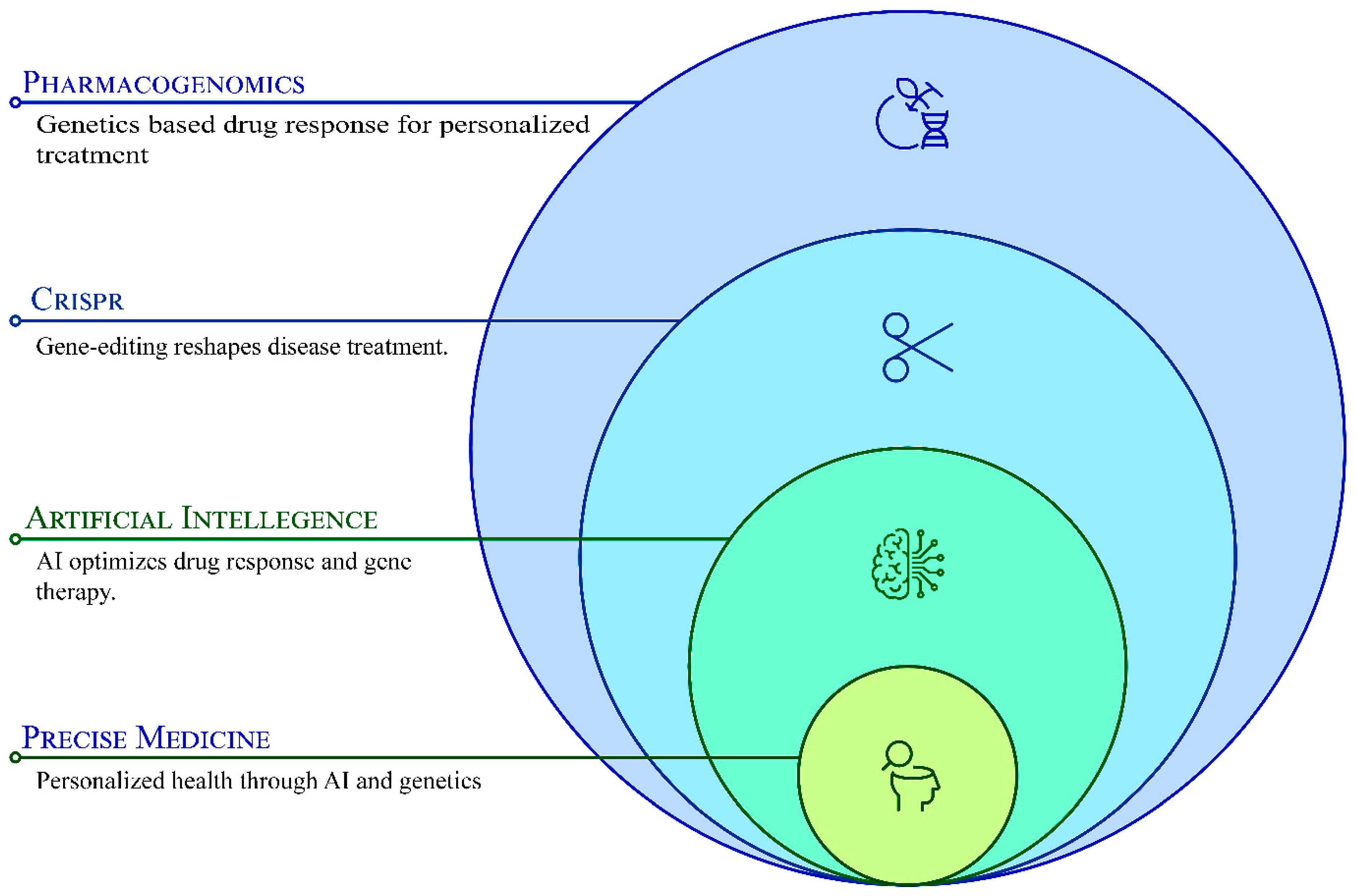
Transforming Pharmacogenomics and CRISPR Gene Editing with the Power of Artificial Intelligence for Precision Medicine
Abstract
1. Introduction
2. Pharmacogenomics: The Foundation of Precision Medicine
2.1. Genetic Variability and Its Role in Drug Response
2.2. Pharmacogenetic Markers: Genetic Variants Influencing Drug Metabolism
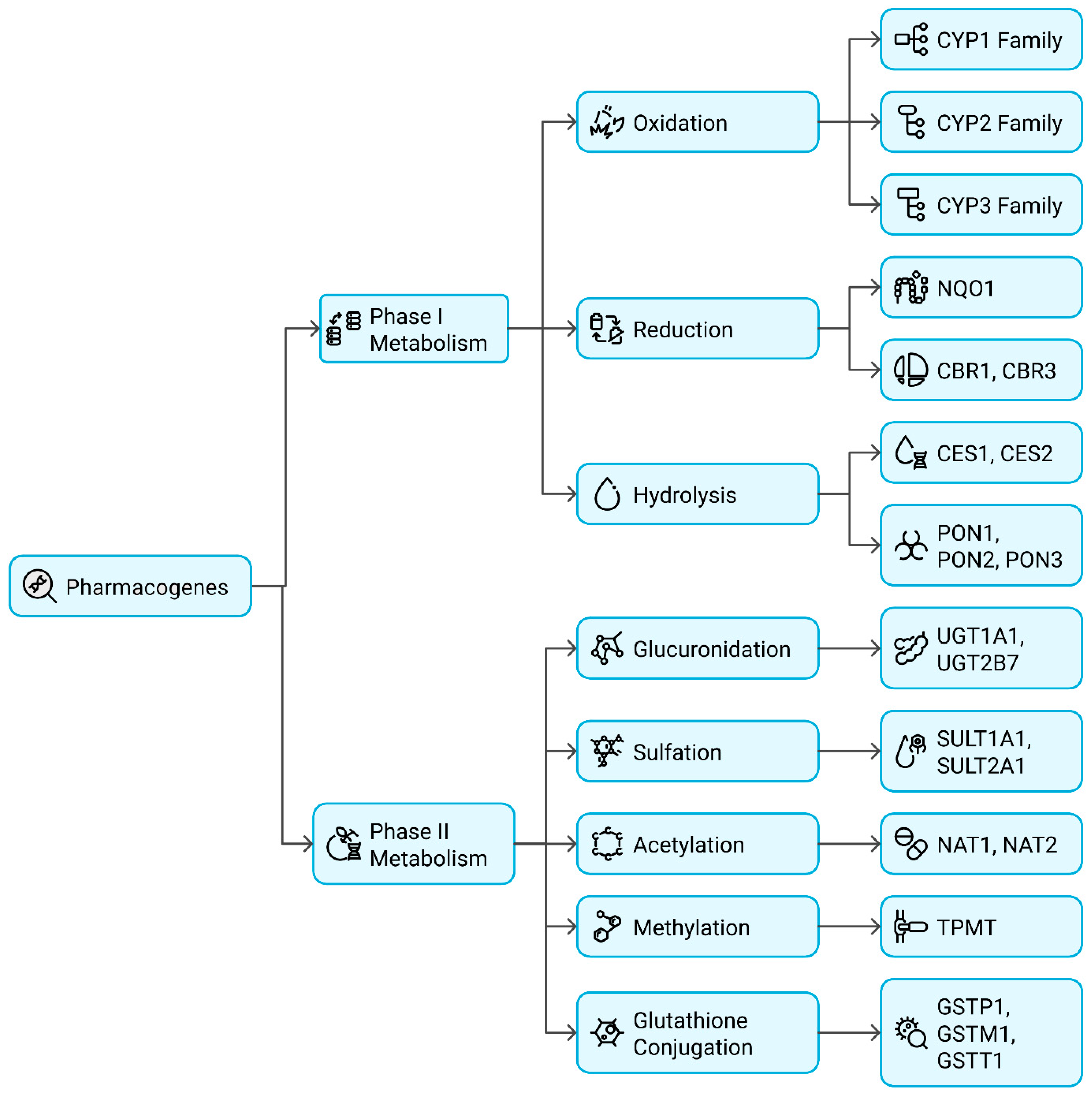
2.2.1. Functional Classification of Pharmacogenes
Phase I Metabolism Genes (Oxidation, Reduction, Hydrolysis)
Phase II Metabolism Genes (Conjugation Reactions)
2.3. Mechanistic Insights into Pharmacogenomic Variations
2.3.1. Functional Consequences of Single Nucleotide Polymorphisms (SNPs)
Pharmacogenes: Synonymous vs. Non-Synonymous SNPs
Drug Response Splice Site and Frameshift Mutations
2.3.2. Copy Number Variations (CNVs) in Drug Metabolism
2.3.3. Epigenetic Modifications and Drug Reactivity
MiRNA Control and DNA Methylation of Drug-Metabolizing Enzymes
Gene Expression and Histone Modifications in Pharmacogenomics
3. CRISPR Genome Editing: A Revolutionary Tool in Personalized Medicine
3.1. CRISPR and Its Role in Precision Medicine
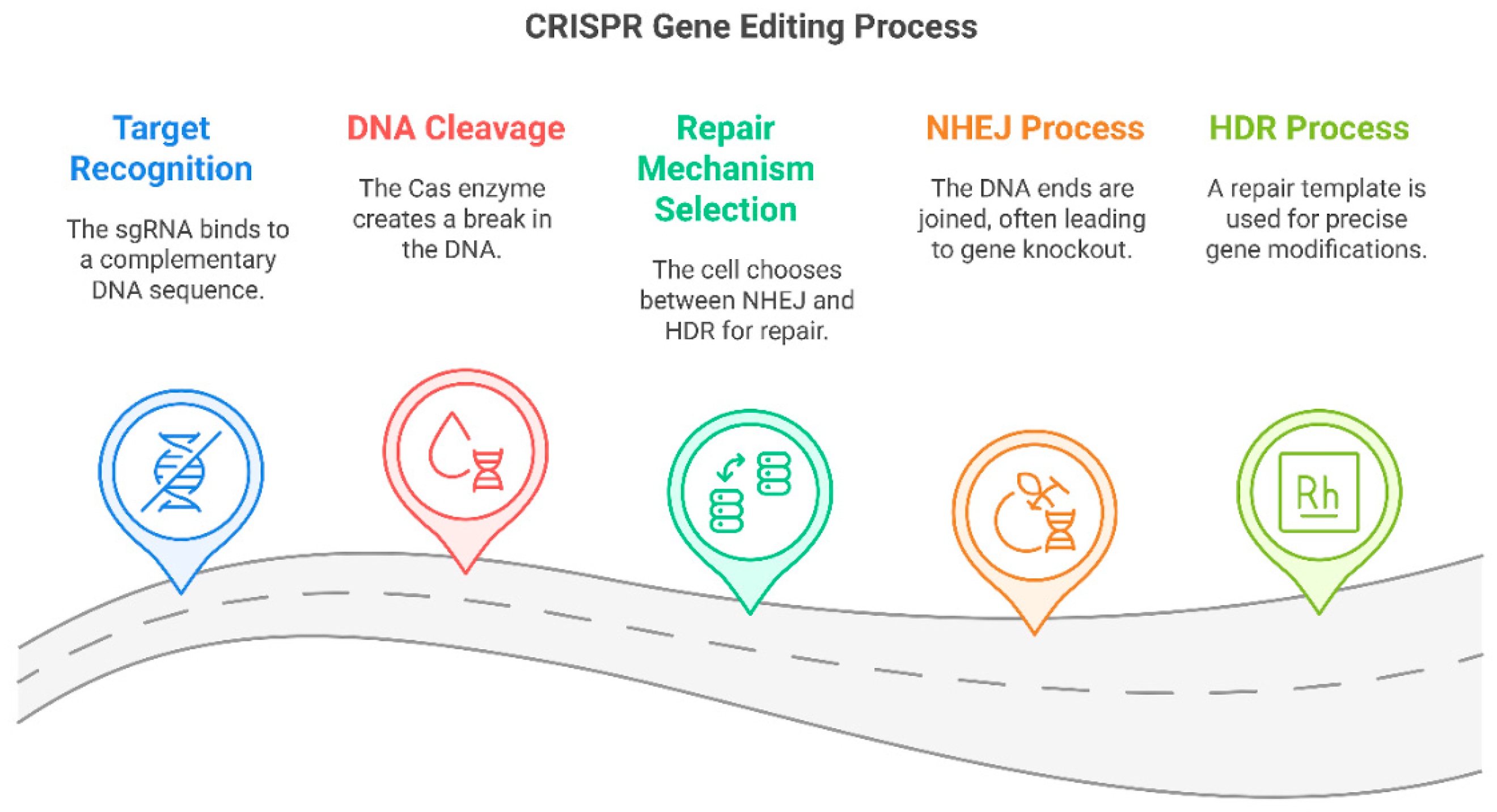
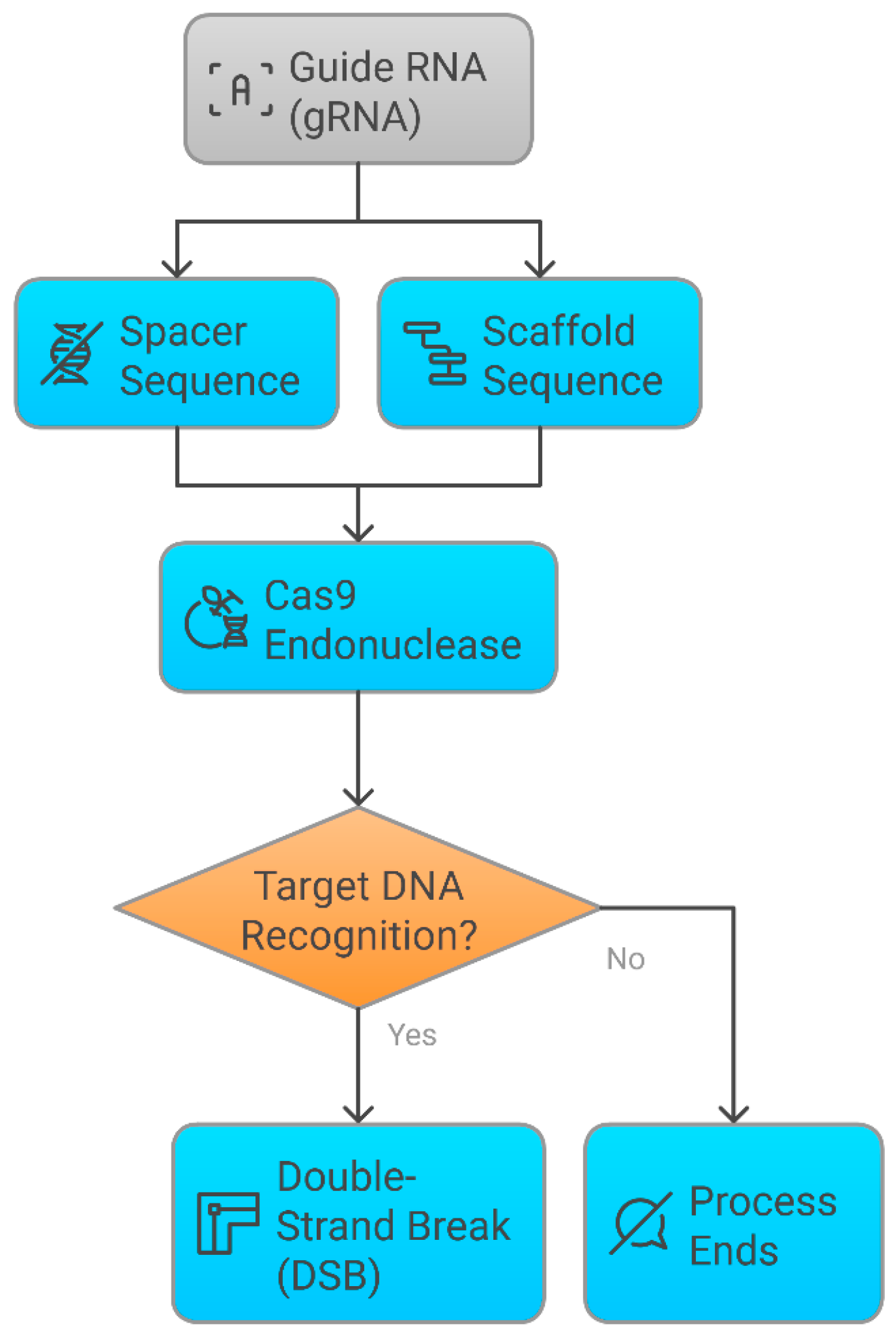
3.2. Mechanistic Insights into CRISPR Genome Editing
3.2.1. Molecular Mechanism of CRISPR-Cas9
3.2.2. Limitations of Subcellular Delivery in CRISPR Genome Editing
3.2.3. Base Editing and Prime Editing: Next-Generation CRISPR Approaches
3.3. CRISPR in Cancer Therapy and Precision Oncology
3.3.1. Targeting Oncogenic Mutations for Personalized Cancer Treatment
3.3.2. CRISPR-Modified CAR-T Cells in Immunotherapy
3.4. CRISPR for Neurological and Metabolic Disorders in Precision Medicine
3.5. CRISPR in Metabolic Disorders
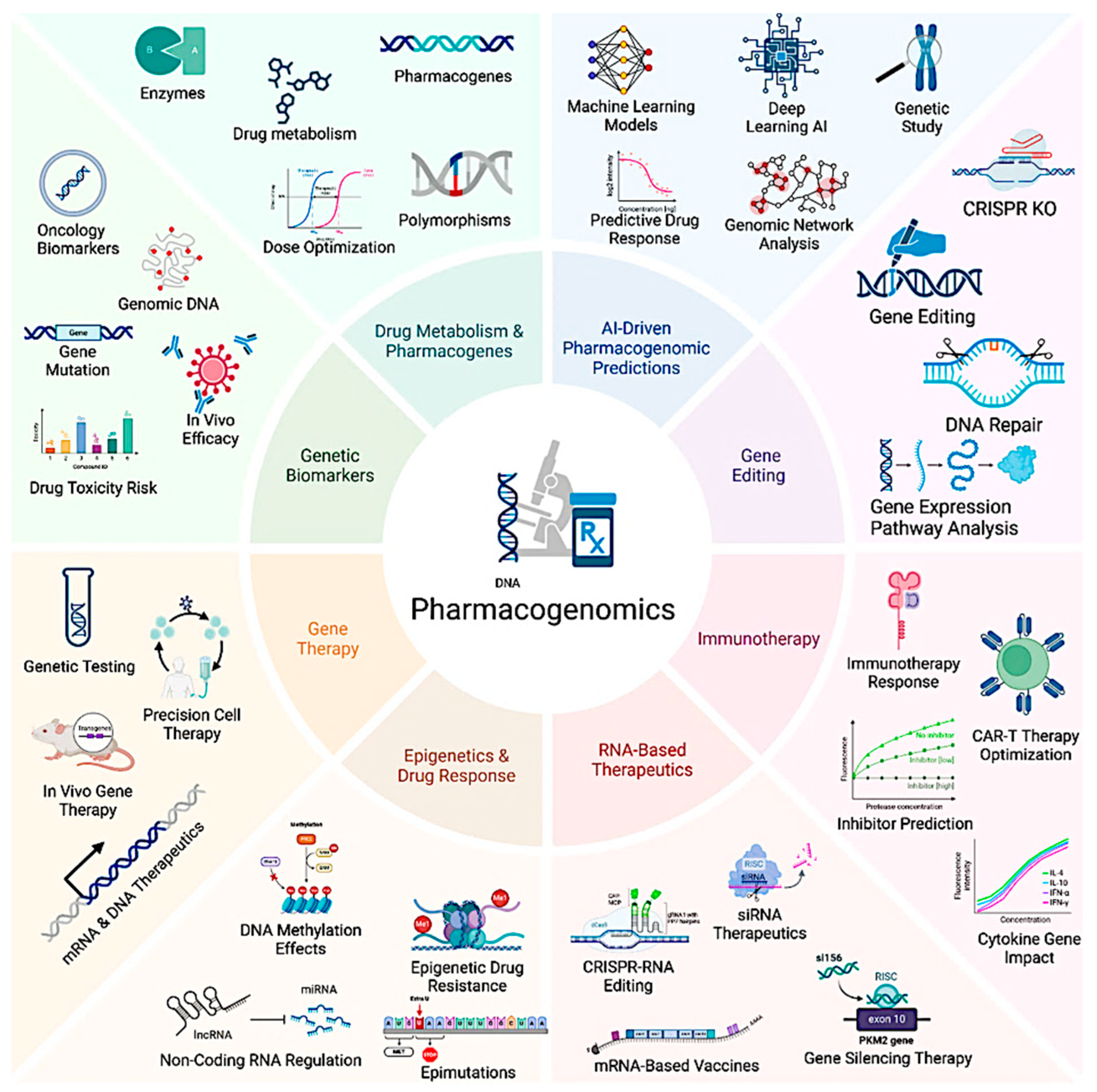
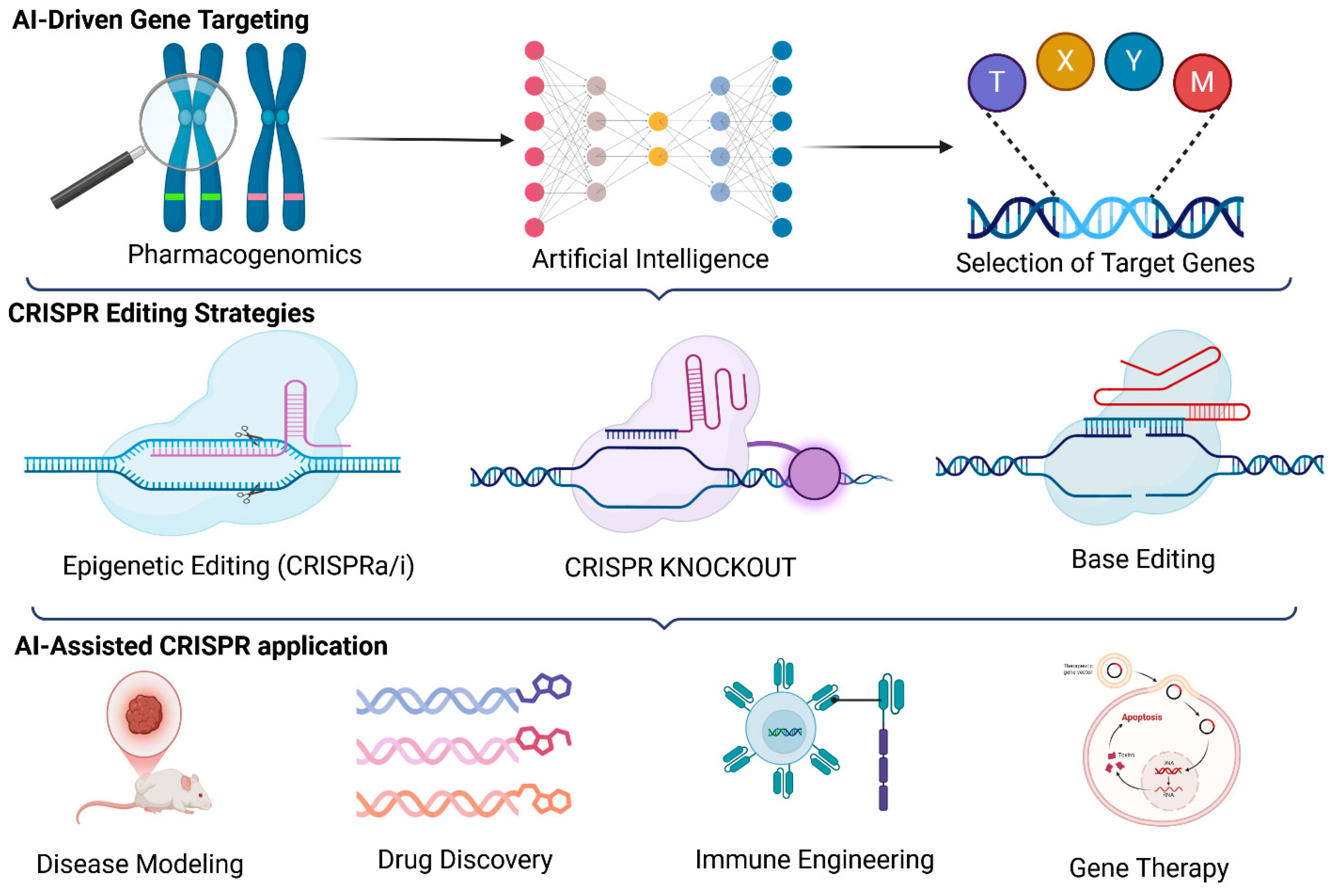
4. Integrating AI, CRISPR, and Pharmacogenomics for Precision Medicine
4.1. The Convergence of AI, CRISPR, and Pharmacogenomics in Precision Medicine
4.2. AI-Enhanced Pharmacogenomic Analysis for Personalized Medicine
4.3. AI-Optimized CRISPR Genome Editing for Precision Therapy
4.4. AI-Guided Drug Repurposing and CRISPR-Driven Functional Genomics
4.5. AI, CRISPR, and Pharmacogenomics in Immunotherapy and Oncology
4.6. AI-CRISPR Integration for Multi-Omics Data Interpretation in Precision Medicine
5. Ethical, Regulatory, and Implementation Challenges
6. Future Directions
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AI | Artificial Intelligence |
| CRISPR | Clustered Regularly Interspaced Short Palindromic Repeats |
