Towards Explainable Computational Toxicology: Linking Antitargets to Rodent Acute Toxicity
Abstract
1. Introduction
2. Materials and Methods
2.1. Dataset Preprocessing
2.2. Physicochemical Properties Analysis
2.3. Chemical Space Visualization
2.4. Molecular Docking
2.4.1. Ligand Preparation
2.4.2. Target Preparation
2.4.3. Molecular Docking Software
2.5. Orthologous Protein Sequence Alignment
2.6. Butina Clustering
2.7. Statistical Analysis
2.7.1. Mann–Whitney U-Test
2.7.2. Spearman Correlations
3. Results
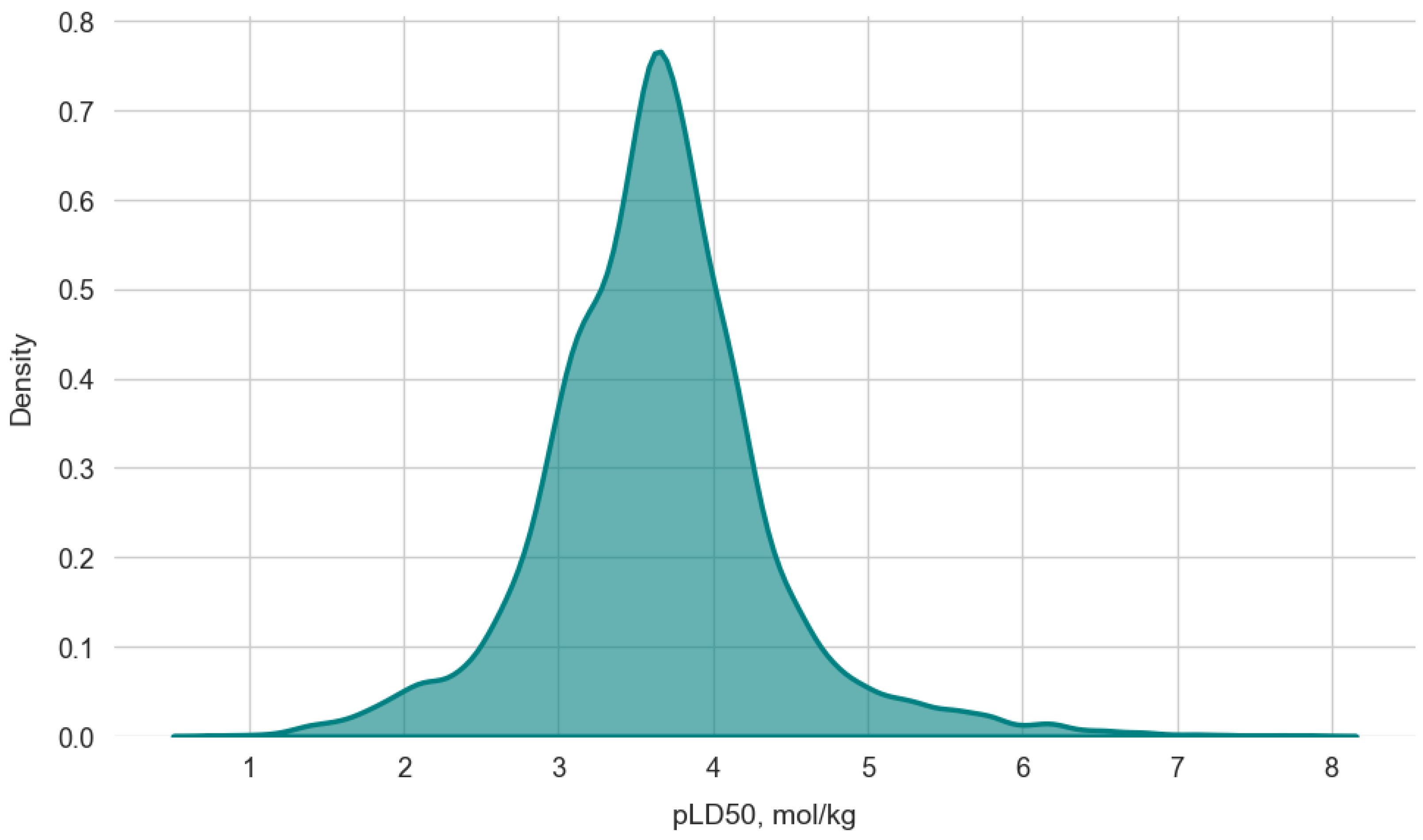
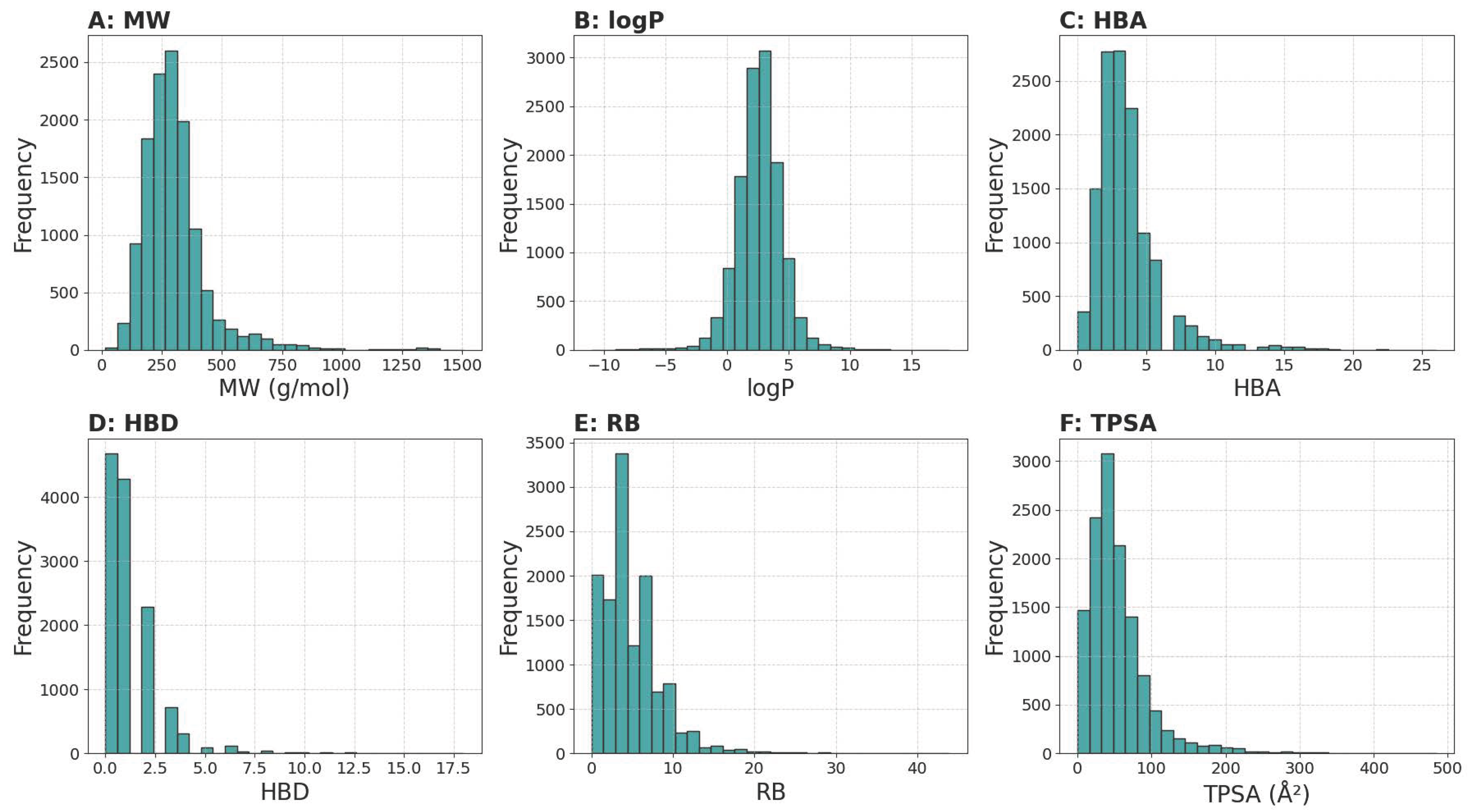
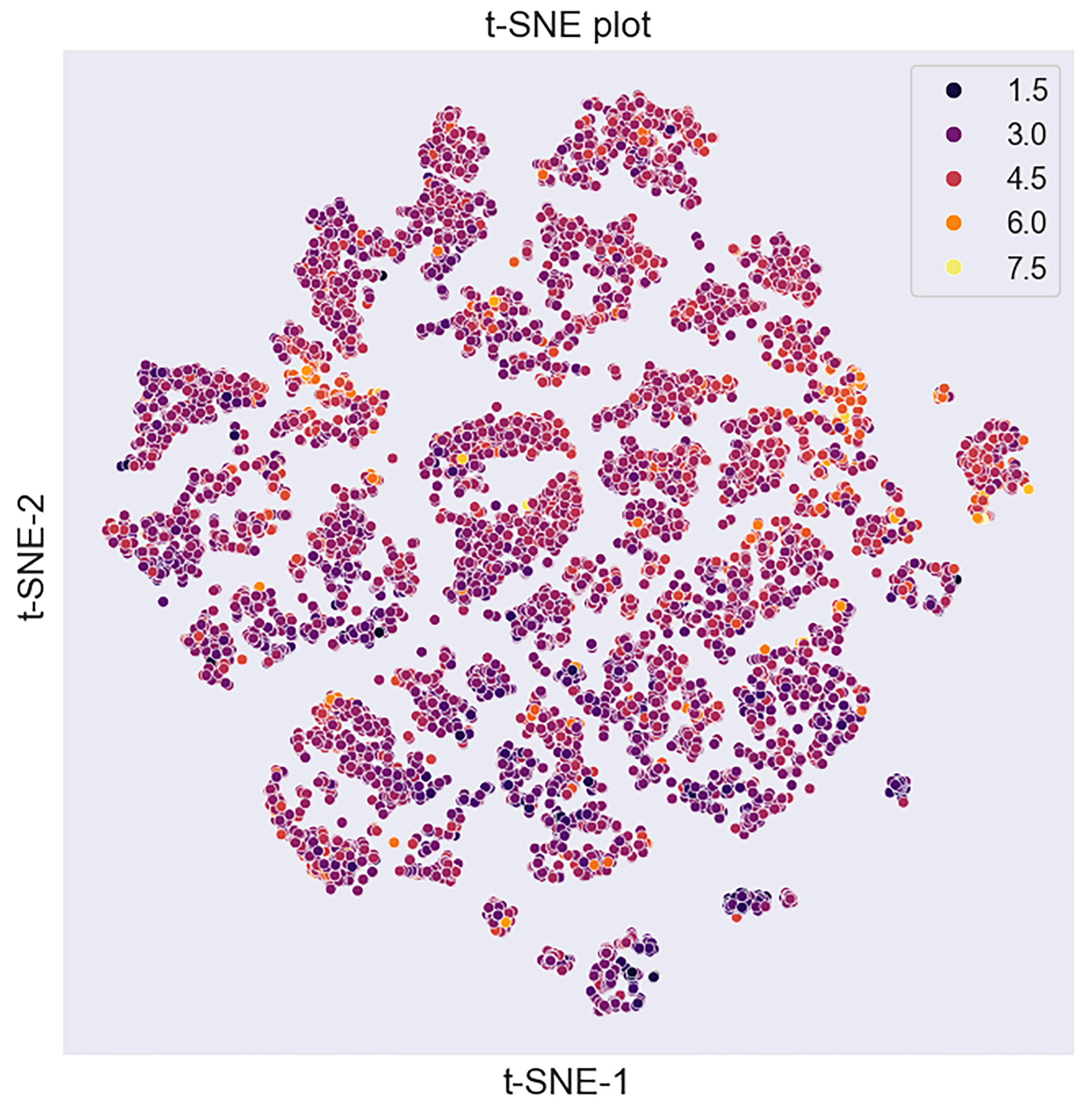
3.1. Data Overview
3.1.1. Physicochemical Properties Analysis
3.1.2. Visualisation of the Dataset’s Chemical Space
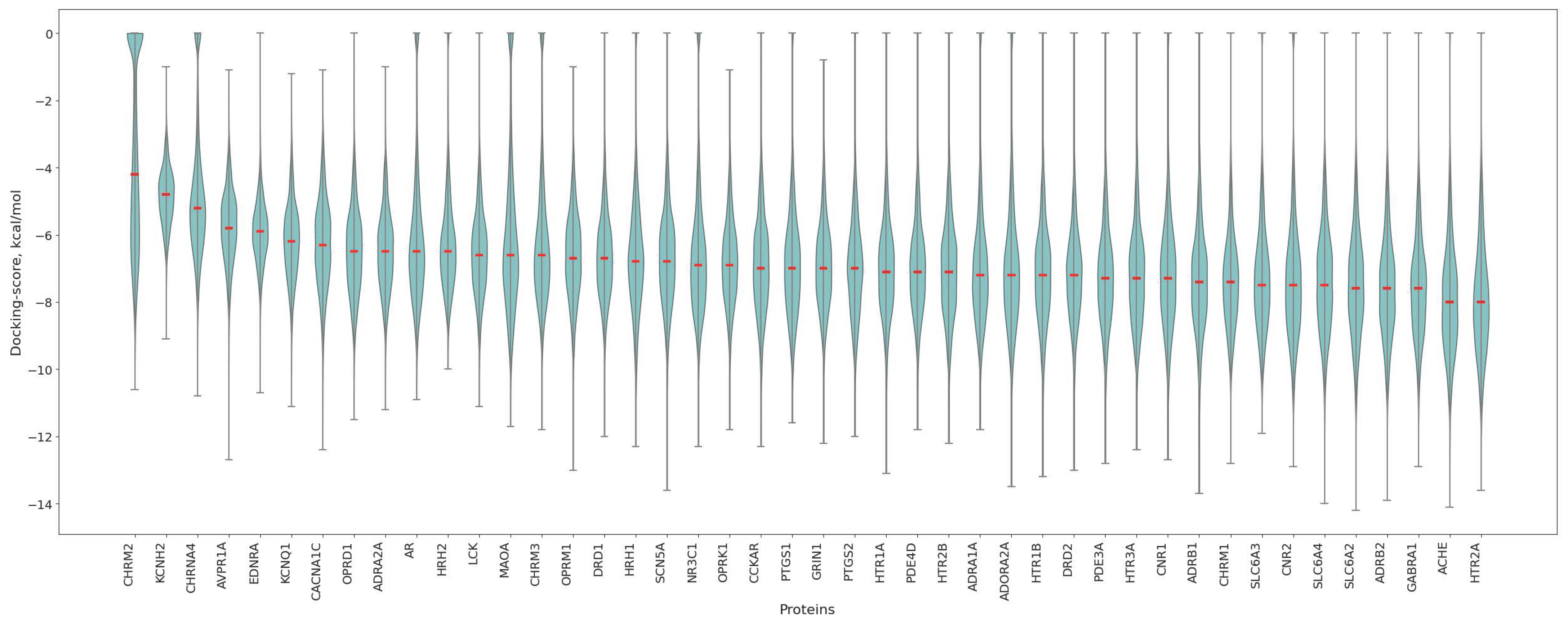
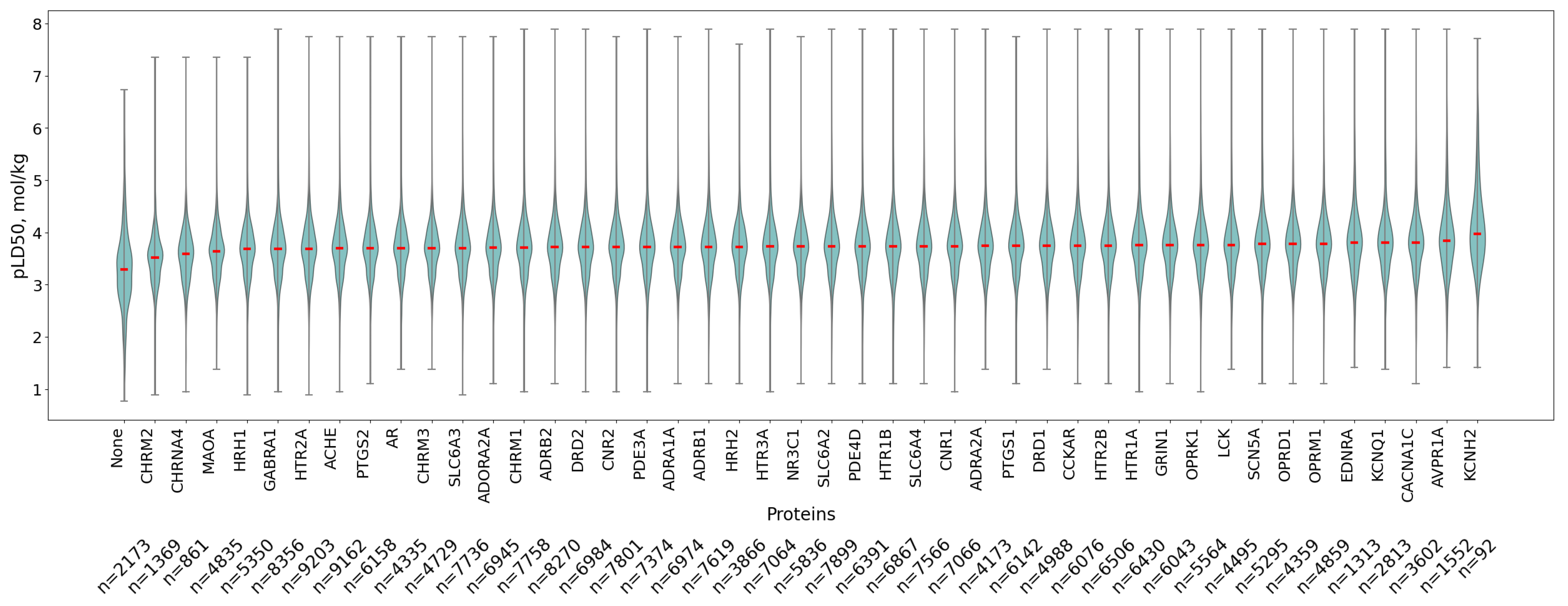
3.2. Protein Affinity Profiles
