Abstract
Introduction: Gene therapy using siRNA is a current area of research in oncology. Although siRNA formulations have not yet been approved for cancer therapy, numerous studies have demonstrated their therapeutic potential for tumor remission. Objective: To provide an overview of the formulations designed and developed to date based on synthetic siRNA for systemic administration to silence cancer genes. Methodology: A thorough search was conducted using the keywords “siRNA”, “therapy”, and “cancer”, with further classification of the resulting works into the various topics addressed in this review. Results: This review encompasses a wide range of aspects, from the design of siRNA using bioinformatics tools to the primary cellular signals and mechanisms targeted for inhibition in cancer therapy. It describes the primary chemical modifications made to siRNA chains to enhance stability, improve bioavailability, and ensure their binding to nanocarrier systems. siRNA formulations ranging from simple conjugates with biomolecules and small molecules to organic, inorganic, and hybrid nanoparticles, which are examined focusing on their advantages and disadvantages. The significance of nanosystems in dual therapy, including siRNA, for developing personalized treatments that achieve better outcomes is emphasized. Conclusions: Personalized cancer therapy appears to be the preferred approach for oncological treatments. To progress, strategies need to be tailored to the patient’s genetic profile. siRNA therapies provide a flexible platform for targeting and inhibiting critical oncogenes, enhancing the prospects of genomics-guided, patient-specific therapies.
1. Introduction
Small interfering RNAs (siRNAs) are double-stranded (sense and antisense) non-coding RNA molecules composed of 19–21 nucleotides with two overhanging nucleotides at the 3′ end of the antisense strand. They are naturally generated from a long RNA. They inhibit (silence) the expression of target genes by cleaving complementary messenger RNA (mRNA) into two halves. They are found in eukaryotic cells but can also be obtained artificially. In principle, any gene can be silenced by a siRNA with a complementary sequence, making siRNAs specific and potent drugs for the treatment of diseases. siRNAs also contribute to transcriptional gene silencing by promoting long-lasting epigenetic changes that are faithfully inherited during cell division [1,2,3,4,5]. It has been suggested that two siRNAs, one targeting the promoter and the other targeting exon 1 of the transcript, can strongly repress the target gene. Weinberg and Morris mention examples of mammalian genes transcriptionally regulated by siRNA (e.g., eukaryotic translation elongation factor 1α, nitric oxide synthase, E-cadherin, among others) [6].
Treatment with siRNA to silence genes that contribute to tumor growth, survival, metastasis, and resistance to other therapies is very promising. The latter is demonstrated in various clinical trials currently underway [7,8]. Despite these studies, the clinical use of siRNA remains limited. So far, of the six drugs approved for commercial use, five have subcutaneous administration [8]. In oncology, however, systemic administration is more convenient as it is more effective and allows for the treatment of several types of cancer with a single product. As naked siRNA presents difficulties for systemic administration, strategies are continually being developed to protect and transport it to target cells, ensuring cell uptake and intracellular release. This review provides an overview of the general steps involved in the development of siRNA formulations, with a focus on those specifically designed for oncology therapy. It covers the design of siRNA molecules using computer tools, the main chemical modifications to the strands, the production of conjugates and transporter nanosystems, and some of the siRNA target genes currently in clinical research.
Although several recent reviews have been published on the use of siRNA as a therapeutic agent, these primarily describe formulations that have reached the clinical trial phase or are already approved for clinical use [9,10,11]. This work takes another approach. It is an overview that compiles and groups the different non-viral delivery systems that have been designed, prepared, and evaluated regardless of whether they have reached clinical evaluation or not, for cancer therapy. This compilation enables the visualization of the variety of systems and nanosystems that have been explored, as well as the extensive research behind a therapeutic strategy that has reached the clinical evaluation phase.
The keywords “cancer”, “therapy”, and “siRNA” were used to perform the search. The results were then grouped by the different siRNA pathways to reach cancer cells. This review contains a general description of siRNA biological mechanism, the bioinformatics tools for designing synthetic siRNAs, the chemical modifications to siRNA strands, siRNA-biomolecules, siRNA conjugate to small molecules, siRNA nanocarrier systems, multimodal nanocarrier systems for oncology therapy, and examples of key siRNA targets in tumor-promoting pathways and their roles in mono- and combination therapies. A notable feature of this work is that it highlights the primary studies in which siRNA has been investigated as part of multimodal cancer therapies, an emerging area of research.
2. Biological Mechanism of siRNA
siRNA degrades the target mRNA without the need to integrate into the genome or enter the nucleus; it only needs to reach the cytosol to associate with the multi-protein RNA-induced silencing complex (RISC) [11,12]. This complex is a ribonucleoprotein family whose ATP-dependent helicase activity unwinds the siRNA double helix, separating it into its two strands. The core protein of the RISC, Argonaute-2 (Ago2), thermodynamically selects the guide or antisense strand and cleaves the passenger strand into fragments. TRBP and PACT, among other proteins present in the RISC, have been shown to influence guide strand selection [11,13]. Studies suggest that endonucleases such as C3PO can degrade the passenger or sense strand.
C3PO is a Mg2+-dependent endonuclease that consists of six Translin/TB-RBP subunits, which confer siRNA-binding specificity, and two TRAX subunits harboring nuclease activity. Evidence indicates that hC3PO functions at the RISC assembly stage rather than in RISC-mediated mRNA cleavage. During this process, the pre-RISC (Ago2/siRNA duplex) is converted into an active RISC (Ago2/guide strand) following Ago2-mediated cleavage of the passenger strand. While the mechanism underlying dissociation of the passenger strand remains unresolved, studies suggest that the intrinsic RNase activity of hC3PO accelerates RISC activation by degrading the Ago2-cleaved passenger strand, liberating the guide strand for specific mRNA recognition and subsequent degradation [14,15].
RISC binds to the guide strand at the 5′-phosphate end, recognizes the complementary sequence of the target mRNA by Watson–Crick base pairing. This pairing, which occurs at the position opposite nucleotide 10 from the 5′-end of the guide strand, leads to the cleavage or recruitment of proteins that mediate repression and/or translational mRNA destabilization, via Ago proteins [12,16]. In this way, gene expression is inhibited. A schematic of the process is shown in Figure 1.
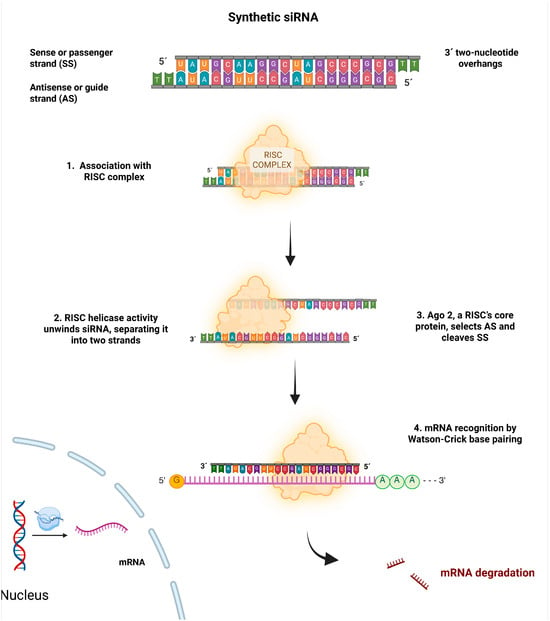
Figure 1.
Biological mechanism of siRNA action. The siRNA associates with the RISC, and Ago-2 degrades the sense strand (SS). The remaining antisense strand (AS) serves as a guide to recognize the target mRNA after Argo-2 degrades it. This decreases or suppresses gene expression.
3. Fundamental Principles for siRNA Design
Gene silencing therapy utilizes rationally designed synthetic siRNAs to degrade complementary messenger RNA (mRNA) selectively. The canonical structure of siRNA consists of a duplex of approximately 19 nucleotides, with two unpaired nucleotides at the 3′ end of each strand, forming two 3′ overhangs [17,18]. Effective siRNA design requires precise optimization of target selection, duplex length, nucleotide composition, sequence specificity, duplex asymmetry, mRNA accessibility, and delivery modality [17,19,20].
The first step in siRNA design is to identify the target gene and select regions within the mRNA that are unique and specific [19,20]. The antisense strand of the siRNA is designed to complement a functional site in the target mRNA. Proper sequence design is crucial for achieving specific gene silencing while minimizing off-target effects. Regarding siRNA length, variations can lead to different outcomes. For example, duplexes longer than 30 nucleotides activate the interferon (IFN) pathway via PKR, causing nonspecific mRNA degradation and apoptosis, which can induce off-target effects [21], whereas duplexes shorter than 15 nucleotides fail to engage the RISC efficiently.
Considering that the RISC enzyme complex uses the thermodynamic properties of the two strands to decide which strand will serve as the functional guide and which will be discarded, strand design is affected by the thermodynamic asymmetry of the 5′ ends, a principle called the “less stable 5′ end rule” [22]. According to this rule, the strand with lower base-pairing stability at its 5′ end is more likely to be incorporated as the guide, while the more stable strand is chosen as the passenger [13,19,22]. For instance, a duplex with an A–U pair (ΔG ≈ −1.8 kcal/mol, less stable) at one 5′ end and a G–C pair (ΔG ≈ −2.7 kcal/mol, more stable) at the other favors the incorporation of the A–U-containing strand as the guide. This example shows how RISC uses thermodynamic differences to enable efficient gene silencing. For guide strand selection, it is also important that the minimum free energy (MFE) at the 5′ end of the strand is higher than that of the opposite strand, and that the ΔMFE of the first three nucleotides of each strand exceeds 1 kcal/mol. These criteria help identify siRNAs with a higher chance of being incorporated into the RISC [22].
The interaction of siRNA with the RISC is heavily influenced by sequence composition and thermodynamic stability. An optimal GC content ranges from approximately 30% to 64%, as too much GC can hinder duplex unwinding, while a region of instability between nucleotides 9 and 14—known as the “energy valley”—helps promote efficient RISC-mediated cleavage [19,23]. Low internal stability at the 5′ end of the antisense strand—fostered by at least four A/U bases within the first seven nucleotides—assists in strand incorporation into RISC. Conversely, high stability at the 3′ end or sequences of G/C bases increase the likelihood of secondary structure formation [19]. Figure 2 illustrates general nucleotide preferences for siRNA design.
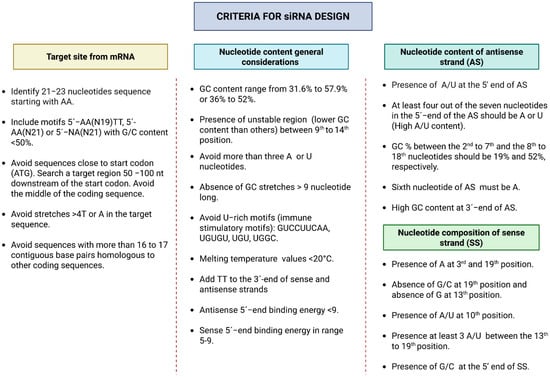
Figure 2.
General considerations for functional siRNA design.
Bioinformatics tools are used to predict potential target sites based on sequence complementarity, thermodynamic stability, and the absence of off-target effects, among other factors. Based on these factors, different variants of molecules are chosen, and possible secondary structures are evaluated. The candidates obtained are experimentally validated to confirm their efficacy and specificity [19,20]. Table 1 shows some of the tools used in the design of siRNAs, as well as their binding directions.

Table 1.
Some bioinformatics tools used in the design of siRNAs.
Tools listed in Table 1 serve different functions. For example, BLOCK-iT™ RNAi Designer is an easy-to-use web tool from Thermo-Fisher that creates RNAi sequences (such as siRNA, shRNA, and miRNA) for any organism using cDNA or GenBank accession numbers. The tool is very effective, with the top three to five designed siRNAs generally achieving over 70% mRNA knockdown under ideal transfection conditions. SMEPred, on the other hand, is a machine learning–based web server that estimates the effectiveness of chemically modified siRNAs. It helps advance siRNA therapeutics by modeling up to 30 chemical modifications, considering both nucleotide composition and modification locations—particularly in the antisense region—to improve silencing efficiency, stability, and minimize off-target effects [27].
Different organizations, such as the RNAi Consortium (TRC) and library manufacturers, offer general standards for siRNA design. There are public databases such as siRNAmod [28], which provide collections of sequences and associated experimental data. This database provides chemically modified siRNAs at various positions, along with experimentally validated permutations and combinations. It also incorporates important information on a specific sequence, including the chemical modification, its respective number and position, structure, simplified canonical input system for molecular data (SMILES), efficacy, cell line, experimental methods, references, and more.
4. siRNA Modifications and Delivery
The main goal in siRNA therapy is to ensure that siRNA reaches the cytosol of the target cell. In vivo siRNA delivery can be administered locally, systemically, or by a combination of both. Local administration (intranasal, intraocular, intrathecal, intratumoral) provides direct access to the target tissue; however, in oncology, the systemic route, particularly intravenous injection, is more convenient as it allows for reaching the target tissue more efficiently. However, naked siRNA is chemically unstable, rapidly cleared by the mononuclear phagocytic system (MPS) and nucleases, exhibits poor cell uptake and endosomal escape, and induces an immune response [12,29]. These drawbacks have led to the development of different delivery strategies, some more clinically viable than others, depending on the tissues to be targeted. The delivery strategies most used are: (i) cell membrane permeabilization by physical methods (beyond the scope of this review), which is little used in the clinic due to its limited effectiveness, although valuable for in vitro studies; (ii) chemical modification of siRNA strands; (iii) developing transport and delivery systems. The latter strategy is the most widely used, mainly with lipidic nanocarriers such as liposomal systems, which are the most prevalent.
4.1. Chemical Modifications to siRNA Strands
Early research aimed to improve the specificity, stability in blood, and cell internalization, as well as decrease the toxicity of naked siRNA, focused on chemical modifications to the strands. These modifications depend on the characteristics of each siRNA and the desired target. They are performed on: (i) the rest ribose; (ii) the ribose-phosphate backbone; (iii) the purine and pyrimidine bases. Many of them have been performed to obtain siRNA conjugates with other molecules or to incorporate them into nanoparticles (NPs). The vast majority do not significantly affect the main characteristics of the siRNA [16,30], although some authors have stressed the importance of performing them with great care so as not to compromise the efficacy of silencing and off-target effects [31].
- Modifications to the rest of the ribose. Ribose modifications, particularly at the 2′ position, have been the most widely used to protect siRNA from attack by ribonucleases, improve affinity for the target mRNA, and decrease the immune response. They have the advantage of not altering backbone conformation or gene silencing efficiency. The most common is the substitution of the 2′-OH group with a less nucleophilic group. The most used group is 2′-O-methyl (2′-O-Me), from which derivatives such as 2′-O-methoxyethyl (2′-O-MOE) and 2′-deoxy-2′-fluoro (2′-F) have been developed [32,33,34]. Substitutions can be made at different sites on the double strand or in the central part of the antisense strand [35]. The best results are obtained by alternating 2′-O-Me and 2′-F substitutions [36,37].
Both 2′-O-methyl (2′-O-Me) and 2′-fluoro (2′-F) modifications enhance the chemical and enzymatic stability of siRNA duplexes by stabilizing the 3′-endo conformation of the ribose sugar. This conformation promotes the characteristic A-form helical geometry of native RNA, which is essential for efficient recognition and loading by the RNA-induced silencing complex (RISC). Additionally, the A-form structure provides greater resistance to nucleases, particularly RNases, which typically target the reactive 2′-OH group of unmodified ribose. Of these modifications, 2′-F most closely resembles the electronic and steric properties of the native 2′-OH, ensuring high compatibility with RISC. Strategically placing 2′-O-Me modifications on the passenger strand promotes efficient RISC loading and can further improve strand selection by enhancing thermodynamic asymmetry and duplex stability [38,39,40]. These modifications also mitigate the innate immune response by preventing recognition by toll-like receptors, thereby reducing immune-stimulant effects, such as TNFα production [41,42,43,44].
Modifications have also been made to 2′-C and 4′-C, as well as to the complete ribose ring, resulting in siRNAs with various physicochemical and biological characteristics [32,33,34,45]. Most of these modified siRNA molecules start by: (i) connecting the 2′-C and 4′-C positions via a methyl bridge to block the sugar residue at the 3′-C end and form so-called locked nucleic acids (LNAs) whose rigid conformation substantially improves the stability of the siRNA [45,46]; (ii) disconnect the 2′-C and 3′-C positions to form unblocked nucleic acids (UNA) to obtain a wide range of derivatives [45,47,48].
- Modifications to the ribose-phosphate backbone. These are designed to minimize off-target effects and immune responses, enhance cell uptake, and increase the bioavailability of siRNA. The most used is the replacement of a non-bridging oxygen in the phosphodiester group linking two consecutive riboses with a sulfur to form a phosphorothioate (PS) [36,49,50,51], a boron to form a borane-phosphate (PB) [52,53], or an acetate to form a phosphonoacetate [54]. These modifications are used to link the siRNA to different molecules [55]. The most prevalent is the formation of PS, where the sulfur atom preserves the negative charge of the siRNA. The principal benefits of this modification are the improvement of siRNA resistance to nucleases, hydrophobicity, stability, and affinity to plasma proteins, resulting in a longer circulation time [56,57]. However, the number of PS should be limited. This can reduce the silencing effect and induce cytotoxicity [49,58,59]. The best PS substitution variant is at the end of the strands. Some authors have synthesized siRNAs with more than one simultaneous modification, with attractive properties for in vivo use [60]. Alternatives with different phosphate derivatives include phosphorodithioate, PS2 [50,61,62], methylphosphonate [58], 5′-(E)-vinyl-phosphonate [63,64,65]. The phosphotriester groups have also been modified [66].
- Modifications to nucleobases. To a lesser extent than ribose and ribose-phosphate backbone modifications, modifications to uridine, cytidine, and adenosine are made to improve thermal stability, nuclease resistance, cell uptake, and reducing immune response [67,68,69,70]. However, concerns exist about the safety of metabolizing modified siRNAs via this route. The modifications described are used to obtain siRNAs with different structures, conjugates, and nanosystems for transport and delivery. There are numerous recent publications that summarize these modifications and their impact on siRNA properties [9,11,39,71]. Table 2 summarizes the advantages and disadvantages of the modifications described, as well as their applications. A common disadvantage is that, depending on the site and extent of the modification, the physicochemical and biological properties of the siRNA may deteriorate.
 Table 2. Major modifications to siRNA strands.
Table 2. Major modifications to siRNA strands.
Simultaneous modifications to siRNA and its carrier can optimize the nanocarrier system. Such modifications can be achieved through a carefully balanced approach that enhances stability, delivery efficiency, and safety without compromising efficacy. siRNA chemical modifications such as 2′-O-Me, 2′-F, phosphorothioate (PS) linkages, and locked nucleic acids (LNAs) can improve siRNA stability, reduce immune stimulation, and enhance specificity without compromising RNA interference activity. For example, the combination of 2′-O-Me and PS at the termini increases nuclease resistance while maintaining silencing activity [11,72]. Optimization of siRNA-carriers should be engineered to protect siRNA from degradation, facilitate cellular uptake, and promote endosomal escape while minimizing toxicity.
In both cases, siRNA and carrier modifications need to be iteratively screened to monitor safety (e.g., immunogenicity, toxicity) and efficacy (target gene silencing, biodistribution). Preclinical tests should also be conducted to evaluate whether modifications to siRNA or the carrier affect siRNA loading or promote off-target effects. Therefore, experimental designs should be followed, including redesign and continuous evaluation of the effectiveness of the forms. This balanced approach enables the development of siRNA therapeutics with optimized stability, delivery, and biocompatibility, which are essential for clinical success in oncology [73].
4.2. siRNA Transporters and Delivery Nanosystems
The most used route for in vivo administration of siRNA is through nanosystems for transport and delivery. These systems are necessary to overcome the primary obstacles associated with the in vivo administration of naked siRNA, namely its high instability and poor pharmacokinetic properties. The phosphodiester bond in siRNA is vulnerable to RNases and phosphatases, which rapidly degrade it and reduce the accumulation of intact siRNA in the desired tissue [11]. These systems can be categorized into two broad groups: viral and non-viral. Viral systems are modified viruses that transport and deliver siRNA into cells. There are several types: (i) adenoviruses; (ii) adeno-associated adenoviruses; (iii) lentiviruses and retroviruses [74,75]. All have high transfection efficiency but have drawbacks such as: (i) limited loading capacity; (ii) off-target effects; (iii) immunogenicity and toxicity. Adeno-associated adenoviruses and lentiviruses infect both dividing and non-dividing cells and are therefore very useful in cancer therapy. Lentiviruses and retroviruses can induce mutagenesis, disrupt essential genes, and activate oncogenes [75].
Non-viral systems are more suitable. They have advantages and disadvantages that need to be evaluated in relation to specific therapy. They can be divided into two main groups: (i) siRNA conjugates with biomolecules or small molecules and (ii) nanocarrier systems.
4.2.1. siRNA Conjugates with Biomolecules and Small Molecules
Although chemical modifications to siRNA strands improve stability, they may negatively affect cellular uptake, intracellular internalization, and interaction with the RNA-induced silencing complex (RISC), ultimately reducing silencing efficiency. Conjugation to stable and biocompatible molecules significantly improves in vivo efficacy. These conjugates have a simple molecular structure and a well-defined ratio. They allow: (i) improved pharmacokinetic profiles; (ii) tissue-specific targeting; and (iii) improved endosomal escape without affecting silencing capacity, among other advantages [76,77,78]. The production process can be regulated in terms of homogeneity and reproducibility. Depending on the type of molecule to be conjugated and the specific characteristics of each siRNA, binding can be done by covalent or non-covalent bonding. Some conjugates spontaneously form nanoparticles (NPs). Most can be incorporated into carrier NPs of different natures by relatively simple processes.
Although siRNA has four terminal phosphate groups available for conjugation, the groups most commonly used for this purpose are the 5′ and 3′ of the sense strand and the 3′of the antisense strand, previously modified (Section 4.1).
- (i)
- siRNA Conjugates with Biomolecules
This section provides an overview of the main conjugates of siRNA biomolecules. These conjugates have the characteristic that they do not require an additional transport system, as they serve as their own delivery systems. Biomolecule siRNA conjugates consist of the biomolecule, the siRNA, and usually a bifunctional connector for binding. In some cases, spacers are added to improve the stability of the conjugate and prevent the formation of NPs. These siRNA conjugates penetrate the cell through various mechanisms, ranging from passive diffusion to receptor-mediated transport [76]. The most used biomolecules for siRNA conjugation include antibodies (Abs) and their fragments (Fabs), peptides, and aptamers (Figure 3).
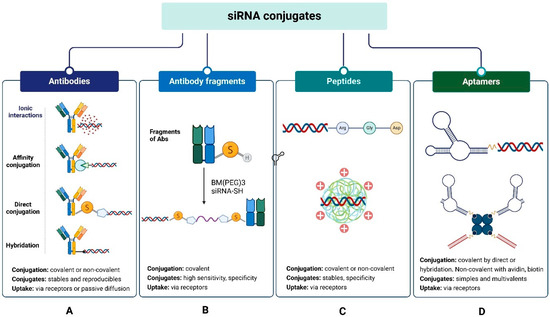
Figure 3.
siRNA conjugates with biomolecules: (A) antibodies (Ab), (B) antibody fragments (Fab), (C) peptides, (D) aptamers.
- Ab-siRNA and FAb-siRNA conjugates. Abs are ideal carrier systems due to their high affinity and specificity, long half-life in blood, and relatively low immunogenicity. Experience gained with antibody-drug conjugates (ADCs) has enabled the development of antibody-siRNA conjugates (ARCs), which combine the precision of siRNA for target gene silencing with the high sensitivity and specificity of Antibodies for binding to their antigens/receptors on the cell surface. They are an advantageous option for targeting siRNA to antigen-expressing tumor cells because they accumulate and internalize more readily than naked siRNA [76,79,80].
Conjugation of Abs to siRNA occurs by covalent and non-covalent coupling (Figure 3A). Covalent docking is a standard method of linking biomolecules. It can be performed using chemical and biological processes, providing stable and reproducible conjugates; however, it sometimes yields low yields and products with varying Ab:siRNA ratios [80]. Additionally, the upstream modifications required for Ab and siRNA binding are relatively expensive [80,81]. Coupling is performed by two variants: (i) direct conjugation and (ii) conjugation by hybridization of the double strand. Non-covalent coupling is simpler and can be performed in a single step; however, the conjugates are less stable and tend to form heterogeneous aggregates and nanoparticles [82]. It is also carried out by two variants: (i) electrostatic interactions and (ii) affinity bonding. For covalent siRNA-Ab/siRNA-Fab coupling by direct conjugation, different reactions can be used, such as (i) disulfide bridge formation; (ii) thiol-maleimide reaction; (iii) carbodiimide-amine reaction; (iv) amino ring opening and β-lactam; (v) azide-alkyne reaction. The bifunctional connector is usually added to the sense strand of the siRNA, and then the connector-siRNA adduct is linked to the Ab or Fab (Figure 3A,B). The method has the advantage that it allows the use of many standard connectors commonly used to obtain ADCs.