| PGx | Pharmacogenomics |
| CAR-T | Chimeric Antigen Receptor T-cell |
| NLP | Natural Language Processing |
| GWAS | Genome-Wide Association Study |
| XAI | Explainable AI |
| TMB | Tumor Mutational Burden |
| miRNA | microRNA |
| lncRNA | Long Non-Coding RNA |
| HDR | Homology-Directed Repair |
| NHEJ | Non-Homologous End Joining |
| iPSC | Induced Pluripotent Stem Cells |
| TCR | T-Cell Receptor |
| FDA | Food and Drug Administration |
| TP53 | Tumor Protein p53 |
| CYP | Cytochrome P450 |
| HLA | Human Leukocyte Antigen |
| NGS | Next-Generation Sequencing |
| PAM | Protospacer Adjacent Motif |
References
- Marques, L.; Costa, B.; Pereira, M.; Silva, A.; Santos, J.; Saldanha, L.; Silva, I.; Magalhães, P.; Schmidt, S.; Vale, N. Advancing Precision Medicine: A Review of Innovative In Silico Approaches for Drug Development, Clinical Pharmacology and Personalized Healthcare. Pharmaceutics 2024, 16, 332. [Google Scholar] [CrossRef] [PubMed]
- Krzyszczyk, P.; Acevedo, A.; Davidoff, E.J.; Timmins, L.M.; Marrero-Berrios, I.; Patel, M.; White, C.; Lowe, C.; Sherba, J.J.; Hartmanshenn, C.; et al. The growing role of precision and personalized medicine for cancer treatment. Technology 2018, 6, 79–100. [Google Scholar] [CrossRef]
- Serrano, D.R.; Luciano, F.C.; Anaya, B.J.; Ongoren, B.; Kara, A.; Molina, G.; Ramirez, B.I.; Sánchez-Guirales, S.A.; Simon, J.A.; Tomietto, G.; et al. Artificial Intelligence (AI) Applications in Drug Discovery and Drug Delivery: Revolutionizing Personalized Medicine. Pharmaceutics 2024, 16, 1328. [Google Scholar] [CrossRef] [PubMed]
- Ho, D.; Quake, S.R.; McCabe, E.R.B.; Chng, W.J.; Chow, E.K.; Ding, X.; Gelb, B.D.; Ginsburg, G.S.; Hassenstab, J.; Ho, C.M.; et al. Enabling Technologies for Personalized and Precision Medicine. Trends Biotechnol. 2020, 38, 497–518. [Google Scholar] [CrossRef] [PubMed]
- Bhat, A.A.; Nisar, S.; Mukherjee, S.; Saha, N.; Yarravarapu, N.; Lone, S.N.; Masoodi, T.; Chauhan, R.; Maacha, S.; Bagga, P.; et al. Integration of CRISPR/Cas9 with artificial intelligence for improved cancer therapeutics. J. Transl. Med. 2022, 20, 534. [Google Scholar] [CrossRef]
- Taherdoost, H.; Ghofrani, A. AI’s role in revolutionizing personalized medicine by reshaping pharmacogenomics and drug therapy. Intell. Pharm. 2024, 2, 643–650. [Google Scholar] [CrossRef]
- Ahmed, S.; Zhou, Z.; Zhou, J.; Chen, S.Q. Pharmacogenomics of Drug Metabolizing Enzymes and Transporters: Relevance to Precision Medicine. Genom. Proteom. Bioinform. 2016, 14, 298–313. [Google Scholar] [CrossRef]