3.3. Association of Antitargets with
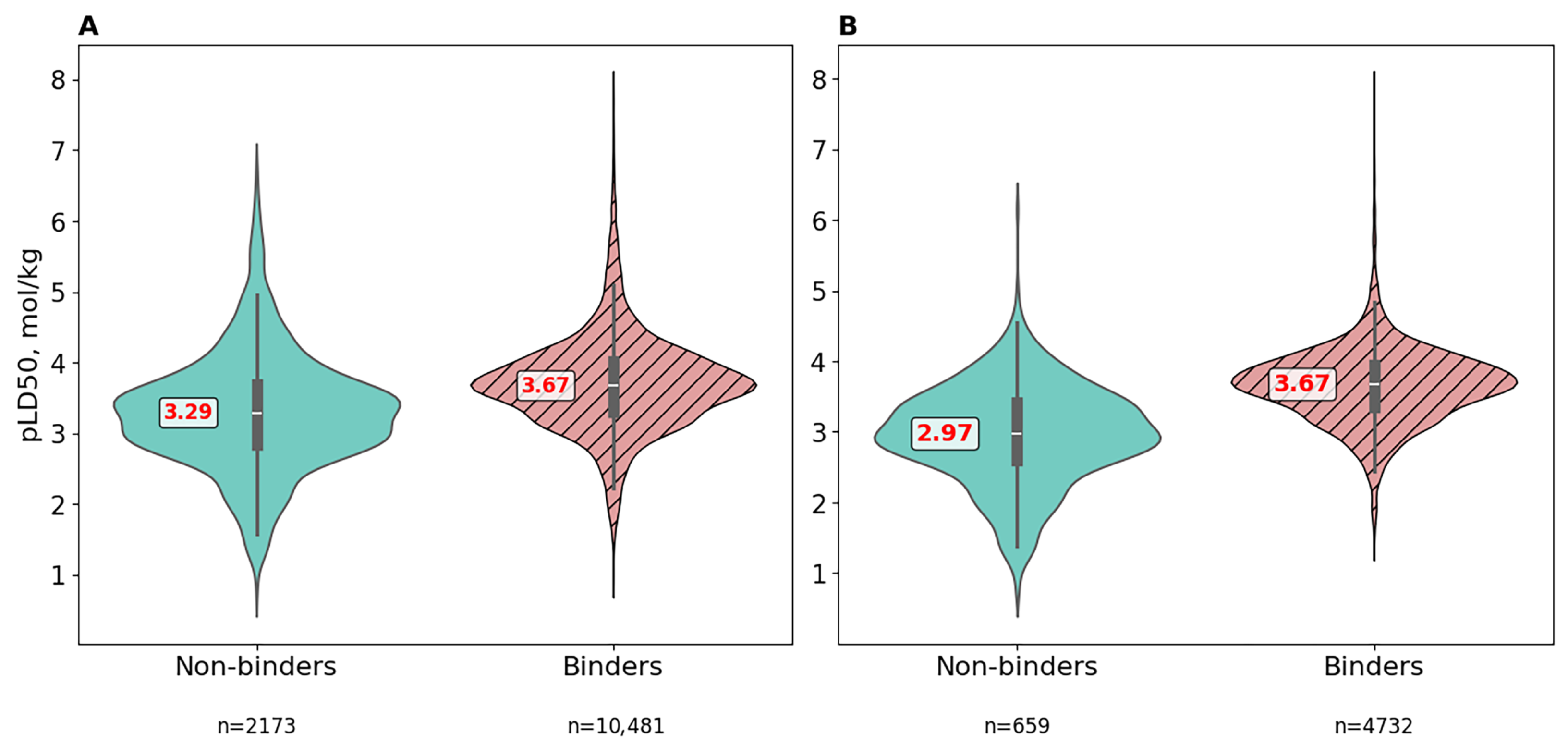
3.4. Comparative Toxicity Analysis of Binders and Non-Binders Molecules
3.4.1. Raw Data
3.4.2. Preprocessed Data Using Molecular Filters
- NIH filterDeveloped by the National Institutes of Health, these filters flag functional groups and scaffolds associated with pan-assay interference compounds (PAINS), reactive electrophiles, redox-active motifs, and other liabilities [47].
