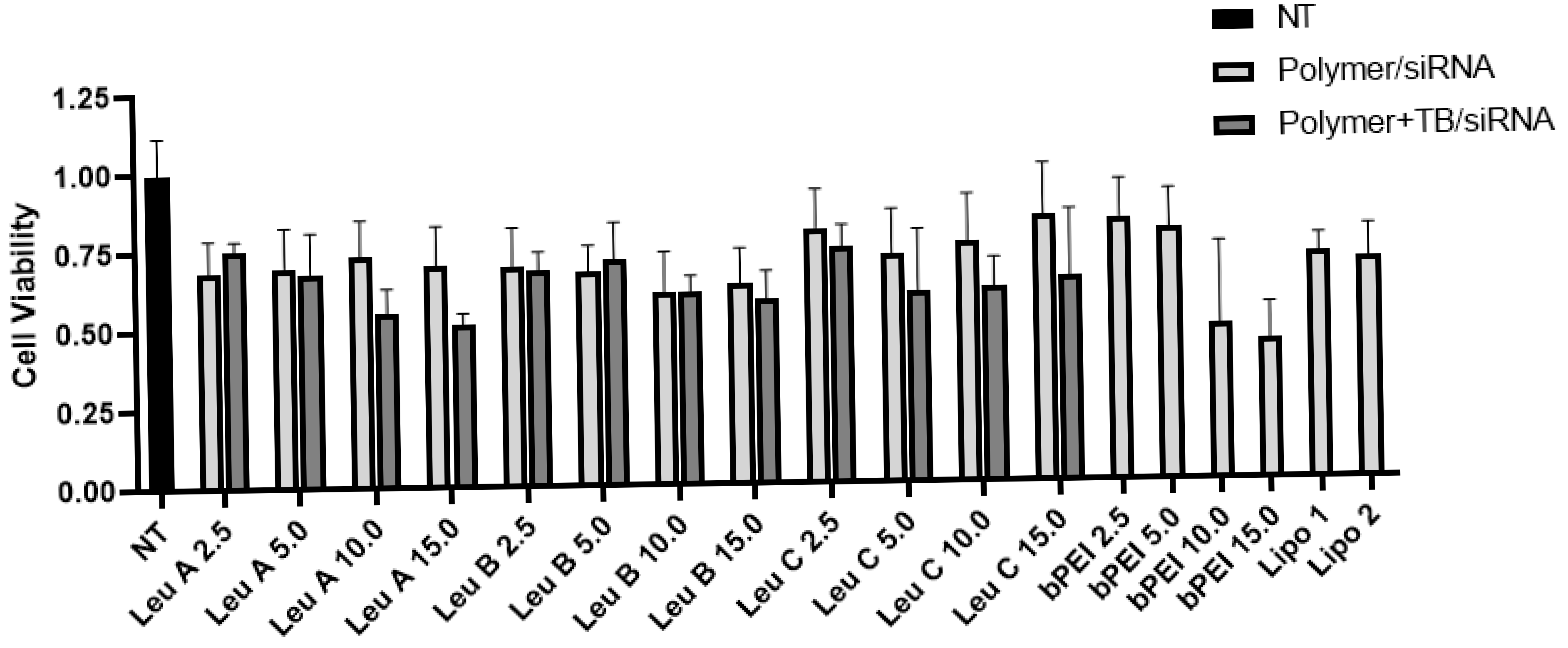
3.1. Leu-Fect Series of Transfection Reagents Exhibit Low Cellular Toxicity
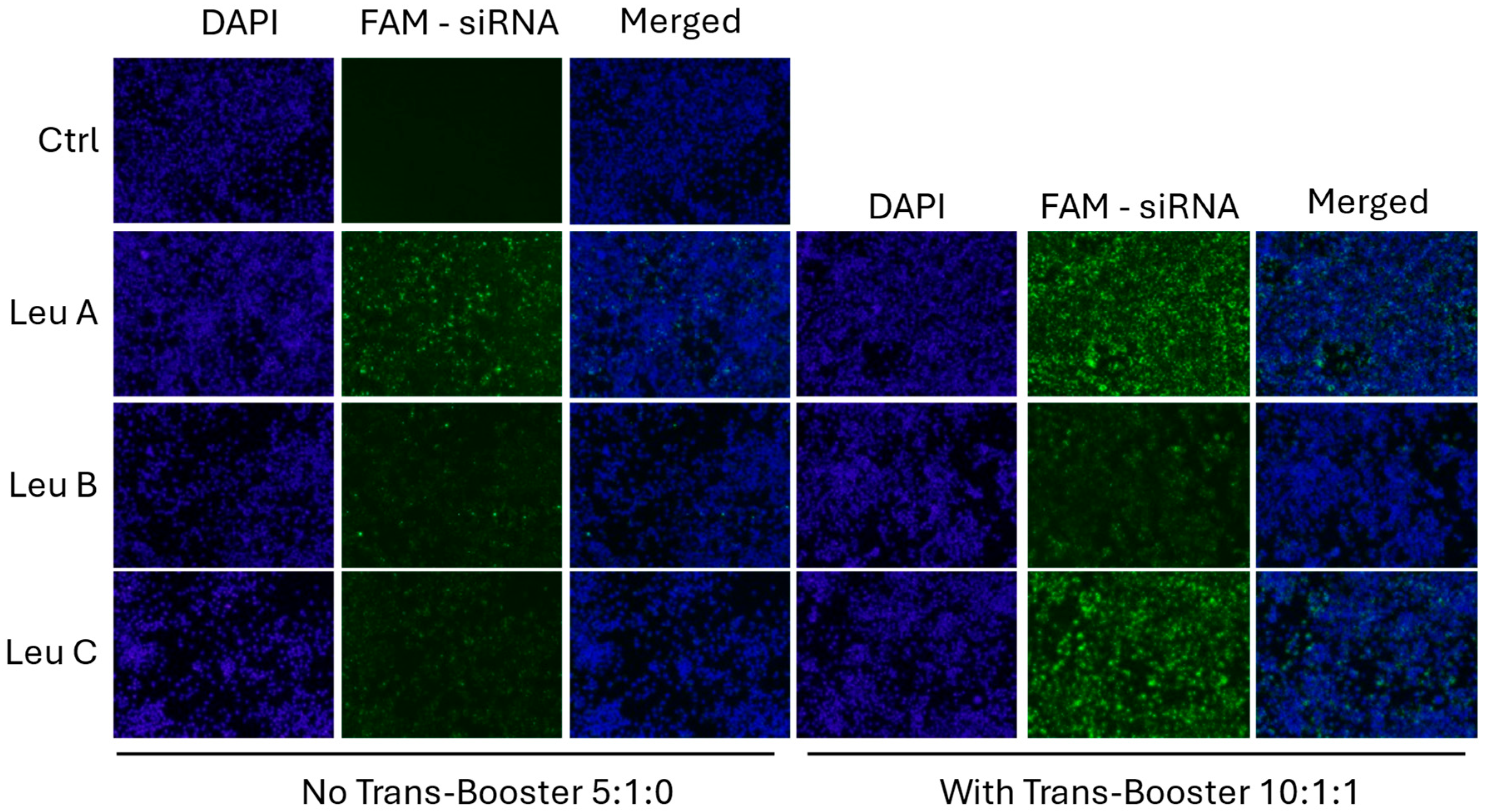
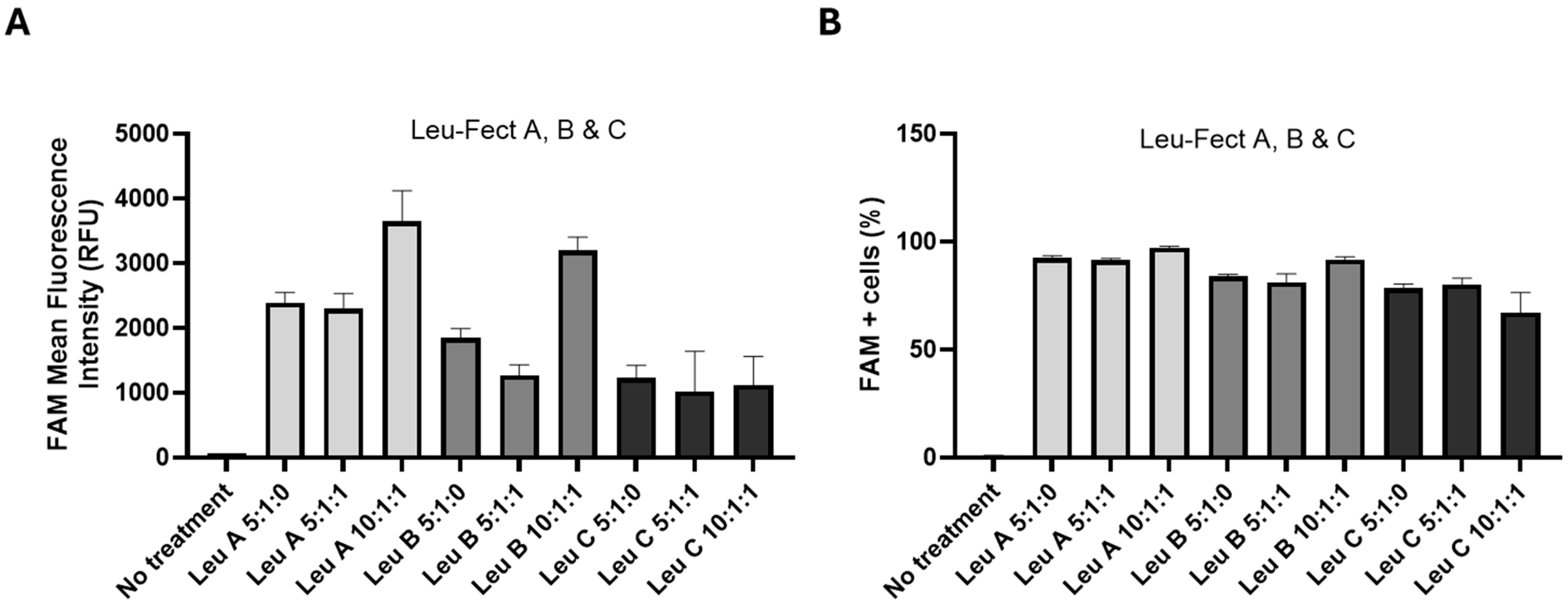
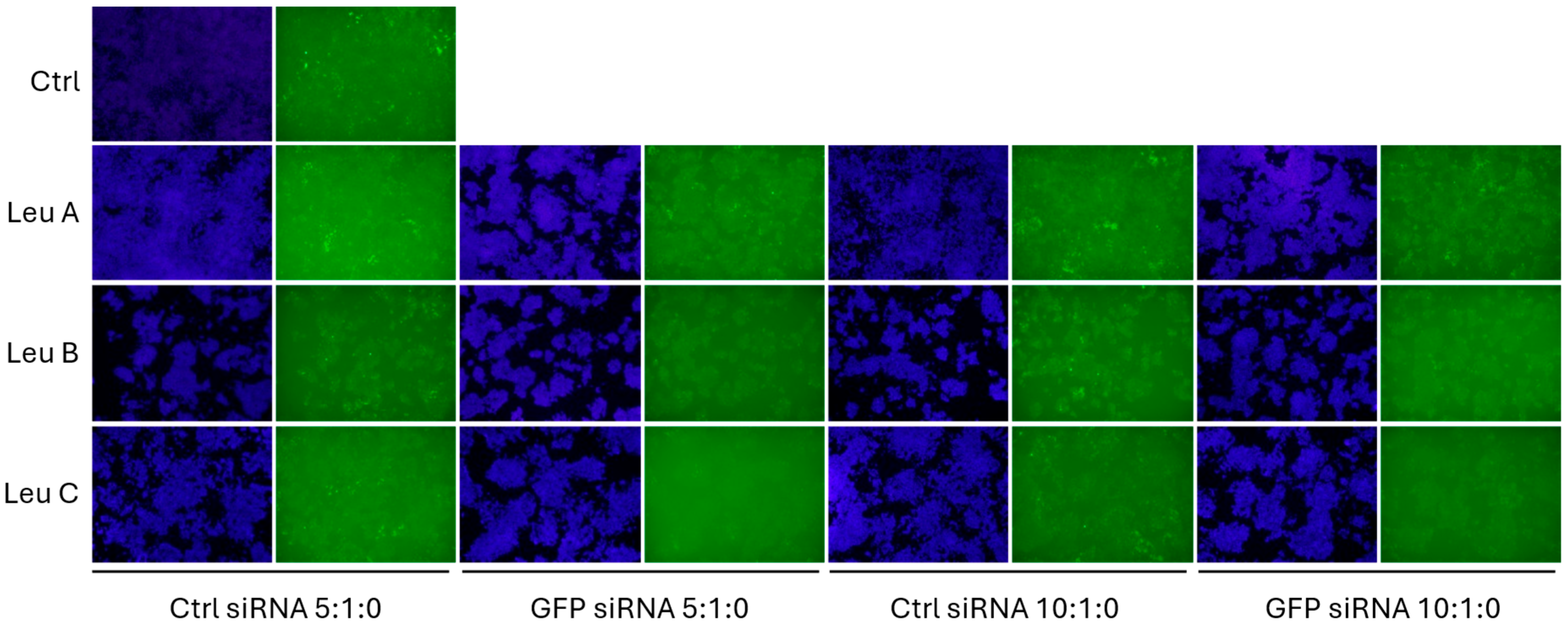
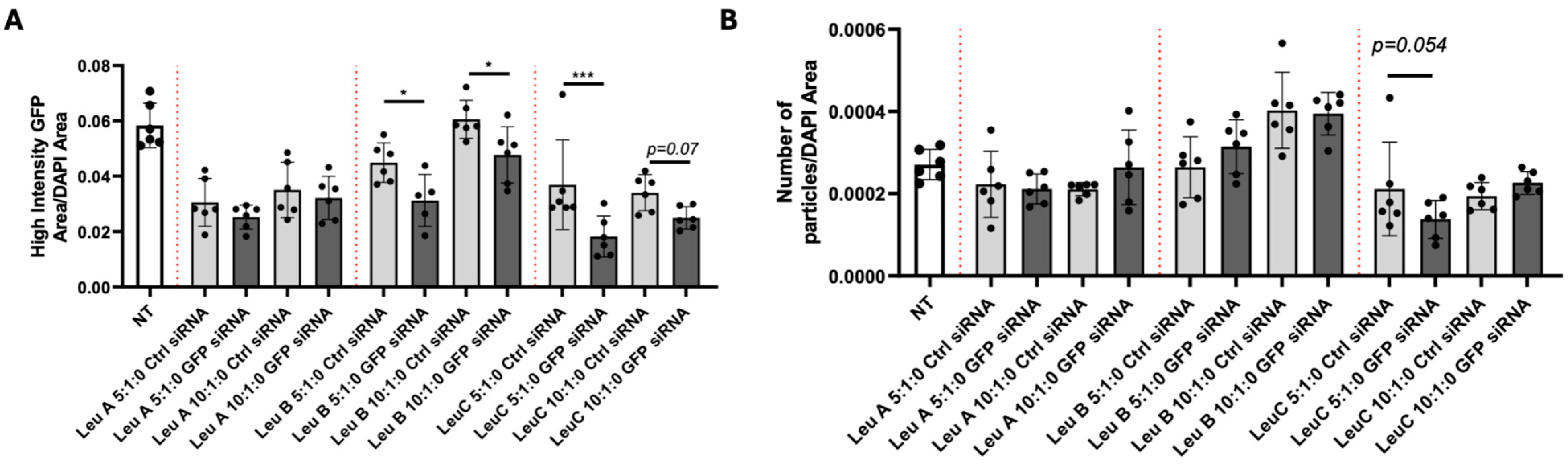
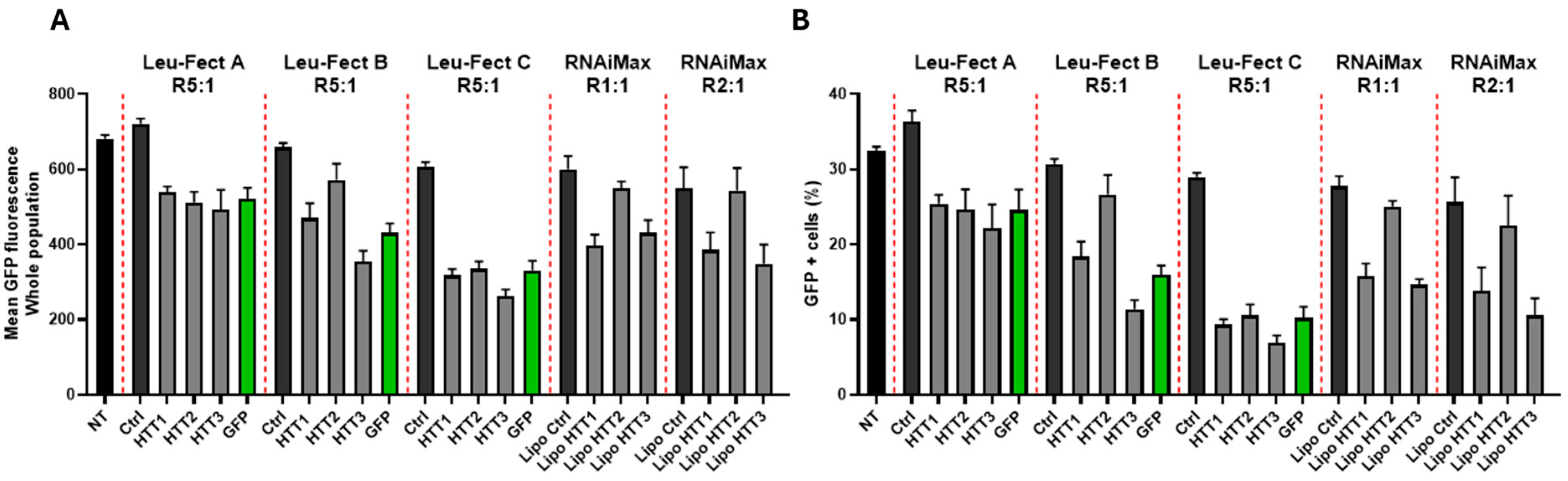
3.2. Leu-Fect Transfection Reagent Effectively Deliver siRNA into Neuronal Cells
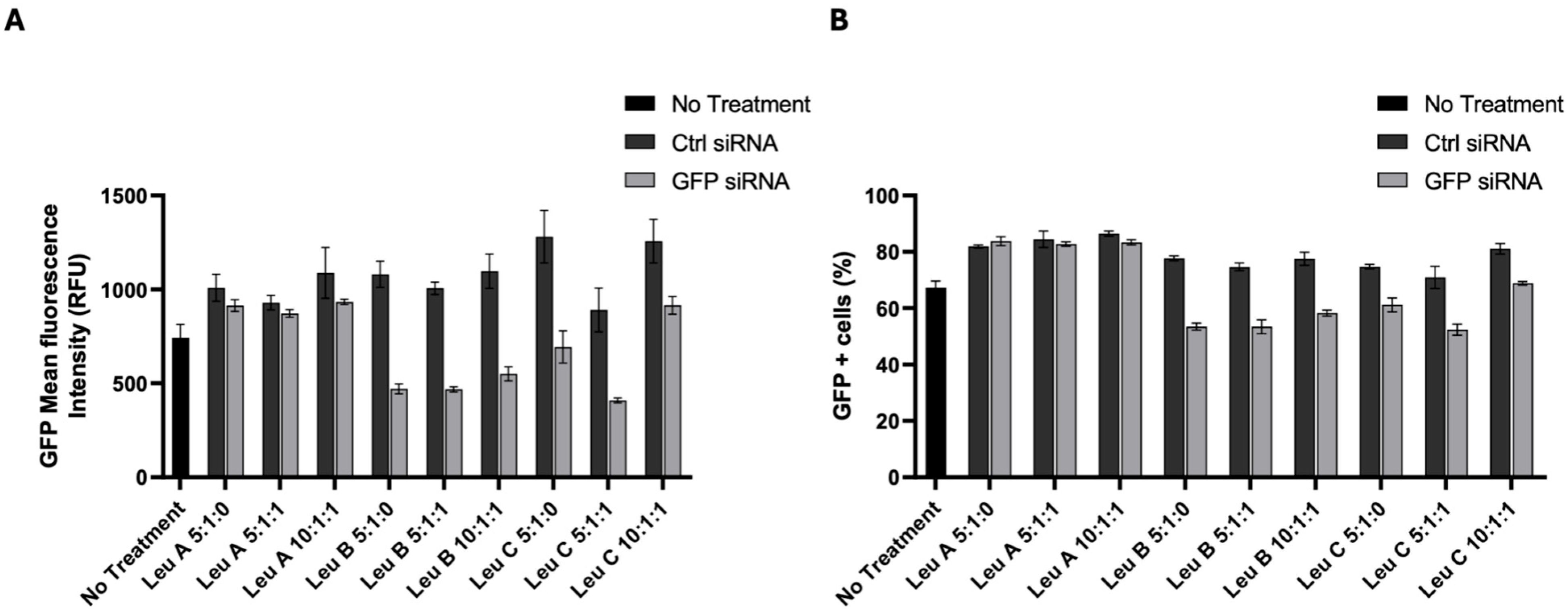
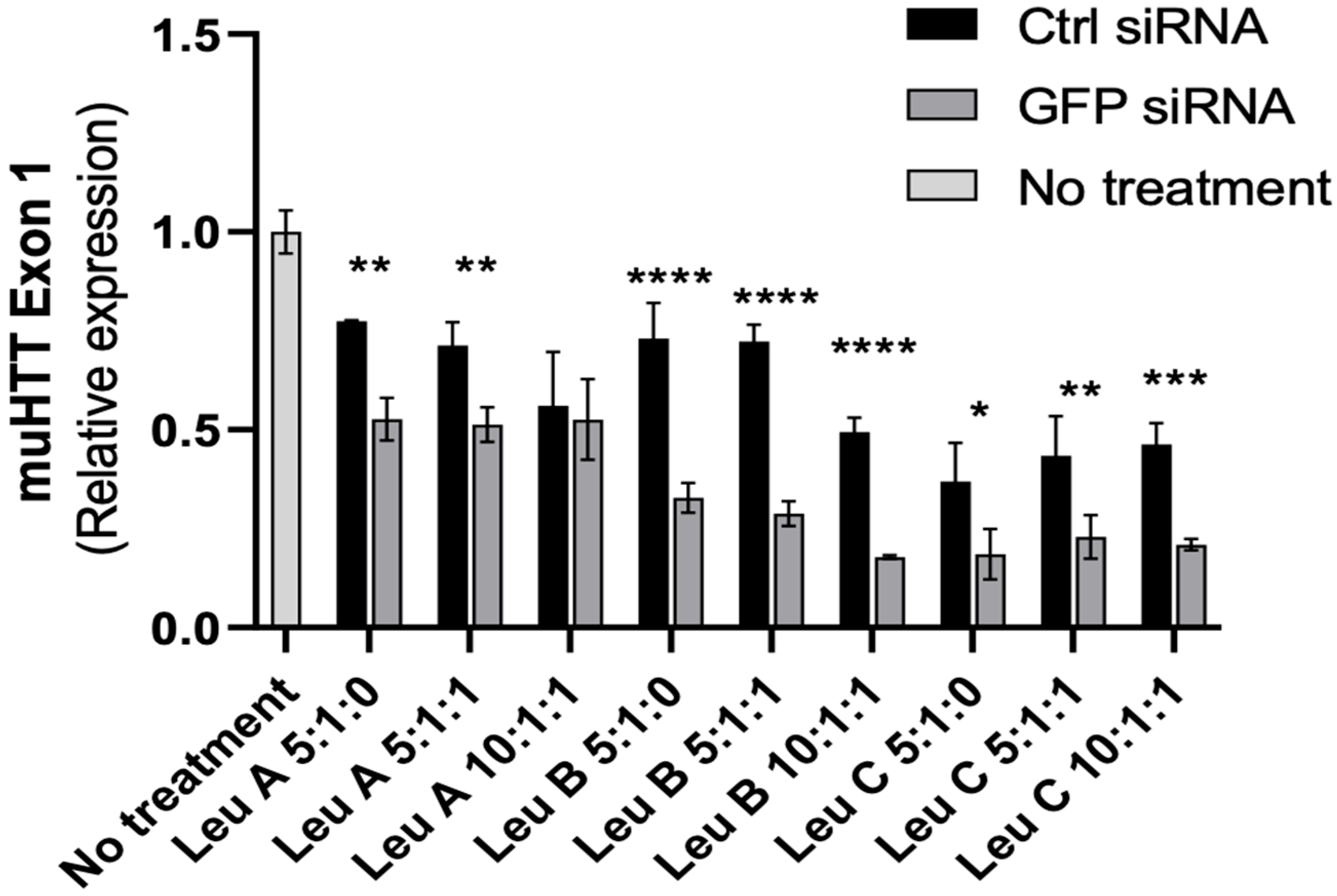
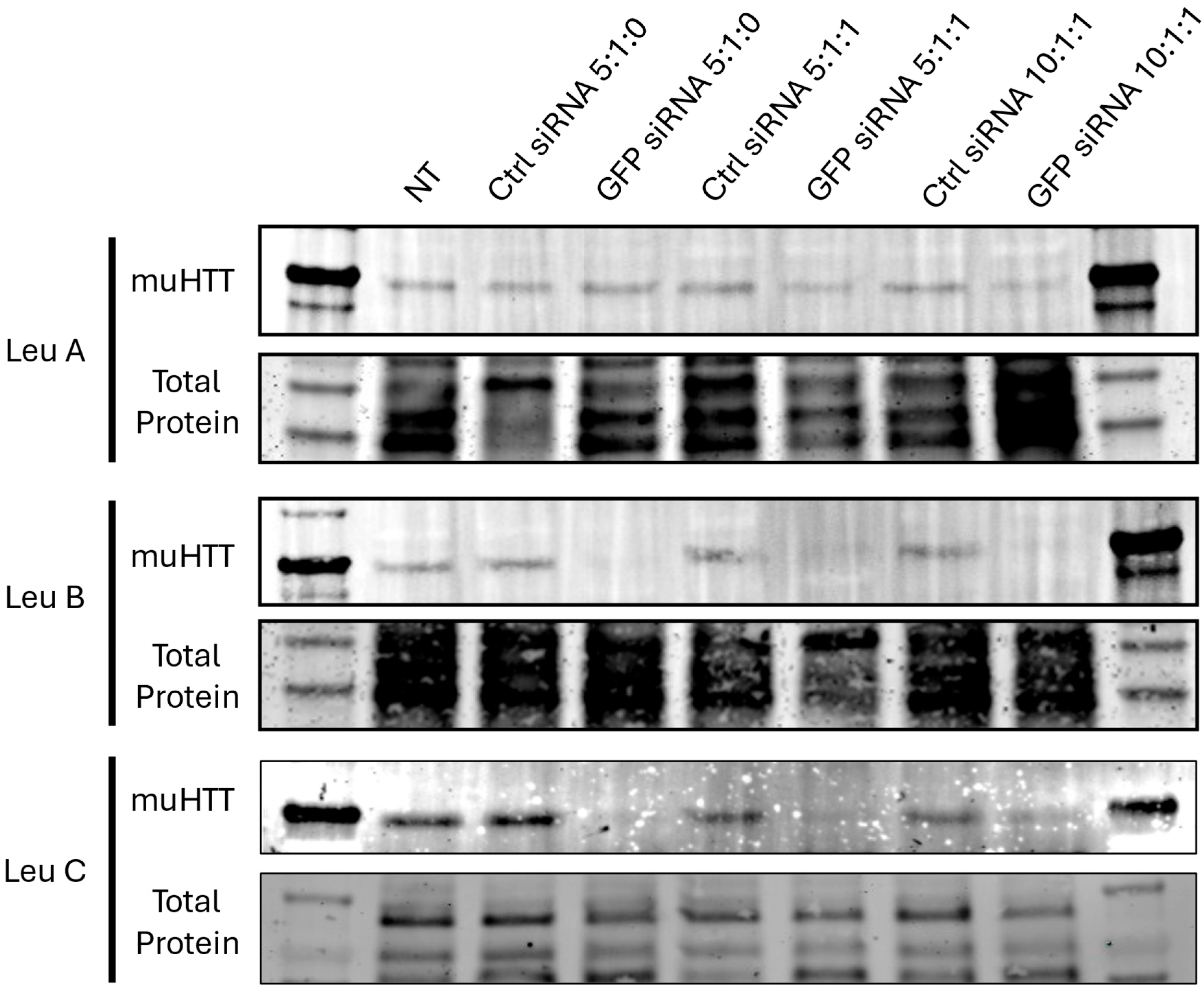
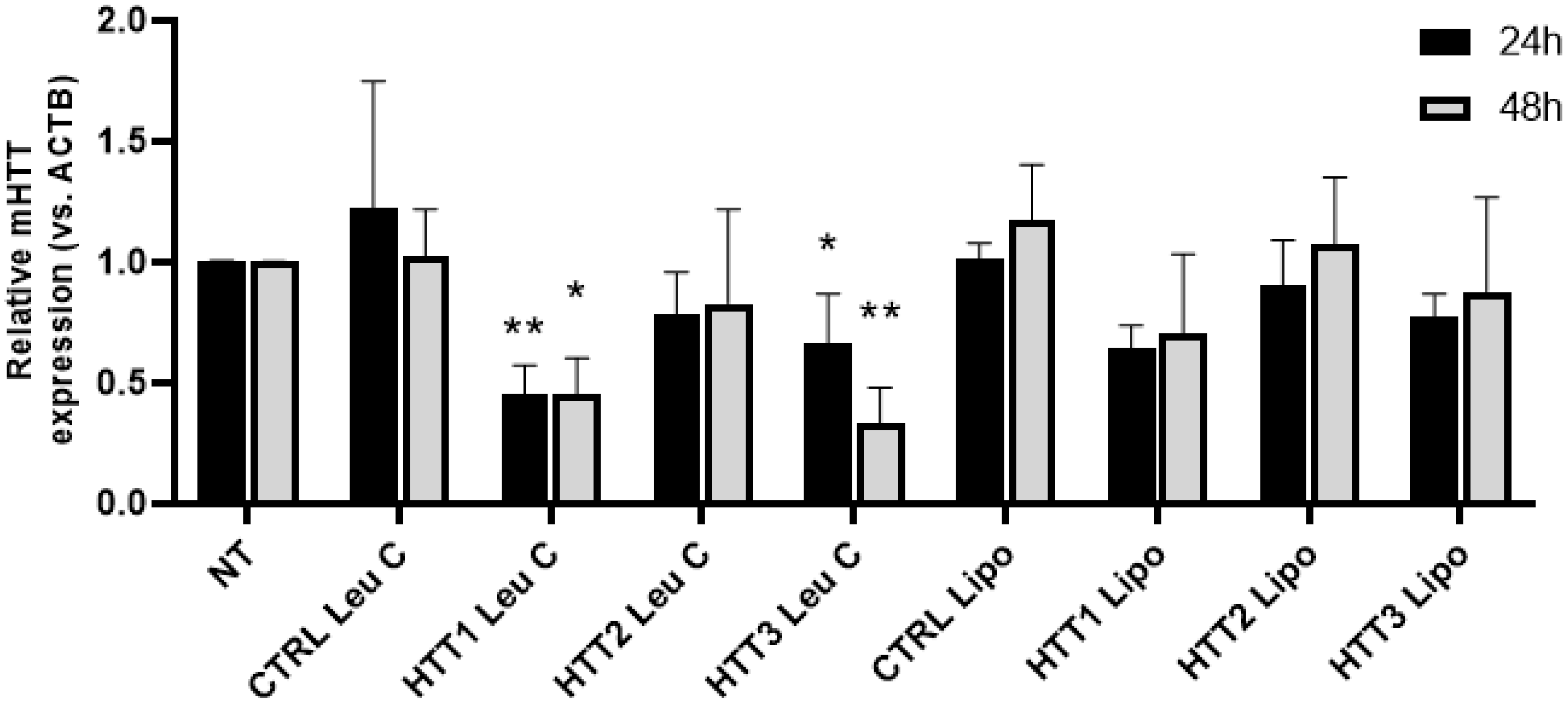
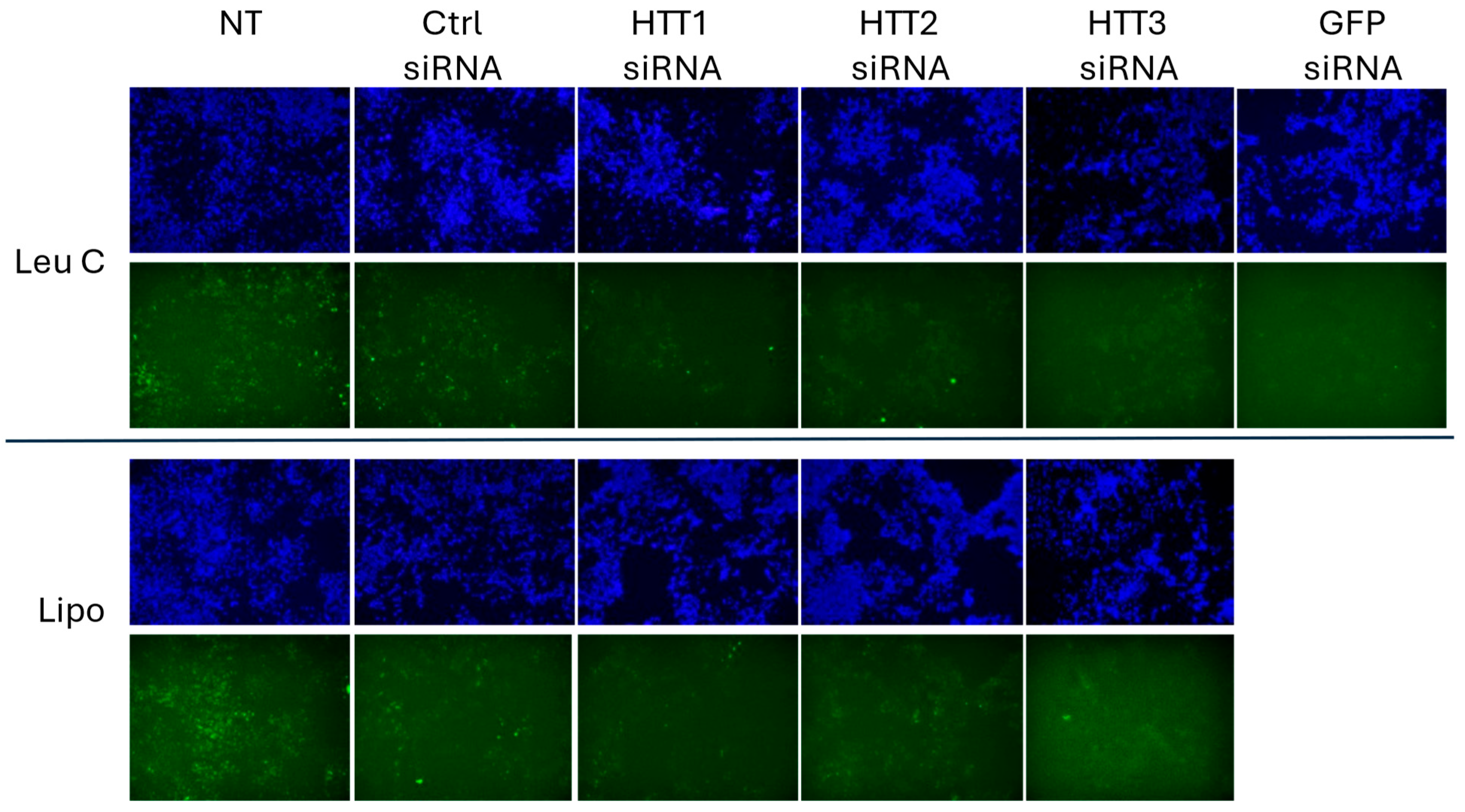
3.3. Leu-Fect/GFP siRNA Complexes Reduced muHTT Transcript and Chimeric Protein Levels
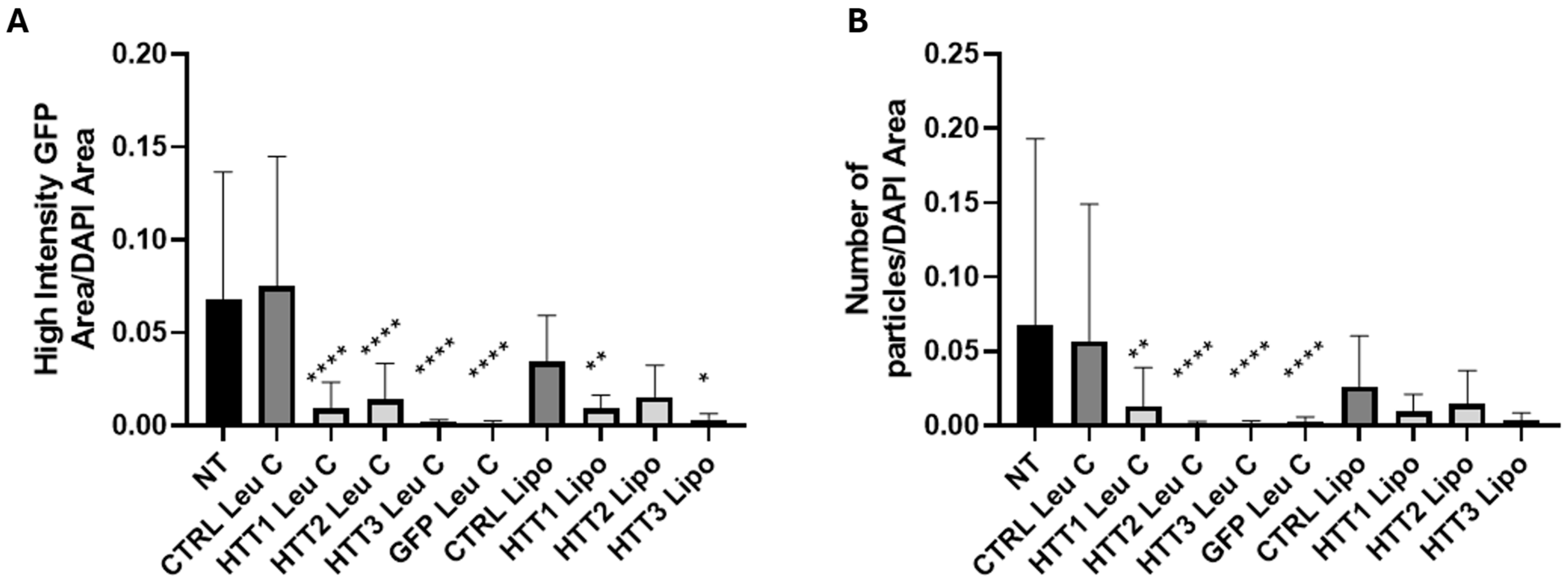
3.4. Leu-Fect C/HTT siRNAs Complexes Reduce muHTT Burden in Neuronal Cells
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dumas, E.M.; van den Bogaard, S.J.; Middelkoop, H.A.; Roos, R.A. A review of cognition in Huntington’s disease. Front. Biosci. Sch. Ed. 2013, 5, 1–18. [Google Scholar]
- Kim, S.D.; Fung, V.S. An update on Huntington’s disease: From the gene to the clinic. Curr. Opin. Neurol. 2014, 27, 477–483. [Google Scholar] [CrossRef]
- Tabrizi, S.J.; Langbehn, D.R.; Leavitt, B.R.; Roos, R.A.; Durr, A.; Craufurd, D.; Kennard, C.; Hicks, S.L.; Fox, N.C.; Scahill, R.I.; et al. Biological and clinical manifestations of Huntington’s disease in the longitudinal TRACK-HD study: Cross-sectional analysis of baseline data. Lancet Neurol. 2009, 8, 791–801. [Google Scholar] [CrossRef]
- Zuccato, C.; Cattaneo, E. Huntington’s disease. In Handbook of Experimental Pharmacology; Springer: Berlin/Heidelberg, Germany, 2014; Volume 220, pp. 357–409. [Google Scholar] [CrossRef]
- MacDonald, M.E.; Strobel, S.A.; Draths, K.M.; Wales, J.L.; Dervan, P.; The Huntington’s Disease Collaborative Research Group. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell 1993, 72, 971–983. [Google Scholar] [CrossRef]