- Nahid, N.A.; Johnson, J.A. CYP2D6 pharmacogenetics and phenoconversion in personalized medicine. Expert Opin. Drug Metab. Toxicol. 2022, 18, 769–785. [Google Scholar] [CrossRef]
- Sissung, T.M.; Goey, A.K.; Ley, A.M.; Strope, J.D.; Figg, W.D. Pharmacogenetics of membrane transporters: A review of current approaches. Methods Mol. Biol. 2014, 1175, 91–120. [Google Scholar] [CrossRef]
- Principi, N.; Petropulacos, K.; Esposito, S. Impact of Pharmacogenomics in Clinical Practice. Pharmaceuticals 2023, 16, 1596. [Google Scholar] [CrossRef]
- Adithan, C.; Subathra, A. NAT2 gene polymorphism: Covert drug interaction causing phenytoin toxicity. Indian J. Med. Res. 2016, 143, 542–544. [Google Scholar] [CrossRef] [PubMed]
- Urbančič, D.; Jukič, M.; Šmid, A.; Gobec, S.; Jazbec, J.; Mlinarič-Raščan, I. Thiopurine S-methyltransferase—An important intersection of drug-drug interactions in thiopurine treatment. Biomed. Pharmacother. 2025, 184, 117893. [Google Scholar] [CrossRef]
- Unissa, A.N.; Sukumar, S.; Hanna, L.E. The Role of N-Acetyl Transferases on Isoniazid Resistance from Mycobacterium tuberculosis and Human: An In Silico Approach. Tuberc. Respir. Dis. 2017, 80, 255–264. [Google Scholar] [CrossRef]
- Roden, D.M.; Wilke, R.A.; Kroemer, H.K.; Stein, C.M. Pharmacogenomics: The genetics of variable drug responses. Circulation 2011, 123, 1661–1670. [Google Scholar] [CrossRef] [PubMed]
- Ran, F.A.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Li, L.; Jiang, J.; Wu, M.; Lin, P. Applications and challenges of CRISPR-Cas gene-editing to disease treatment in clinics. Precis. Clin. Med. 2021, 4, 179–191. [Google Scholar] [CrossRef]
- Aljabali, A.A.A.; El-Tanani, M.; Tambuwala, M.M. Principles of CRISPR-Cas9 technology: Advancements in genome editing and emerging trends in drug delivery. J. Drug Deliv. Sci. Technol. 2024, 92, 105338. [Google Scholar] [CrossRef]
- Xue, C.; Greene, E.C. DNA Repair Pathway Choices in CRISPR-Cas9-Mediated Genome Editing. Trends Genet. 2021, 37, 639–656. [Google Scholar] [CrossRef]
- Shaikh, T.H. Copy Number Variation Disorders. Curr. Genet. Med. Rep. 2017, 5, 183–190. [Google Scholar] [CrossRef]
- Bhatt, D.K.; Basit, A.; Zhang, H.; Gaedigk, A.; Lee, S.B.; Claw, K.G.; Mehrotra, A.; Chaudhry, A.S.; Pearce, R.E.; Gaedigk, R.; et al. Hepatic Abundance and Activity of Androgen- and Drug-Metabolizing Enzyme UGT2B17 Are Associated with Genotype, Age, and Sex. Drug Metab. Dispos. 2018, 46, 888–896. [Google Scholar] [CrossRef]
- Loureiro, A.; da Silva, G.J. CRISPR-Cas: Converting A Bacterial Defence Mechanism into A State-of-the-Art Genetic Manipulation Tool. Antibiotics 2019, 8, 18. [Google Scholar] [CrossRef]