For disulfide bridging, both siRNA and Ab/Fab are modified with thiol groups, and then both molecules react [83]. A variant is to add connectors with thiol groups to Ab/Fab and siRNA and then link the two adducts together [84].
The thiol-maleimide reaction is the most used method for direct siRNA-Ab conjugation [76,85,86]. For this conjugation, the interchain disulfide bonds of Ab (4-linked) are reduced to thiol groups using mild reductants such as dithiothreitol (DTT), 2-mercaptoethanol (2ME), or tris [2-carboxyethyl] phosphine (TCEP). The thiol groups are deprotonated and react, by nucleophilic addition or substitution, with electrophilic connectors without affecting the intrachain disulfide bonds (Figure 3A). The most used connectors are maleimide and its derivatives, including SMCC [45,85], N-succinimidyl 3-(2-pyridyldithio)-propionate (SPDP) [45,85,87], N-succinimidyl S acetylthioacetate (SATA) [85], N-succinimidyl 4-(2′-pyridyldithio) pentanoate (SPP) [87], and N-succinimidyl 3-maleimidopropionate (BMPS) [87]. Peptides have also been used as connectors [88]. The connector first binds to siRNA, and then the siRNA-connector adduct is conjugated to Ab [77,80,86,89]. The main drawback of the reaction is the unspecificity of the binding site. To improve this, cysteine is inserted at specific positions in Ab (via protein engineering). After obtaining the thiol groups from the cysteine, the reaction with the siRNA-connector adduct occurs [82,89]. This variant allows the use of THIOMAB platforms. THIOMABs are Abs with cysteine residues in determined positions. They were developed to obtain ADCs but are also successful for ARCs. When THIOMABs are used, the siRNA is modified to obtain an amine group on the sense strand. This amine group is attached to the amide-thiol linker. Finally, the siRNA-connector adduct reacts with THIOMAB thiols [85].
Direct conjugation via the thiol-maleimide reaction can also be performed by first attaching the linker to the Ab and then to the modified siRNA. Under this variant, Ab-siRNA conjugates were produced with cationic gelatin as a linker. Cationic gelatin is obtained by modifying carboxyl groups that bind to lysine residues of Ab through amide bonds. Subsequently, thiol-modified siRNA is conjugated to gelatin [90].
In carbodiimide-amine conjugation, the carboxyl groups either naturally present or chemically introduced into the siRNA are activated with 1-ethyl-3-(3-dimethyl aminopropyl) carbodiimide (EDC). Upon activation, the siRNA forms an intermediate that reacts with primary amine groups on the Ab, typically those on lysine residues, to form a very stable amide bond. The method has the disadvantage that the binding is nonspecific since the linker can also bind to the amine groups of tyrosine and cysteine [89]. Some authors conjugate the EDC to carboxyl groups of Ab and then bind them with siRNA, previously modified with amine groups [80]. The addition of N-hydroxysuccinimide (NHS) enhances the reaction efficiency [91].
Conjugation via the amino ring opening reaction and β-lactam is performed by functionalizing the siRNA with β-lactam. Then, the siRNA-β-lactam adduct reacts with the amino groups of the lysine residues [92]. For direct azide-alkyne conjugation, siRNA is modified with an azide (N3) or an alkyne at the ribose moiety. Ab is conjugated to a linker containing alkyne or azide groups [93,94,95]. The linker can also be conjugated to siRNA [95].
Double-stranded hybridization involves covalently conjugating a single-stranded oligonucleotide to the Ab and then adding the complementary strand to form the double-stranded siRNA (Figure 3A) [82].
Non-covalent coupling by electrostatic interactions allows for the easy obtaining of Ab-siRNA conjugates with the following advantages: (i) no chemical modification of siRNA is required; (ii) the complex formed is easily internalized and promotes endosomal escape. Cationic peptides that bind siRNA by electrostatic interactions are used, for example, 9-arginines [96] and protamines [97,98,99].
The method consists of first coupling arginine or protamine to the Ab by a covalent bond and subsequently to the siRNA. This spontaneously forms a peptide-siRNA complex (Figure 3A). The positive charges of the 9-residue of arginine and protamine interact with the negative charges (~40) of siRNA phosphates. For covalent binding of siRNA to arginine or protamine, bifunctional linkers such as those mentioned are used (e.g., SMCC) [97,98,99]. DPDPB (1,4-di-(3′-[2′pyridyldithio]-propionamido) butane) is also used [80]. The choice of linker depends on the coupling site. EDC, for example, couples the carboxyl groups of the Ab to the amine groups of 9-arginine, and SPDP couples the thiol groups of the Ab to the amine groups of 9-arginine. If a 9-arginine derivative with a cysteine residue is used, then its thiol groups can be coupled to the thiols of the Ab using DPDPB as the linker [80]. SMCC is used to couple the amino terminus of protamines to the thiol groups of Ab [97,98,99]. There are commercial kits that allow these couplings to be made easily.
Ab-siRNA electrostatic complexes have been used to transfect siRNAs into different cell types. However, the method has two significant drawbacks: (i) depending on the reaction conditions, aggregates and superstructures can form [99]; (ii) since the interactions are reversible, they can dissociate under certain pH and salt concentrations [82].
Non-covalent affinity coupling (Figure 3A) is based on spontaneous affinity reactions between avidin and biotin [100,101]. It has the advantage that it does not require purification. Some authors, however, consider that it is not helpful for therapeutic applications.
Avidin-biotin conjugates can be obtained by (i) modifying the siRNA with a thiol to conjugate it to the previously maleimide-linked avidin. Biotin is also modified and binds to the primary amines of the Ab. The siRNA-avidin adduct is then bound to the biotinylated Ab [100,101]; (ii) linking the Ab to avidin and the siRNA to biotin, using tetraethylene glycol (TEG) as a connector [79,100]. Since avidin has four binding sites for biotin, some authors biotinylate both the siRNA and the Ab and subsequently bind them to avidin [89]. The high affinity between avidin and biotin (KD = 10−15 M and t1/2 (disoc) = 89 days) [101] enables the formation of conjugates with high stability [80,101]. Similar methods obtain Fab/siRNA conjugates.
Ab-siRNA/Fab-siRNA conjugates present several limitations: (i) in some cases, multiple tissues express the same antigens and/or cell receptors, leading to potential nonspecific targeting; (ii) some conjugates exhibit low levels of internalization, particularly in solid tumors; (iii) others may interfere with the immune system or induce toxicity [78,80,82,89].
- Peptide-siRNA conjugates. The small size of peptides, their relatively low molecular weight, ease of synthesis, and low cost of production have facilitated the development of peptide-siRNA conjugates (Figure 3C) with lower immunogenicity and toxicity, better pharmacokinetic properties, high cellular uptake, and more efficient endocytosis than Ab-siRNA conjugates [102,103]. Amino acids (AA) with acidic, hydrophilic, hydrophobic, or aromatic residues can be combined to generate several peptides with siRNA delivery potential, called cell-penetrating peptides (CPP) or membrane transduction peptides (MTP). These peptides are generally composed of <30 AA, often including lysine, histidine, and arginine. They readily cross the anionic surface of cell membranes and reach intracellular compartments without interacting with receptors or altering their functions [102,104,105]. They can be chemically modified to enhance endosomal escape and decrease the influence of endocytic proteases [105,106]. They have been used to internalize, through passive diffusion or endocytosis, various types of macromolecules, including siRNA. Some are internalized by both mechanisms, depending on factors such as the AA sequence and its structure, the CPP/siRNA concentration ratio, cell line and others [102,107,108].
The first CPPs used in siRNA delivery were natural peptides such as (i) the gene transcription transactivator (TAT, 49–57), with high arginine content, derived from HIV-1 TAT protein and (ii) penetratin, peptide derived from the third helix of the Antennapedia homeodomain [109,110,111]. The key functional component of these peptides is the guanidine group of arginine, which is capable of forming hydrogen bonds with various elements of the cell membrane [104,112]. Some chimeric and synthetic peptides can interact with cell membrane phosphates, carboxylates, and phosphonates [104]. Among the most used CPPs is a chimeric peptide of 27 AA in length that contains 12 functional AA from the amino terminus of galanin and mastoparan at the carboxyl terminus, connected through a lysine [108,113], as well as low molecular weight protamine derivatives (LMWP) [107,114]. Currently, many CPPs with different AA sequences are available, which differ in length, charge, hydrophobicity, structure, and flexibility. These peptides can penetrate a wide variety of cells, with low toxicity and high transfection efficiency.
Some CPPs also enhance endosomal escape and increase the accumulation in the cytosol of the peptides captured by endocytosis and remain trapped in endosomes for a long time. They are referred to as endosome-disrupting peptides [104,110,115]. These peptides are obtained by introducing histidine residues into the CPPs, which destabilize the endosomal membrane due to their sensitivity to pH changes [116,117]. They are especially lethal to drug-resistant cells, making their application in combination therapies beneficial [115]. However, they require careful evaluation of the balance between endosomolytic activity and toxicity.
Other CPPs are modified to specifically target tumor cell-associated receptors (integrin, somatostatin, gastrin, neurotensin, folate receptors, and others). They are then called targeted peptides. The delivery of siRNA with targeted peptides enhances cell uptake and internalization [118,119,120]. In some cases, they do not require additional agents for transfection [110]. Among the most studied targeting peptides are the cyclic peptides Arg-Gly-Asp (RGD) [121,122]. The RGD receptor, integrin alpha V/β, is ubiquitously expressed on tumor endothelial cells, making it a very favorable target. Some CPPs combine endosomal escape and uptake via receptors and are therefore referred to as multifunctional CPPs [123].
Like Abs, peptides are coupled to siRNA by covalent and non-covalent bonds. The choice of one or the other method depends on the sequence and modification of the side chains of the AAs that make up the peptide, as well as its structure. Conjugation by covalent bonds produces better-defined conjugates that are very stable in circulation [114,121,124]. It is usually performed by direct conjugation, in a similar manner to that described for direct Ab-siRNA conjugation. The most common method is the thiol-maleimide reaction, which involves the amine of the lysine or cysteine residue in the peptide [114,124,125]. The previously modified peptide is bound to the amide-thiol linker, and subsequently, the peptide-linker adduct is reacted with the siRNA, which is also modified with thiol groups. The conjugates obtained are monomeric [114,124,125] and easily release siRNA into the cytoplasm because the disulfide bridges that are formed are easily reduced in this environment [114,124].
When peptides with a high cationic charge are used, it is recommended to place spacers between the peptide and the connector to prevent the interaction between positive charges and negative charges of siRNA, and to form aggregates [111,124,125]. The addition of spacers has been shown to improve cell internalization compared to systems without spacers. One of the most used spacers is PEG, which also contributes to reducing the immune response [102,114].
Another strategy for covalently linking peptides to siRNA is the azide-alkyne reaction. The AA sequences of the peptide are modified to decrease positive charges, a crucial step in preventing aggregation. Then, an azide group is introduced into the terminal chain of the peptide. The siRNA is modified with an alkyl group and finally reacts with the azide-modified peptide. The inertness of the alkyne and azide towards other functional groups, such as amines, carboxylates, thiols, alcohols, and esters, makes the binding very selective and regiospecific. The reaction is also insensitive to oxygen and water. It allows the preparation of conjugates with high stability in serum but requires an additional transfection agent [94,126,127]. The reaction is also used to obtain peptide-siRNA conjugates by hybridization [128].
Similar to Ab-siRNA conjugates, covalent peptide-siRNA conjugation requires many steps and a high level of purification to exclude the effects of the free peptide. Furthermore, the formation of covalent bonds between the peptide and siRNA can affect their biological activity [78,108,127]. Some conjugates may remain trapped in the endocytic pathway, while others trigger immune responses.
Obtaining peptide-siRNA conjugates by non-covalent bonds is simpler. Electrostatic interactions primarily drive it. To form these conjugates, low-molecular-weight cationic peptides with a high charge density of lysine or arginine (+7 to +10) are used. Among the most used are octaarginine (R8), nonaarginine (R9), and their derivatives [120,125,129,130]. The drawback of the method is that to obtain an appropriate conjugate that easily penetrates the cell, an excess of peptide is required. This can lead to the formation of unwanted aggregates with complex secondary and tertiary structures [120,125], and increase conjugate toxicity [108,116,131]. Some tend to be localized in the nucleus rather than the cytosol [124].
AA sequence and peptide structure are significant factors in the formation of electrostatic peptide-siRNA conjugates. Some authors consider that the best way to obtain stable electrostatic conjugates is to use amphipathic peptides [116,125,132,133]. The peptide’s cationic part forms an electrostatic complex with siRNA, and the hydrophobic part contributes to structural stability and cell internalization [134]. Amphipathic peptides with high silencing capacity and low toxicity can be obtained by the addition of tryptophan (aromatic) residues at one end and arginine and lysin [132,135]. However, there is evidence that amphipathic peptides tend to produce off-target effects and toxicity [134].
Although siRNA-polypeptide conjugates are typically delivered using carrier systems rather than conjugates, polypeptides are included here due to their structural similarities with peptides. siRNA-polypeptide conjugates face several challenges: (i) they interact with blood proteins and form aggregates that accumulate in the lungs, liver, and spleen [29,136]; (ii) they tend to induce off-target effects [137]; (iii) some exhibit low encapsulation efficiency [138,139,140]; (iv) others fail to promote efficient endosomal escape or cause toxicity [29,140]. To address these issues, various strategies have been developed, such as (i) modifying the surface of the siRNA-polypeptide conjugate by coating with PEG [141,142] and/or conjugating with molecules that target cell receptors [143,144]; (ii) incorporating the conjugate into inorganic or organic nanostructures to create hybrid nanosystems; (iii) adding histidine and arginine residues; histidine allows pH-responsive siRNA release and endosomal escape [145], while arginine enhances membrane penetration via receptor-mediated endocytosis [146]; (iv) improving siRNA release by attaching non-cationic components to the cationic carrier, which triggers charge conversion to reduce binding affinity at the target site, such as the cytosol [147]. One of the most used polypeptides is polylysine (PLL).
PLL-siRNA conjugates employ PLL, a widely used polypeptide for siRNA delivery. They are characterized by biocompatibility and biodegradability; however, they tend to exhibit instability in vivo and may pose toxicity concerns [148,149]. The facile chemical modification of PLL facilitates the creation of conjugates with various spatial configurations [149,150,151], which can be incorporated into alternative nanosystems [139,148]. Conventionally, PLL is modified through the addition of histidine and arginine residues [152].
- Aptamer-siRNA conjugates: Aptamers are single-stranded DNA or RNA molecules known for their high affinity and specificity for cell receptors overexpressed in tumors. Their large-scale production and low cost have driven the development of aptamer-siRNA conjugates (Figure 3D), also known as chimeras, as alternatives to Ab-siRNA and peptide-siRNA conjugates [153,154]. Unlike antibodies and peptides, aptamers maintain their biological activity when linked with siRNA. Since aptamers are smaller (6–30 kDa) compared to antibodies (150 kDa), their conjugates with siRNA are more easily internalized, and their endosomal escape is more efficient [155,156,157].
The broad chemical versatility of aptamers enables the synthesis of multivalent and simple aptamer-siRNA conjugates through both covalent and non-covalent interactions [153,158]. Covalent attachment is accomplished via direct methods or hybridization. In both methods, the aptamer and siRNA strands are modified, and then linked through reactions such as: (i) carbodiimide; (ii) thiol-maleimide; (iii) azide-alkyne, as previously described [158,159,160]. Some researchers also use a technique called oxidative coupling [153].
The hybridization process is similar to that used for Abs and CPPs. It begins with in vitro transcription to produce a siRNA strand bound to the aptamer, followed by the addition of the complementary strand [157,161,162]. Another method involves the so-called “universal adhesive bridge” conjugation, where the 3′-end of the aptamer is joined to the 5′-end of the sense strand of the siRNA via a U-U-U connector acting as a bridge. The complementary strand of the siRNA is then added (Figure 3D) [163,164,165].
An advantage of hybridization conjugation is that multivalent siRNA-aptamer conjugates can be obtained. For example, two different siRNAs can be linked with a bivalent aptamer to simultaneously silence two different genes [166,167]. Multimeric conjugates called comb-type conjugates can also be obtained [155,160]. These conjugates are formed by the hybridization of antisense chains of the siRNA with sense chains that are previously bound to the aptamer (Figure 3D).
Aptamer-siRNA conjugates by covalent means are expensive [168], so many authors prefer non-covalent binding by affinity conjugation using the avidin-biotin system [159,165]. The siRNA binds to biotin and the aptamer to avidin (or vice versa). Similarly, two biotinylated siRNAs and two biotinylated aptamers can be used and bound to avidin [169]. To ensure the release of siRNA into the cytosol, a cleavable linker can be added between the siRNA and the biotin. This allows the preparation of multivalent conjugates with a greater internalization capacity compared to the usual conjugates, although silencing does not improve significantly [168,169].
Aptamers are easily degraded in the biological environment. To increase the blood circulation time and silencing efficiency, different strategies are used such as: (i) truncating the aptamer [161,170]; (ii) adding overhanging nucleotides to the antisense strand of the siRNA [161,171]; (iii) add PEG to the sense strand of siRNA [161,170]; (iv) mixing (i)–(ii) [169]; (v) link the aptamer-siRNA conjugate to nanocarrier systems of different nature. The latter is one of the most widely used strategies [155,156,158,166,172].
- Toxins-siRNA conjugates. Toxins are poisonous substances produced by living organisms such as plants, animals, and microorganisms. The AB-type toxin has a domain that can be engineered to reduce (attenuate) toxicity, a translocation domain that enables endosomal escape, and a receptor domain that induces endocytosis mediated by cell receptors [173]. Diphtheria and anthrax are examples of AB-type toxins, which have been used to create conjugates for delivering siRNA in vitro to cancer cells [173,174]. siRNA conjugates with attenuated AB-toxins become a promising delivery system because they address some of the challenges of siRNA delivery, especially endosomal escape, and improve upon some disadvantages of polycationic delivery systems. Although these engineered modified toxins have been used for protein delivery [175], few studies have reported on siRNA delivery. This field is likely to grow in the future as methods for modifying toxins and conjugating siRNA continue to advance.
- (ii)
- siRNA Conjugates with Small Molecules
The binding of siRNA to small molecules has led to the formation of conjugates with low toxicity and immunogenicity, which can easily penetrate the tumor microenvironment [39,55]. Among these molecules, the most used in oncology therapy are folic acid (FA), cholesterol, fatty acids, and calcium complexes. These molecules can be conjugated directly to the siRNA or used in a packaged delivery vehicle. Anisamides and α-tocopherol have been used to a lesser extent. N-acetyl galactosamine (GalNAc) siRNA conjugates, which are widely used in clinical practice, are only mentioned here as they are not used in oncology. These conjugates with small molecules employ different targeting mechanisms, such as receptor recognition or enhancing cell internalization through hydrophobicity.
Conjugating siRNA to small molecules has been achieved by direct covalent binding. Copper-catalyzed azide-alkyne cycloaddition (CuAAC) is one of the most widely used routes [176,177]. The reaction is based on modifying the sense strand of the siRNA with one or more alkyne groups that are introduced either at the ends or in central positions. The small molecule is modified with the azide and attached to a linker. Depending on the molecule to be linked, hydrophobic or hydrophilic azides are used. Finally, the parts are reacted to form the conjugate. The reaction can be carried out either in the liquid phase or on solid supports, allowing the molecule to be conjugated to the siRNA with simple purification. This reaction is widely used to obtain RNA conjugates with folic acid (FA), cholesterol, long-chain fatty acids, oligoamines, and carbohydrates [176,177,178]. Some siRNA conjugates with small molecules are listed below.
- FA-siRNA conjugates. FA is a crucial component for cell growth and proliferation, serving as a transporter for several therapeutic agents. It is especially useful in oncology because FA receptors (FR), particularly FRα and FRβ, are overexpressed in many tumor cells, while they are expressed at very low or no levels in normal tissues [179,180]. The uptake of FA via FR is specific and exhibits high affinity (KD 10−9 M) [181]. The small size of FA-siRNA conjugates allows them to easily reach solid tumors [179,180]. These conjugates are produced by direct covalent conjugation or by hybridization [177]. However, direct conjugation has limitations. Since folate conjugates bind to FRs through the pteridine residue, conjugation must occur through the glutamate residue, which, having two carboxyl groups, results in mixtures of α and γ isomers [182]. Although folate can be incorporated into the 3′ or 5′ ends of the sense strand of siRNA, many researchers prefer central positions [179]. Common covalent conjugation methods include: (i) linking siRNA to a bifunctional amide thiol linker followed by attachment of folate [181]; (ii) connecting the 5′-end of an oligodeoxynucleotide (ODN) to a folate molecule (ODN-FA), then extending the sense strand of the siRNA to couple it with the ODN-FA adduct [183]; and azide-alkyne reactions with various variants [177,184]. The same reactions are used for conjugation via hybridization [183].
For these covalent FA-siRNA reactions, a wide diversity of linkers/spacers is available. For example, the most used are PEG, NHS, N, N′-dicyclohexylcarbodiimide (DCC), polyethylene imine (PEI), N-propargyl-diethanolamine, and dibenzo cyclooctyl amine (DBCO) [179,181,184].
- Cholesterol-siRNA conjugates were developed to increase hydrophobicity and improve siRNA cell uptake [185,186,187]. They are created through direct covalent bonding or hybridization. The most commonly used reactions are thiol-maleimide and azide-alkyne. For the linkage, the 3′ or 5′ ends of the sense strand are modified, followed by adding cholesterol bound to a linker [188,189,190]. The common reaction methods include: (i) attaching the cholesterol connector to a terminal -OH group of the sense strand of the siRNA [188]; (ii) performing PS modifications at the 3′-end of both strands, adding modified nucleotides (2′-OMe) to the antisense strand, and attaching the cholesterol linker to the sense strand [188,190]; (iii) similar to the previous method but alternating modifications with 2′-OMe and 2′F, then attaching nucleotides to the antisense strand [191]; (iv) truncating one or both strands to produce short asymmetric siRNAs, then adding nucleotides with PS modifications to the antisense strand and attaching the linker-cholesterol adduct to the sense strand [189]. These variants enhance thermodynamic stability, pharmacokinetics, internalization, and silencing efficiency. The linker’s nature and the siRNA binding site significantly influence the biological properties of the conjugates [185,189,192]. The most mentioned connectors include triethylene glycol (TEG) [190,192], 2-amino butyl-1-3-propanediol (C7) [188], trans-4-hydroxyprolinol [78,190], and hexamethylenediamine [189].
siRNA-cholesterol conjugates tend to remain trapped in the cell membrane. To improve internalization, so-called tagged siRNA has been obtained. A tagged siRNA is obtained by chelating the siRNA with six cholesterol conjugates. Each cholesterol conjugate is formed by a chelator, a ligand, and a cholesterol molecule. One of the cholesterol conjugates is captured by the cell membrane, and the remaining five “push” the siRNA inwards. The pushing force disappears when the siRNA is released into the cytosol. Ethidium [193] and zinc (II)-dipicolylamine (Zn/DPA) [194] are used as chelators.
The application of cholesterol-siRNA conjugates is primarily carried out locally (in the skin, eyes, and brain [185,195]. The application in oncological therapy (and by systemic route) is used less since a large percentage (80%) accumulates in the liver [196] and in organs that overexpress LDL/HDL receptors [77,185,189]. Some conjugates can also induce cytotoxicity at high concentrations [196,197]. For these reasons, cholesterol-siRNA conjugates are preferred for incorporation into lipid nanosystems for oncology therapy.
- Fatty acid conjugates with siRNA. siRNA has been conjugated to fatty acids such as docosanoic acid (DCA, C22H44O2), docosahexaenoic acid (DHA, C22H32O2), lithocholic acid (LCA, C24H40O3), palmitic acid (C16H32O2), among others [198,199]. These conjugates are obtained by methods similar to those described for cholesterol-siRNA conjugates. Their physicochemical properties, biodistribution, and silencing capacity depend on the specific characteristics of each fatty acid [198,199,200,201,202]. However, their direct use is also limited. For this reason, it is preferred to associate them with lipid nanosystems.
- Calcium-siRNA complexes. The spontaneous formation of calcium ion with siRNA produces nanocomplexes that are stable, have a uniform size (~100 nm), and carry a negative surface charge (−8 mV). The reversible nature of the electrostatic interactions between Ca2+ and siRNA has been effectively used for in vitro siRNA transfection [203]. These complexes serve as the foundation for developing highly efficient siRNA nanocarrier systems [204].