- Harding, R.J.; Tong, Y.F. Proteostasis in Huntington’s disease: Disease mechanisms and therapeutic opportunities. Acta Pharmacol. Sin. 2018, 39, 754–769. [Google Scholar] [CrossRef]
- Tabrizi, S.J.; Scahill, R.I.; Durr, A.; Roos, R.A.; Leavitt, B.R.; Jones, R.; Landwehrmeyer, G.B.; Fox, N.C.; Johnson, H.; Hicks, S.L.; et al. Biological and clinical changes in premanifest and early stage Huntington’s disease in the TRACK-HD study: The 12-month longitudinal analysis. Lancet Neurol. 2011, 10, 31–42. [Google Scholar] [CrossRef]
- Sipione, S.; Rigamonti, D.; Valenza, M.; Zuccato, C.; Conti, L.; Pritchard, J.; Kooperberg, C.; Olson, J.M.; Cattaneo, E. Early transcriptional profiles in huntingtin-inducible striatal cells by microarray analyses. Hum. Mol. Genet. 2002, 11, 1953–1965. [Google Scholar] [CrossRef]
- Sipione, S.; Cattaneo, E. Modeling brain pathologies using neural stem cells. Methods Mol. Biol. 2002, 198, 245–262. [Google Scholar] [CrossRef]
- Tabrizi, S.J.; Estevez-Fraga, C.; van Roon-Mom, W.M.C.; Flower, M.D.; Scahill, R.I.; Wild, E.J.; Muñoz-Sanjuan, I.; Sampaio, C.; E Rosser, A.; Leavitt, B.R. Potential disease-modifying therapies for Huntington’s disease: Lessons learned and future opportunities. Lancet Neurol. 2022, 21, 645–658. [Google Scholar] [CrossRef]
- Thomson, S.B.; Stam, A.; Brouwers, C.; Fodale, V.; Bresciani, A.; Vermeulen, M.; Mostafavi, S.; Petkau, T.L.; Hill, A.; Yung, A.; et al. AAV5-miHTT-mediated huntingtin lowering improves brain health in a Huntington’s disease mouse model. Brain J. Neurol. 2023, 146, 2298–2315. [Google Scholar] [CrossRef]
- Caron, N.S.; Southwell, A.L.; Brouwers, C.C.; Cengio, L.D.; Xie, Y.; Black, H.F.; Anderson, L.M.; Ko, S.; Zhu, X.; van Deventer, S.J.; et al. Potent and sustained huntingtin lowering via AAV5 encoding miRNA preserves striatal volume and cognitive function in a humanized mouse model of Huntington disease. Nucleic Acids Res. 2020, 48, 36–54. [Google Scholar] [CrossRef]