- Asmamaw, M.; Zawdie, B. Mechanism and Applications of CRISPR/Cas-9-Mediated Genome Editing. Biologics 2021, 15, 353–361. [Google Scholar] [CrossRef] [PubMed]
- Sioson, V.A.; Kim, M.; Joo, J. Challenges in delivery systems for CRISPR-based genome editing and opportunities of nanomedicine. Biomed. Eng. Lett. 2021, 11, 217–233. [Google Scholar] [CrossRef] [PubMed]
- Alamillo, J.M.; López, C.M.; Martínez Rivas, F.J.; Torralbo, F.; Bulut, M.; Alseekh, S. Clustered regularly interspaced short palindromic repeats/CRISPR-associated protein and hairy roots: A perfect match for gene functional analysis and crop improvement. Curr. Opin. Biotechnol. 2023, 79, 102876. [Google Scholar] [CrossRef]
- Xu, C.L.; Ruan, M.Z.C.; Mahajan, V.B.; Tsang, S.H. Viral Delivery Systems for CRISPR. Viruses 2019, 11, 28. [Google Scholar] [CrossRef] [PubMed]
- Yoon, H.; Shaw, J.L.; Haigis, M.C.; Greka, A. Lipid metabolism in sickness and in health: Emerging regulators of lipotoxicity. Mol. Cell 2021, 81, 3708–3730. [Google Scholar] [CrossRef]
- Tsuchida, C.A.; Wasko, K.M.; Hamilton, J.R.; Doudna, J.A. Targeted nonviral delivery of genome editors in vivo. Proc. Natl. Acad. Sci. USA 2024, 121, e2307796121. [Google Scholar] [CrossRef]
- Mehta, A.; Merkel, O.M. Immunogenicity of Cas9 Protein. J. Pharm. Sci. 2020, 109, 62–67. [Google Scholar] [CrossRef]
- Saeed, S.; Bonnefond, A.; Froguel, P. Obesity: Exploring its connection to brain function through genetic and genomic perspectives. Mol. Psychiatry 2025, 30, 651–658. [Google Scholar] [CrossRef]
- Molla, G.; Bitew, M. Revolutionizing Personalized Medicine: Synergy with Multi-Omics Data Generation, Main Hurdles, and Future Perspectives. Biomedicines 2024, 12, 2750. [Google Scholar] [CrossRef]
- Chakravarthi, B.V.; Nepal, S.; Varambally, S. Genomic and Epigenomic Alterations in Cancer. Am. J. Pathol. 2016, 186, 1724–1735. [Google Scholar] [CrossRef] [PubMed]
- Kalinin, A.A.; Higgins, G.A.; Reamaroon, N.; Soroushmehr, S.; Allyn-Feuer, A.; Dinov, I.D.; Najarian, K.; Athey, B.D. Deep learning in pharmacogenomics: From gene regulation to patient stratification. Pharmacogenomics 2018, 19, 629–650. [Google Scholar] [CrossRef] [PubMed]
- Askr, H.; Elgeldawi, E.; Aboul Ella, H.; Elshaier, Y.A.M.M.; Gomaa, M.M.; Hassanien, A.E. Deep learning in drug discovery: An integrative review and future challenges. Artif. Intell. Rev. 2023, 56, 5975–6037. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Ma, X.; Gao, F.; Guo, Y. Off-target effects in CRISPR/Cas9 gene editing. Front. Bioeng. Biotechnol. 2023, 11, 1143157. [Google Scholar] [CrossRef]