- Anisamide-siRNA conjugates. One and two receptors are polypeptide chains located in the endoplasmic reticulum and as transmembrane proteins in nervous system cells. They have neuromodulatory and ion-channeling functions and bind to a wide range of psychoactive drugs [205]. These receptors are also expressed in several tumor cell types [205] and exhibit high affinity for anisamide (2-(4′-methoxy benzamido) ethyleneamide) and its derivatives [206]. Anisamide conjugates with drugs demonstrate high internalization efficiency via endocytosis mediated by these receptors [207,208], which has driven the development of mono- and multivalent anisamide-siRNA conjugates. Monovalent conjugates are formed by direct covalent bonding between an anisamide phosphoramidate and the 5′ end of the sense strand of the siRNA, which has been previously modified with 2′O-Me. For multivalent conjugation, multifunctional linkers are used to first bind to the siRNA and then to the modified anisamide. These conjugates show a high transfection capacity in prostate cancer cells [206]. However, the use of anisamide-siRNA conjugates remains limited, likely due to concerns about potential side effects. Some have been incorporated into nanocarrier systems.
- N-acetyl galactosamine-siRNA conjugates. These are designed to silence specific mRNAs of liver proteins by binding to asialoglycoprotein receptors ASGP-R [209,210]. These conjugates consist of three molecules of N-acetyl galactosamine (GaINaC) linked via a spacer to the siRNA. By using different spacers and modifications of the siRNA, a wide variety of these conjugates have been developed, and they are incorporated into nanocarrier systems. Some of these formulations have received approval for treating various diseases [8].
Other small molecule siRNA conjugates used in oncology therapy include anandamide-siRNA for immune cell treatment [211] and α-tocopherol-siRNA for liver disorders and pancreatic cancer therapy [212,213].
Although small-molecule siRNA conjugates provide a simple and promising method for targeted delivery, especially to specific tissues like the liver (e.g., GalNAc–siRNA), many of these conjugates show poor pharmacokinetics, which restricts their effectiveness in vivo. These issues include low stability, inadequate delivery to target tissues, and potential off-target effects. To overcome these problems, small-molecule siRNA conjugates are often incorporated into nanocarrier systems. Although more complex, nanocarriers offer improved stability, better biodistribution, and the ability to co-deliver therapeutic agents, making them a more versatile and effective platform for a broader range of therapeutic uses.
4.2.2. siRNA Nanocarrier Systems
Although modifications to the strands and conjugations with biomolecules and small molecules help avoid many problems linked to naked siRNA, nanocarrier systems are the best choice for systemic siRNA delivery in cancer therapy. These nanocarriers are tiny structures (NP) that have a therapeutic effect similar to or greater than free drugs, with fewer side effects.
The essential physicochemical properties to consider in a nanosystem for transporting therapeutic agents (including siRNAs) are size, shape, surface charge, hydrophobicity, and surface modifications [214,215]. Emerging evidence indicates that mechanical properties such as deformability, stiffness, and elasticity are equally important in affecting biological outcomes [216,217]. Sizes larger than 10 nm are preferred because they are not easily eliminated by the kidneys, and sizes less than 200 nm are favored because they can penetrate the tumor neovasculature through the fenestrations of endothelial cells in tumor blood vessels (EPR effect). A size smaller than 200 nm also prevents them from being cleared from the bloodstream (which could lead to accumulation in the liver and spleen and activate the immune system) [218], allowing them to accumulate in the tumor.
Describing the ideal nanocarrier can be challenging. Usually, their size is around 100 nm, especially for rigid nanoparticles like inorganic ones and some polymeric variants. Soft particles larger than 200 nm, such as lipid nanoparticles, micelles, liposomes, and certain polymers, can be internalized by tumors because they are capable of deforming and flattening. However, these bigger particles are more susceptible to clearance from the bloodstream. Spherical shapes are generally preferred for improved cellular uptake [219], although they tend to activate the immune system [220]. For longer blood circulation, neutral or negatively charged surfaces at physiological pH are ideal, as they minimize nonspecific serum protein interactions and off-target tissue uptake, thereby increasing specificity [214,215]. Nonetheless, membrane electrostatic repulsion makes their internalization into target cells less efficient [220]. Additionally, the nanoparticle pKa affects off-target organ uptake in systemic siRNA therapy, with an optimal pKa around 5.4–6.5 to ensure effective endosomal escape. In acidic endosomes, neutral or anionic nanocarriers become protonated, which disrupts membranes and releases siRNA into the cytoplasm [221].
For siRNA transport, the overall surface charge of the carrier depends on the chemical state of the siRNA. If naked siRNA is transported on the nanoparticle surface, the nanoparticle should have a positive charge. Conversely, if siRNA forms a complex, it can be transported inside or on the surface of a neutral or negatively charged nanoparticle. Nanoparticles with hydrophilic surfaces are preferred to reduce the corona effect and are often coated to improve colloidal stability [214,215]. On the other hand, hydrophobic domains stabilize nanoparticles and facilitate cellular uptake, also decreasing toxicity; however, they may increase the corona effect [16]. The nanoparticle surface should be modified with a molecule that enables specific recognition and targeting to the tumor site.
Generally, the relationship between nanoparticle size and immune response is as follows. Nanoparticles smaller than 500 nm are most effectively recognized and taken up by dendritic cells and macrophages. Specifically, those between 100 and 200 nm are internalized via endocytosis, which triggers CD4 and CD8 responses as well as Th1-type immune responses (cellular-based immunity). These smaller nanoparticles elicit a stronger immune response than larger ones. Particles larger than 500 nm are absorbed through phagocytosis and micropinocytosis by macrophages specialized in taking up bigger particles. They stimulate a humoral adaptive response (antibody-mediated immunity) [222].
All of the above factors must be considered when designing siRNA nanocarrier systems. Although the mentioned characteristics improve tumor biodistribution and accumulation, each must be studied thoroughly for every carrier used. Nanocarrier systems are classified into two main categories: “organic” and “inorganic and hybrid”. The choice between them depends on the specific application. Each type has its advantages and disadvantages that influence silencing effectiveness.
- (i)
- Organic Nanocarriers
Organic nanocarriers are derived from natural, synthetic, or semi-synthetic sources. To date, the most studied siRNA transporters are polymeric and lipid-based (Figure 4). Cell derivatives have been examined to a lesser extent. Some authors include carbon derivatives in this group, but in this work, they were classified as inorganic nanocarriers.
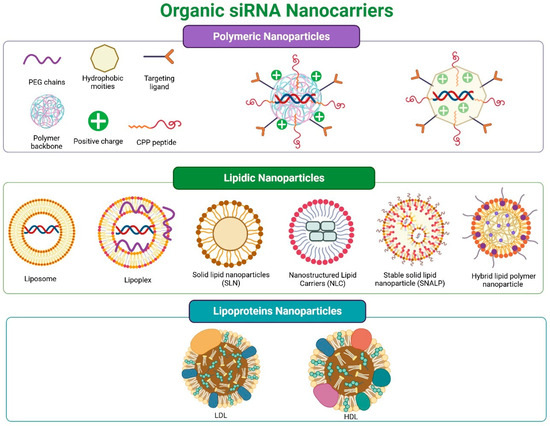
Figure 4.
Organic siRNA nanocarriers: (top panel) polymeric; (middle panel) lipidic: lipoplexes, liposomes, solid lipid nanoparticles (SLN), nanostructured lipids (NLC), stabilized nucleic acid lipid particles (SNALP), lipid-polymer hybrid nanoparticles; (bottom panel) lipoproteins: LDL and HDL.
- (a)
- Polymeric Nanosystems
Polymeric nanosystems carrying siRNA (polymeric NPs) consist of polymers and siRNA, which may be attached to other molecules and include auxiliary components (see Figure 4 (top panel)). These NPs are highly durable, exhibit low polydispersity, and possess high stability. They are easy to customize for various applications and are cost-effective [223,224]. They can be cationic, anionic, or amphiphilic, whether natural or synthetic, but all share qualities of biodegradability, biocompatibility, and low immunogenicity [224,225].
Among the natural polymers most used in siRNA transport, chitosan (CS) stands out [141,226,227]. To a lesser extent, alginates, cellulose, gelatin, atelocollagen, and hyaluronic acid have been used [228,229]. The diversity of synthetic polymers is extensive and complex to cover. Among the most used are polyethylene imine (PEI) and poly(β-amino esters) (PBAE) [136,148,149,230,231]. Polylysine (PLL) can also be considered as a synthetic polymer, but in this work, it was included in the previous subsection “Peptide-siRNA conjugate”, because it is also a polypeptide.
A useful way to classify polymeric nanosystems is based on the morphology of the nanoparticle formed. Polyplexes are created from cationic polymers (e.g., PEI, CS, PBAE). A polyplex is a complex formed through spontaneous electrostatic interactions between the polymer’s positive charge and the negative charge of the siRNA. Nanogels, polymersomes, or polymeric micelles are formed from amphiphilic polymers. A positively charged polymer binds to siRNA, and hydrophobic residues compact this polyplex within the nanosystem. Nanospheres are primarily formed through self-assembly of hydrophobic polymers and copolymers (e.g., PLA, PLGA, and PCL), where hydrophobic segments aggregate to create an inner core, while hydrophilic portions form a stabilizing outer shell in aqueous environments. PLGA (poly-L-lactic-co-glycolic acid) is a specific anionic polymer that is “cationized” by adding functional groups, mainly positively charged amines. All these structures are mostly spherical [223,225,232,233], which carry siRNA in a condensed or complexed form [137,138,139,223,234].
The physicochemical and biological characteristics of polymeric carriers are influenced by factors such as molecular weight, charge density, and the spatial configuration of the polymer (whether linear or branched). Additional determinants include the size of the siRNA, ionic strength of the preparation medium, concentrations of both polymer and siRNA, the ratio between the amines of the polymer and the phosphates of the siRNA (N/P ratio), order of reagent addition, among others [16,29,223,225,232]. Multiple techniques are employed for synthesis, including emulsification, nanoprecipitation, ionic gelation, and the use of supercritical fluids [139,232]. Depending on the chosen preparation method, siRNA can be encapsulated within the core, entrapped within the polymeric matrix, covalently conjugated to the polymer, or adsorbed onto the surface. This allows for the utilization of both naked and conjugated siRNA [139,232]
Cationic polymers are the most commonly used in polymeric nanoparticles for siRNA delivery [29,143,223,235,236]. These polymers can be surface-modified, being linked to peptides, receptor ligands, polysaccharides, exosomes, or cancer cell membranes. Such modifications aim to enhance siRNA delivery to cancer cells. A recent review covers these modifications [237]. In this work, only the most representative polyplexes—PEI-siRNA, CS-siRNA, and PBAE-siRNA—are described below.
- PEI-siRNA polyplexes. PEI is a widely used cationic polymer which forms polyplexes with siRNA because of its high cation density and ability to buffer protons across a broad pH range [238,239]. Its structure includes repetitive ethylenimine groups, which confer extensive buffering capacity within the pH range of the endosomal/lysosomal pathway [240,241]. Chemically, it is highly versatile, easily functionalized and branched [136,240,242]. The transfection efficiency of PEI-siRNA polyplexes is high because, under acidic conditions, their amines facilitate endosomal escape, helping siRNA reach the cytoplasm [136,241]. Branched PEI generally outperforms linear PEI in transfection efficiency [241,243,244]. Additionally, some PEIs can form covalent complexes with siRNA [241,245].
The main drawback of PEI-siRNA polyplexes is the toxicity linked to the high molecular weight of PEI. To lower toxicity, strategies similar to those used for reducing toxicity in PLL have been employed, such as: (i) decreasing the molecular weight to around 25 kDa [240,244]; (ii) reducing the positive Z-potential of the polyplex by partially neutralizing the peripheral amino groups [136,240,244]; (iii) adding hydrophobic residues [246]; (iv) coating the surface of the polyplexes with PEG [238,244,247]; (v) using copolymers [136,244]; (vi) embedding the polyplex in more complex nanosystems [136,139,233,236,241,248].
- CS-siRNA polyplexes. Chitosan (CS) is the most widely used natural polymer for preparing polyplexes that directly deliver siRNA to cells or form part of other nanosystems. It is a linear polysaccharide derived by deacetylation of chitin, composed of repeated units of N-acetyl-D-glucosamine and D-glucosamine linked via β-1,4 bonds. The proportion of these units determines the polymer’s degree of deacetylation (DD) [141,227]. CS is biocompatible, biodegradable, non-toxic, and easily modified [227,249,250]. siRNA can be incorporated into CS through: (i) encapsulation; (ii) adsorption; or (iii) electrostatic interactions [249,251]. The most common method is electrostatic complexation, as the high positive charge of CS facilitates binding with the negatively charged phosphate groups of siRNA [137,227,250].
CS-siRNA polyplexes are stable, their endosomal escape is effective, and they do not cause significant changes in the biological activity of siRNA [141,252,253,254]. The size, shape, and charge of these polyplexes heavily depend on the molecular weight of the CS and the molar ratio between both components (N/P ratio), which then influence transfection efficiency [251,253,255,256].
Despite these qualities, the effectiveness of polyplexes can be hindered by certain physicochemical factors, which include: (i) low stability, especially in those using low molecular weight CS [251,257]; (ii) a tendency to aggregate [257]; (iii) limited cell internalization [250,258]. To improve stability, it is advisable to: (i) use CS with a molecular weight over 10 kDa [249,250,251]; (ii) chemically modify CS [249,250]; (iii) coat the polyplex surface with polymers [137,141,256]; (iv) add CPP or targeting ligands [250,258,259]; (v) incorporate the polyplex into other nanocarrier systems [260,261].
- PBAE-siRNA polyplexes involve polymers prepared by polymerizing diacrylate and amino compounds. These polymers were designed to enhance biodegradability and reduce cytotoxicity compared to PEI and PLL [262,263,264]. They come in diverse structures and shapes, such as linear, spherical, and multi-layered films [231,265]. Their chemical modifications allow for control over size, surface charge, hydrophobicity, degradability, and stimulus responsiveness, influenced by factors like chain length, structure, end groups, solution pH, and N/P ratio [12,231,262,266]. Typically, these polyplexes are serum-stable and readily taken up by cells [12,262]. Inside the cell, they quickly degrade, releasing siRNA and PBAE is hydrolyzed into biocompatible products [12,231]. Their transfection efficiency is influenced by terminal groups, while toxicity relates to hydrophobicity levels and the spacing of amino groups [267]. Adding groups like amines or hydroxyls to the PBAE end chains notably enhances transfection, and the alkyl chain length and end-group hydrophobicity directly impact toxicity [264].
Amphiphilic polymers, also known as polysaccharide peptide conjugates, are utilized for siRNA delivery [268]. These complex molecules are prepared by covalently attaching a polysaccharide, a carbohydrate polymer, to a polypeptide. Various architectures, such as polysaccharide–short cationic peptide and polysaccharide–cationic polypeptide, can be designed for siRNA conjugates. They outperform peptides and polysaccharides alone, being fully biodegradable, offering better biocompatibility than other polymers, and facilitating enhanced endosomal escape due to amino acid residues with proton groups (H+). Additionally, they can recognize cell receptors depending on their amino acid sequence. Although not yet widely used in siRNA delivery, they represent a promising platform, especially since they have demonstrated superior response compared to the well-established PEI polymer.
Other polymeric nanoparticles used to deliver siRNA in cancer treatment include siRNA polyplexes with (i) poly(lactic acid co-glycolic acid) (PLGA) [269]; (ii) α, β-poly(N-2-hydroxyethyl)-D,L-aspartamide (PHEA) [139]; (iii) poly(amidoamine) (PAMAM) [270]; and (iv) Poly(2-dimethylaminoethyl methacrylate) (PDMAEMA) [236], among others.
- (b)
- Lipid Nanosystems
Lipid nanosystems are currently the most commonly used vehicles for siRNA delivery. They naturally enhance cell uptake, protect siRNA from nuclease degradation, and reduce renal clearance. Their key properties, such as size, structure, surface charge, and transfection efficiency, depend on the specific lipids used, allowing their biological functions to be tuned. These nanosystems are biocompatible, biodegradable, less immunogenic than polymeric alternatives, highly effective in delivering siRNA, and are easily and inexpensively produced. They are taken up through various endocytosis pathways. However, a drawback is their tendency to become trapped in the liver [223,271,272,273,274]. They are mainly categorized into two groups: lipid nanosystems (LNPs) and lipoprotein transporters.
LNPs consist of acylglycerides, fatty acids, phospholipids, and steroids, forming mostly spherical structures. Based on their composition and structure, they can be categorized into lipoplexes, liposomes, solid lipid nanoparticles (SLN), nanostructured lipids (NLC), stabilized nucleic acid lipid nanoparticles (SNALP), and lipid-polymer hybrid nanoparticles (see Figure 4 middle panel). Notably, self-nanoemulsifying drug delivery systems (SNEDDS), which are taken orally, and cubosomes, regarded as the third generation of LNPs, are also critical.
Lipoplexes. There are different perspectives on how to define and structure lipoplexes. Many authors refer to all LNPs as lipoplexes, while others consider only liposomes as lipoplexes. In this work, a lipoplex is defined as a supramolecular structure formed by a lipid and siRNA (Figure 4 middle panel). These lipoplexes form spontaneously through electrostatic interactions between the negatively charged siRNA and the positively charged polar heads of the lipid [275,276]. Therefore, lipids with a permanent positive charge (cationic lipids) or zwitterionic lipids that are neutral at physiological pH (7.4) but become positively charged at lower pH (5.5–6.5) due to protonation of their amino groups (ionizable lipids) are used to prepare lipoplexes. Anionic lipids are rarely used for this purpose because they require a cationic mediator, have low cell penetration capacity, and can trigger an immune response [277]. Lipoplexes can also be produced by covalent linkage to the 3′ end of the sense strand of siRNA, but this approach is less common [196].
The electrostatic interaction between the cationic head group of the lipid (either cationic or ionizable) and siRNA results in the formation of thermodynamically stable particles with various morphologies, sizes, stabilities, and interactions with cell membranes. These properties depend on the specific characteristics of the lipid and the preparation conditions [275,278,279]. Cationic lipids used for producing polyplexes include: (i) N-(1-[2,3-dioleoyloxyloxy]-propyl)-N,N,N-trimethylammonium chloride (DOTMA) [272,280,281,282]; (ii) 2,3-dioleoloxy-N-[2-(sperminacarboxamido)ethyl]-N,N-dimethyl-1-propaniminium trifluoroacetate (or chloride) (DOSPA) [276,283,284]; (iii) N,N-dioleoyl-N,N-dimethyl ammonium chloride (DODAC) [272]; (iv) dimethyl-dioctadecyl ammonium bromide (DDAB) [285,286]; (v) N-(N′, N′-dimethylamino ethane)-carbamoyl cholesterol (DC-cholesterol) [283]; (vi) 1,2-dioleoyl-3-trimethylammonium propane sulfate (DOTAP) [195,285,287]. These lipids consist of three parts: (i) a polar head containing one or more amine groups, which can be primary, secondary, tertiary, or quaternary; (ii) two hydrophobic chains, which may be saturated or unsaturated; and (iii) a connecting group linking parts (i) and (ii). The amine groups are responsible for complexing with siRNA, the hydrophobic chains aid in packaging, and the connector enhances the chemical and enzymatic stability of the nanosystem [275,281,285].
The transfection ability of lipoplexes is high. Their overall positive charge helps them interact electrostatically with the negatively charged lipids of cell membranes, allowing entry into the cell through endocytosis [198,288]. However, cationic lipoplexes are cleared from the plasma very quickly and tend to form aggregates and micelles, which are absorbed by macrophages in the lungs, liver, and spleen. In contrast, their uptake in tumors and inflammation sites is lower, leading to reduced silencing efficiency [198,272,289]. To address these issues, additional polymers and other components are incorporated to create liposomes and more advanced lipid nanosystems [290].
Ionizable pH-dependent lipids are preferable to permanent cationic lipids. Lipoplexes with ionizable lipids are prepared at low pH to ensure electrostatic interaction with siRNA [281,289]. Because they are neutral (pKa < 7) at physiological pH, their interaction with blood cells is less than that of cationic lipids [237,272,278]. Once inside the cell, they protonate again, interact with the negatively charged endosomal membrane, and destabilize it, allowing the siRNA to be released easily into the cytosol. Ionizable lipids used for preparing siRNA lipoplexes include: (i) dioleoyl-phosphatidyl ethanolamine (DOPE) [273,290,291,292]; (ii) 1,2-dioleoyl-3-dimethylammonium propane (DODAP) [272,293]; and (iii) 1,2-dilinoleyloxy-3-dimethylaminopropane (DLin-DMA) and its derivatives [16,196,271,272,273,294]. All of these are used to formulate various lipid nanosystems.
The silencing ability of lipoplexes depends on the pKa of the amino group, the unsaturation of the acyl chains, and the structure of the connectors [272,281,289]. Ionizable lipid-like molecules, called lipidoids, are now being synthesized, with their structure designed through specific combinations of the three lipid components to enhance the in vivo efficacy and safety of LPNs. Among the most commonly used are 98N12-5, C12-200, OF-02, L3-19 [16,272,295,296]. Patrisiran was the first siRNA formulation approved for clinical use [237]. Although it was not developed for cancer therapy, it is a carrier nanosystem based on ionizable lipids. The advantages of ionizable lipids as targeted delivery systems for siRNA were recently reviewed [237].
Liposomes. Liposomes are the most commonly used LNPs for siRNA transport and delivery. They are spherical vesicular structures composed of one or more phospholipid bilayers (natural or synthetic) that self-assemble around a central aqueous core when dispersed in water (Figure 4 middle panel). Hydrophobic interactions and Van der Waals forces stabilize them. The phospholipid bilayers are separated by aqueous compartments. This structure enables the encapsulation of hydrophilic molecules in aqueous compartments, while molecules with different characteristics can be trapped or attached to the lipid bilayer. Molecules with intermediate log P are retained in both the aqueous compartments and the lipid bilayer. These features enable liposomes to carry multiple types of molecules simultaneously, making them helpful in developing theranostic and multimodal therapy systems. The physicochemical and biological characteristics of liposomes—such as structure, size, lipid composition, surface charge, bilayer fluidity, and interaction with cell membranes—depend on their component combinations and production methods [297,298,299,300]. For example, the number of bilayers influences size and encapsulation efficiency [271,289,300,301]. For medical applications, single bilayer (unilamellar) liposomes with sizes of 50–150 nm are preferred [300].
Key components of siRNA-carrying liposomes include (i) phospholipids that form lipoplexes with the siRNA and promote its endosomal escape; (ii) hydrophobic lipids that provide stability to the liposome; (iii) polymeric molecules that coat the liposome to enhance pharmacokinetics; and often (iv) ligands targeting cell receptors [271,276,289,299,300]. The hydrophilic head of the phospholipids (positive, negative, or zwitterionic) and their hydrophobic tail define the amphipathic nature of the bilayer and the surface charge of the liposome [276,300].
The most commonly used natural phospholipids are phosphatidic acid (PA), phosphatidyl ethanolamine (PEA), phosphatidyl glycerol (PG), phosphatidyl serine (PS), phosphatidyl choline (PC), and phosphatidyl inositol (PI) [300,302]. These phospholipids are modified to obtain synthetic derivatives with improved characteristics, such as DOPE, DOTAP, DOTMA, dilauryl phosphatidyl choline (DLPC), 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC), 1,2-dipalmitoyl-sn-glycero-e-phosphatidylcholine (DPPC), 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC), dilauryl phosphatidyl ethanolamine (DLPE), dilauryl phosphatidyl glycerol (DLPG), dioleoyl phosphatidyl serine (DOPS), 1,2-dioleoyl-sn-glycero-3-phosphatidylcholine (DOPC), among others [272,278,283].
To improve bilayer properties and in vivo stability, hydrophobic lipids (co-lipids) are added. These lipids, usually non-cationic, include sterols, phospholipids, and glycerolipids. The most used is cholesterol and its derivatives [283,291,300], although DSPC, DOPE, and DOPC are also used and contribute to fusion with cell membranes [275,283,299,300,303]. Cholesterol fills the empty spaces between phospholipid molecules, inserts with the -OH groups facing the aqueous phase, and the aliphatic chain aligned parallel to the acyl chains of the hospholipids. In this way, it contributes to the rigidity of the bilayer, reduces the permeability of hydrophilic compounds through the bilayer and the tendency of liposomes to aggregate while increasing the absolute Z-potential, the blood lifetime, the permeability of hydrophobic compounds, and promotes interaction with cell membranes [272,276,291,299,300,304].