- Sogorb-Gonzalez, M.; Landles, C.; Caron, N.S.; Stam, A.; Osborne, G.; Hayden, M.R.; Howland, D.; Van Deventer, S.; Bates, G.P.; Vallès, A.; et al. Exon 1-targeting miRNA reduces the pathogenic exon 1 HTT protein in Huntington’s disease models. Brain 2024, 147, 4043–4055. [Google Scholar] [CrossRef]
- Zhang, L.; Wu, T.; Shan, Y.; Li, G.; Ni, X.; Chen, X.; Hu, X.; Lin, L.; Li, Y.; Guan, Y.; et al. Therapeutic reversal of Huntington’s disease by in vivo self-assembled siRNAs. Brain J. Neurol. 2021, 144, 3421–3435. [Google Scholar] [CrossRef]
- Kingwell, K. Double setback for ASO trials in Huntington disease. Nat. Rev. Drug Discov. 2021, 20, 412–413. [Google Scholar] [CrossRef]
- Attia, N.; Mashal, M.; Puras, G.; Pedraz, J.L. Mesenchymal Stem Cells as a Gene Delivery Tool: Promise, Problems, and Prospects. Pharmaceutics 2021, 13, 843. [Google Scholar] [CrossRef]
- Dezfouli, S.A.; Michailides, M.E.; Uludag, H. Delivery Aspects for Implementing siRNA Therapeutics for Blood Diseases. Biochemistry 2024, 63, 3059–3077. [Google Scholar] [CrossRef]
- Funhoff, A.M.; van Nostrum, C.F.; Koning, G.A.; Schuurmans-Nieuwenbroek, N.M.E.; Crommelin, D.J.A.; Hennink, W.E. Endosomal escape of polymeric gene delivery complexes is not always enhanced by polymers buffering at low pH. Biomacromolecules 2004, 5, 32–39. [Google Scholar] [CrossRef]
- Aliabadi, H.M.; Landry, B.; Bahadur, R.K.; Neamnark, A.; Suwantong, O.; Uludağ, H. Impact of lipid substitution on assembly and delivery of siRNA by cationic polymers. Macromol. Biosci. 2011, 11, 662–672. [Google Scholar] [CrossRef]
- Remant Bahadur, K.C.; Kucharski, C.; Uludağ, H. Additive nanocomplexes of cationic lipopolymers for improved non-viral gene delivery to mesenchymal stem cells. J. Mater. Chem. B 2015, 3, 3972–3982. [Google Scholar] [CrossRef]
- Dezfouli, S.A.; Rajendran, A.P.; Claerhout, J.; Uludag, H. Designing Nanomedicines for Breast Cancer Therapy. Biomolecules 2023, 13, 1559. [Google Scholar] [CrossRef]