- Erdoğan, S. Integration of Artificial Intelligence and Genome Editing System for Determining the Treatment of Genetic Disorders. Balk. Med. J. 2024, 41, 419–420. [Google Scholar] [CrossRef]
- Chehelgerdi, M.; Chehelgerdi, M.; Khorramian-Ghahfarokhi, M.; Shafieizadeh, M.; Mahmoudi, E.; Eskandari, F.; Rashidi, M.; Arshi, A.; Mokhtari-Farsani, A. Comprehensive review of CRISPR-based gene editing: Mechanisms, challenges, and applications in cancer therapy. Mol. Cancer 2024, 23, 9. [Google Scholar] [CrossRef] [PubMed]
- Bock, C.; Datlinger, P.; Chardon, F.; Coelho, M.A.; Dong, M.B.; Lawson, K.A.; Lu, T.; Maroc, L.; Norman, T.M.; Song, B.; et al. High-content CRISPR screening. Nat. Rev. Methods Primers 2022, 2, 8. [Google Scholar] [CrossRef]
- Tao, R.; Han, X.; Bai, X.; Yu, J.; Ma, Y.; Chen, W.; Zhang, D.; Li, Z. Revolutionizing cancer treatment: Enhancing CAR-T cell therapy with CRISPR/Cas9 gene editing technology. Front. Immunol. 2024, 15, 1354825. [Google Scholar] [CrossRef]
- Subramanian, I.; Verma, S.; Kumar, S.; Jere, A.; Anamika, K. Multi-omics Data Integration, Interpretation, and Its Application. Bioinform. Biol. Insights 2020, 14, 1177932219899051. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Oikonomou, P.; Tavazoie, S. Comprehensive Genome-wide Perturbations via CRISPR Adaptation Reveal Complex Genetics of Antibiotic Sensitivity. Cell 2020, 180, 1002–1017.E31. [Google Scholar] [CrossRef]
- Ayanoğlu, F.B.; Elçin, A.E.; Elçin, Y.M. Bioethical issues in genome editing by CRISPR-Cas9 technology. Turk. J. Biol. 2020, 44, 110–120. [Google Scholar] [CrossRef] [PubMed]
- Gershon, E.S.; Alliey-Rodriguez, N.; Grennan, K. Ethical and public policy challenges for pharmacogenomics. Dialogues Clin. Neurosci. 2014, 16, 567–574. [Google Scholar] [CrossRef]
- Minssen, T.; Gerke, S.; Aboy, M.; Price, N.; Cohen, G. Regulatory responses to medical machine learning. J. Law Biosci. 2020, 7, lsaa002. [Google Scholar] [CrossRef] [PubMed]
- Lafi, Z.; Ata, T.; Asha, S. CRISPR in clinical diagnostics: Bridging the gap between research and practice. Bioanalysis 2025, 17, 281–290. [Google Scholar] [CrossRef]
- Johnson, K.B.; Wei, W.Q.; Weeraratne, D.; Frisse, M.E.; Misulis, K.; Rhee, K.; Zhao, J.; Snowdon, J.L. Precision Medicine, AI, and the Future of Personalized Health Care. Clin. Transl. Sci. 2021, 14, 86–93. [Google Scholar] [CrossRef]
- Bhinder, B.; Gilvary, C.; Madhukar, N.S.; Elemento, O. Artificial Intelligence in Cancer Research and Precision Medicine. Cancer Discov. 2021, 11, 900–915. [Google Scholar] [CrossRef] [PubMed]