To increase the lifetime in blood, decrease MPS recognition and toxicity, it is common practice to coat liposomes with different molecules. The most used is PEG, natural or modified [272,273,305,306]. In addition to improving pharmacokinetics, PEG allows antibodies, antibody fragments, peptides, aptamers, radionuclides, and ligands to be conjugated to the liposome surface, enabling the targeting of specific cell receptors [307,308,309]. As natural PEG has the disadvantage of negatively influencing cell uptake and endosomal escape of siRNA, and may cause unwanted toxicity, derivatives have been developed to improve both properties and increase transfection capacity [305,306].
The method of liposome preparation influences their physicochemical and biological properties [271,289,299,302,308]. Depending on the method, liposomes with different structural and surface properties can be obtained, which interact with cells by multiple mechanisms [299,300,306,310].
The siRNA can be inserted into the aqueous core (encapsulation) or into the lipid bilayer of liposomes. The site of insertion influences subsequent biodistribution [298,305,311]. Encapsulation is preferred as it ensures greater stability in vivo. Encapsulation can be carried out during the preparation [298,312,313,314,315] or after liposome preparation [303,310,316,317]. If the encapsulation is done during the preparation (passive encapsulation), naked siRNA is used [297] or in the form of positively charged complexes [274,293]. Suppose the encapsulation is performed after the liposome has been prepared (active encapsulation). In that case, the naked siRNA is not suitable because if the liposome contains PEG, the addition of the siRNA may form a very large nanosystem or the siRNA may adhere to the liposome surface and be released as soon as it enters the bloodstream [297,298].
The insertion of siRNA into the lipid bilayer depends on the phospholipids, the preparation conditions, and the N/P ratio [271,289,298]. The simplest variant of incorporation into single-layer liposomes is to pre-conjugate the siRNA to hydrophobic molecules such as cholesterol and fatty acids [289,306,318,319]. Binding by adhesion to the surface can be done either with naked siRNA (cationic liposomes) or in the form of conjugates [298].
Lipofectamine 2000 is the most used commercial liposome for siRNA transfection, especially in vitro. It consists of a cationic liposome made from a 3:1 mix of DOSPA and DOPE, enabling high-efficiency and reproducible transfection across various cell lines [272]. However, a limitation of liposomes is their instability during storage, which has prompted the development of more stable nanosystems.
Solid lipid nanoparticles (SLN) and nanostructured lipids (LNC) represent a more advanced generation of lipid-based carriers than liposomes, with a stiffer morphology, higher physical stability, and greater loading efficiency [272,274,276]. They are easy to produce, stable during freeze-drying, do not require organic solvents for preparation, and can release encapsulated siRNA in a controlled manner. SLNs consist of a solid lipid core surrounded by a layer of non-ionic surfactant [274,314,320]. They may or may not be coated with additional materials [321]. Typical lipids used in SLNs include fatty acids, fatty alcohols, glycerides, and waxes with a melting point above 40 °C [274,276]. Surfactants are employed to reduce interfacial tension between the lipid and aqueous phases, enhance formulation stability, and influence the release kinetics of the encapsulated siRNA [321,322]. Many combinations of lipids and surfactants are available to create SLNs with different physicochemical and biological properties, which can incorporate siRNA in either hydrophilic or lipophilic forms. To load siRNA into the SLNs’ lipophilic core, it must first be conjugated, preferably as lipoplexes [323].
LNCs are modified SLNs (Figure 4 middle panel). They are considered as some of the most advanced lipid carriers for siRNA delivery [272,314]. They consist of a mixture of solid and liquid lipids, to which surfactants are also added [274,314,324]. They exhibit higher loading efficiency and stability than SLNs, with similar toxicity levels. Their capacity to internalize tumor tissue is also high [314]. Both SLNs and LNCs are produced using several methods summarized in different publications [324], although all methods involve incorporating the siRNA into melted or liquid lipids before mixing with the surfactant solution.
Stabilized nucleic acid lipid particles (SNALP) can be considered a specific type of liposomes (Figure 4, middle panel) [325]. They have a size of approximately 100 nm and a neutral charge. They consist of four main components: (i) a lipid that can be cationic or ionizable to form the lipid-siRNA lipoplex that is encapsulated in the nucleus; (ii) one or more ionizable lipids forming the membrane and contribute to the siRNA endosomal escape and cytosolic release; (iii) one or more high-transition-temperature ancillary lipids that stabilize the lipid layer; (iv) a PEG lipid derivative to enhance pharmacokinetics and target cell surface molecules [326,327,328]. Cationic and ionizable lipids are the ones previously mentioned (see lipoplexes). Generally, auxiliary lipids such as cholesterol, DSPC, and DSPE are used [16,321,326,327].
The most common PEG derivatives are diacylglycerols like PEG-DSG and PEG-DMG [327,329]. Unlike liposomes, SNALPs have a micelle-like structure. In these micelles, the hydrophobic core contains the lipoplex, the bilayer is composed of the ionizable neutral lipid, cholesterol, and auxiliary phospholipids, and the surface features the PEG lipid conjugate, which may or may not have targeted molecules attached [328]. Several methods are used to prepare SNALPs, but the most common involves microfluidic mixing of lipids and siRNA in ethanol followed by dialysis [297,327]. After mixing and prior to dialysis, connectors are added.
Lipid-polymer hybrid nanoparticles (LPHNPs) combine the benefits of lipid and polymeric nanoparticles. Their pharmacokinetic properties, biocompatibility, bioavailability, shelf stability, and ability for controlled release are superior to those of lipid or polymeric nanoparticles alone, while they exhibit lower immunogenicity and toxicity [183,330,331,332]. Their main components include siRNA, a lipid, and a biodegradable polymer. These nanoparticles are divided into three sections: (i) the polymeric core, where siRNA is efficiently encapsulated as a polyplex; (ii) an inner lipid monolayer formed by a cationic lipid that surrounds and protects the core, promotes biocompatibility, and enables controlled siRNA release; (iii) an outer layer composed of a hydrophilic lipid-PEG (or lipid-PVP) conjugate that enhances stability and pharmacokinetics and allows attachment of molecules targeting specific cell receptors (Figure 4 middle panel) [183,331]. Due to their multimodal loading capacity, they are ideal for designing theranostic systems [333,334]. Additionally, siRNA can be incorporated into the lipid bilayer through covalent bonding [335].
The polymers used to prepare the LPHNPs core include those described earlier. Commonly used ones are PEI, PLGA, polycaprolactone (PCL), poly-lactic acid (PLA), poly-hydroxyethyl methacrylate (PHEMA), PBAE, and CS [183,242,331,333,336,337,338,339,340]. Depending on the polymer used to form the core polyplex, the inner lipid monolayer may consist of cationic, anionic, or neutral lipids. If the core is formed with a charged polyplex, then the inner layer lipids are oppositely charged. If the polyplex is hydrophobic, then the interaction with the lipids occurs through the hydrophobic tails of the lipids [341].
The list of lipids used to form the inner and outer lipid layers is extensive. Among the most common are myristic acid, phosphatidylcholine, cholesterol, stearic acid, 1,2-dipalmitoyl-sn-glycero-3-phosphocholine, DOTAP, DSPG, DOPC, DSPE, and glycerylmonooleate (GMO) [330,338,342,343,344]. The variety of lipid/polymer combinations results in LPHNPs with diverse physicochemical, structural, and functional properties [335,340,345]. A table listing some of these combinations can be found in [346].
Although there are different methods of preparing LPHNP, they are all based on mixing its components in one or more stages [264,274,331,335,340]. There are also hybrid nanoparticles that are prepared with inorganic/organic components.
Self-emulsifying drug delivery systems (SNEDDS) are lipid-based vehicles suitable for oral siRNA transport. They are thin oil-in-water (o/w) nanoemulsions smaller than 100 nm, consisting of an isotropic and stable mix of lipids, surfactants, cosurfactants, and cosolvents, in which active substances are dissolved or suspended [347,348]. These systems undergo self-transformation in situ when their oily phase contacts the aqueous environment of the intestinal tract (GIT). GIT motility creates agitation, allowing spontaneous formation [347,349]. They protect against enzymatic hydrolysis, have a fast onset of action, higher loading efficiency compared to other lipid systems, and can be formulated in liquid or solid forms. They are easy to manufacture and scale up. Additionally, they can incorporate both hydrophilic and lipophilic molecules and are therefore an alternative for siRNA delivery [348,349].
Other lipid siRNA transport systems, including cubosomes or cuboplexes, are at earlier stages of pharmaceutical development [350,351]. Cubosomes are LNPs with an internal bicontinuous cubic symmetry and have higher transfection efficiency than liposomes, probably because of their different mechanisms of interaction with cells [352]. They are considered the third generation of LNPs.
- (c)
- Lipoprotein-Based Nanosystems
Lipoproteins are complex assemblies of lipids and proteins with varying sizes and densities, present in blood serum. They are crucial for transporting cholesterol, triglycerides, and other lipids, and also participate in processes such as coagulation, tissue repair, and immune responses [353]. Their hydrophobic core, consisting of triglycerides and non-polar cholesterol esters, is covered by a monolayer of phospholipids and free cholesterol, into which specialized proteins called apolipoproteins are intercalated to contribute to the stability and facilitate recognition by cell receptors [308,354]. Free cholesterol, located between the alkyl chains of phospholipids, helps maintain the outer layer’s rigidity [355]. These lipoproteins are classified by size, composition, and density into: (i) chylomicrons (CM, >100 nm), (ii) very low-density lipoproteins (VLDL, 30–80 nm), (iii) intermediate-density lipoproteins (IDL, 30–50 nm), (iv) low-density lipoproteins (LDL, ~25–35 nm), and (v) high-density lipoproteins (HDL, 5–12 nm). In living organisms, they can convert from one type to another based on the current lipid-to-protein ratios [353,354].
LDL and HDL (Figure 4 bottom panel) are employed as drug carrier systems due to: (i) they are not recognized by the MPS; (ii) they have long blood circulation times; (iii) they are non-immunogenic; and (iv) they can transport hydrophobic, hydrophilic, and amphiphilic molecules without significantly altering the structure or biological properties of the transported molecule. They can be used in native forms (isolated from plasma) or in synthetic forms. The native form is less practical because isolation, purification, and quantification involve lengthy and costly procedures that are not very scalable and are hard to standardize [353,354,355]. It is preferable to synthesize them from their natural or synthetic components (reconstituted, rLDL, and rHDL). Reconstituted lipoproteins, where the apolipoproteins are replaced by mimetic peptides, are called mimetics (mLDL and mHDL) [354,356,357]. These synthetic lipoproteins offer the advantage that their component ratios can be adjusted to create nanoparticles with specific physico-chemical properties, without compromising the biological functions of native lipoproteins. They are easy to produce and well tolerated by patients [308,354,358]. Compared to liposomes, they are smaller and can directly interact with targeted cell receptors.
Synthetic LDL and HDL NPs are typically spherical, though they can also be discoidal [357,359]. They can attach biomolecules and ligands to their surfaces, allowing them to target their own cell receptors as well as other receptors of interest on the cell membrane. They are internalized in cells by both passive and active transport [357,360]. Their multimodal loading capacity supports the development of theranostic systems [360,361,362] and multimodal therapy systems [363,364]. They break down into recyclable units of cholesterol, fatty acids, and amino acids and do not require PEG coating to extend their blood circulation times [353,355]. These advantages have contributed to their use in diagnosing and treating various types of tumors.
LDL-siRNA nanosystems. LDL is the primary cholesterol transporter to different extrahepatic tissues and plays a crucial role in their homeostasis and steroid synthesis. Native LDL is a diverse group of particles that vary in size, density, and composition (Figure 4 bottom panel). Apo B-100 is embedded in the amphiphilic monolayer of phospholipids and free cholesterol [353,355,365]. It is among the largest proteins known (>500 kDa, 4536 amino acids). Apo B-100 encircles the LDL surface like a belt, contributing to its stability and mediating receptor (LDLR) binding [355,365]. LDL and HDL compositions are detailed in Table 3.

Table 3.
Main characteristics of LDL and HDL.
The tissue with the highest expression of LDLR is the liver, but the adrenal glands, reproductive organs, skeletal muscle, lymphocytes, kidneys, and intestines also express these receptors, although in smaller amounts [353,355]. Some tumors overexpress LDLRs because they need lipids for growth and proliferation [355,359,365,366,367,368,369,370,371]. Cholesterol is also a precursor to steroid hormones, which are important in the development of breast and prostate cancer [367,372]. These facts have motivated the use of LDL for cancer diagnosis and therapy.
Although HDL is more commonly used than LDL for siRNA delivery, cholesterol-siRNA conjugates can be effectively loaded into LDL [355,368,373]. The main pathway for internalizing cholesterol-siRNA conjugates via LDL involves ApoB-100 binding to its receptors (LDLR) [353,368,373]. After endocytosis, LDL merges with lysosomes, where it is hydrolyzed, releasing its contents into the cytosol. LDLR is then recycled back to the cell surface, while its components are degraded, and the siRNA is freed from cholesterol to bind to RISC [353].
Various methods have been developed for rLDL preparation [355,365], which utilize natural or synthetic lipids to which Apo-B 100 is added [353]. Some researchers also include Apo E [374,375]. A challenge with ApoB-100 is that it is difficult to isolate and tends to aggregate, making mimetic peptides a more practical option [375].
Naked siRNA is not naturally incorporated into LDL, so it must be conjugated with other molecules. For example, lipophilic conjugates such as cholesterol-siRNA and fatty acids-siRNA are used [368,370,373,376]. Amphiphilic conjugates designed to integrate into the phospholipid layer are also employed. The amino acids of ApoB100 allow covalent attachment to various molecules, including peptides, proteins, antibodies, antibody fragments, and aptamers.
Lipophilic cholesterol-siRNA and fatty acid-siRNA conjugates are incorporated into the LDL core during nanosystem preparation through exchange with core lipids [355,362,373]. Alternatively, some researchers prefer to perform this exchange after preparing rLDL [368,374]. These lipophilic conjugates, encapsulated within the rLDL core, are mainly absorbed by the liver, with limited uptake in other LDLR-expressing organs [368,373], although brain uptake has also been observed [374].
siRNA conjugates are incorporated into rLDL phospholipids through both covalent binding via phosphodiester bonds and non-covalent interactions by embedding conjugate hydrophobic tails, which orient hydrophilic heads toward the aqueous environment. In this way, the conjugate is inserted via Van der Waals forces, hydrogen bonds, and ionic interactions [355,362]. The balance and strength of Van der Waals forces between the inserted molecule and phospholipids, along with the hydrogen bonds and ionic interactions, determine the efficiency and stability [355,362]. However, this method has the drawback that some molecules tend to escape from rLDL.
For siRNA binding to Apo B100, specific residues such as lysine, arginine, tyrosine, and cysteine are utilized to attach molecules that target cellular receptors other than LDLR [355,362,377]. For example, FA can be attached to ApoB-100 by pre-modifying lysine residues [377]. Although this reduces ApoB-100’s ability to bind LDLR, it allows rLDL to bind to FR, which is expressed in various cancers. Alternatively, surface modification of rLDL with cationic polymers can create electrostatic complexes with siRNA, providing attaching to rLDL [365].
HDL-siRNA nanosystems use HDL, the smallest and densest lipoprotein, which is crucial for reverse cholesterol transport. Similar to LDL, HDL contains various NP subpopulations that affect its functions in vivo [372,378]. It differs from LDL in its component ratios and protein composition (see Table 3). Its main protein, Apo A-1, accounts for about 70%, influencing HDL’s size and appearance, allowing its binding to SR-B1 receptors, which are often overexpressed in tumor cells [358,372,379,380]. Additionally, Apo A-1 has anti-tumor effects [381].
Physicochemical properties of rHDL are similar to those of native HDL [361,382]. When rHDL interacts with SR-B1, a hydrophobic structure forms. This enables the direct release of transported molecules into the cytosol [383,384]. This interaction is especially beneficial when SR-B1 is overexpressed, as it enables selective delivery of therapeutic molecules [385].
rHDL prevents siRNA endolysosomal degradation and offers high transfection efficiency [373,380,386,387]. Similar to rLDL, siRNA can be incorporated into both the core and the outer layer. To incorporate siRNA into the rHDL core, hydrophobic conjugates with an octanol:water partition coefficient, expressed as Log P > 3, are required. Conjugates with Log P > 1 can also be used. Encapsulation can occur during preparation [361,379,387] or after the rHDL is formed [374,388,389]. In the first approach, the molecule is exchanged or previously bound to one of the core components. In the second, the desired agent is introduced after the rHDL has been prepared.
Cholesterol is commonly used as a hydrophobic agent to bind siRNA to rHDL [202,374,390,391,392]. Oligosine [379], fatty acids [202,373], α-tocopherol-siRNA [388] are also used. These conjugates are encapsulated in the core or inserted into the bilayer depending on their physicochemical properties and preparation method. Some authors have coupled siRNA to the rHDL surface through electrostatic interactions. In this case, cationic polymers or lipids are bound to rHDL surface to form polyplexes or lipoplexes with siRNA [356,379,389]. Covalent binding by hybridization has also been used [356]. The siRNA chemical structure, together with the preparation method of rHDL/siRNA complexes, critically affects NP–receptor interactions and transfection efficiency [393]. These variables are considered required.
Similar to rLDL, the insertion of molecules into the rHDL surface occurs through: (i) covalent bonding to the head groups of phospholipids [308,360,362] or to the lysine, arginine, tyrosine, or cysteine residues of Apo A-1 [362,394,395], and (ii) non-covalent intercalation of conjugates with membrane components via Van der Waals forces, hydrogen bonds, or ionic interactions [308,357,362,396].
An advantage of rHDL is that it can simultaneously transport several molecules, which enables the design of theranostic systems and multimodal therapy [361,397,398]. However, it has the disadvantage that it is taken up by the liver, kidneys, stomach, steroidogenic tissues, and smooth muscle [379,380], which could cause unwanted off-target effects.
- (d)
- Cell-Derived Nanosystems
Organic nanosystems derived from cells, called extracellular vesicles, are in the pharmaceutical development stages. These nanosystems are small vesicles (30–200 nm) secreted by various types of cells, including tumor cells. They play different roles in cell communication, the immune system, tissue repair, and facilitate the transport of bioactive molecules [399,400]. They have numerous advantages because they easily cross biological membranes, have long circulation times in the blood, and are non-toxic [400]. There are different subtypes of extracellular microvesicles, but the most studied as siRNA transporters are exosomes [320,399,401,402]. Its main drawback is the low siRNA loading capacity and high hepatic uptake [402]. Efforts are currently focused on developing strategies to overcome these challenges.
- (ii)
- Inorganic and Hybrid Nanocarriers
Over the past twenty years, inorganic nanosystems (metallic and non-metallic) have been developed for siRNA delivery, offering better mechanical properties than organic nanoparticles, sufficient bioavailability, low toxicity, and high stability in serum, among other beneficial qualities. The surface of these inorganic nanoparticles, which vary in morphology and physicochemical characteristics, can be easily modified through covalent conjugation with organic ligands or amphiphilic polymers, electrostatic interactions, or coating with lipid systems [384]. They are useful in designing theranostic systems and multimodal therapies [278,403,404]. These are also called multifunctional nanoparticles and hybrid nanoparticles [384,405]. Their great diversity enables various strategies for coupling siRNA through both covalent and non-covalent bonds [384,404]. The most commonly used include: (i) gold nanoparticles; (ii) magnetic nanoparticles; (iii) inorganic semiconductor crystals (quantum dots); (iv) carbon-based nanoparticles; and (v) mesoporous silica nanoparticles. This category also includes calcium phosphate-siRNA complexes encapsulated in lipid systems.
- Au nanoparticles. These are colloidal suspensions of Au with unique chemical and physicochemical properties that have been used in biomedicine since ancient times. They absorb light and X-rays, and their absorption maxima can be adjusted during synthesis. They can disperse and convert absorbed light into heat, enhance the Raman spectra of molecules near their surface, produce fluorescence, and carry different molecules with high loading capacity. They are bioinert, low in toxicity, and easy to obtain [406,407,408]. They easily conjugate to biomolecules due to their high affinity for thiols, disulfides, and amine groups [384,409,410]. They efficiently transfect siRNA without needing additional transfection agents.
siRNA binds to AuNPs through covalent bonding or electrostatic interactions. For covalent attachment, AuNPs are functionalized with amine, cysteamine, or azide groups [407,411,412,413], and then the modified siRNA is linked via the bioconjugation reactions already described. Alternatively, siRNA can be directly attached to the surface of AuNPs after modification with thiol groups, forming an Au-S bond [404,414]. This method also allows hybridization-based binding [415]. Some researchers attach PEG functionalized with amine groups or thiols to the AuNP and then connect the siRNA to the PEG using a linker [414]. Others conjugate siRNA to PEG first and then attach the siRNA-PEG complex to the AuNP [412].
For AuNP-siRNA binding through electrostatic interactions, the AuNP is positively charged before adding the siRNA, which can be naked or conjugated to a linker. Cationic polymers and peptides are used to positively charge the AuNP. One of the most common is PEI, due to its dual role as a reducing agent and stabilizer of Au+ ions, as well as its strong electrostatic interaction with siRNA [416,417,418]. However, its toxicity is a drawback, which is why some researchers prefer CS [419,420,421]. Among cationic peptides, protamine is the most used, though TAT and RGD have also been employed [410,422]. Electrostatic bonding enables layer-by-layer assembly [417,420].
Since the physicochemical properties of AuNPs depend on their shape and size, they are prepared in various forms—such as spheres, rods, stars, cubes, and plates—to suit different applications. This variety broadens the range of therapeutic options [409,423]. Spherical AuNPs are used to produce so-called spherical nucleic acids (SNAs).
AuNP/siRNA coated with cationic carboxylane dendrimers has demonstrated its therapeutic efficacy for silencing antiapoptotic proteins of the BCL family, which are overexpressed in different tumors [424]. Other systems, where AuNP/siRNA are coated with PEG-acrylate and folate monomers, have been used to promote tumor vessel normalization, enhance immunotherapy, and inhibit metastasis formation [425].
- Magnetic nanoparticles are used to develop NMR theranostic systems and for gene silencing. They consist of materials containing ferromagnetic, paramagnetic, diamagnetic, antiferromagnetic, and ferrimagnetic elements, with sizes ranging from 10 to 20 nm. These particles exhibit different responses to external magnetic fields [426,427]. Superparamagnetic iron oxide nanoparticles (Fe3O4 and γ-Fe2O3), known as SPIONs, are the most commonly used in medicine due to their versatility, biocompatibility, strong magnetization, and low production costs [428,429,430].
Like other inorganic systems, magnetic NPs are functionalized with different molecules, and siRNA is assembled using both covalent bonds and electrostatic interactions. This results in systems with diverse morphology, size, surface charge, pharmacokinetics, and magnetic properties [405,427,429,431,432]. The methods employed for siRNA coupling are similar to those used for AuNPs.
Among the most commonly used molecules to attach siRNA to magnetic NPs are cationic polymers, with PEI standing out [405,428]. Cationic peptides have also been utilized [433], along with slightly anionic polymers such as PEG and its derivatives [429,430,434], polysaccharides [427], and combinations of molecules [435,436]. The concurrent binding of siRNA and molecules targeting specific cellular receptors enhances the efficiency of these nanosystems [437].
An advantage of magnetic NPs is that siRNA can be concentrated in a given tissue by applying an external magnetic field. The method, called magnetofection, depends on the interaction of many variables including NP composition, the applied magnetic field, the strength of the siRNA-NP bond, the route of administration, and the location and depth of the target tissue [426,438]. It has been shown, however, that it offers the possibility of internalizing siRNA in difficult-to-transfect cells such as neuronal cells [437,438].
- Inorganic semiconductor crystals, known as quantum dots (QDs). They are nanocrystals ranging from 2 to 20 nm that are created from binary combinations such as CdSe, CdTe, CdS, ZnS, ZnHgSe, PbS, CdHgTe, and CdxPb1-xTe alloys. Their optical properties vary with changes in composition, size, and shape [439]. These QDs offer several advantages: (i) tunable emission; (ii) high fluorescence quantum yield; (iii) resistance to photobleaching; (iv) a high surface-to-volume ratio; and (v) ease of functionalization, which facilitates the transport of molecules of diverse types [440]).