- Brenk filter
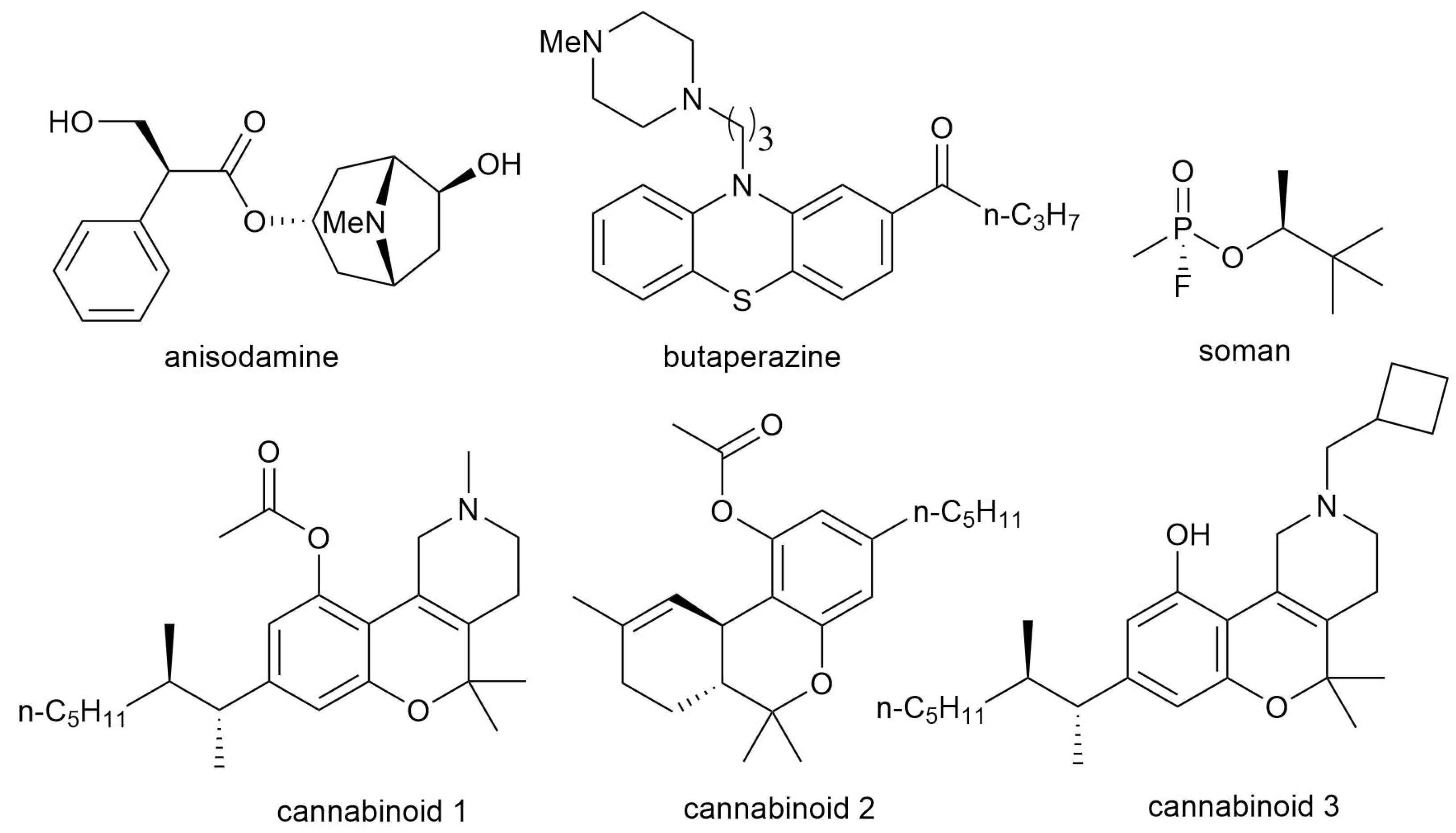
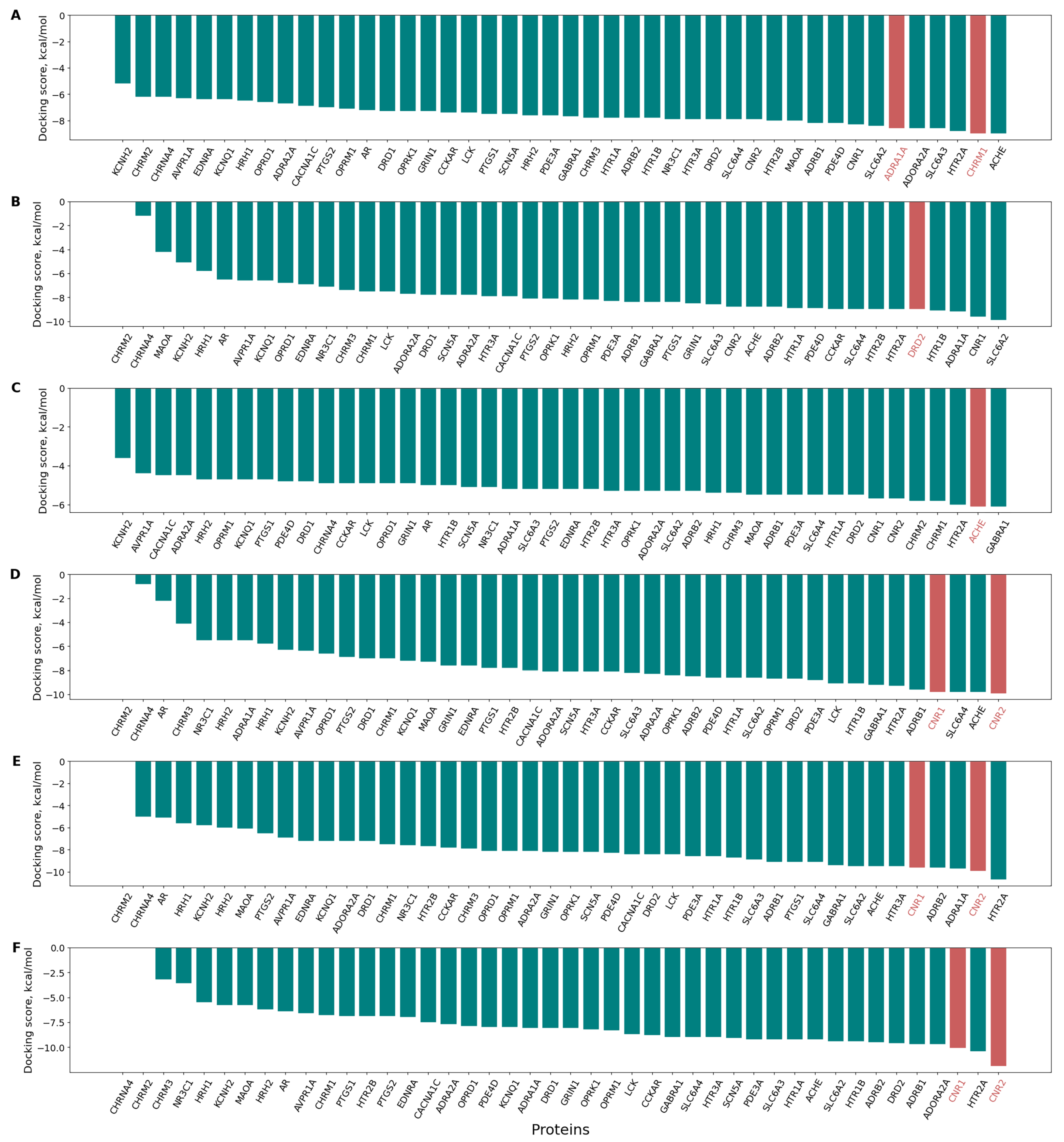
3.5. Confirmation of the Pharmaco- and Toxicodynamics of Known Bioactive Molecules via Inverse Docking
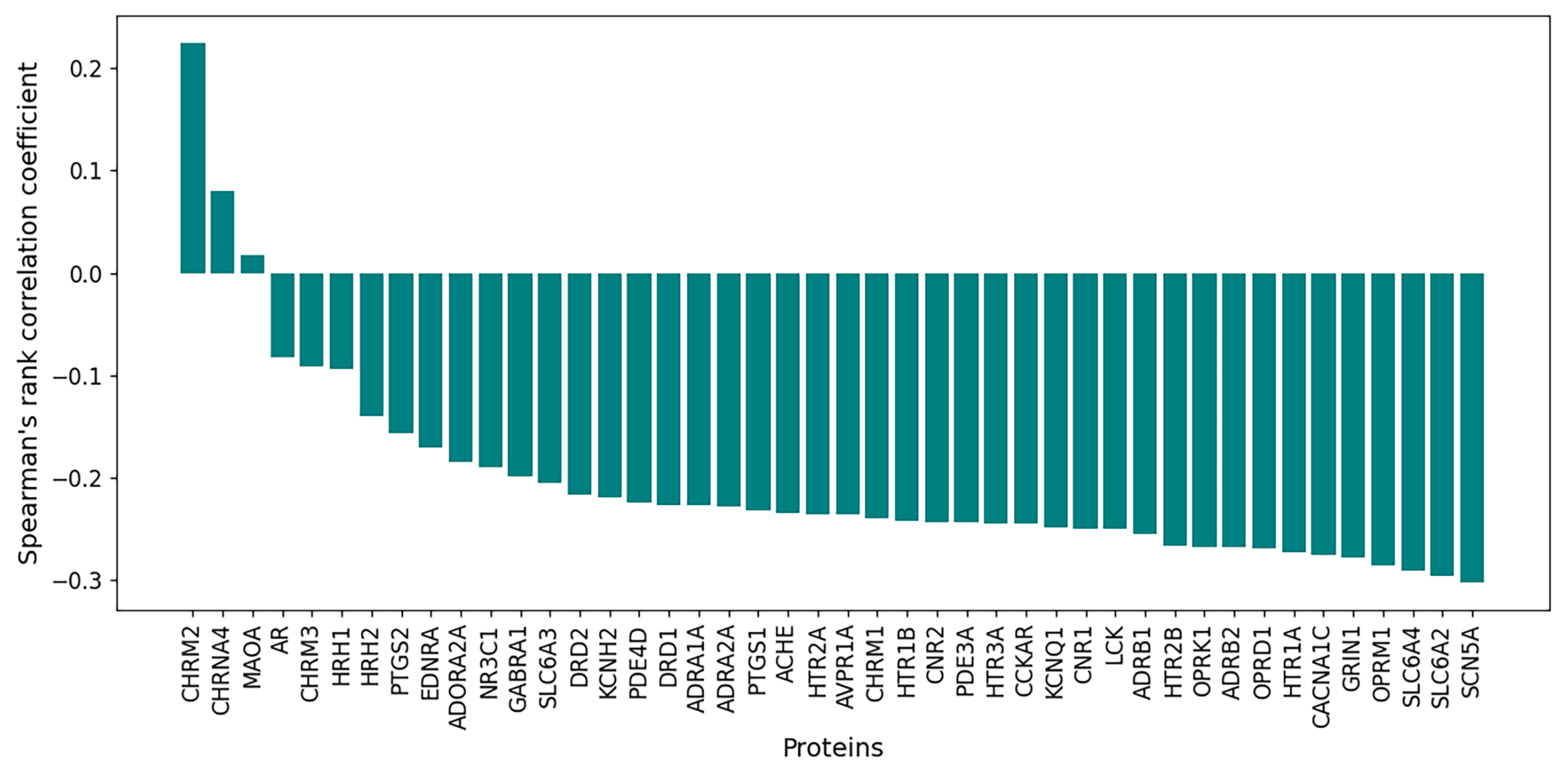
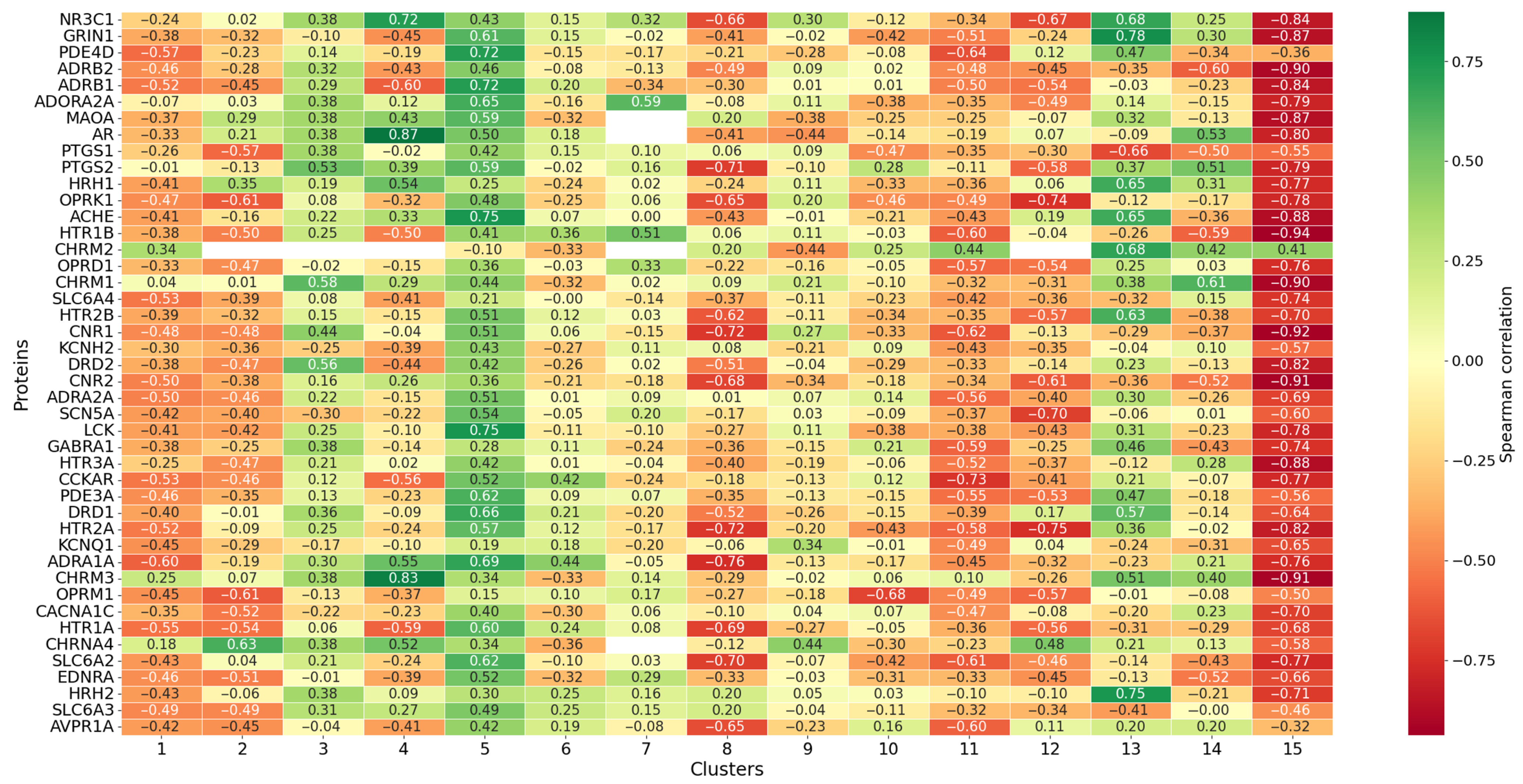
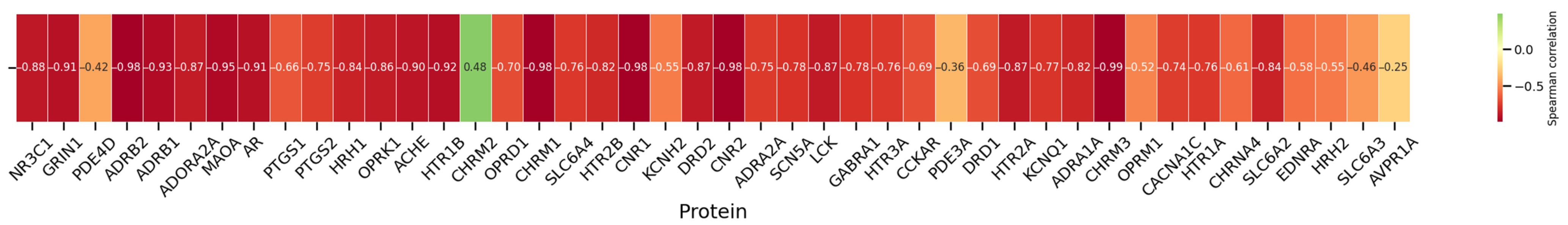
3.6. Correlations Between and Protein Binding Affinity
3.6.1. Initial Dataset
3.6.2. Cluster Analysis
4. Discussion
Challenges and Prospects
- Mechanistic domainThe panel-based approach is relevant when ligand toxicity is driven by interaction with one of the listed proteins. Therefore, it is crucial to properly filter out nonspecific toxicants. Moreover, the panel of 44 proteins is not exhaustive and requires expansion. Toxicity targets may also include other biopolymers (such as DNA), not just proteins.
- Binding affinity prediction methodsIn this study, we used molecular docking to assess affinities. However, this method involves several assumptions and limitations [58,59,60,61]. In particular, an accurate three-dimensional structure of the target protein is essential, as it enables the prediction of ligand-protein interactions by modeling binding poses and affinities. However, the selected protein structure may be incorrectly chosen due to factors such as incomplete experimental data or modeling errors. For example, while full-length protein structures are preferable for comprehensive analysis, sometimes only individual domains are available. A promising direction is to use consensus approaches that combine multiple in silico tools (QSAR, pharmacophore search, etc.) for affinity prediction.