- Kazantsev, A.; Preisinger, E.; Dranovsky, A.; Goldgaber, D.; Housman, D. Insoluble detergent-resistant aggregates form between pathological and nonpathological lengths of polyglutamine in mammalian cells. Proc. Natl. Acad. Sci. USA 1999, 96, 11404–11409. [Google Scholar] [CrossRef]
- DiFiglia, M.; Sena-Esteves, M.; Chase, K.; Sapp, E.; Pfister, E.; Sass, M.; Yoder, J.; Reeves, P.; Pandey, R.K.; Rajeev, K.G.; et al. Therapeutic silencing of mutant huntingtin with siRNA attenuates striatal and cortical neuropathology and behavioral deficits. Proc. Natl. Acad. Sci. USA 2007, 104, 17204–17209. [Google Scholar] [CrossRef]
- Godinho, B.M.D.C.; McCarthy, D.J.; Torres-Fuentes, C.; Beltrán, C.J.; McCarthy, J.; Quinlan, A.; Ogier, J.R.; Darcy, R.; O’Driscoll, C.M.; Cryan, J.F. Differential nanotoxicological and neuroinflammatory liabilities of non-viral vectors for RNA interference in the central nervous system. Biomaterials 2014, 35, 489–499. [Google Scholar] [CrossRef]
- Postigo, A.; Cross, J.R.; Downward, J.; Way, M. Interaction of F1L with the BH3 domain of Bak is responsible for inhibiting vaccinia-induced apoptosis. Cell Death Differ. 2006, 13, 1651–1662. [Google Scholar] [CrossRef]
- Valencia-Serna, J.; Aliabadi, H.M.; Manfrin, A.; Mohseni, M.; Jiang, X.; Uludag, H. siRNA/lipopolymer nanoparticles to arrest growth of chronic myeloid leukemia cells in vitro and in vivo. Eur. J. Pharm. Biopharm. 2018, 130, 66–70. [Google Scholar] [CrossRef]
- Wang, H.; Liu, X.; Ai, X.; Remant-Bahadur, K.C.; Dick, T.A.; Yan, B.; Lu, T.; Zhou, X.; Luo, R.; Liu, M.; et al. Safe and Effective Delivery of mRNA Using Modified PEI-Based Lipopolymers. Pharmaceutics 2023, 15, 410. [Google Scholar] [CrossRef]
- Yotsomnuk, P.; Rajendran, A.P.; Sundaram, D.N.M.; Morales, L.C.; Kucharski, C.; Nasrullah, M.; Skolpap, W.; Jiang, X.; Gibson, S.B.; Brandwein, J.; et al. Lipopolymers as the Basis of Non-Viral Delivery of Therapeutic siRNA Nanoparticles in a Leukemia (MOLM-13) Model. Biomolecules 2025, 15, 115. [Google Scholar] [CrossRef]