The physicochemical properties of QDs depend on the method of production, which can be physical, chemical or biological [440,441]. However, QDs offer the possibility of simultaneously coupling molecules of different nature and obtaining high contrast images, which is useful for theranostic devices [442,443,444,445]. They have the advantage of being easily integrated into LNP.
siRNA binds to QDs through both covalent and non-covalent interactions. The QD surface is functionalized with molecules containing terminal groups such as carboxyl, amine, thiol, hydrazine, or diol [384,441,446]. siRNA is then attached via established reactions, including (i) disulfide bridges, (ii) carbodiimide-amine coupling, (iii) thiol-maleimide reaction, and (iv) azide-alkyne click chemistry. Non-covalent coupling occurs through affinity and electrostatic interactions. For affinity coupling, streptavidin is attached to the QD followed by biotinylated siRNA [441]. Electrostatic coupling involves cationic molecules first attaching to the QD, then linking siRNA. Common functionalization molecules include PEG and derivatives [446,447,448], PEI [447,449,450], peptides [451], saccharides [443], and amino acids [442]. To enhance biocompatibility and transfection efficiency, some researchers use combinations of these molecules [445,447,448] or more advanced nanosystems [452,453]. Recently, carbon QDs (carbon dots, CD) have emerged as an alternative.
- Carbon-based nanoparticles leverage the chemical properties of carbon to create carrier NPs with sizes akin to many biological structures. Hybridization of sp2 carbon enables the production of various graphitic materials—such as nanodiamonds (3D), graphene (2D), nanotubes (1D), fullerenes, and quantum dots (0D)—which are used to fabricate NPs with diverse structures. These materials offer exceptional mechanical strength, high stability, a large surface area-to-volume ratio, unique optical features, excellent thermal and caloric conductivity, biocompatibility, ease of functionalization, and antibacterial properties [454,455]. For siRNA coupling, the primary options include: (i) carbon nanotubes, (ii) graphene oxide nanosheets, (iii) fullerenes, and (iv) carbon quantum dots.
Carbon nanotubes (CNTs). These are elongated tubular nanostructures, measuring 50–100 nm in length, made from graphite sheets derived from carbon atoms arranged in a hexagonal pattern. They exhibit high biocompatibility, excellent electrical and thermal conductivity, and are straightforward to synthesize and functionalize [455,456]. Their optical properties support combined applications in gene and photothermal therapy. CNTs can be produced as either single-walled (SWCNT) or multi-walled (MWCNT) varieties [428].
SWCNTs are primarily employed for siRNA delivery, with their surfaces functionalized through covalent bonding, hydrophobic interactions, or van der Waals forces [428,455,457,458,459]. The specific molecule used for CNT functionalization determines whether siRNA is attached via covalent or electrostatic bonds [457,458,459,460]. Amphiphilic PEG derivatives are commonly utilized because they also facilitate the attachment of various biomolecules for receptor recognition [458,461]. The techniques for siRNA coupling have already been detailed.
CNTs have a high loading capacity, and their tubular structure enables them to easily penetrate the plasma membrane, releasing siRNA into the cytosol through a mechanism that does not rely on endocytosis [455,459]. An example of effective CNT/siRNA application is the system developed by Siu et al. [458], who use SWCNTs covalently bonded with PEI-modified lipids, allowing siRNA to bind via electrostatic interactions. However, these systems have been shown to potentially cause cytotoxicity [461].
Graphene oxide nanoparticles (GON). These are nanosheets derived from graphene or graphite, with atoms in a hexagonal sp2-hybridized configuration. They also contain domains of sp3-hybridized carbon atoms [338]. GONs are produced through various methods, with the most common being chemical or thermal exfoliation of graphite oxide, followed by dispersion in water to form stable colloids [462]. Their physicochemical properties are unique, with hydrophilicity and biocompatibility surpassing those of graphene. Additionally, they exhibit high specific surface area, thermal and electrical conductivity, and optical transmittance [338,455,456], enabling the development of multimodal therapy and theranostic systems [445,463,464,465]. The surface of GONs can be easily functionalized to attach siRNA, as hydroxyl, carboxyl, epoxy, and carbonyl groups are highly accessible. Functionalization occurs via both covalent and non-covalent bonding [455,465,466,467,468,469]. Covalent bonding involves various reactions, with nucleophilic substitution of epoxy and carboxyl groups by molecules containing primary amines being one of the most common methods [466,468,469]. Non-covalent attachment primarily occurs through electrostatic interactions using positively charged polymers and peptides [468,470]. The process of coupling siRNA to the functionalized GONs follows the methods previously described.
GONs offer lower toxicity and higher loading capacity than other inorganic nanocarriers, and they can be integrated into various nanosystems to prepare hybrid nanoparticles [468].
Fullerenes. They are three-dimensional, mostly spherical structures composed of large, complex, and highly durable carbon molecules arranged in an alternating pattern of hexagons and pentagons. The most common fullerene is C60, formed by 60 carbon atoms connected by single bonds (12 pentagons) and double bonds (20 hexagons). Their size and non-polar nature enable them to easily penetrate cell membranes [456,471], making them highly suitable for drug delivery. Fullerenes can also be functionalized with molecules of different nature [341,471,472,473]. Additionally, their photoelectrochemical properties make them excellent candidates for photodynamic therapy (PDT) [341,456]. siRNA primarily binds to fullerenes through electrostatic interactions, which requires pre-cationizing the fullerene with molecules containing amine groups [341,473,474,475].
Carbon quantum dots (CDs). These are nanoparticles less than 10 nm in size, exhibiting optoelectronic properties similar to traditional quantum dots while offering greater biocompatibility and lower toxicity [445,469,476]. They are ideal for siRNA delivery. CDs consist of a carbon skeleton with sp2/sp3 hybrid [477,478]. Their solubility and biocompatibility surpass those of CNTs, GONs, and fullerenes, making them better suited for in-vivo applications [477]. They are produced through diverse low-cost methods, either by top-down techniques that break down larger NPs or by bottom [476,478,479]. Depending on the core structure—crystalline or amorphous—and surface functional groups, they are categorized into four groups. Graphene quantum dots (GQDs), derived from graphene or graphite, have a crystalline structure composed of one or more graphene sheets (up to 10), connected by chemical groups at the edges, and are rich in hydroxyl, carboxyl, carbonyl, and epoxy groups like GONs [469,480]. Carbon quantum dots (CQDs) are produced from small organic molecules or CNTs, feature a crystalline core, and are always spherical [480]. Carbon nanodots (CNDs), obtained from graphite, have an amorphous core and spherical shape, with a high degree of carbonization [478,480]. Carbonized polymer dots (CPDs) are hybrid nanoparticles formed by incomplete carbonization of polymers, with attached functional groups [480].
Like other carbon NPs, CDs are functionalized with groups such as amine, carboxyl, and hydroxyl to enable covalent and non-covalent linking of molecules. These molecules then bind to siRNA, primarily through electrostatic interactions [481,482,483]. The most commonly used molecules have already been described. Their choice for CD functionalization aimed at siRNA binding depends on the intended application—whether theranostic, multimodal, or combined with gene silencing [482,484].
- Mesoporous silica nanoparticles (MSNP). These are silica nanoparticles with various morphologies and numerous pores. Their structure, shape, nanoparticle size (50–200 nm), and pore size (2–50 nm) can vary depending on the production method [485,486,487]. They are highly stable under physiological conditions, biocompatible, biodegradable, low in toxicity, and considered safe for in vivo applications by the FDA [488,489]. Adjustable pore sizes enable the loading of small molecules into small nanoparticles [486]. Surface silanol groups (Si-OH) facilitate functionalization with amines, thiols, chlorides, phosphates, carboxyl groups, and others, allowing the attachment of a variety of molecules [488,489,490,491]. This versatility supports the development of controlled release systems [486], targeted therapy [489,490], stimuli-responsive systems [492,493], theranostics [494,495], and multimodal therapy [496,497,498]. They can be either conventional (MSN) or hollow (HMS), with different synthesis methods utilized depending on the specific application, as summarized in several publications [487,499].
siRNA can be incorporated into MSNPs either during their synthesis or afterward, with the latter being more common [81,486]. This can happen inside the pores or on the surface of the MSNPs. For pore incorporation, the MSNPs need to have large pores to allow siRNA, conjugated with various molecules, to enter [489]. However, surface coupling is generally preferred and can be achieved either covalently or via electrostatic interactions, using the reactions and conjugates previously described [489].
4.2.3. siRNA as a Tool for Multimodal Cancer Treatment
An important feature of siRNA carrier systems is their ability to support multimodal therapy, which is one of the most promising strategies in cancer treatment. The synergistic effect of combining two or more treatments is more effective than using them separately, as one therapy often boosts the other. This enables lower drug doses, reducing tissue and systemic toxicity while limiting resistance to chemotherapy and radiotherapy [81,492,500]. Current multimodal approaches combined with siRNA-based gene silencing include chemotherapy, photothermal therapy (PTT), photodynamic therapy (PDT), and radiotherapy. In this context, siRNA-mediated gene silencing targets multiple cancer hallmarks, such as cell cycle progression (PLK-1, KSP), evasion of apoptosis (Survivin, Bcl-2), DNA repair (MGMT, RAD51), tumor microenvironment remodeling (CXCR4, nerve growth factor), and immune evasion (PD-L1), collectively enhancing the antitumor effect.
An example of such approaches is the use of AuNP/siRNA NPs for gene therapy/PTT [501,502], cationic photosensitizing polymers conjugated with siRNA for gene therapy/PDT [503], as well as siRNA/radionuclide/SNEEDS nanosystems for radiotherapy/gene silencing/PDT [261]. Table 4 shows some delivery systems used for this purpose.

Table 4.
Representative siRNA carriers targeting tumor-promoting pathways in monotherapy or combination therapy.
Even when nanocarriers possess their “ideal” traits and demonstrate strong in vitro results, their success in clinical cancer treatment is not guaranteed. Several factors limit their therapeutic potential beyond laboratory conditions. These include: (i) the effects of the reticuloendothelial system and barriers like blood vessel endothelium and extracellular matrix [214]; (ii) the stability of siRNA nanocarriers and their degradation by nucleases and enzymes in the bloodstream; (iii) clearance by the mononuclear phagocytic system (MPS) and kidneys, which decreases tumor delivery [134,515]; (iv) toxicity from off-target gene silencing caused by nonspecific distribution; and (v) the challenge of targeting tumors specifically due to the tumor microenvironment’s complexity and receptor variety [134,516]. Additionally, technical issues like complex manufacturing processes that involve multiple synthesis and purification steps hamper scalability and regulatory approval. Variability between batches also poses obstacles for clinical translation compared to more controlled in vitro settings.
Some strategies for designing theranostic siRNA systems that combine effective gene silencing with the ability to monitor treatment in real time could involve functionalizing siRNA carriers with imaging agents. The nanocarrier system should respond to stimuli and act as a modular nanoplatform, allowing the incorporation of advanced imaging techniques and targeting ligand carriers [517].
In our view, the most clinically promising delivery system—offering the greatest potential to overcome the significant challenges of toxicity and immunogenicity in systemic siRNA delivery—is a sophisticated, theranostic platform that integrates several key features. This system must be capable of encapsulating siRNA (whether naked or in complexes) within its core while housing the imaging agent in its coating, ensuring maximum protection of the siRNA and high loading efficiency. Importantly, this design guarantees that the siRNA and imaging agent do not compete for space, optimizing functionality. Small inorganic nanoparticles can be used in this platform, but mainly as an additional therapeutic agent or imaging probe.
Furthermore, such platform should be able to recognize tumor cells with high internalization rates. Its size should be meticulously maintained between 100 and 200 nm, and it should be inherently “soft” to promote cellular uptake, making organic nanosystems the most promising choice. Combining lipid cores—capable of carrying other hydrophobic therapeutic agents—with biodegradable polymeric components on the surface creates a highly effective platform. These hybrid designs can incorporate pH-sensitive or membrane-disruptive features like the proton sponge effect to maximize endosomal escape, which is critical for efficacy.
A hydrophilic polymer surface minimizes undesirable corona effects and stabilizes colloidal suspension, while also allowing for a slightly positive net charge that enhances siRNA loading—even in complex form—and facilitates tumor cell uptake. This surface can be functionalized with specific groups to enable precise tumor targeting via a vector, further boosting therapeutic effectiveness.
Finally, the platform should be spheroidal, ideally a distorted sphere, as this shape ensures superior internalization and elicits a weaker immune response compared to more spherical forms. Overall, hybrid lipid/polymer organic systems stand out as the most compelling and versatile option, poised to revolutionize systemic siRNA delivery and dramatically improve clinical outcomes.
Ultimately, all variables involved in preparing a nanocarrier system need to be validated through repeated preclinical testing. Moreover, dose optimization offers the greatest potential for safe and effective systemic siRNA therapy [518].
5. siRNA-Targeted Genes Under Clinical Investigation
Cancer is a complex, heterogeneous disease caused by deregulated pathways that promote tumor initiation, progression, metastasis, and therapy resistance. Several candidates are currently being evaluated in clinical trials to inhibit key regulators, including serine/threonine kinases, receptor tyrosine kinases, oncoproteins, and signaling mediators. Table 5 lists representative therapeutic targets and their associated siRNA formulations in clinical trials, highlighting their biological functions, alterations in cancer, and the therapeutic benefits of targeting them.

Table 5.
siRNA therapeutic targets in clinical trials: biological roles and effects of inhibition.
As shown in Table 5, siRNA-based cancer therapies in clinical trials are designed either as monotherapies or in combination strategies. These approaches include (i) inhibition of tumor survival, (ii) inhibition of angiogenesis, and (iii) induction of apoptosis [551,552]. The most common strategy in siRNA formulations is targeting proteins that act as key effectors in signaling pathways that regulate tumor growth, survival, and invasion (e.g., PLK1, PKN3, EphA2, KRAS, TGF-β1, COX-2, BCL2L12, VEGF, GSTP) [553]. Notably, many of these targets are directly linked to therapy resistance, including PLK1 (TKM-080301 candidate), KRAS (siG12D LODER candidate), TGF-β1 and COX-2 (STP-705 candidate), BCL2L12 (NU-0129 candidate), VEGF and KSP (ALN-VSP02 candidate), and GSTP (NBF-06 candidate).
In cancers, oncogene overexpression, loss of tumor suppressors, and alterations in their regulatory networks promote dedifferentiation and tumor heterogeneity [554], which decreases therapeutic effectiveness due to compensatory signaling and ultimately worsens clinical outcomes [555,556]. Significant molecular barriers include mutations or deletions in the PI3K tumor suppressor phosphatase, mutations in receptor tyrosine kinase (RTK) pathways (e.g., EGFR, HER2, MET), amplification of oncogenic signals (such as mutant K-Ras activation and SMAD4 inactivation via TGFβ), buildup of transcription factors (c-Jun, Myc, Elk, ETS, ATF), and activation of the PI3K/AKT/mTOR and MAPK/ERK pathways. For example, blocking PI3K pathway activators can trigger compensatory signaling that promotes resistance or partial response to inhibitors [556].
Due to its intrinsic heterogeneity and molecular differences among patients, it cannot be effectively treated with single-pathway monotherapies, which often lead to resistance, recurrence, activation of compensatory net [557,558]. These issues highlight the need for combination therapies that target multiple signaling pathways simultaneously [558]. For example, therapeutic candidates such as STP-075 and ALN-VSP02 demonstrate this approach by targeting distinct molecular pathways. STP-075 inhibits TGF-β1 and COX-2, while ALN-VSP02 targets VEGF and KSP simultaneously (Table 5). Such strategies are poised to enhance personalized cancer treatment by tailoring therapies to a patient’s genetic profile. Integrating genomic profiling with siRNA-based therapies offers a compelling approach in precision oncology.
6. Conclusions
The increasing interest in siRNAs, driven by their success in treating various diseases including cancer, has spurred multidisciplinary research efforts. These range from using bioinformatics to design siRNAs that target specific genes to developing delivery systems that are both safe and effective for siRNA conjugates. Despite numerous clinical trials, no siRNA-based formulation has yet received approval for cancer treatment. Creating formulations that are both effective and safe for systemic cancer therapy is challenging, as it involves chemical modifications to both the siRNA and its carrier. These modifications must ensure high stability in blood, efficient cellular uptake, and successful endosomal escape of siRNA. Additionally, the system needs to be highly specific to avoid off-target effects, while minimizing toxicity and immunogenicity. Many candidates with promising in vitro results fail to reach in vivo viability due to these stringent requirements. Nonetheless, ongoing clinical trials suggest that in the near future, viable siRNA formulations for oncology will become available.
The current findings confirm that siRNA therapy holds considerable promise for cancer treatment. Evidence from combined therapies that enhance treatment outcomes, along with the development of theranostic systems for disease monitoring, indicates that siRNA as a treatment option is a promising field that will likely play an even greater role in future cancer therapies. Nevertheless, several critical challenges remain to be addressed. Ongoing research aims to streamline the production of nanosystems, enhance specificity to prevent uptake in non-target organs, and boost siRNA internalization and endosomal escape—all crucial for effective gene silencing. Although challenges remain with stability, specificity, and delivery, progress in formulation techniques, combination therapies, theranostics and patient’s genetic profile (for personalized medicine) boosts confidence that siRNAs will soon play a crucial role in precision oncology.