- Structures of orthologous proteinsOne limitation of this study is the reliance on orthologous proteins 3D-structures. This approach may introduce inaccuracies in docking predictions, as subtle structural variations between orthologs—such as differences in active site conformations—could alter ligand binding affinities and lead to misestimation of toxicodynamics. Furthermore, the absence of comprehensive species-specific crystallographic data for many antitarget proteins necessitates this approximation, potentially limiting the generalizability of the findings to real-world interspecies extrapolations in toxicological assessments.
- Affinity
- Binding siteAffinity assessments of small molecules for proteins usually focus on the orthosteric site. However, molecules may also bind to allosteric sites, acting as either positive or negative allosteric modulators (PAMs, NAMs) [64].
- Absorption and bioavailabilityMolecules administered into the body are characterized by bioavailability—the percentage of the substance that reaches the systemic circulation. In our study, we chose the intravenous route of administration, ensuring 100% bioavailability for all compounds [65]. However, applying this concept to other datasets may be challenging. Another important aspect of bioavailability is formulation. Even with the same route of administration and substance, bioavailability may vary significantly depending on formulation. Various methods for increasing bioavailability are known, such as micronization, solid dispersion and so on [66,67]. This issue also relates more broadly to data aggregation. It is important to pay close attention to experimental protocols when compiling datasets.
- DistributionDocking assumes inevitable ligand–protein encounters. However, xenobiotics administered to the body do not necessarily reach every protein, due to distribution. For example, for proteins located in the CNS, a ligand must cross the blood–brain barrier [68]. To enhance the informativeness of binding data in toxicity prediction, information about protein localization should be considered.
- MetabolismIn the body, xenobiotics undergo biotransformation reactions that alter their structures, primarily via CYP450 family enzymes [69,70]. A single xenobiotic may yield dozens or even hundreds of metabolites, which can differ markedly in bioactivity. For example, prodrugs are a class whose therapeutic effect depends on the formation of active metabolites. Sofosbuvir is a prodrug that is metabolized to the active antiviral agent GS-461203 (2’-deoxy-2’--fluoro--C-methyluridine-5’-triphosphate), which acts as a defective substrate for the NS5B protein, a viral RNA polymerase [71]. In our study, docking was performed only for the parent compounds. Explicitly accounting for metabolism is quite challenging, as it requires predicting metabolite structures, which is still subject to uncertainty. Moreover, this would significantly increase the scale of ligand–protein docking calculations.
- ExcretionExcretion is a key pharmacokinetic process determining the elimination of xenobiotics and their metabolites from the body through routes such as renal filtration, biliary secretion, or pulmonary exhalation. Variability in excretion rates and pathways can greatly influence systemic exposure and toxicity outcomes due to accumulation [72].
- Constraints of the model interpretability approachAs discussed above in Section 3.6.2, a strong correlation may sometimes be misinterpreted. It is important to recognize that in such cases, the observed relationship does not necessarily reflect a true mechanism of action but rather a correlation with other descriptors (such as logP). Moreover, molecular docking can produce false-positive results, which in turn may lead to misleading interpretations of the underlying biological mechanism.
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pognan, F.; Beilmann, M.; Boonen, H.C.; Czich, A.; Dear, G.; Hewitt, P.; Mow, T.; Oinonen, T.; Roth, A.; Steger-Hartmann, T.; et al. The evolving role of investigative toxicology in the pharmaceutical industry. Nat. Rev. Drug Discov. 2023, 22, 317–335. [Google Scholar] [CrossRef]
- Nabholz, J.V. Environmental hazard and risk assessment under the United States Toxic Substances Control Act. Sci. Total Environ. 1991, 109–110, 649–665. [Google Scholar] [CrossRef] [PubMed]
- Fairhurst, S. Hazard and risk assessment of industrial chemicals in the occupational context in europe: Some current issues. Food Chem. Toxicol. 2003, 41, 1453–1462. [Google Scholar] [CrossRef] [PubMed]
- Taylor, K.; Alvarez, L.R. An Estimate of the Number of Animals Used for Scientific Purposes Worldwide in 2015. Altern. Lab. Anim. 2019, 47, 196–213. [Google Scholar] [CrossRef] [PubMed]
- European Commission. REACH Regulation—Environment—European Commission. Available online: https://environment.ec.europa.eu/topics/chemicals/reach-regulation_en (accessed on 13 October 2025).
- Chinen, K.; Malloy, T. QSAR Use in REACH Analyses of Alternatives to Predict Human Health and Environmental Toxicity of Alternative Chemical Substances. Integr. Environ. Assess. Manag. 2020, 16, 745–760. [Google Scholar] [CrossRef]
- Vaillancourt, E.; Mohamed, A.A.; Ansell, J.; Ashikaga, T.; Ayad, A.; Ayman, H.; Campos, M.V.; Chien, H.S.; Giusti, A.; Hatao, M.; et al. Highlighting best practices to advance next-generation risk assessment of cosmetic ingredients. NAM J. 2025, 1, 100020. [Google Scholar] [CrossRef]
- Hristozov, D.; Badetti, E.; Bigini, P.; Brunelli, A.; Dekkers, S.; Diomede, L.; Doak, S.H.; Fransman, W.; Gajewicz-Skretna, A.; Giubilato, E.; et al. Next Generation Risk Assessment approaches for advanced nanomaterials: Current status and future perspectives. NanoImpact 2024, 35, 100523. [Google Scholar] [CrossRef]
- Sewell, F.; Alexander-White, C.; Brescia, S.; Currie, R.A.; Roberts, R.; Roper, C.; Vickers, C.; Westmoreland, C.; Kimber, I. New approach methodologies (NAMs): Identifying and overcoming hurdles to accelerated adoption. Toxicol. Res. 2024, 13, tfae044. [Google Scholar] [CrossRef]
- Vora, L.K.; Gholap, A.D.; Jetha, K.; Thakur, R.R.S.; Solanki, H.K.; Chavda, V.P. Artificial Intelligence in Pharmaceutical Technology and Drug Delivery Design. Pharmaceutics 2023, 15, 1916. [Google Scholar] [CrossRef]
- GHS Classification Summary—PubChem. Available online: https://pubchem.ncbi.nlm.nih.gov/ghs/ (accessed on 13 October 2025).
- eCFR::40 CFR 156.62—Toxicity Category. Available online: https://www.ecfr.gov/current/title-40/chapter-I/subchapter-E/part-156/subpart-D/section-156.62 (accessed on 13 October 2025).