- Abdalla, M.M.I. Advancing diabetes management: Exploring pancreatic beta-cell restoration’s potential and challenges. World J. Gastroenterol. 2024, 30, 4339–4353. [Google Scholar] [CrossRef]
- Karpov, D.S.; Sosnovtseva, A.O.; Pylina, S.V.; Bastrich, A.N.; Petrova, D.A.; Kovalev, M.A.; Shuvalova, A.I.; Eremkina, A.K.; Mokrysheva, N.G. Challenges of CRISPR/Cas-Based Cell Therapy for Type 1 Diabetes: How Not to Engineer a “Trojan Horse”. Int. J. Mol. Sci. 2023, 24, 17320. [Google Scholar] [CrossRef]
- Dixit, S.; Kumar, A.; Srinivasan, K.; Vincent, P.; Ramu Krishnan, N. Advancing genome editing with artificial intelligence: Opportunities, challenges, and future directions. Front. Bioeng. Biotechnol. 2023, 11, 1335901. [Google Scholar] [CrossRef]
| Feature | CRISPR-Cas9 | CRISPR-Cas12 | CRISPR-Cas13 |
|---|---|---|---|
| Target Type | Double-stranded DNA | Single-stranded and double-stranded DNA | Single-stranded RNA |
| Cleavage Mechanism | Creates double-strand breaks (DSBs) | Creates single-strand staggered cuts | Cleave RNA molecules |
| Guide RNA (gRNA) | Single guide RNA (sgRNA) | Single guide RNA (sgRNA) | CRISPR RNA (crRNA) |
| Recognition Motif (PAM or PFS) | PAM sequence (5′-NGG-3′) | PAM sequence (5′-TTTV-3′) | PFS (Protospacer Flanking Site) instead of PAM |
| Editing Precision | High, but off-target effects possible | Higher specificity than Cas9 | High specificity for RNA editing |
| Primary Applications | Gene knockout, gene correction, genome-wide screening | Genome editing, diagnostics (e.g., SHERLOCK) | RNA interference, viral RNA targeting, transcriptome regulation |
| Advantages | Efficient for genome editing, widely studied, well-characterized | Higher specificity, useful for diagnostics and collateral activity, enables detection applications | Can target RNA without affecting genome, potential for antiviral therapies |
| Limitations | Potential for off-target mutations requires PAM sequence | Less well-characterized than Cas9, requires PAM sequence | Collateral RNA degradation limits specificity and is not useful for DNA editing |
| AI, CRISPR, and Pharmacogenomics Concept | Key Focus | Applications | AI Techniques Used | Challenges |
|---|---|---|---|---|
| Drug Repurposing and Functional Genomics | AI-powered drug repurposing and CRISPR functional genomics to identify new therapeutic applications of existing drugs and novel drug targets. | Oncology, rare diseases, metabolic disorders, identification of synthetic lethal interactions. | Neural networks, AI-driven drug-target interaction prediction, deep learning-based functional genomics screening. | Validation of AI-identified drug repurposing candidates, ethical considerations in genetic screening. |