Author Contributions
Conceptualization: L.A.-L.; investigation: L.A.-L., A.E.-C., M.T.-N., E.M.-A., B.O.-G., R.O.-P., V.S.-M. and K.I.-O.; data curation: L.A.-L., M.T.-N. and K.I.-O.; writing—original draft preparation: L.A.-L., A.E.-C., M.T.-N., E.M.-A., B.O.-G., R.O.-P., V.S.-M. and K.I.-O.; writing—review and editing: L.A.-L., M.T.-N. and K.I.-O.; visualization: L.A.-L.; supervision, M.T.-N. and K.I.-O.; funding acquisition, L.A.-L. and K.I.-O. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Secretaría de Ciencias de la Universidad Autónoma del Estado de México/Secretariat of Science of the Autonomous University of the State of Mexico, grant number 7151/2024ECON.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
This work was carried out as a part of the activities of the national research network “Research on Pharmacy and Theranostics/Investigation en Farmacia y teranóstica” registered at Universidad Autónoma del Estado de México (UAEMex-6406/REDP2021). The critiques and comments on this work and the scientific discussions with Eunice Olivé-Álvarez were also very appreciated.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Morris, K.V. SiRNA-Mediated Transcriptional Gene Silencing: The Potential Mechanism and a Possible Role in the Histone Code. Cell Mol. Life Sci. 2005, 62, 3057–3066. [Google Scholar] [CrossRef]
- Morris, K.V.; Chan, S.W.L.; Jacobsen, S.E.; Looney, D.J. Small Interfering RNA-Induced Transcriptional Gene Silencing in Human Cells. Science (1979) 2004, 305, 1289–1292. [Google Scholar] [CrossRef]
- Ting, A.H.; Schuebel, K.E.; Herman, J.G.; Baylin, S.B. Short Double-Stranded RNA Induces Transcriptional Gene Silencing in Human Cancer Cells in the Absence of DNA Methylation. Nat. Genet. 2005, 37, 906–910. [Google Scholar] [CrossRef]
- Zhang, X.; Rossi, J.J. Phylogenetic Comparison of Small RNA-Triggered Transcriptional Gene Silencing. J. Biol. Chem. 2011, 286, 29443–29448. [Google Scholar] [CrossRef]
- Zhang, K.; Wang, Q.; Xie, Y.; Mor, G.; Sega, E.; Low, P.S.; Huang, Y. Receptor-Mediated Delivery of SiRNAs by Tethered Nucleic Acid Base-Paired Interactions. RNA 2008, 14, 577–583. [Google Scholar] [CrossRef]
- Weinberg, M.S.; Morris, K.V. Transcriptional Gene Silencing in Humans. Nucleic. Acids. Res. 2016, 44, 6505–6517. [Google Scholar] [CrossRef] [PubMed]
- Motamedi, H.; Ari, M.M.; Alvandi, A.; Abiri, R. Principle, Application and Challenges of Development SiRNA-Based Therapeutics against Bacterial and Viral Infections: A Comprehensive Review. Front. Microbiol. 2024, 15, 1393646. [Google Scholar] [CrossRef] [PubMed]
- Guo, F.; Li, Y.; Yu, W.; Fu, Y.; Zhang, J.; Cao, H. Recent Progress of Small Interfering RNA Delivery on the Market and Clinical Stage. Mol. Pharm. 2024, 21, 2081–2096. [Google Scholar] [CrossRef]
- Friedrich, M.; Aigner, A. Therapeutic SiRNA: State-of-the-Art and Future Perspectives. BioDrugs 2022, 36, 549–571. [Google Scholar] [CrossRef]
- Ebenezer, O.; Oyebamiji, A.K.; Olanlokun, J.O.; Tuszynski, J.A.; Wong, G.K.-S. Recent Update on SiRNA Therapeutics. Int. J. Mol. Sci. 2025, 26, 3456. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Zhong, L.; Weng, Y.; Peng, L.; Huang, Y.; Zhao, Y.; Liang, X.-J. Therapeutic SiRNA: State of the Art. Signal Transduct. Target. Ther. 2020, 5, 101. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Chen, B.; Gan, C.; Sun, H.; Zhang, J.; Feng, L. A Comprehensive Review of Small Interfering RNAs (SiRNAs): Mechanism, Therapeutic Targets, and Delivery Strategies for Cancer Therapy. Int. J. Nanomed. 2023, 18, 7605–7635. [Google Scholar] [CrossRef] [PubMed]
- Lisowiec-Wąchnicka, J.; Bartyś, N.; Pasternak, A. A Systematic Study on the Influence of Thermodynamic Asymmetry of 5′-Ends of SiRNA Duplexes in Relation to Their Silencing Potency. Sci. Rep. 2019, 9, 255. [Google Scholar] [CrossRef]
- Ye, X.; Huang, N.; Liu, Y.; Paroo, Z.; Huerta, C.; Li, P.; Chen, S.; Liu, Q.; Zhang, H. Structure of C3PO and Mechanism of Human RISC Activation. Nat. Struct. Mol. Biol. 2011, 18, 650–657. [Google Scholar] [CrossRef]
- Sahu, S.; Williams, L.; Perez, A.; Philip, F.; Caso, G.; Zurawsky, W.; Scarlata, S. Regulation of the Activity of the Promoter of RNA-Induced Silencing, C3PO. Protein Sci. 2017, 26, 1807–1818. [Google Scholar] [CrossRef]
- Dong, Y.; Siegwart, D.J.; Anderson, D.G. Strategies, Design, and Chemistry in SiRNA Delivery Systems. Adv. Drug Deliv. Rev. 2019, 144, 133–147. [Google Scholar] [CrossRef]
- Elbashir, S.M. Functional Anatomy of SiRNAs for Mediating Efficient RNAi in Drosophila Melanogaster Embryo Lysate. EMBO J. 2001, 20, 6877–6888. [Google Scholar] [CrossRef]
- Martinelli, D.D. Machine Learning for SiRNA Efficiency Prediction: A Systematic Review. Health Sci. Rev. 2024, 11, 100157. [Google Scholar] [CrossRef]
- Fakhr, E.; Zare, F.; Teimoori-Toolabi, L. Precise and Efficient SiRNA Design: A Key Point in Competent Gene Silencing. Cancer Gene Ther. 2016, 23, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Setten, R.L.; Rossi, J.J.; Han, S. The Current State and Future Directions of RNAi-Based Therapeutics. Nat. Rev. Drug Discov. 2019, 18, 421–446. [Google Scholar] [CrossRef]
- Lam, J.K.-W.; Liang, W.; Chan, H.-K. Pulmonary Delivery of Therapeutic SiRNA. Adv. Drug Deliv. Rev. 2012, 64, 1–15. [Google Scholar] [CrossRef]
- Lück, S.; Kreszies, T.; Strickert, M.; Schweizer, P.; Kuhlmann, M.; Douchkov, D. SiRNA-Finder (Si-Fi) Software for RNAi-Target Design and Off-Target Prediction. Front. Plant Sci. 2019, 10, 1023. [Google Scholar] [CrossRef]
- Birmingham, A.; Anderson, E.; Sullivan, K.; Reynolds, A.; Boese, Q.; Leake, D.; Karpilow, J.; Khvorova, A. A Protocol for Designing SiRNAs with High Functionality and Specificity. Nat. Protoc. 2007, 2, 2068–2078. [Google Scholar] [CrossRef]
- Piekna-Przybylska, D.; DiChiacchio, L.; Mathews, D.H.; Bambara, R.A. A Sequence Similar to TRNA3Lys Gene Is Embedded in HIV-1 U3–R and Promotes Minus-Strand Transfer. Nat. Struct. Mol. Biol. 2010, 17, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Mysara, M.; Garibaldi, J.M.; ElHefnawi, M. Mysirna-Designer: A Workflow for Efficient SiRNA Design. PLoS ONE 2011, 6, e25642. [Google Scholar] [CrossRef] [PubMed]
- Ayyagari, V.S. Design of SiRNA Molecules for Silencing of Membrane Glycoprotein, Nucleocapsid Phosphoprotein, and Surface Glycoprotein Genes of SARS-CoV2. J. Genet. Eng. Biotechnol. 2022, 20, 113. [Google Scholar] [CrossRef]
- Dar, S.A.; Gupta, A.K.; Thakur, A.; Kumar, M. SMEpred Workbench: A Web Server for Predicting Efficacy of Chemicallymodified SiRNAs. RNA Biol. 2016, 13, 1144–1151. [Google Scholar] [CrossRef]
- Dar, S.A.; Thakur, A.; Qureshi, A.; Kumar, M. SiRNAmod: A Database of Experimentally Validated Chemically Modified SiRNAs. Sci. Rep. 2016, 6, 20031. [Google Scholar] [CrossRef] [PubMed]
- Shtykalova, S.; Deviatkin, D.; Freund, S.; Egorova, A.; Kiselev, A. Non-Viral Carriers for Nucleic Acids Delivery: Fundamentals and Current Applications. Life 2023, 13, 903. [Google Scholar] [CrossRef]
- Paul, A.; Muralidharan, A.; Biswas, A.; Kamath, B.V.; Joseph, A.; Alex, A.T. SiRNA Therapeutics and Its Challenges: Recent Advances in Effective Delivery for Cancer Therapy. OpenNano 2022, 7, 100063. [Google Scholar] [CrossRef]
- Davis, S.M.; Sousa, J.; Vangjeli, L.; Hassler, M.R.; Echeverria, D.; Knox, E.; Turanov, A.A.; Alterman, J.F.; Khvorova, A. 2′-O-Methyl at 20-Mer Guide Strand 3′ Termini May Negatively Affect Target Silencing Activity of Fully Chemically Modified SiRNA. Mol. Ther. Nucleic. Acids. 2020, 21, 266–277. [Google Scholar] [CrossRef]
- Basila, M.; Kelley, M.L.; Smith, A.V.B. Minimal 2′-O-Methyl Phosphorothioate Linkage Modification Pattern of Synthetic Guide RNAs for Increased Stability and Efficient CRISPR-Cas9 Gene Editing Avoiding Cellular Toxicity. PLoS ONE 2017, 12, e0188593. [Google Scholar] [CrossRef]
- Gore, K.R.; Nawale, G.N.; Harikrishna, S.; Chittoor, V.G.; Pandey, S.K.; Höbartner, C.; Patankar, S.; Pradeepkumar, P.I. Synthesis, Gene Silencing, and Molecular Modeling Studies of 4′C-Aminomethyl-2′-O-Methyl Modified Small Interfering RNAs. J. Org. Chem. 2012, 77, 3233–3245. [Google Scholar] [CrossRef]
- Harp, J.M.; Guenther, D.C.; Bisbe, A.; Perkins, L.; Matsuda, S.; Bommineni, G.R.; Zlatev, I.; Foster, D.J.; Taneja, N.; Charisse, K.; et al. Structural Basis for the Synergy of 4ʹ- and 2ʹ-Modifications on SiRNA Nuclease Resistance, Thermal Stability and RNAi Activity. Nucleic. Acids. Res. 2018, 46, 8090–8104. [Google Scholar] [CrossRef]
- Song, X.; Wang, X.; Ma, Y.; Liang, Z.; Yang, Z.; Cao, H. Site-Specific Modification Using the 2′-Methoxyethyl Group Improves the Specificity and Activity of SiRNAs. Mol. Ther. Nucleic. Acids. 2017, 9, 242–250. [Google Scholar] [CrossRef] [PubMed]
- Ly, S.; Echeverria, D.; Sousa, J.; Khvorova, A. Single-Stranded Phosphorothioated Regions Enhance Cellular Uptake of Cholesterol-Conjugated SiRNA but Not Silencing Efficacy. Mol. Ther. Nucleic. Acids. 2020, 21, 991–1005. [Google Scholar] [CrossRef] [PubMed]
- Hassler, M.R.; Turanov, A.A.; Alterman, J.F.; Haraszti, R.A.; Coles, A.H.; Osborn, M.F.; Echeverria, D.; Nikan, M.; Salomon, W.E.; Roux, L.; et al. Comparison of Partially and Fully Chemically-Modified SiRNA in Conjugate-Mediated Delivery in Vivo. Nucleic. Acids. Res. 2018, 46, 2185–2196. [Google Scholar] [CrossRef]
- Czauderna, F.; Fechtner, M.; Dames, S.; Aygün, H.; Klippel, A.; Pronk, G.J.; Giese, K.; Kaufmann, J. Structural Variations and Stabilising Modifications of Synthetic SiRNAs in Mammalian Cells. Nucleic. Acids. Res. 2003, 31, 2705–2716. [Google Scholar] [CrossRef] [PubMed]
- Chernikov, I.V.; Vlassov, V.V.; Chernolovskaya, E.L. Current Development of SiRNA Bioconjugates: From Research to the Clinic. Front. Pharmacol. 2019, 10, 444. [Google Scholar] [CrossRef]
- Prakash, T.P.; Allerson, C.R.; Dande, P.; Vickers, T.A.; Sioufi, N.; Jarres, R.; Baker, B.F.; Swayze, E.E.; Griffey, R.H.; Bhat, B. Positional Effect of Chemical Modifications on Short Interference RNA Activity in Mammalian Cells. J. Med. Chem. 2005, 48, 4247–4253. [Google Scholar] [CrossRef]
- Gangopadhyay, S.; Gore, K.R. Advances in SiRNA Therapeutics and Synergistic Effect on SiRNA Activity Using Emerging Dual Ribose Modifications. RNA Biol. 2022, 19, 452–467. [Google Scholar] [CrossRef] [PubMed]
- Esparza-Garrido, R.; Velázquez-Flores, M. Activation of Toll-like Receptors by Non-coding RNAs and Their Fragments (Review). Mol. Med. Rep. 2025, 32, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Pallan, P.S.; Greene, E.M.; Jicman, P.A.; Pandey, R.K.; Manoharan, M.; Rozners, E.; Egli, M. Unexpected Origins of the Enhanced Pairing Affinity of 2′-Fluoro-Modified RNA. Nucleic. Acids. Res. 2011, 39, 3482–3495. [Google Scholar] [CrossRef]
- Ge, Q.; Dallas, A.; Ilves, H.; Shorenstein, J.; Behlke, M.A.; Johnston, B.H. Effects of Chemical Modification on the Potency, Serum Stability, and Immunostimulatory Properties of Short ShRNAs. RNA 2010, 16, 118–130. [Google Scholar] [CrossRef][Green Version]
- Cochran, M.; Arias, D.; Burke, R.; Chu, D.; Erdogan, G.; Hood, M.; Kovach, P.; Kwon, H.W.; Chen, Y.; Moon, M.; et al. Structure-Activity Relationship of Antibody-Oligonucleotide Conjugates: Evaluating Bioconjugation Strategies for Antibody-SiRNA Conjugates for Drug Development. J. Med. Chem. 2024, 67, 14852–14867. [Google Scholar] [CrossRef]
- Langkjær, N.; Pasternak, A.; Wengel, J. UNA (Unlocked Nucleic Acid): A Flexible RNA Mimic That Allows Engineering of Nucleic Acid Duplex Stability. Bioorg Med. Chem. 2009, 17, 5420–5425. [Google Scholar] [CrossRef]
- Snead, N.M.; Escamilla-Powers, J.R.; Rossi, J.J.; McCaffrey, A.P. 5′ Unlocked Nucleic Acid Modification Improves SiRNA Targeting. Mol. Ther. Nucleic. Acids. 2013, 2, e103. [Google Scholar] [CrossRef]
- Laursen, M.B.; Pakula, M.M.; Gao, S.; Fluiter, K.; Mook, O.R.; Baas, F.; Langklær, N.; Wengel, S.L.; Wengel, J.; Kjems, J.; et al. Utilization of Unlocked Nucleic Acid (UNA) to Enhance SiRNA Performance in Vitro and in Vivo. Mol. Biosyst. 2010, 6, 862–870. [Google Scholar] [CrossRef]
- Crooke, S.T.; Seth, P.P.; Vickers, T.A.; Liang, X. The Interaction of Phosphorothioate-Containing RNA Targeted Drugs with Proteins Is a Critical Determinant of the Therapeutic Effects of These Agents. J. Am. Chem. Soc. 2020, 142, 14754–14771. [Google Scholar] [CrossRef]
- Yang, X.; Sierant, M.; Janicka, M.; Peczek, L.; Martinez, C.; Hassell, T.; Li, N.; Li, X.; Wang, T.; Nawrot, B. Gene Silencing Activity of SiRNA Molecules Containing Phosphorodithioate Substitutions. ACS Chem. Biol. 2012, 7, 1214–1220. [Google Scholar] [CrossRef] [PubMed]
- Zharkov, T.D.; Markov, O.V.; Zhukov, S.A.; Khodyreva, S.N.; Kupryushkin, M.S. Influence of Combinations of Lipophilic and Phosphate Backbone Modifications on Cellular Uptake of Modified Oligonucleotides. Molecules 2024, 29, 452. [Google Scholar] [CrossRef]
- Hall, A.H.S.; Wan, J.; Spesock, A.; Sergueeva, Z.; Shaw, B.R.; Alexander, K.A. High Potency Silencing by Single-Stranded Boranophosphate SiRNA. Nucleic. Acids. Res. 2006, 34, 2773–2781. [Google Scholar] [CrossRef]
- Hall, A.H.S. RNA Interference Using Boranophosphate SiRNAs: Structure-Activity Relationships. Nucleic. Acids. Res. 2004, 32, 5991–6000. [Google Scholar] [CrossRef]
- Ryan, D.E.; Diamant-Levi, T.; Steinfeld, I.; Taussig, D.; Visal-Shah, S.; Thakker, S.; Lunstad, B.D.; Kaiser, R.J.; McCaffrey, R.; Ortiz, M.; et al. Phosphonoacetate Modifications Enhance the Stability and Editing Yields of Guide RNAs for Cas9 Editors. Biochemistry 2023, 62, 3512–3520. [Google Scholar] [CrossRef]
- Hawner, M.; Ducho, C. Cellular Targeting of Oligonucleotides by Conjugation with Small Molecules. Molecules 2020, 25, 5963. [Google Scholar] [CrossRef]
- Berk, C.; Civenni, G.; Wang, Y.; Steuer, C.; Catapano, C.V.; Hall, J. Pharmacodynamic and Pharmacokinetic Properties of Full Phosphorothioate Small Interfering RNAs for Gene Silencing in Vivo. Nucleic. Acid. Ther. 2021, 31, 237–244. [Google Scholar] [CrossRef]
- Crooke, S.T.; Wang, S.; Vickers, T.A.; Shen, W.; Liang, X. Cellular Uptake and Trafficking of Antisense Oligonucleotides. Nat. Biotechnol. 2017, 35, 230–237. [Google Scholar] [CrossRef]
- Migawa, M.T.; Shen, W.; Wan, W.B.; Vasquez, G.; Oestergaard, M.E.; Low, A.; De Hoyos, C.L.; Gupta, R.; Murray, S.; Tanowitz, M.; et al. Site-Specific Replacement of Phosphorothioate with Alkyl Phosphonate Linkages Enhances the Therapeutic Profile of Gapmer ASOs by Modulating Interactions with Cellular Proteins. Nucleic. Acids. Res. 2019, 47, 5465–5479. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.; De Hoyos, C.L.; Migawa, M.T.; Vickers, T.A.; Sun, H.; Low, A.; Bell, T.A.; Rahdar, M.; Mukhopadhyay, S.; Hart, C.E.; et al. Chemical Modification of PS-ASO Therapeutics Reduces Cellular Protein-Binding and Improves the Therapeutic Index. Nat. Biotechnol. 2019, 37, 640–650. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Sato, K.; Wada, T. Solid-Phase Synthesis of Boranophosphate/Phosphorothioate/Phosphate Chimeric Oligonucleotides and Their Potential as Antisense Oligonucleotides. J. Org. Chem. 2022, 87, 3895–3909. [Google Scholar] [CrossRef] [PubMed]
- Duschmalé, J.; Schäublin, A.; Funder, E.; Schmidt, S.; Kiełpiński, Ł.J.; Nymark, H.; Jensen, K.; Koch, T.; Duschmalé, M.; Koller, E.; et al. Investigating Discovery Strategies and Pharmacological Properties of Stereodefined Phosphorodithioate LNA Gapmers. Mol. Ther. Nucleic. Acids. 2022, 29, 176–188. [Google Scholar] [CrossRef]
- Wu, S.Y.; Yang, X.; Gharpure, K.M.; Hatakeyama, H.; Egli, M.; McGuire, M.H.; Nagaraja, A.S.; Miyake, T.M.; Rupaimoole, R.; Pecot, C.V.; et al. 2′-OMe-Phosphorodithioate-Modified SiRNAs Show Increased Loading into the RISC Complex and Enhanced Anti-Tumour Activity. Nat. Commun. 2014, 5, 3455. [Google Scholar] [CrossRef]
- Elkayam, E.; Parmar, R.; Brown, C.R.; Willoughby, J.L.; Theile, C.S.; Manoharan, M.; Joshua-Tor, L. SiRNA Carrying an (E)-Vinylphosphonate Moiety at the 5′ End of the Guide Strand Augments Gene Silencing by Enhanced Binding to Human Argonaute-2. Nucleic. Acids. Res. 2017, 45, 3528–3536. [Google Scholar] [CrossRef] [PubMed]
- Parmar, R.; Willoughby, J.L.S.; Liu, J.; Foster, D.J.; Brigham, B.; Theile, C.S.; Charisse, K.; Akinc, A.; Guidry, E.; Pei, Y.; et al. 5′-(E)-Vinylphosphonate: A Stable Phosphate Mimic Can Improve the RNAi Activity of SiRNA-GalNAc Conjugates. ChemBioChem 2016, 17, 985–989. [Google Scholar] [CrossRef]
- Parmar, R.G.; Brown, C.R.; Matsuda, S.; Willoughby, J.L.S.; Theile, C.S.; Charissé, K.; Foster, D.J.; Zlatev, I.; Jadhav, V.; Maier, M.A.; et al. Facile Synthesis, Geometry, and 2′-Substituent-Dependent in Vivo Activity of 5′-(E)- and 5′-(Z)-Vinylphosphonate-Modified SiRNA Conjugates. J. Med. Chem. 2018, 61, 734–744. [Google Scholar] [CrossRef]
- Tsubaki, K.; Hammill, M.L.; Varley, A.J.; Kitamura, M.; Okauchi, T.; Desaulniers, J.-P. Synthesis and Evaluation of Neutral Phosphate Triester Backbone-Modified SiRNAs. ACS Med. Chem. Lett. 2020, 11, 1457–1462. [Google Scholar] [CrossRef] [PubMed]
- Sipa, K.; Sochacka, E.; Kazmierczak-Baranska, J.; Maszewska, M.; Janicka, M.; Nowak, G.; Nawrot, B. Effect of Base Modifications on Structure, Thermodynamic Stability, and Gene Silencing Activity of Short Interfering RNA. RNA 2007, 13, 1301–1316. [Google Scholar] [CrossRef] [PubMed]
- Peacock, H.; Kannan, A.; Beal, P.A.; Burrows, C.J. Chemical Modification of SiRNA Bases to Probe and Enhance RNA Interference. J. Org. Chem. 2011, 76, 7295–7300. [Google Scholar] [CrossRef]
- Shinohara, F.; Oashi, T.; Harumoto, T.; Nishikawa, T.; Takayama, Y.; Miyagi, H.; Takahashi, Y.; Nakajima, T.; Sawada, T.; Koda, Y.; et al. SiRNA Potency Enhancement via Chemical Modifications of Nucleotide Bases at the 5 ′-End of the SiRNA Guide Strand. RNA 2021, 27, 163–173. [Google Scholar] [CrossRef]
- Valenzuela, R.A.P.; Suter, S.R.; Ball-Jones, A.A.; Ibarra-Soza, J.M.; Zheng, Y.; Beal, P.A. Base Modification Strategies to Modulate Immune Stimulation by an SiRNA. ChemBioChem 2015, 16, 262–267. [Google Scholar] [CrossRef]
- Li, Q.; Dong, M.; Chen, P. Advances in Structural-Guided Modifications of SiRNA. Bioorg. Med. Chem. 2024, 110, 117825. [Google Scholar] [CrossRef]
- Chernikov, I.V.; Ponomareva, U.A.; Chernolovskaya, E.L. Structural Modifications of SiRNA Improve Its Performance In Vivo. Int. J. Mol. Sci. 2023, 24, 956. [Google Scholar] [CrossRef]
- Li, Z.; Wang, X.; Zhou, X.; Wang, J.; Guan, Z.; Yang, Z. Optimization in Chemical Modification of Single-Stranded SiRNA Encapsulated by Neutral Cytidinyl/Cationic Lipids. Front. Chem. 2022, 10, 843181. [Google Scholar] [CrossRef]
- Bulcha, J.T.; Wang, Y.; Ma, H.; Tai, P.W.L.; Gao, G. Viral Vector Platforms within the Gene Therapy Landscape. Signal Transduct. Target. Ther. 2021, 6, 53. [Google Scholar] [CrossRef]
- Lundstrom, K. Viral Vectors in Gene Therapy: Where Do We Stand in 2023? Viruses 2023, 15, 698. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.W.; Choi, J.; Kim, E.H.; Choi, J.; Kim, S.H.; Yang, Y. Design of SiRNA Bioconjugates for Efficient Control of Cancer-Associated Membrane Receptors. ACS Omega 2023, 8, 36435–36448. [Google Scholar] [CrossRef]
- Lee, J.W.; Choi, J.; Choi, Y.; Kim, K.; Yang, Y.; Kim, S.H.; Yoon, H.Y.; Kwon, I.C. Molecularly Engineered SiRNA Conjugates for Tumor-Targeted RNAi Therapy. J. Control. Release 2022, 351, 713–726. [Google Scholar] [CrossRef] [PubMed]
- Tai, W. Current Aspects of SiRNA Bioconjugate for In Vitro and In Vivo Delivery. Molecules 2019, 24, 2211. [Google Scholar] [CrossRef]
- Im, S.-W.; Pravinsagar, P.; Im, S.-R.; Jang, Y.-J. Variable Heavy Chain Domain Derived from a Cell-Penetrating Anti-DNA Monoclonal Antibody for the Intracellular Delivery of Biomolecules. Immunol. Investig. 2017, 46, 500–517. [Google Scholar] [CrossRef] [PubMed]
- Cao, W.; Li, R.; Pei, X.; Chai, M.; Sun, L.; Huang, Y.; Wang, J.; Barth, S.; Yu, F.; He, H. Antibody–SiRNA Conjugates (ARC): Emerging SiRNA Drug Formulation. Med. Drug Discov. 2022, 15, 100128. [Google Scholar] [CrossRef]