- Nasrullah, M.; Kc, R.; Nickel, K.; Parent, K.; Kucharski, C.; Sundaram, D.N.M.; Rajendran, A.P.; Jiang, X.; Brandwein, J.; Uludağ, H. Lipopolymer/siRNA Nanoparticles Targeting the Signal Transducer and Activator of Transcription 5A Disrupts Proliferation of Acute Lymphoblastic Leukemia. ACS Pharmacol. Transl. Sci. 2024, 7, 2840–2855. [Google Scholar] [CrossRef]
- Morales, L.C.; Rajendran, A.; Ansari, A.; Kc, R.; Nasrullah, M.; Kiti, K.; Yotsomnuk, P.; Kulka, M.; Sundaram, D.N.M.; Uludağ, H. Biodistribution of Therapeutic Small Interfering RNAs Delivered with Lipid-Substituted Polyethylenimine-Based Delivery Systems. Mol. Pharm. 2024, 21, 1436–1449. [Google Scholar] [CrossRef]
- Ansari, A.S.; Remant, K.C.; Morales, L.C.; Nasrullah, M.; Sundaram, D.N.M.; Kucharski, C.; Jiang, X.; Brandwein, J.; Uludağ, H. Lipopolymer mediated siRNA delivery targeting aberrant oncogenes for effective therapy of myeloid leukemia in preclinical animal models. J. Control. Release 2024, 367, 821–836. [Google Scholar] [CrossRef]
- Santadkha, T.; Skolpap, W.; Remant, K.C.; Ansari, A.; Kucharski, C.; Uludağ, H. Improved delivery of Mcl-1 and survivin siRNA combination in breast cancer cells with additive siRNA complexes. Investig. New Drugs 2022, 40, 962–976. [Google Scholar] [CrossRef]
- Wang, S.; Frappier, L. Nucleosome assembly proteins bind to Epstein-Barr virus nuclear antigen 1 and affect its functions in DNA replication and transcriptional activation. J. Virol. 2009, 83, 11704–11714. [Google Scholar] [CrossRef]
- Piao, M.J.; Kang, K.A.; Lee, I.K.; Kim, H.S.; Kim, S.; Choi, J.Y.; Choi, J.; Hyun, J.W. Silver nanoparticles induce oxidative cell damage in human liver cells through inhibition of reduced glutathione and induction of mitochondria-involved apoptosis. Toxicol. Lett. 2011, 201, 92–100. [Google Scholar] [CrossRef]
- Hoogeveen, A.T.; Willemsen, R.; Meyer, N.; de Rooij, K.E.; Roos, R.A.; van Ommen, G.J.; Galjaard, H. Characterization and localization of the Huntington disease gene product. Hum. Mol. Genet. 1993, 2, 2069–2073. [Google Scholar] [CrossRef]