| Immunotherapy and Oncology | Optimization of cancer immunotherapy through AI-driven patient stratification, CRISPR-enhanced CAR-T cell therapy, and immune checkpoint modulation. | Tumor microenvironment analysis, AI-powered immune profiling, CRISPR-engineered immunotherapies. | AI-based single-cell sequencing analysis, AI-guided CRISPR knockout studies, immune response prediction. | Patient-specific immunotherapy optimization, overcoming tumor immune evasion mechanisms. |
| Multi-Omics Data Interpretation | AI-driven multi-omics data integration for understanding disease mechanisms, drug responses, and personalized treatment strategies. | Personalized multi-omics analysis, single-cell transcriptomics, and disease pathway modeling. | Integrating AI with genomics, transcriptomics, proteomics, and metabolomics data analysis. | Standardization of multi-omics data interpretation, regulatory challenges in AI-driven precision medicine. |
| Neurodegenerative Disease Therapy | CRISPR-based correction of neurodegenerative disease mutations, AI-guided identification of genetic targets for personalized therapy. | Alzheimer’s, Parkinson’s, Huntington’s disease, CRISPR-based neuroprotection. | Machine learning-based biomarker discovery, AI-guided CRISPR target selection, and deep learning for disease progression modeling. | Ethical concerns in CRISPR neuro-editing, ensuring AI accuracy in neurogenomic predictions. |
| Metabolic Disorder Management | Precision genome editing for diabetes, obesity, and cholesterol management, AI-driven analysis of metabolic pathways. | CRISPR-driven insulin sensitivity modulation, lipid metabolism optimization, personalized treatment for metabolic disorders. | AI-enhanced metabolic profiling, deep learning for CRISPR-mediated genetic corrections. | Long-term safety of CRISPR-based metabolic modifications, AI-driven identification of unintended gene interactions. |
| AI-Driven Pharmacovigilance | AI-driven real-time monitoring of adverse drug reactions (ADRs), CRISPR validation of pharmacogenomic safety markers. | Early detection of ADRs, pharmacogenomic safety monitoring, and AI-enhanced drug safety assessments. | Natural language processing (NLP) for pharmacovigilance data, AI-assisted safety screening of CRISPR modifications. | Regulatory approval of AI-powered pharmacovigilance systems, balancing AI automation with clinical oversight. |