- Li, G.; Moellering, R.E. A Concise, Modular Antibody–Oligonucleotide Conjugation Strategy Based on Disuccinimidyl Ester Activation Chemistry. ChemBioChem 2019, 20, 1599–1605. [Google Scholar] [CrossRef]
- Dugal-Tessier, J.; Thirumalairajan, S.; Jain, N. Antibody-Oligonucleotide Conjugates: A Twist to Antibody-Drug Conjugates. J. Clin. Med. 2021, 10, 838. [Google Scholar] [CrossRef]
- Bäumer, N.; Berdel, W.E.; Bäumer, S. Immunoprotein-Mediated SiRNA Delivery. Mol. Pharm. 2017, 14, 1339–1351. [Google Scholar] [CrossRef]
- Ma, Y.; Kowolik, C.M.; Swiderski, P.M.; Kortylewski, M.; Yu, H.; Horne, D.A.; Jove, R.; Caballero, O.L.; Simpson, A.J.G.; Lee, F.T.; et al. Humanized Lewis-Y Specific Antibody Based Delivery of STAT3 SiRNA. ACS Chem. Biol. 2011, 6, 962–970. [Google Scholar] [CrossRef]
- Cuellar, T.L.; Barnes, D.; Nelson, C.; Tanguay, J.; Yu, S.-F.; Wen, X.; Scales, S.J.; Gesch, J.; Davis, D.; van Brabant Smith, A.; et al. Systematic Evaluation of Antibody-Mediated SiRNA Delivery Using an Industrial Platform of THIOMAB–SiRNA Conjugates. Nucleic. Acids. Res. 2015, 43, 1189–1203. [Google Scholar] [CrossRef] [PubMed]
- Hammond, S.M.; Abendroth, F.; Goli, L.; Stoodley, J.; Burrell, M.; Thom, G.; Gurrell, I.; Ahlskog, N.; Gait, M.J.; Wood, M.J.A.; et al. Antibody-Oligonucleotide Conjugate Achieves CNS Delivery in Animal Models for Spinal Muscular Atrophy. JCI Insight 2022, 7, e154142. [Google Scholar] [CrossRef] [PubMed]
- Sugo, T.; Terada, M.; Oikawa, T.; Miyata, K.; Nishimura, S.; Kenjo, E.; Ogasawara-Shimizu, M.; Makita, Y.; Imaichi, S.; Murata, S.; et al. Development of Antibody-SiRNA Conjugate Targeted to Cardiac and Skeletal Muscles. J. Control. Release 2016, 237, 1–13. [Google Scholar] [CrossRef]
- Yu, Z.; Zhang, X.; Pei, X.; Cao, W.; Ye, J.; Wang, J.; Sun, L.; Yu, F.; Wang, J.; Li, N.; et al. Antibody-SiRNA Conjugates (ARCs) Using Multifunctional Peptide as a Tumor Enzyme Cleavable Linker Mediated Effective Intracellular Delivery of SiRNA. Int. J. Pharm. 2021, 606, 120940. [Google Scholar] [CrossRef] [PubMed]
- Dovgan, I.; Koniev, O.; Kolodych, S.; Wagner, A. Antibody–Oligonucleotide Conjugates as Therapeutic, Imaging, and Detection Agents. Bioconjug. Chem. 2019, 30, 2483–2501. [Google Scholar] [CrossRef]
- Sreedurgalakshmi, K.; Srikar, R.; Harikrishnan, K.; Srinivasan, L.; Rajkumari, R. Cetuximab–SiRNA Conjugate Linked Through Cationized Gelatin Knocks Down KRAS G12C Mutation in NSCLC Sensitizing the Cells Toward Gefitinib. Technol. Cancer. Res. Treat. 2021, 20, 15330338211041453. [Google Scholar] [CrossRef]
- Asiaei, S.; Smith, B.; Nieva, P. Enhancing Conjugation Rate of Antibodies to Carboxylates: Numerical Modeling of Conjugation Kinetics in Microfluidic Channels and Characterization of Chemical over-Exposure in Conventional Protocols by Quartz Crystal Microbalance. Biomicrofluidics 2015, 9, 024122. [Google Scholar] [CrossRef]
- Nanna, A.R.; Kel’in, A.V.; Theile, C.; Pierson, J.M.; Voo, Z.X.; Garg, A.; Nair, J.K.; Maier, M.A.; Fitzgerald, K.; Rader, C. Generation and Validation of Structurally Defined Antibody-SiRNA Conjugates. Nucleic. Acids. Res. 2020, 48, 5281–5293. [Google Scholar] [CrossRef] [PubMed]
- Satake, N.; Duong, C.; Yoshida, S.; Oestergaard, M.; Chen, C.; Peralta, R.; Guo, S.; Seth, P.P.; Li, Y.; Beckett, L.; et al. Novel Targeted Therapy for Precursor B-Cell Acute Lymphoblastic Leukemia: Anti-CD22 Antibody-MXD3 Antisense Oligonucleotide Conjugate. Mol. Med. 2016, 22, 632–642. [Google Scholar] [CrossRef]
- Honcharenko, D.; Druceikaite, K.; Honcharenko, M.; Bollmark, M.; Tedebark, U.; Strömberg, R. New Alkyne and Amine Linkers for Versatile Multiple Conjugation of Oligonucleotides. ACS Omega 2021, 6, 579–593. [Google Scholar] [CrossRef]
- Perrone, D.; Marchesi, E.; Preti, L.; Navacchia, M.L. Modified Nucleosides, Nucleotides and Nucleic Acids via Click Azide-Alkyne Cycloaddition for Pharmacological Applications. Molecules 2021, 26, 3100. [Google Scholar] [CrossRef]
- Kumar, P.; Ban, H.-S.; Kim, S.-S.; Wu, H.; Pearson, T.; Greiner, D.L.; Laouar, A.; Yao, J.; Haridas, V.; Habiro, K.; et al. T Cell-Specific SiRNA Delivery Suppresses HIV-1 Infection in Humanized Mice. Cell 2008, 134, 577–586. [Google Scholar] [CrossRef] [PubMed]
- Bäumer, N.; Rehkämper, J.; Appel, N.; Terheyden, L.; Hartmann, W.; Wardelmann, E.; Buchholz, F.; Müller-Tidow, C.; Berdel, W.E.; Bäumer, S. Downregulation of PIK3CA via Antibody-EsiRNA-Complexes Suppresses Human Xenograft Tumor Growth. PLoS ONE 2018, 13, e0200163. [Google Scholar] [CrossRef]
- Shi, S.-J.; Wang, L.-J.; Han, D.-H.; Wu, J.-H.; Jiao, D.; Zhang, K.-L.; Chen, J.-W.; Li, Y.; Yang, F.; Zhang, J.-L.; et al. Therapeutic Effects of Human Monoclonal PSMA Antibody-Mediated TRIM24 SiRNA Delivery in PSMA-Positive Castration-Resistant Prostate Cancer. Theranostics 2019, 9, 1247–1263. [Google Scholar] [CrossRef]
- Bäumer, N.; Tiemann, J.; Scheller, A.; Meyer, T.; Wittmann, L.; Suburu, M.E.G.; Greune, L.; Peipp, M.; Kellmann, N.; Gumnior, A.; et al. Targeted SiRNA Nanocarrier: A Platform Technology for Cancer Treatment. Oncogene 2022, 41, 2210–2224. [Google Scholar] [CrossRef] [PubMed]
- Xia, C.F.; Boado, R.J.; Pardridge, W.M. Antibody-Mediated Targeting of SiRNA via the Human Insulin Receptor Using Avidin-Biotin Technology. Mol. Pharm. 2009, 6, 747–751. [Google Scholar] [CrossRef]
- Xia, C.F.; Zhang, Y.; Zhang, Y.; Boado, R.J.; Pardridge, W.M. Intravenous SiRNA of Brain Cancer with Receptor Targeting and Avidin-Biotin Technology. Pharm. Res. 2007, 24, 2309–2316. [Google Scholar] [CrossRef]
- Falato, L.; Gestin, M.; Langel, Ü. Cell-Penetrating Peptides Delivering SiRNAs: An Overview. In Design and Delivery of SiRNA Therapeutics; Methods in Molecular Biology; Humana Press: New York, NY, USA, 2021; Volume 2282, pp. 329–352. [Google Scholar]
- Lopuszynski, J.; Agrawal, V.; Zahid, M. Tissue-Specific Cell Penetrating Peptides for Targeted Delivery of Small Interfering RNAs. Med. Res. Arch. 2022, 10, 2998. [Google Scholar] [CrossRef]
- Walrant, A.; Cardon, S.; Burlina, F.; Sagan, S. Membrane Crossing and Membranotropic Activity of Cell-Penetrating Peptides: Dangerous Liaisons? Acc. Chem. Res. 2017, 50, 2968–2975. [Google Scholar] [CrossRef] [PubMed]
- Sajid, M.I.; Mandal, D.; El-Sayed, N.S.; Lohan, S.; Moreno, J.; Tiwari, R.K. Oleyl Conjugated Histidine-Arginine Cell-Penetrating Peptides as Promising Agents for SiRNA Delivery. Pharmaceutics 2022, 14, 881. [Google Scholar] [CrossRef]
- Jagrosse, M.L.; Baliga, U.K.; Jones, C.W.; Russell, J.J.; García, C.I.; Najar, R.A.; Rahman, A.; Dean, D.A.; Nilsson, B.L. Impact of Peptide Sequence on Functional SiRNA Delivery and Gene Knockdown with Cyclic Amphipathic Peptide Delivery Agents. Mol. Pharm. 2023, 20, 6090–6103. [Google Scholar] [CrossRef] [PubMed]
- Ruseska, I.; Zimmer, A. Internalization Mechanisms of Cell-Penetrating Peptides. Beilstein J. Nanotechnol. 2020, 11, 101–123. [Google Scholar] [CrossRef]
- Beloor, J.; Zeller, S.; Choi, C.S.; Lee, S.-K.; Kumar, P. Cationic Cell-Penetrating Peptides as Vehicles for SiRNA Delivery. Ther. Deliv. 2015, 6, 491–507. [Google Scholar] [CrossRef]
- Moschos, S.A.; Jones, S.W.; Perry, M.M.; Williams, A.E.; Erjefalt, J.S.; Turner, J.J.; Barnes, P.J.; Sproat, B.S.; Gait, M.J.; Lindsay, M.A. Lung Delivery Studies Using SiRNA Conjugated to TAT(48-60) and Penetratin Reveal Peptide Induced Reduction in Gene Expression and Induction of Innate Immunity. Bioconjug. Chem. 2007, 18, 1450–1459. [Google Scholar] [CrossRef]
- Tai, W.; Gao, X. Functional Peptides for SiRNA Delivery. Adv. Drug Deliv. Rev. 2017, 110–111, 157–168. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Huang, Y.; Liu, Y.; Chen, Y.; Jin, H.; Zheng, Y.; Du, Q.; Yang, Z.; Zhang, L. Synthesis and Biological Evaluation of Peptide-SiRNA Conjugates with Phosphodiester Unit as Linker. Sci. China Chem. 2013, 56, 1542–1549. [Google Scholar] [CrossRef]
- Desale, K.; Kuche, K.; Jain, S. Cell-Penetrating Peptides (CPPs): An Overview of Applications for Improving the Potential of Nanotherapeutics. Biomater. Sci. 2021, 9, 1153–1188. [Google Scholar] [CrossRef]
- Muratovska, A.; Eccles, M.R. Conjugate for Efficient Delivery of Short Interfering RNA (SiRNA) into Mammalian Cells. FEBS Lett. 2004, 558, 63–68. [Google Scholar] [CrossRef]
- Yu, Z.; Ye, J.; Pei, X.; Sun, L.; Liu, E.; Wang, J.; Huang, Y.; Lee, S.J.; He, H. Improved Method for Synthesis of Low Molecular Weight Protamine–SiRNA Conjugate. Acta Pharm. Sin. B 2018, 8, 116–126. [Google Scholar] [CrossRef]
- Lin, L.; Chi, J.; Yan, Y.; Luo, R.; Feng, X.; Zheng, Y.; Xian, D.; Li, X.; Quan, G.; Liu, D.; et al. Membrane-Disruptive Peptides/Peptidomimetics-Based Therapeutics: Promising Systems to Combat Bacteria and Cancer in the Drug-Resistant Era. Acta Pharm. Sin. B 2021, 11, 2609–2644. [Google Scholar] [CrossRef] [PubMed]
- Pan, R.; Xu, W.; Yuan, F.; Chu, D.; Ding, Y.; Chen, B.; Jafari, M.; Yuan, Y.; Chen, P. A Novel Peptide for Efficient SiRNA Delivery in Vitro and Therapeutics in Vivo. Acta Biomater. 2015, 21, 74–84. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.; Dussouillez, C.; Padilla, B.; Frisch, B.; Mason, A.J.; Kichler, A. Design of a New Cell Penetrating Peptide for DNA, SiRNA and MRNA Delivery. J. Gene Med. 2022, 24, e3401. [Google Scholar] [CrossRef]
- He, S.; Cen, B.; Liao, L.; Wang, Z.; Qin, Y.; Wu, Z.; Liao, W.; Zhang, Z.; Ji, A. A Tumor-Targeting CRGD-EGFR SiRNA Conjugate and Its Anti-Tumor Effect on Glioblastoma in Vitro and in Vivo. Drug Deliv. 2017, 24, 471–481. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Xu, S.; Xiang, T.; Liu, H.; Chen, L.; Jiang, B.; Yao, J.; Zhu, H.; Hu, R.; Chen, Z. Multifunctional Building Elements for the Construction of Peptide Drug Conjugates. Eng. Regen. 2022, 3, 92–109. [Google Scholar] [CrossRef]
- Zeller, S.; Choi, C.S.; Uchil, P.D.; Ban, H.-S.; Siefert, A.; Fahmy, T.M.; Mothes, W.; Lee, S.-K.; Kumar, P. Attachment of Cell-Binding Ligands to Arginine-Rich Cell-Penetrating Peptides Enables Cytosolic Translocation of Complexed SiRNA. Chem. Biol. 2015, 22, 50–62. [Google Scholar] [CrossRef]
- Cen, B.; Wei, Y.; Huang, W.; Teng, M.; He, S.; Li, J.; Wang, W.; He, G.; Bai, X.; Liu, X.; et al. An Efficient Bivalent Cyclic RGD-PIK3CB SiRNA Conjugate for Specific Targeted Therapy against Glioblastoma In Vitro and In Vivo. Mol. Ther. Nucleic. Acids. 2018, 13, 220–232. [Google Scholar] [CrossRef]
- Huang, Y.; Wang, X.; Huang, W.; Cheng, Q.; Zheng, S.; Guo, S.; Cao, H.; Liang, X.J.; Du, Q.; Liang, Z. Systemic Administration of SiRNA via CRGD-Containing Peptide. Sci. Rep. 2015, 5, 12458. [Google Scholar] [CrossRef]
- Juretić, D. Designed Multifunctional Peptides for Intracellular Targets. Antibiotics 2022, 11, 1196. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Liu, E.; Gong, J.; Wang, J.; Huang, Y.; He, H.; Yang, V.C. High-Yield Synthesis of Monomeric LMWP(CPP)-SiRNA Covalent Conjugate for Effective Cytosolic Delivery of SiRNA. Theranostics 2017, 7, 2495–2508. [Google Scholar] [CrossRef] [PubMed]
- Park, J.W.; Bang, E.-K.; Jeon, E.M.; Kim, B.H. Complexation and Conjugation Approaches to Evaluate SiRNA Delivery Using Cationic, Hydrophobic and Amphiphilic Peptides. Org. Biomol. Chem. 2012, 10, 96–102. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, X.F.; Chen, Y.; Zhang, L.H.; Yang, Z.J. A Solid-Phase Method for Peptide-SiRNA Covalent Conjugates Based on Click Chemistry. Medchemcomm 2012, 3, 506–511. [Google Scholar] [CrossRef]
- Matsubara, M.; Honda, K.; Ozaki, K.; Kajino, R.; Kakisawa, Y.; Maeda, Y.; Ueno, Y. Synthesis of SiRNAs Incorporated with Cationic Peptides R8G7 and R8A7 and the Effect of the Modifications on SiRNA Properties. RSC Adv. 2020, 10, 34815–34824. [Google Scholar] [CrossRef]
- Smidt, J.M.; Lykke, L.; Stidsen, C.E.; Pristovšek, N.; Gothelf, K.V. Synthesis of Peptide–SiRNA Conjugates via Internal Sulfonylphosphoramidate Modifications and Evaluation of Their in Vitro Activity. Nucleic. Acids. Res. 2024, 52, 49–58. [Google Scholar] [CrossRef]
- Ando, Y.; Nakazawa, H.; Miura, D.; Otake, M.; Umetsu, M. Enzymatic Ligation of an Antibody and Arginine 9 Peptide for Efficient and Cell-Specific SiRNA Delivery. Sci. Rep. 2021, 11, 10035. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.W.; Kim, N.Y.; Choi, Y.B.; Park, S.H.; Yang, J.M.; Shin, S. RNA Interference in Vitro and in Vivo Using an Arginine Peptide/SiRNA Complex System. J. Control. Release 2010, 143, 335–343. [Google Scholar] [CrossRef]
- Maeda, Y.; Iwata Hara, R.; Nishina, K.; Yoshida-Tanaka, K.; Sakamoto, T.; Yokota, T.; Wada, T. Artificial Cationic Peptides That Increase Nuclease Resistance of SiRNA without Disturbing RNAi Activity. Nucleosides Nucleotides Nucleic Acids 2019, 38, 338–348. [Google Scholar] [CrossRef] [PubMed]
- Crombez, L.; Aldrian-Herrada, G.; Konate, K.; Nguyen, Q.N.; McMaster, G.K.; Brasseur, R.; Heitz, F.; Divita, G. A New Potent Secondary Amphipathic Cell-Penetrating Peptide for SiRNA Delivery into Mammalian Cells. Mol. Ther. 2009, 17, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Crowet, J.M.; Lins, L.; Deshayes, S.; Divita, G.; Morris, M.; Brasseur, R.; Thomas, A. Modeling of Non-Covalent Complexes of the Cell-Penetrating Peptide CADY and Its SiRNA Cargo. Biochim. Biophys. Acta Biomembr. 2013, 1828, 499–509. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ali Zaidi, S.S.; Fatima, F.; Ali Zaidi, S.A.; Zhou, D.; Deng, W.; Liu, S. Engineering SiRNA Therapeutics: Challenges and Strategies. J. Nanobiotechnol. 2023, 21, 381. [Google Scholar] [CrossRef]
- Crombez, L.; Divita, G. A Non-Covalent Peptide-Based Strategy for SiRNA Delivery. In Methods in Molecular Biology; Humana Press Inc.: Totowa, NJ, USA, 2011; Volume 683, pp. 349–360. [Google Scholar]
- Wang, F.; Gao, L.; Meng, L.Y.; Xie, J.M.; Xiong, J.W.; Luo, Y. A Neutralized Noncharged Polyethylenimine-Based System for Efficient Delivery of SiRNA into Heart without Toxicity. ACS Appl. Mater. Interfaces 2016, 8, 33529–33538. [Google Scholar] [CrossRef]
- Farra, R.; Musiani, F.; Perrone, F.; Čemažar, M.; Kamenšek, U.; Tonon, F.; Abrami, M.; Ručigaj, A.; Grassi, M.; Pozzato, G.; et al. Polymer-Mediated Delivery of SiRNAs to Hepatocellular Carcinoma: Variables Affecting Specificity and Effectiveness. Molecules 2018, 23, 777. [Google Scholar] [CrossRef] [PubMed]
- Patil, Y.; Panyam, J. Polymeric Nanoparticles for SiRNA Delivery and Gene Silencing. Int. J. Pharm. 2009, 367, 195–203. [Google Scholar] [CrossRef]
- Cavallaro, G.; Sardo, C.; Craparo, E.F.; Porsio, B.; Giammona, G. Polymeric Nanoparticles for SiRNA Delivery: Production and Applications. Int. J. Pharm. 2017, 525, 313–333. [Google Scholar] [CrossRef]
- Zhang, L.; Lou, W.; Wang, J. Advances in Nucleic Acid Therapeutics: Structures, Delivery Systems, and Future Perspectives in Cancer Treatment. Clin. Exp. Med. 2024, 24, 200. [Google Scholar] [CrossRef]
- Guţoaia, A.; Schuster, L.; Margutti, S.; Laufer, S.; Schlosshauer, B.; Krastev, R.; Stoll, D.; Hartmann, H. Fine-Tuned PEGylation of Chitosan to Maintain Optimal SiRNA-Nanoplex Bioactivity. Carbohydr. Polym. 2016, 143, 25–34. [Google Scholar] [CrossRef]
- Jiang, Z.; Thayumanavan, S. Noncationic Material Design for Nucleic Acid Delivery. Adv. Ther. 2020, 3, 1900206. [Google Scholar] [CrossRef]
- Zamboni, C.G.; Kozielski, K.L.; Vaughan, H.J.; Nakata, M.M.; Kim, J.; Higgins, L.J.; Pomper, M.G.; Green, J.J. Polymeric Nanoparticles as Cancer-Specific DNA Delivery Vectors to Human Hepatocellular Carcinoma. J. Control. Release 2017, 263, 18–28. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Mundra, V.; Peng, Y.; Wang, Y.; Tan, C.; Mahato, R.I. Pharmacokinetics and Biodistribution of Polymeric Micelles Containing MiRNA and Small-Molecule Drug in Orthotopic Pancreatic Tumor-Bearing Mice. Theranostics 2018, 8, 4033–4049. [Google Scholar] [CrossRef]
- Ahmad, A.; Khan, J.M. PH-Sensitive Endosomolytic Peptides in Gene and Drug Delivery: Endosomal Escape and Current Challenges. J. Drug Deliv. Sci. Technol. 2022, 76, 103786. [Google Scholar] [CrossRef]
- Xie, L.; Liu, R.; Chen, X.; He, M.; Zhang, Y.; Chen, S. Micelles Based on Lysine, Histidine, or Arginine: Designing Structures for Enhanced Drug Delivery. Front. Bioeng. Biotechnol. 2021, 9, 744657. [Google Scholar] [CrossRef]
- Dutta, K.; Das, R.; Medeiros, J.; Kanjilal, P.; Thayumanavan, S. Charge-Conversion Strategies for Nucleic Acid Delivery. Adv. Funct. Mater. 2021, 31, 2011103. [Google Scholar] [CrossRef]
- Guo, J.; Cheng, W.P.; Gu, J.; Ding, C.; Qu, X.; Yang, Z.; O’Driscoll, C. Systemic Delivery of Therapeutic Small Interfering RNA Using a PH-Triggered Amphiphilic Poly-l-Lysine Nanocarrier to Suppress Prostate Cancer Growth in Mice. Eur. J. Pharm. Sci. 2012, 45, 521–532. [Google Scholar] [CrossRef]
- Gorzkiewicz, M.; Kopeć, O.; Janaszewska, A.; Konopka, M.; Pędziwiatr-Werbicka, E.; Tarasenko, I.I.; Bezrodnyi, V.V.; Neelov, I.M.; Klajnert-Maculewicz, B. Poly(Lysine) Dendrimers Form Complexes with Sirna and Provide Its Effcient Uptake by Myeloid Cells: Model Studies for Therapeutic Nucleic Acid Delivery. Int. J. Mol. Sci. 2020, 21, 3138. [Google Scholar] [CrossRef]
- Liyanage, W.; Wu, T.; Kannan, S.; Kannan, R.M. Dendrimer–SiRNA Conjugates for Targeted Intracellular Delivery in Glioblastoma Animal Models. ACS Appl. Mater. Interfaces 2022, 14, 46290–46303. [Google Scholar] [CrossRef]
- Osipova, O.; Zakharova, N.; Pyankov, I.; Egorova, A.; Kislova, A.; Lavrentieva, A.; Kiselev, A.; Tennikova, T.; Korzhikova-Vlakh, E. Amphiphilic PH-Sensitive Polypeptides for SiRNA Delivery. J. Drug Deliv. Sci. Technol. 2022, 69, 103135. [Google Scholar] [CrossRef]
- Egorova, A.A.; Kiselev, A.V.; Tarasenko, I.I.; Il’ina, P.L.; Pankova, G.A.; Il’ina, I.E.; Baranov, V.C.; Vlasov, G.P. Hyperbranched Polylysines Modified with Histidine and Arginine: The Optimization of Their DNA Compacting and Endosomolytic Properties. Russ. J. Bioorg. Chem. 2009, 35, 437–445. [Google Scholar] [CrossRef] [PubMed]
- Kruspe, S.; Giangrande, P. Aptamer-SiRNA Chimeras: Discovery, Progress, and Future Prospects. Biomedicines 2017, 5, 45. [Google Scholar] [CrossRef]
- Mahmoudian, F.; Ahmari, A.; Shabani, S.; Sadeghi, B.; Fahimirad, S.; Fattahi, F. Aptamers as an Approach to Targeted Cancer Therapy. Cancer Cell Int. 2024, 24, 108. [Google Scholar] [CrossRef]
- Sivakumar, P.; Kim, S.; Kang, H.C.; Shim, M.S. Targeted SiRNA Delivery Using Aptamer-siRNA Chimeras and Aptamer-conjugated Nanoparticles. WIREs Nanomed. Nanobiotechnol. 2019, 11, e1543. [Google Scholar] [CrossRef]
- Safarzadeh Kozani, P.; Safarzadeh Kozani, P.; Rahbarizadeh, F. Aptamer-Assisted Delivery of Nucleotides with Tumor-Suppressing Properties for Targeted Cancer Therapies. Trends Med. Sci. 2021, 1, 114909. [Google Scholar] [CrossRef]
- McNamara, J.O.; Andrechek, E.R.; Wang, Y.; Viles, K.D.; Rempel, R.E.; Gilboa, E.; Sullenger, B.A.; Giangrande, P.H. Cell Type–Specific Delivery of SiRNAs with Aptamer-SiRNA Chimeras. Nat. Biotechnol. 2006, 24, 1005–1015. [Google Scholar] [CrossRef]
- Odeh, F.; Nsairat, H.; Alshaer, W.; Ismail, M.A.; Esawi, E.; Qaqish, B.; Al Bawab, A.; Ismail, S.I. Aptamers Chemistry: Chemical Modifications and Conjugation Strategies. Molecules 2019, 25, 3. [Google Scholar] [CrossRef]
- Lai, W.Y.; Wang, W.Y.; Chang, Y.C.; Chang, C.J.; Yang, P.C.; Peck, K. Synergistic Inhibition of Lung Cancer Cell Invasion, Tumor Growth and Angiogenesis Using Aptamer-SiRNA Chimeras. Biomaterials 2014, 35, 2905–2914. [Google Scholar] [CrossRef]
- Yoo, H.; Jung, H.; Kim, S.A.; Mok, H. Multivalent Comb-Type Aptamer–SiRNA Conjugates for Efficient and Selective Intracellular Delivery. Chem. Commun. 2014, 50, 6765–6767. [Google Scholar] [CrossRef] [PubMed]
- Dassie, J.P.; Liu, X.Y.; Thomas, G.S.; Whitaker, R.M.; Thiel, K.W.; Stockdale, K.R.; Meyerholz, D.K.; McCaffrey, A.P.; McNamara, J.O.; Giangrande, P.H. Systemic Administration of Optimized Aptamer-SiRNA Chimeras Promotes Regression of PSMA-Expressing Tumors. Nat. Biotechnol. 2009, 27, 839–846. [Google Scholar] [CrossRef] [PubMed]