- Thompson, P.D.; Bhatia, K.P.; Brown, P.; Davis, M.B.; Pires, M.; Quinn, N.P.; Luthert, P.; Honovar, M.; O’Brien, M.D.; Marsden, C.D.; et al. Cortical myoclonus in Huntington’s disease. Mov. Disord. 1994, 9, 633–641. [Google Scholar] [CrossRef]
- Mangiarini, L.; Sathasivam, K.; Seller, M.; Cozens, B.; Harper, A.; Hetherington, C.; Lawton, M.; Trottier, Y.; Lehrach, H.; Davies, S.W.; et al. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell 1996, 87, 493–506. [Google Scholar] [CrossRef]
- Meade, C.A.; Deng, Y.-P.; Fusco, F.R.; Del Mar, N.; Hersch, S.; Goldowitz, D.; Reiner, A. Cellular localization and development of neuronal intranuclear inclusions in striatal and cortical neurons in R6/2 transgenic mice. J. Comp. Neurol. 2002, 449, 241–269. [Google Scholar] [CrossRef]
- Orth, M.; Cooper, J.M.; Bates, G.P.; Schapira, A.H.V. Inclusion formation in Huntington’s disease R6/2 mouse muscle cultures. J. Neurochem. 2003, 87, 1–6. [Google Scholar] [CrossRef]
- Carter, R.J.; Lione, L.A.; Humby, T.; Mangiarini, L.; Mahal, A.; Bates, G.P.; Dunnett, S.B.; Morton, A.J. Characterization of progressive motor deficits in mice transgenic for the human Huntington’s disease mutation. J. Neurosci. Off. J. Soc. Neurosci. 1999, 19, 3248–3257. [Google Scholar] [CrossRef]
- Sundaram, D.N.M.; Plianwong, S.; Kc, R.; Ostergaard, H.; Uludağ, H. In Vitro Cytotoxicity and Cytokine Production by Lipid-Substituted Low Molecular Weight Branched PEIs Used for Gene Delivery. Acta Biomater. 2022, 148, 279–297. [Google Scholar] [CrossRef]
- Belgrad, J.; Tang, Q.; Hildebrand, S.; Summers, A.; Sapp, E.; Echeverria, D.; O’Reilly, D.; Luu, E.; Bramato, B.; Allen, S.; et al. A programmable dual-targeting siRNA scaffold supports potent two-gene modulation in the central nervous system. Nucleic Acids Res. 2024, 52, 6099–6113. [Google Scholar] [CrossRef] [PubMed]
- Mathews, E.W.; Coffey, S.R.; Gärtner, A.; Belgrad, J.; Bragg, R.M.; O’Reilly, D.; Cantle, J.P.; McHugh, C.; Summers, A.; Fentz, J.; et al. Suppression of Huntington’s Disease Somatic Instability by Transcriptional Repression and Direct CAG Repeat Binding. BioRxiv 2024. [Google Scholar] [CrossRef]