| Synthetic Biology and AI-CRISPR Integration | AI-powered synthetic biology tools for CRISPR-based genome engineering, designing gene circuits for therapeutic applications. | AI-designed gene circuits for cell therapy, CRISPR-driven biosynthetic pathway modifications. | AI-assisted protein engineering, machine learning-driven pathway modeling, CRISPR gene circuit optimization. | Ethical concerns in AI-driven synthetic biology, potential risks of unintended gene expression alterations. |
| Gene Therapy Optimization | Improving gene therapy efficiency using AI-optimized CRISPR delivery mechanisms and patient-specific therapeutic modeling. | Rare genetic disease therapy, AI-enhanced vector selection for CRISPR delivery, CRISPR-based ex vivo gene correction. | AI-powered vector design, deep learning for optimizing CRISPR efficiency, AI-driven prediction of therapeutic outcomes. | Minimizing CRISPR off-target effects in gene therapy, regulatory challenges in AI-assisted gene therapy development. |
| Personalized Regenerative Medicine | AI-CRISPR-engineered stem cells for regenerative medicine developed personalized tissue and organ repair strategies. | Tissue regeneration, CRISPR-based iPSC therapies, AI-powered stem cell differentiation modeling. | AI-driven stem cell fate prediction, machine learning-guided tissue engineering, CRISPR-based genome stability analysis. | AI-driven stem cell differentiation reliability, CRISPR safety in regenerative medicine, patient-specific variability. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Srivastav, A.K.; Mishra, M.K.; Lillard, J.W., Jr.; Singh, R. Transforming Pharmacogenomics and CRISPR Gene Editing with the Power of Artificial Intelligence for Precision Medicine. Pharmaceutics 2025, 17, 555. https://doi.org/10.3390/pharmaceutics17050555
Srivastav AK, Mishra MK, Lillard JW Jr., Singh R. Transforming Pharmacogenomics and CRISPR Gene Editing with the Power of Artificial Intelligence for Precision Medicine. Pharmaceutics. 2025; 17(5):555. https://doi.org/10.3390/pharmaceutics17050555
Chicago/Turabian StyleSrivastav, Amit Kumar, Manoj Kumar Mishra, James W. Lillard, Jr., and Rajesh Singh. 2025. "Transforming Pharmacogenomics and CRISPR Gene Editing with the Power of Artificial Intelligence for Precision Medicine" Pharmaceutics 17, no. 5: 555. https://doi.org/10.3390/pharmaceutics17050555
APA StyleSrivastav, A. K., Mishra, M. K., Lillard, J. W., Jr., & Singh, R. (2025). Transforming Pharmacogenomics and CRISPR Gene Editing with the Power of Artificial Intelligence for Precision Medicine. Pharmaceutics, 17(5), 555. https://doi.org/10.3390/pharmaceutics17050555