- Mansouri, K.; Karmaus, A.L.; Fitzpatrick, J.; Patlewicz, G.; Pradeep, P.; Alberga, D.; Alepee, N.; Allen, T.E.; Allen, D.; Alves, V.M.; et al. CATMoS: Collaborative Acute Toxicity Modeling Suite. Environ. Health Perspect. 2021, 129, 047013. [Google Scholar] [CrossRef]
- Shkil, D.O.; Muhamedzhanova, A.A.; Petrov, P.I.; Skorb, E.V.; Aliev, T.A.; Steshin, I.S.; Tumanov, A.V.; Kislinskiy, A.S.; Fedorov, M.V. Expanding Predictive Capacities in Toxicology: Insights from Hackathon-Enhanced Data and Model Aggregation. Molecules 2024, 29, 1826. [Google Scholar] [CrossRef] [PubMed]
- Allen, T.E.; Goodman, J.M.; Gutsell, S.; Russell, P.J. Defining Molecular Initiating Events in the Adverse Outcome Pathway Framework for Risk Assessment. Chem. Res. Toxicol. 2014, 27, 2100–2112. [Google Scholar] [CrossRef] [PubMed]
- Allen, T.E.; Goodman, J.M.; Gutsell, S.; Russell, P.J. A History of the Molecular Initiating Event. Chem. Res. Toxicol. 2016, 29, 2060–2070. [Google Scholar] [CrossRef] [PubMed]
- Cronin, M.T.; Basiri, H.; Chrysochoou, G.; Enoch, S.J.; Firman, J.W.; Spînu, N.; Madden, J.C. The predictivity of QSARs for toxicity: Recommendations for improving model performance. Comput. Toxicol. 2025, 33, 100338. [Google Scholar] [CrossRef]
- Gissi, A.; Tcheremenskaia, O.; Bossa, C.; Battistelli, C.L.; Browne, P. The OECD (Q)SAR Assessment Framework: A tool for increasing regulatory uptake of computational approaches. Comput. Toxicol. 2024, 31, 100326. [Google Scholar] [CrossRef]
- Ancuceanu, R.; Lascu, B.E.; Drăgănescu, D.; Dinu, M. In Silico ADME Methods Used in the Evaluation of Natural Products. Pharmaceutics 2025, 17, 1002. [Google Scholar] [CrossRef]
- Allen, C.H.; Mervin, L.H.; Mahmoud, S.Y.; Bender, A. Leveraging heterogeneous data from GHS toxicity annotations, molecular and protein target descriptors and Tox21 assay readouts to predict and rationalise acute toxicity. J. Cheminform. 2019, 11, 36. [Google Scholar] [CrossRef]
- Škuta, C.; Cortés-Ciriano, I.; Dehaen, W.; Kříž, P.; Westen, G.J.V.; Tetko, I.V.; Bender, A.; Svozil, D. QSAR-derived affinity fingerprints (part 1): Fingerprint construction and modeling performance for similarity searching, bioactivity classification and scaffold hopping. J. Cheminform. 2020, 12, 39. [Google Scholar] [CrossRef]
- Liu, J.; Gui, Y.; Rao, J.; Sun, J.; Wang, G.; Ren, Q.; Qu, N.; Niu, B.; Chen, Z.; Sheng, X.; et al. In silico off-target profiling for enhanced drug safety assessment. Acta Pharm. Sin. B 2024, 14, 2927–2941. [Google Scholar] [CrossRef]
- Zakharov, A.V.; Lagunin, A.A.; Filimonov, D.A.; Poroikov, V.V. Quantitative Prediction of Antitarget Interaction Profiles for Chemical Compounds. Chem. Res. Toxicol. 2012, 25, 2378–2385. [Google Scholar] [CrossRef]
- Füzi, B.; Mathai, N.; Kirchmair, J.; Ecker, G.F. Toxicity prediction using target, interactome, and pathway profiles as descriptors. Toxicol. Lett. 2023, 381, 20–26. [Google Scholar] [CrossRef] [PubMed]
- Rao, M.; Nassiri, V.; Srivastava, S.; Yang, A.; Brar, S.; McDuffie, E.; Sachs, C. Artificial Intelligence and Machine Learning Models for Predicting Drug-Induced Kidney Injury in Small Molecules. Pharmaceuticals 2024, 17, 1550. [Google Scholar] [CrossRef] [PubMed]
- Bowes, J.; Brown, A.J.; Hamon, J.; Jarolimek, W.; Sridhar, A.; Waldron, G.; Whitebread, S. Reducing safety-related drug attrition: The use of in vitro pharmacological profiling. Nat. Rev. Drug Discov. 2012, 11, 909–922. [Google Scholar] [CrossRef] [PubMed]
- ncats/LD50 -Multitask: Official Repository for Multitask Deep Learning Models. Available online: https://github.com/ncats/ld50-multitask (accessed on 13 October 2025).
- Landrum, G. RDKit: Open-Source Cheminformatics Software. Available online: https://www.rdkit.org/ (accessed on 13 October 2025).
- Sorkun, M.C.; Mullaj, D.; Koelman, J.M.A.; Er, S. ChemPlot, a Python Library for Chemical Space Visualization**. Chem.-Methods 2022, 2, e202200005. [Google Scholar] [CrossRef]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An Open chemical toolbox. J. Cheminform. 2011, 3, 33. [Google Scholar] [CrossRef]
- Halgren, T.A. Merck molecular force field. I. Basis, form, scope, parameterization, and performance of MMFF94. J. Comput. Chem. 1996, 17, 490–519. [Google Scholar] [CrossRef]
- Gasteiger, J.; Marsili, M. Iterative partial equalization of orbital electronegativity—A rapid access to atomic charges. Tetrahedron 1980, 36, 3219–3228. [Google Scholar] [CrossRef]
- RCSB Protein Data Bank. Available online: https://www.rcsb.org (accessed on 13 October 2025).
- X5D2B0|SWISS-MODEL Repository. Available online: https://swissmodel.expasy.org/repository/uniprot/X5D2B0 (accessed on 13 October 2025).
- Delano, W. PyMOL An Open-Source Molecular Graphics Tool—References—Scientific Research Publishing. Available online: https://www.scirp.org/reference/referencespapers?referenceid=2323923 (accessed on 13 October 2025).
- Martins, D.S.; He, Y.; Eberhardt, J.; Sharma, P.; Bruciaferri, N.; Holcomb, M.; Llanos, M.A.; Hansel-Harris, A.; Barkdull, A.P.; Tillack, A.F.; et al. Meeko: Molecule parameterization and software interoperability for docking and beyond. ChemRxiv 2025. [Google Scholar] [CrossRef]
- Ding, J.; Tang, S.; Mei, Z.; Wang, L.; Huang, Q.; Hu, H.; Ling, M.; Wu, J. Vina-GPU 2.0: Further Accelerating AutoDock Vina and Its Derivatives with Graphics Processing Units. J. Chem. Inf. Model. 2023, 63, 1982–1998. [Google Scholar] [CrossRef]
- Shen, C.; Zhang, X.; Deng, Y.; Gao, J.; Wang, D.; Xu, L.; Pan, P.; Hou, T.; Kang, Y. Boosting Protein–Ligand Binding Pose Prediction and Virtual Screening Based on Residue–Atom Distance Likelihood Potential and Graph Transformer. J. Med. Chem. 2022, 65, 10691–10706. [Google Scholar] [CrossRef]
- Ye, J.; McGinnis, S.; Madden, T.L. BLAST: Improvements for better sequence analysis. Nucleic Acids Res. 2006, 34, W6–W9. [Google Scholar] [CrossRef]
- Butina, D. Unsupervised Data Base Clustering Based on Daylight’s Fingerprint and Tanimoto Similarity: A Fast and Automated Way To Cluster Small and Large Data Sets. J. Chem. Inf. Comput. Sci. 1999, 39, 747–750. [Google Scholar] [CrossRef]
- Dankwa, B.; Broni, E.; Enninful, K.S.; Kwofie, S.K.; Wilson, M.D. Consensus docking and MM-PBSA computations identify putative furin protease inhibitors for developing potential therapeutics against COVID-19. Struct. Chem. 2022, 33, 2221–2241. [Google Scholar] [CrossRef] [PubMed]
- Wong, F.; Krishnan, A.; Zheng, E.J.; Stärk, H.; Manson, A.L.; Earl, A.M.; Jaakkola, T.; Collins, J.J. Benchmarking AlphaFold-enabled molecular docking predictions for antibiotic discovery. Mol. Syst. Biol. 2022, 18, e11081. [Google Scholar] [CrossRef] [PubMed]
- Sanguinetti, M.C.; Tristani-Firouzi, M. hERG potassium channels and cardiac arrhythmia. Nature 2006, 440, 463–469. [Google Scholar] [CrossRef]
- Vandenberg, J.I.; Perry, M.D.; Perrin, M.J.; Mann, S.A.; Ke, Y.; Hill, A.P. hERG K+ Channels: Structure, Function, and Clinical Significance. Physiol. Rev. 2012, 92, 1393–1478. [Google Scholar] [CrossRef]
- Rampe, D.; Brown, A.M. A history of the role of the hERG channel in cardiac risk assessment. J. Pharmacol. Toxicol. Methods 2013, 68, 13–22. [Google Scholar] [CrossRef]
- Brown, A.M. Drugs, hERG and sudden death. Cell Calcium 2004, 35, 543–547. [Google Scholar] [CrossRef]
- Getting Started with the RDKit in Python—The RDKit 2025.03.6 Documentation. Available online: https://www.rdkit.org/docs/GettingStartedInPython.html#jadhav (accessed on 13 October 2025).