- Hirunagi, T.; Sahashi, K.; Tachikawa, K.; Leu, A.I.; Nguyen, M.; Mukthavaram, R.; Karmali, P.P.; Chivukula, P.; Tohnai, G.; Iida, M.; et al. Selective suppression of polyglutamine-expanded protein by lipid nanoparticle-delivered siRNA targeting CAG expansions in the mouse CNS. Mol. Ther. Nucleic Acids 2021, 24, 1–10. [Google Scholar] [CrossRef]
- Aliabadi, H.M.; Totonchy, J.; Mahdipoor, P.; Parang, K.; Uludağ, H. Suppression of Human Coronavirus 229E Infection in Lung Fibroblast Cells via RNA Interference. Front. Nanotechnol. 2021, 3, 670543. [Google Scholar] [CrossRef]
- Mantha, N.; Das, S.K.; Das, N.G. RNAi-based therapies for Huntington’s disease: Delivery challenges and opportunities. Ther. Deliv. 2012, 3, 1061–1076. [Google Scholar] [CrossRef]
- Scacheri, P.C.; Rozenblatt-Rosen, O.; Caplen, N.J.; Wolfsberg, T.G.; Umayam, L.; Lee, J.C.; Hughes, C.M.; Shanmugam, K.S.; Bhattacharjee, A.; Meyerson, M.; et al. Short interfering RNAs can induce unexpected and divergent changes in the levels of untargeted proteins in mammalian cells. Proc. Natl. Acad. Sci. USA 2004, 101, 1892–1897. [Google Scholar] [CrossRef]
- Fedorov, Y.; Anderson, E.M.; Birmingham, A.; Reynolds, A.; Karpilow, J.; Robinson, K.; Leake, D.; Marshall, W.S.; Khvorova, A. Off-target effects by siRNA can induce toxic phenotype. RNA 2006, 12, 1188–1196. [Google Scholar] [CrossRef]
- Wood, T.E.; Barry, J.; Yang, Z.; Cepeda, C.; Levine, M.S.; Gray, M. Mutant huntingtin reduction in astrocytes slows disease progression in the BACHD conditional Huntington’s disease mouse model. Hum. Mol. Genet. 2019, 28, 487–500. [Google Scholar] [CrossRef]
- Zheng, M.; Tao, W.; Zou, Y.; Farokhzad, O.C.; Shi, B. Nanotechnology-Based Strategies for siRNA Brain Delivery for Disease Therapy. Trends Biotechnol. 2018, 36, 562–575. [Google Scholar] [CrossRef]