- Brenk, R.; Schipani, A.; James, D.; Krasowski, A.; Gilbert, I.H.; Frearson, J.; Wyatt, P.G. Lessons Learnt from Assembling Screening Libraries for Drug Discovery for Neglected Diseases. ChemMedChem 2008, 3, 435–444. [Google Scholar] [CrossRef]
- Skipper, P.L.; Kim, M.Y.; Sun, H.L.P.; Wogan, G.N.; Tannenbaum, S.R. Monocyclic aromatic amines as potential human carcinogens: Old is new again. Carcinogenesis 2010, 31, 50–58. [Google Scholar] [CrossRef]
- Cui, L.; Sun, H.L.; Wishnok, J.S.; Tannenbaum, S.R.; Skipper, P.L. Identification of Adducts Formed by Reaction of N-Acetoxy-3,5-dimethylaniline with DNA. Chem. Res. Toxicol. 2007, 20, 1730–1736. [Google Scholar] [CrossRef] [PubMed]
- Lorent, J.H.; Quetin-Leclercq, J.; Mingeot-Leclercq, M.P. The amphiphilic nature of saponins and their effects on artificial and biological membranes and potential consequences for red blood and cancer cells. Org. Biomol. Chem. 2014, 12, 8803–8822. [Google Scholar] [CrossRef] [PubMed]
- Sudji, I.R.; Subburaj, Y.; Frenkel, N.; García-Sáez, A.J.; Wink, M. Membrane Disintegration Caused by the Steroid Saponin Digitonin Is Related to the Presence of Cholesterol. Molecules 2015, 20, 20146–20160. [Google Scholar] [CrossRef] [PubMed]
- Böttger, S.; Hofmann, K.; Melzig, M.F. Saponins can perturb biologic membranes and reduce the surface tension of aqueous solutions: A correlation? Bioorg. Med. Chem. 2012, 20, 2822–2828. [Google Scholar] [CrossRef]
- Cereto-Massagué, A.; Ojeda, M.J.; Valls, C.; Mulero, M.; Pujadas, G.; Garcia-Vallve, S. Tools for in silico target fishing. Methods 2015, 71, 98–103. [Google Scholar] [CrossRef]
- Jenkins, J.L.; Bender, A.; Davies, J.W. In silico target fishing: Predicting biological targets from chemical structure. Drug Discov. Today Technol. 2006, 3, 413–421. [Google Scholar] [CrossRef]
- Zhang, Y.; Zou, J.; Wan, F.; Peng, F.; Peng, C. Update on the sources, pharmacokinetics, pharmacological action, and clinical application of anisodamine. Biomed. Pharmacother. 2023, 161, 114522. [Google Scholar] [CrossRef]
- Pruksaphon, K.; Tanid, K.Y.; Tan, C.H.; Simsiriwong, P.; Gutiérrezid, J.M.; Ratanabanangkoonid, K. An in vitro a-neurotoxin—nAChR binding assay correlates with lethality and in vivo neutralization of a large number of elapid neurotoxic snake venoms from four continents. PLoS Neglected Trop. Dis. 2020, 14, e0008581. [Google Scholar] [CrossRef]
- Pantsar, T.; Poso, A. Binding Affinity via Docking: Fact and Fiction. Molecules 2018, 23, 1899. [Google Scholar] [CrossRef]
- Elokely, K.M.; Doerksen, R.J. Docking Challenge: Protein Sampling and Molecular Docking Performance. J. Chem. Inf. Model. 2013, 53, 1934–1945. [Google Scholar] [CrossRef]
- Chen, Y.C. Beware of docking! Trends Pharmacol. Sci. 2015, 36, 78–95. [Google Scholar] [CrossRef] [PubMed]
- Pinzi, L.; Rastelli, G. Molecular Docking: Shifting Paradigms in Drug Discovery. Int. J. Mol. Sci. 2019, 20, 4331. [Google Scholar] [CrossRef] [PubMed]
- Berg, K.A.; Clarke, W.P. Making Sense of Pharmacology: Inverse Agonism and Functional Selectivity. Int. J. Neuropsychopharmacol. 2018, 21, 962. [Google Scholar] [CrossRef] [PubMed]
- Weiss, D.R.; Ahn, S.; Sassano, M.F.; Kleist, A.; Zhu, X.; Strachan, R.; Roth, B.L.; Lefkowitz, R.J.; Shoichet, B.K. Conformation Guides Molecular Efficacy in Docking Screens of Activatedb-2 Adrenergic G Protein Coupled Receptor. ACS Chem. Biol. 2013, 8, 1018–1026. [Google Scholar] [CrossRef]
- Chen, C.J.; Jiang, C.; Yuan, J.; Chen, M.; Cuyler, J.; Xie, X.Q.; Feng, Z. How Do Modulators Affect the Orthosteric and Allosteric Binding Pockets? ACS Chem. Neurosci. 2022, 13, 959–977. [Google Scholar] [CrossRef]
- Katzung, B.G.; Masters, S.B.; Trevor, A.J. Basic & Clinical Pharmacology, 9th ed.; McGraw-Hill: New York, NY, USA, 2004. [Google Scholar]
- Sahu, R.K.; Khan, J. Formulation strategies to improve the bioavailability of poorly absorbed drugs. In Advances and Challenges in Pharmaceutical Technology: Materials, Process Development and Drug Delivery Strategies; Academic Press: Cambridge, MA, USA, 2021; pp. 229–242. [Google Scholar] [CrossRef]
- Bhalani, D.V.; Nutan, B.; Kumar, A.; Chandel, A.K.S. Bioavailability Enhancement Techniques for Poorly Aqueous Soluble Drugs and Therapeutics. Biomedicines 2022, 10, 2055. [Google Scholar] [CrossRef]
- Wu, D.; Chen, Q.; Chen, X.; Han, F.; Chen, Z.; Wang, Y. The blood–brain barrier: Structure, regulation, and drug delivery. Signal Transduct. Target. Ther. 2023, 8, 1–27. [Google Scholar] [CrossRef]
- Esteves, F.; Rueff, J.; Kranendonk, M. The Central Role of Cytochrome P450 in Xenobiotic Metabolism—A Brief Review on a Fascinating Enzyme Family. J. Xenobiotics 2021, 11, 94–114. [Google Scholar] [CrossRef]
- Hakkola, J.; Pelkonen, O.; Fasanen, M.; Raunio, H. Xenobiotic-Metabolizing Cytochrome P450 Enzymes in the Human Feto-Placental Unit: Role in Intrauterine Toxicity. Crit. Rev. Toxicol. 1998, 28, 35–72. [Google Scholar] [CrossRef]
- Fung, A.; Jin, Z.; Dyatkina, N.; Wang, G.; Beigelman, L.; Deval, J. Efficiency of Incorporation and Chain Termination Determines the Inhibition Potency of 2’-Modified Nucleotide Analogs against Hepatitis C Virus Polymerase. Antimicrob. Agents Chemother. 2014, 58, 3636–3645. [Google Scholar] [CrossRef]
- Asiri, Y. The Role of Pharmacokinetics in Pharmaceutical Toxicology. J. Pharm. Toxicol. 2023, 6, 54–56. Available online: https://www.openaccessjournals.com/articles/the-role-of-pharmacokinetics-in-pharmaceutical-toxicology-16261.html (accessed on 13 October 2025).
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nikitin, I.; Morgunov, I.; Safronov, V.; Kalyuzhnaya, A.; Fedorov, M. Towards Explainable Computational Toxicology: Linking Antitargets to Rodent Acute Toxicity. Pharmaceutics 2025, 17, 1573. https://doi.org/10.3390/pharmaceutics17121573
Nikitin I, Morgunov I, Safronov V, Kalyuzhnaya A, Fedorov M. Towards Explainable Computational Toxicology: Linking Antitargets to Rodent Acute Toxicity. Pharmaceutics. 2025; 17(12):1573. https://doi.org/10.3390/pharmaceutics17121573
Chicago/Turabian StyleNikitin, Ilia, Igor Morgunov, Victor Safronov, Anna Kalyuzhnaya, and Maxim Fedorov. 2025. "Towards Explainable Computational Toxicology: Linking Antitargets to Rodent Acute Toxicity" Pharmaceutics 17, no. 12: 1573. https://doi.org/10.3390/pharmaceutics17121573
APA StyleNikitin, I., Morgunov, I., Safronov, V., Kalyuzhnaya, A., & Fedorov, M. (2025). Towards Explainable Computational Toxicology: Linking Antitargets to Rodent Acute Toxicity. Pharmaceutics, 17(12), 1573. https://doi.org/10.3390/pharmaceutics17121573