- Burgess, A.; Huang, Y.; Querbes, W.; Sah, D.W.; Hynynen, K. Focused ultrasound for targeted delivery of siRNA and efficient knockdown of Htt expression. J. Control. Release 2012, 163, 125–129. [Google Scholar] [CrossRef]
| siRNA | Sense Strand | Antisense Strand | Region of HTT (Nucleotides) |
|---|---|---|---|
| HTT1 siRNA | UUCAUCAGCUUUUCCAGGGdTdC | CCCUGGAAAAGCUGAUGACdGdG | 8–26 |
| HTT2 siRNA | GCCUUCGAGUCCCUCAAUdCdC | ACUUGAGGGACUGAAGGCdCdT | 28–44 |
| HTT3 siRNA | UUGCUGUUGCUGCUGUUGGdAdA | CCAACAGCAGCAACAGCAAdCdA | 49–71 (in CAG mutation) |
| GFP-22 siRNA | GAACUUCAGGGUCAGCUUGCCG | GCAAGCUGACCCUGAAGUUCAU | 620–644 |
| Ctrl siRNA | AUGCUACCAAUUCAAGUGAAGAUdTdA | UAAUCUUCACUUGAAUUGGUAGCAUUU | N/A |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morales, L.C.; Modi, L.; Abbasi Dezfouli, S.; Rajendran, A.P.; Kc, R.; Kadam, V.; Sipione, S.; Uludağ, H. In Vitro Efficacy of PEI-Derived Lipopolymers in Silencing of Toxic Proteins in a Neuronal Model of Huntington’s Disease. Pharmaceutics 2025, 17, 726. https://doi.org/10.3390/pharmaceutics17060726
Morales LC, Modi L, Abbasi Dezfouli S, Rajendran AP, Kc R, Kadam V, Sipione S, Uludağ H. In Vitro Efficacy of PEI-Derived Lipopolymers in Silencing of Toxic Proteins in a Neuronal Model of Huntington’s Disease. Pharmaceutics. 2025; 17(6):726. https://doi.org/10.3390/pharmaceutics17060726
Chicago/Turabian StyleMorales, Luis C., Luv Modi, Saba Abbasi Dezfouli, Amarnath Praphakar Rajendran, Remant Kc, Vaibhavi Kadam, Simonetta Sipione, and Hasan Uludağ. 2025. "In Vitro Efficacy of PEI-Derived Lipopolymers in Silencing of Toxic Proteins in a Neuronal Model of Huntington’s Disease" Pharmaceutics 17, no. 6: 726. https://doi.org/10.3390/pharmaceutics17060726
APA StyleMorales, L. C., Modi, L., Abbasi Dezfouli, S., Rajendran, A. P., Kc, R., Kadam, V., Sipione, S., & Uludağ, H. (2025). In Vitro Efficacy of PEI-Derived Lipopolymers in Silencing of Toxic Proteins in a Neuronal Model of Huntington’s Disease. Pharmaceutics, 17(6), 726. https://doi.org/10.3390/pharmaceutics17060726