- Puplampu-Dove, Y.; Gefen, T.; Rajagopalan, A.; Muheramagic, D.; Schrand, B.; Gilboa, E. Potentiating Tumor Immunity Using Aptamer-Targeted RNAi to Render CD8+ T Cells Resistant to TGFβ Inhibition. Oncoimmunology 2018, 7, e1349588. [Google Scholar] [CrossRef] [PubMed]
- Esposito, C.L.; Nuzzo, S.; Catuogno, S.; Romano, S.; de Nigris, F.; de Franciscis, V. STAT3 Gene Silencing by Aptamer-SiRNA Chimera as Selective Therapeutic for Glioblastoma. Mol. Ther. Nucleic. Acids. 2018, 10, 398–411. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, A.; Priceman, S.J.; Kujawski, M.; Xin, H.; Cherryholmes, G.A.; Zhang, W.; Zhang, C.; Lahtz, C.; Kowolik, C.; Forman, S.J.; et al. CTLA4 Aptamer Delivers STAT3 SiRNA to Tumor-Associated and Malignant T Cells. J. Clin. Investig. 2014, 124, 2977–2987. [Google Scholar] [CrossRef]
- Zhou, J.; Tiemann, K.; Chomchan, P.; Alluin, J.; Swiderski, P.; Burnett, J.; Zhang, X.; Forman, S.; Chen, R.; Rossi, J. Dual Functional BAFF Receptor Aptamers Inhibit Ligand-Induced Proliferation and Deliver SiRNAs to NHL Cells. Nucleic. Acids. Res. 2013, 41, 4266–4283. [Google Scholar] [CrossRef]
- Liu, H.Y.; Yu, X.; Liu, H.; Wu, D.; She, J.-X. Co-Targeting EGFR and Survivin with a Bivalent Aptamer-Dual SiRNA Chimera Effectively Suppresses Prostate Cancer. Sci. Rep. 2016, 6, 30346. [Google Scholar] [CrossRef]
- Xue, L.; Maihle, N.J.; Yu, X.; Tang, S.-C.; Liu, H.Y. Synergistic Targeting HER2 and EGFR with Bivalent Aptamer-SiRNA Chimera Efficiently Inhibits HER2-Positive Tumor Growth. Mol. Pharm. 2018, 15, 4801–4813. [Google Scholar] [CrossRef]
- Aaldering, L.J.; Tayeb, H.; Krishnan, S.; Fletcher, S.; Wilton, S.D.; Veedu, R.N. Smart Functional Nucleic Acid Chimeras: Enabling Tissue Specific RNA Targeting Therapy. RNA Biol. 2015, 12, 412–425. [Google Scholar] [CrossRef]
- Zhou, J.; Rossi, J.J. Cell-Type-Specific, Aptamer-Functionalized Agents for Targeted Disease Therapy. Mol. Ther. Nucleic. Acids. 2014, 3, e169. [Google Scholar] [CrossRef]
- Pastor, F.; Kolonias, D.; Giangrande, P.H.; Gilboa, E. Induction of Tumour Immunity by Targeted Inhibition of Nonsense-Mediated MRNA Decay. Nature 2010, 465, 227–230. [Google Scholar] [CrossRef] [PubMed]
- Quirico, L.; Orso, F.; Esposito, C.L.; Bertone, S.; Coppo, R.; Conti, L.; Catuogno, S.; Cavallo, F.; de Franciscis, V.; Taverna, D. Axl-148b Chimeric Aptamers Inhibit Breast Cancer and Melanoma Progression. Int. J. Biol. Sci. 2020, 16, 1238–1251. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Yang, J.; Liu, D.; Zhang, H.; Ou, T.; Xiao, L.; Chen, W. Construction of Aptamer-SiRNA Chimera and Glutamine Modified Carboxymethyl-β-Cyclodextrin Nanoparticles for the Combination Therapy against Lung Squamous Cell Carcinoma. Biomed. Pharmacother. 2024, 174, 116506. [Google Scholar] [CrossRef]
- Arnold, A.E.; Smith, L.J.; Beilhartz, G.L.; Bahlmann, L.C.; Jameson, E.; Melnyk, R.A.; Shoichet, M.S. Attenuated Diphtheria Toxin Mediates SiRNA Delivery. Sci. Adv. 2020, 6, 4848. [Google Scholar] [CrossRef]
- Dyer, P.D.R.; Shepherd, T.R.; Gollings, A.S.; Shorter, S.A.; Gorringe-Pattrick, M.A.M.; Tang, C.K.; Cattoz, B.N.; Baillie, L.; Griffiths, P.C.; Richardson, S.C.W. Disarmed Anthrax Toxin Delivers Antisense Oligonucleotides and SiRNA with High Efficiency and Low Toxicity. J. Control. Release 2015, 220, 316–328. [Google Scholar] [CrossRef]
- Zhao, S.; Karp, J.M.; Joshi, N. Toxin-Mediated SiRNA Delivery. Trends Pharmacol. Sci. 2020, 41, 511–513. [Google Scholar] [CrossRef]
- Yamada, T.; Peng, C.G.; Matsuda, S.; Addepalli, H.; Jayaprakash, K.N.; Alam, M.R.; Mills, K.; Maier, M.A.; Charisse, K.; Sekine, M.; et al. Versatile Site-Specific Conjugation of Small Molecules to SiRNA Using Click Chemistry. J. Org. Chem. 2011, 76, 1198–1211. [Google Scholar] [CrossRef]
- Salim, L.; Desaulniers, J.-P. To Conjugate or to Package? A Look at Targeted SiRNA Delivery Through Folate Receptors. Nucleic. Acid. Ther. 2021, 31, 21–38. [Google Scholar] [CrossRef]
- Abdelaal, A.M.; Kasinski, A.L. Ligand-Mediated Delivery of RNAi-Based Therapeutics for the Treatment of Oncological Diseases. NAR Cancer 2021, 3, 030. [Google Scholar] [CrossRef] [PubMed]
- Salim, L.; Islam, G.; Desaulniers, J.-P. Targeted Delivery and Enhanced Gene-Silencing Activity of Centrally Modified Folic Acid–SiRNA Conjugates. Nucleic. Acids. Res. 2020, 48, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Gangopadhyay, S.; Nikam, R.R.; Gore, K.R. Folate Receptor-Mediated SiRNA Delivery: Recent Developments and Future Directions for RNAi Therapeutics. Nucleic. Acid. Ther. 2021, 31, 245–270. [Google Scholar] [CrossRef]
- Thomas, M.; Kularatne, S.A.; Qi, L.; Kleindl, P.; Leamon, C.P.; Hansen, M.J.; Low, P.S. Ligand-Targeted Delivery of Small Interfering RNAs to Malignant Cells and Tissues. Ann. N. Y. Acad. Sci. 2009, 1175, 32–39. [Google Scholar] [CrossRef] [PubMed]
- Boss, S.D.; Betzel, T.; Müller, C.; Fischer, C.R.; Haller, S.; Reber, J.; Groehn, V.; Schibli, R.; Ametamey, S.M. Comparative Studies of Three Pairs of α- and γ-Conjugated Folic Acid Derivatives Labeled with Fluorine-18. Bioconjug Chem. 2016, 27, 74–86. [Google Scholar] [CrossRef]
- Zhang, L.; Chan, J.M.; Gu, F.X.; Rhee, J.W.; Wang, A.Z.; Radovic-Moreno, A.F.; Alexis, F.; Langer, R.; Farokhzad, O.C. Self-Assembled Lipid-Polymer Hybrid Nanoparticles: A Robust Drug Delivery Platform. ACS Nano 2008, 2, 1696–1702. [Google Scholar] [CrossRef]
- Dohmen, C.; Fröhlich, T.; Lächelt, U.; Röhl, I.; Vornlocher, H.P.; Hadwiger, P.; Wagner, E. Defined Folate-PEG-SiRNA Conjugates for Receptor-Specific Gene Silencing. Mol. Ther. Nucleic. Acids. 2012, 1, e7. [Google Scholar] [CrossRef] [PubMed]
- Osborn, M.F.; Khvorova, A. Improving SiRNA Delivery in Vivo Through Lipid Conjugation. Nucleic. Acid. Ther. 2018, 28, 128–136. [Google Scholar] [CrossRef]
- Chen, M.; Chen, M.; He, J. Cancer Cell Membrane Cloaking Nanoparticles for Targeted Co-Delivery of Doxorubicin and PD-L1 SiRNA. Artif. Cells Nanomed. Biotechnol. 2019, 47, 1635–1641. [Google Scholar] [CrossRef]
- Zheng, Y.; Tai, W. Insight into the SiRNA Transmembrane Delivery—From Cholesterol Conjugating to Tagging. WIREs Nanomed. Nanobiotechnol. 2020, 12, e1606. [Google Scholar] [CrossRef]
- Petrova, N.S.; Chernikov, I.V.; Meschaninova, M.I.; Dovydenko, I.S.; Venyaminova, A.G.; Zenkova, M.A.; Vlassov, V.V.; Chernolovskaya, E.L. Carrier-Free Cellular Uptake and the Gene-Silencing Activity of the Lipophilic SiRNAs Is Strongly Affected by the Length of the Linker between SiRNA and Lipophilic Group. Nucleic. Acids. Res. 2012, 40, 2330–2344. [Google Scholar] [CrossRef] [PubMed]
- Chernikov, I.V.; Ponomareva, U.A.; Meschaninova, M.I.; Bachkova, I.K.; Vlassov, V.V.; Zenkova, M.A.; Chernolovskaya, E.L. Cholesterol Conjugates of Small Interfering RNA: Linkers and Patterns of Modification. Molecules 2024, 29, 786. [Google Scholar] [CrossRef] [PubMed]
- Shmushkovich, T.; Monopoli, K.R.; Homsy, D.; Leyfer, D.; Betancur-Boissel, M.; Khvorova, A.; Wolfson, A.D. Functional Features Defining the Efficacy of Cholesterol-Conjugated, Self-Deliverable, Chemically Modified SiRNAs. Nucleic. Acids. Res. 2018, 46, 10905–10916. [Google Scholar] [CrossRef]
- Alterman, J.F.; Hall, L.M.; Coles, A.H.; Hassler, M.R.; Didiot, M.C.; Chase, K.; Abraham, J.; Sottosanti, E.; Johnson, E.; Sapp, E.; et al. Hydrophobically Modified SiRNAs Silence Huntingtin MRNA in Primary Neurons and Mouse Brain. Mol. Ther. Nucleic. Acids. 2015, 4, e266. [Google Scholar] [CrossRef]
- Wada, S.; Yasuhara, H.; Wada, F.; Sawamura, M.; Waki, R.; Yamamoto, T.; Harada-Shiba, M.; Obika, S. Evaluation of the Effects of Chemically Different Linkers on Hepatic Accumulations, Cell Tropism and Gene Silencing Ability of Cholesterol-Conjugated Antisense Oligonucleotides. J. Control. Release 2016, 226, 57–65. [Google Scholar] [CrossRef]
- Tai, W.; Gao, X. Noncovalent Tagging of SiRNA with Steroids for Transmembrane Delivery. Biomaterials 2018, 178, 720–727. [Google Scholar] [CrossRef]
- Kim, J.B.; Lee, Y.M.; Ryu, J.; Lee, E.; Kim, W.J.; Keum, G.; Bang, E.K. Coordinative Amphiphiles as Tunable SiRNA Transporters. Bioconjug. Chem. 2016, 27, 1850–1856. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Li, F.; Xu, Y.; Zhang, W.; Hu, Y.; Fu, Y.; Xu, W.; Ge, S.; Fan, X.; Lu, L. Cholesterol Modification of SDF-1-Specific SiRNA Enables Therapeutic Targeting of Angiogenesis through Akt Pathway Inhibition. Exp. Eye. Res. 2019, 184, 64–71. [Google Scholar] [CrossRef]
- Biscans, A.; Ly, S.; McHugh, N.; Cooper, D.A.; Khvorova, A. Engineered Ionizable Lipid SiRNA Conjugates Enhance Endosomal Escape but Induce Toxicity in Vivo. J. Control. Release 2022, 349, 831–843. [Google Scholar] [CrossRef]
- Østergaard, M.E.; Jackson, M.; Low, A.; Chappell, A.E.; Lee, R.G.; Peralta, R.Q.; Yu, J.; Kinberger, G.A.; Dan, A.; Carty, R.; et al. Conjugation of Hydrophobic Moieties Enhances Potency of Antisense Oligonucleotides in the Muscle of Rodents and Non-Human Primates. Nucleic. Acids. Res. 2019, 47, 6045–6058. [Google Scholar] [CrossRef]
- Biscans, A.; Coles, A.; Echeverria, D.; Khvorova, A. The Valency of Fatty Acid Conjugates Impacts SiRNA Pharmacokinetics, Distribution, and Efficacy in Vivo. J. Control. Release 2019, 302, 116–125. [Google Scholar] [CrossRef]
- Biscans, A.; Caiazzi, J.; McHugh, N.; Hariharan, V.; Muhuri, M.; Khvorova, A. Docosanoic Acid Conjugation to SiRNA Enables Functional and Safe Delivery to Skeletal and Cardiac Muscles. Mol. Ther. 2021, 29, 1382–1394. [Google Scholar] [CrossRef] [PubMed]
- Alexander, P.; Kucera, G.; Pardee, T.S. Improving Nucleoside Analogs via Lipid Conjugation: Is Fatter Any Better? Crit. Rev. Oncol. Hematol. 2016, 100, 46–56. [Google Scholar] [CrossRef] [PubMed]
- Osborn, M.F.; Coles, A.H.; Biscans, A.; Haraszti, R.A.; Roux, L.; Davis, S.; Ly, S.; Echeverria, D.; Hassler, M.R.; Godinho, B.M.D.C.; et al. Hydrophobicity Drives the Systemic Distribution of Lipid-Conjugated SiRNAs via Lipid Transport Pathways. Nucleic. Acids. Res. 2019, 47, 1070–1081. [Google Scholar] [CrossRef] [PubMed]
- Tran, P.; Weldemichael, T.; Liu, Z.; Li, H. Delivery of Oligonucleotides: Efficiency with Lipid Conjugation and Clinical Outcome. Pharmaceutics 2022, 14, 342. [Google Scholar] [CrossRef]
- Ruvinov, E.; Kryukov, O.; Forti, E.; Korin, E.; Goldstein, M.; Cohen, S. Calcium–SiRNA Nanocomplexes: What Reversibility Is All About. J. Control. Release 2015, 203, 150–160. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Li, Z.; Zhao, X.; Keen, L.; Kong, X. Calcium Phosphate Nanoparticles-Based Systems for SiRNA Delivery. Regen. Biomater. 2016, 3, 187–195. [Google Scholar] [CrossRef] [PubMed]
- Oyer, H.M.; Sanders, C.M.; Kim, F.J. Small-Molecule Modulators of Sigma1 and Sigma2/TMEM97 in the Context of Cancer: Foundational Concepts and Emerging Themes. Front. Pharmacol. 2019, 10, 1141. [Google Scholar] [CrossRef]
- Alam, M.R.; Ming, X.; Nakagawa, O.; Jin, J.; Juliano, R.L. Covalent Conjugation of Oligonucleotides with Cell-Targeting Ligands. Bioorg. Med. Chem. 2013, 21, 6217–6223. [Google Scholar] [CrossRef]
- Qu, D.; Jiao, M.; Lin, H.; Tian, C.; Qu, G.; Xue, J.; Xue, L.; Ju, C.; Zhang, C. Anisamide-Functionalized PH-Responsive Amphiphilic Chitosan-Based Paclitaxel Micelles for Sigma-1 Receptor Targeted Prostate Cancer Treatment. Carbohydr. Polym. 2020, 229, 115498. [Google Scholar] [CrossRef]
- Yao, W.; Liu, C.; Wang, N.; Zhou, H.; Chen, H.; Qiao, W. Anisamide-Modified Dual-Responsive Drug Delivery System with MRI Capacity for Cancer Targeting Therapy. J. Mol. Liq. 2021, 340, 116889. [Google Scholar] [CrossRef]
- Springer, A.D.; Dowdy, S.F. GalNAc-SiRNA Conjugates: Leading the Way for Delivery of RNAi Therapeutics. Nucleic. Acid. Ther. 2018, 28, 109–118. [Google Scholar] [CrossRef]
- Biessen, E.A.L.; Van Berkel, T.J.C. N-Acetyl Galactosamine Targeting: Paving the Way for Clinical Application of Nucleotide Medicines in Cardiovascular Diseases. Arterioscler. Thromb. Vasc. Biol. 2021, 41, 2855–2865. [Google Scholar] [CrossRef]
- Willibald, J.; Harder, J.; Sparrer, K.; Conzelmann, K.-K.; Carell, T. Click-Modified Anandamide SiRNA Enables Delivery and Gene Silencing in Neuronal and Immune Cells. J. Am. Chem. Soc. 2012, 134, 12330–12333. [Google Scholar] [CrossRef] [PubMed]
- Nishina, K.; Unno, T.; Uno, Y.; Kubodera, T.; Kanouchi, T.; Mizusawa, H.; Yokota, T. Efficient In Vivo Delivery of SiRNA to the Liver by Conjugation of α-Tocopherol. Mol. Ther. 2008, 16, 734–740. [Google Scholar] [CrossRef]
- Tang, S.; Kapoor, E.; Ding, L.; Yu, A.; Tang, W.; Hang, Y.; Smith, L.M.; Sil, D.; Oupický, D. Effect of Tocopherol Conjugation on Polycation-Mediated SiRNA Delivery to Orthotopic Pancreatic Tumors. Biomater. Adv. 2023, 145, 213236. [Google Scholar] [CrossRef]
- Yagublu, V.; Karimova, A.; Hajibabazadeh, J.; Reissfelder, C.; Muradov, M.; Bellucci, S.; Allahverdiyev, A. Overview of Physicochemical Properties of Nanoparticles as Drug Carriers for Targeted Cancer Therapy. J. Funct. Biomater. 2022, 13, 196. [Google Scholar] [CrossRef]
- Yuan, Z.; Yan, R.; Fu, Z.; Wu, T.; Ren, C. Impact of Physicochemical Properties on Biological Effects of Lipid Nanoparticles: Are They Completely Safe. Sci. Total Environ. 2024, 927, 172240. [Google Scholar] [CrossRef]
- Hui, Y.; Yi, X.; Hou, F.; Wibowo, D.; Zhang, F.; Zhao, D.; Gao, H.; Zhao, C.-X. Role of Nanoparticle Mechanical Properties in Cancer Drug Delivery. ACS Nano 2019, 13, 7410–7424. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Yuk, S.A.; Dieterly, A.M.; Kwon, S.; Park, J.; Meng, F.; Gadalla, H.H.; Cadena, M.J.; Lyle, L.T.; Yeo, Y. Nanosac, a Noncationic and Soft Polyphenol Nanocapsule, Enables Systemic Delivery of SiRNA to Solid Tumors. ACS Nano 2021, 15, 4576–4593. [Google Scholar] [CrossRef]
- Zein, R.; Sharrouf, W.; Selting, K. Physical Properties of Nanoparticles That Result in Improved Cancer Targeting. J. Oncol. 2020, 2020, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, L.; Li, L.; Song, F.; Song, F. Interplay of Nanoparticle Properties during Endocytosis. Crystals 2021, 11, 728. [Google Scholar] [CrossRef]
- Palmieri, V.; Caracciolo, G. Tuning the Immune System by Nanoparticle–Biomolecular Corona. Nanoscale Adv. 2022, 4, 3300–3308. [Google Scholar] [CrossRef]
- Lu, Z.-R.; Sun, D. Mechanism of PH-Sensitive Amphiphilic Endosomal Escape of Ionizable Lipid Nanoparticles for Cytosolic Nucleic Acid Delivery. Pharm. Res. 2025, 42, 1065–1077. [Google Scholar] [CrossRef]
- Oyewumi, M.O.; Kumar, A.; Cui, Z. Nano-Microparticles as Immune Adjuvants: Correlating Particle Sizes and the Resultant Immune Responses. Expert. Rev. Vaccines 2010, 9, 1095–1107. [Google Scholar] [CrossRef]
- Morales-Becerril, A.; Aranda-Lara, L.; Isaac-Olivé, K.; Ocampo-García, B.E.; Morales-ávila, E. Nanocarriers for delivery of sirna as gene silencing mediator. Excli J. 2022, 21, 1028–1052. [Google Scholar] [PubMed]
- Zhang, T.; Yin, H.; Li, Y.; Yang, H.; Ge, K.; Zhang, J.; Yuan, Q.; Dai, X.; Naeem, A.; Weng, Y.; et al. Optimized Lipid Nanoparticles (LNPs) for Organ-Selective Nucleic Acids Delivery in Vivo. iScience 2024, 27, 109804. [Google Scholar] [CrossRef] [PubMed]
- Sinani, G.; Durgun, M.E.; Cevher, E.; Özsoy, Y. Polymeric-Micelle-Based Delivery Systems for Nucleic Acids. Pharmaceutics 2023, 15, 2021. [Google Scholar] [CrossRef] [PubMed]
- Serrano-Sevilla, I.; Artiga, Á.; Mitchell, S.G.; De Matteis, L.; de la Fuente, J.M. Natural Polysaccharides for SiRNA Delivery: Nanocarriers Based on Chitosan, Hyaluronic Acid, and Their Derivatives. Molecules 2019, 24, 2570. [Google Scholar] [CrossRef]
- Cao, Y.; Tan, Y.F.; Wong, Y.S.; Liew, M.W.J.; Venkatraman, S. Recent Advances in Chitosan-Based Carriers for Gene Delivery. Mar. Drugs 2019, 17, 381. [Google Scholar] [CrossRef]
- Oh, Y.-K.; Park, T.G. SiRNA Delivery Systems for Cancer Treatment. Adv. Drug Deliv. Rev. 2009, 61, 850–862. [Google Scholar] [CrossRef]
- Singha, K.; Namgung, R.; Kim, W.J. Polymers in Small-Interfering RNA Delivery. Nucleic. Acid. Ther. 2011, 21, 133–147. [Google Scholar] [CrossRef]
- Ewe, A.; Höbel, S.; Heine, C.; Merz, L.; Kallendrusch, S.; Bechmann, I.; Merz, F.; Franke, H.; Aigner, A. Optimized Polyethylenimine (PEI)-Based Nanoparticles for SiRNA Delivery, Analyzed in Vitro and in an Ex Vivo Tumor Tissue Slice Culture Model. Drug Deliv. Transl. Res. 2017, 7, 206–216. [Google Scholar] [CrossRef]
- Karlsson, J.; Rhodes, K.R.; Green, J.J.; Tzeng, S.Y. Poly(Beta-Amino Ester)s as Gene Delivery Vehicles: Challenges and Opportunities. Expert. Opin. Drug Deliv. 2020, 17, 1395–1410. [Google Scholar] [CrossRef]
- Zielińska, A.; Carreiró, F.; Oliveira, A.M.; Neves, A.; Pires, B.; Venkatesh, D.N.; Durazzo, A.; Lucarini, M.; Eder, P.; Silva, A.M.; et al. Polymeric Nanoparticles: Production, Characterization, Toxicology and Ecotoxicology. Molecules 2020, 25, 3731. [Google Scholar] [CrossRef]
- Fus-Kujawa, A.; Teper, P.; Botor, M.; Klarzyńska, K.; Sieroń, Ł.; Verbelen, B.; Smet, M.; Sieroń, A.L.; Mendrek, B.; Kowalczuk, A. Functional Star Polymers as Reagents for Efficient Nucleic Acids Delivery into HT-1080 Cells. Int. J. Polym. Mater. Polym. Biomater. 2021, 70, 356–370. [Google Scholar] [CrossRef]
- Nguyen, J.; Steele, T.W.J.; Merkel, O.; Reul, R.; Kissel, T. Fast Degrading Polyesters as SiRNA Nano-Carriers for Pulmonary Gene Therapy. J. Control. Release 2008, 132, 243–251. [Google Scholar] [CrossRef]
- Zhu, J.; Qiao, M.; Wang, Q.; Ye, Y.; Ba, S.; Ma, J.; Hu, H.; Zhao, X.; Chen, D. Dual-Responsive Polyplexes with Enhanced Disassembly and Endosomal Escape for Efficient Delivery of SiRNA. Biomaterials 2018, 162, 47–59. [Google Scholar] [CrossRef] [PubMed]
- Bholakant, R.; Qian, H.; Zhang, J.; Huang, X.; Huang, D.; Feijen, J.; Zhong, Y.; Chen, W. Recent Advances of Polycationic SiRNA Vectors for Cancer Therapy. Biomacromolecules 2020, 21, 2966–2982. [Google Scholar] [CrossRef]
- Yao, Z.; Liu, T.; Wang, J.; Fu, Y.; Zhao, J.; Wang, X.; Li, Y.; Yang, X.; He, Z. Targeted Delivery Systems of SiRNA Based on Ionizable Lipid Nanoparticles and Cationic Polymer Vectors. Biotechnol. Adv. 2025, 81, 108546. [Google Scholar] [CrossRef]
- Hibbitts, A.J.; Ramsey, J.M.; Barlow, J.; Macloughlin, R.; Cryan, S.A. In Vitro and in Vivo Assessment of Pegylated Pei for Anti-Il-8/Cxcl-1 Sirna Delivery to the Lungs. Nanomaterials 2020, 10, 1248. [Google Scholar] [CrossRef]
- Cho, S.K.; Dang, C.; Wang, X.; Ragan, R.; Kwon, Y.J. Mixing-Sequence-Dependent Nucleic Acid Complexation and Gene Transfer Efficiency by Polyethylenimine. Biomater. Sci. 2015, 3, 1124–1133. [Google Scholar] [CrossRef]
- Kazemi Oskuee, R.; Dabbaghi, M.; Gholami, L.; Taheri-Bojd, S.; Balali-Mood, M.; Mousavi, S.H.; Malaekeh-Nikouei, B. Investigating the Influence of Polyplex Size on Toxicity Properties of Polyethylenimine Mediated Gene Delivery. Life Sci. 2018, 197, 101–108. [Google Scholar] [CrossRef]
- Höbel, S.; Aigner, A. Polyethylenimines for SiRNA and MiRNA Delivery in Vivo. WIREs Nanomed. Nanobiotechnol. 2013, 5, 484–501. [Google Scholar] [CrossRef]
- Ewe, A.; Panchal, O.; Pinnapireddy, S.R.; Bakowsky, U.; Przybylski, S.; Temme, A.; Aigner, A. Liposome-Polyethylenimine Complexes (DPPC-PEI Lipopolyplexes) for Therapeutic SiRNA Delivery in Vivo. Nanomedicine 2017, 13, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Parhamifar, L.; Larsen, A.K.; Hunter, A.C.; Andresen, T.L.; Moghimi, S.M. Polycation Cytotoxicity: A Delicate Matter for Nucleic Acid Therapy—Focus on Polyethylenimine. Soft Matter. 2010, 6, 4001. [Google Scholar] [CrossRef]
- Hall, A.; Lächelt, U.; Bartek, J.; Wagner, E.; Moghimi, S.M. Polyplex Evolution: Understanding Biology, Optimizing Performance. Mol. Ther. 2017, 25, 1476–1490. [Google Scholar] [CrossRef]
- Liu, S.; Huang, W.; Jin, M.J.; Fan, B.; Xia, G.M.; Gao, Z.G. Inhibition of Murine Breast Cancer Growth and Metastasis by Survivin-Targeted SiRNA Using Disulfide Cross-Linked Linear PEI. Eur. J. Pharm. Sci. 2016, 82, 171–182. [Google Scholar] [CrossRef]
- Zhang, Q.Y.; Ho, P.Y.; Tu, M.J.; Jilek, J.L.; Chen, Q.X.; Zeng, S.; Yu, A.M. Lipidation of Polyethylenimine-Based Polyplex Increases Serum Stability of Bioengineered RNAi Agents and Offers More Consistent Tumoral Gene Knockdown in Vivo. Int. J. Pharm. 2018, 547, 537–544. [Google Scholar] [CrossRef]
- Merkel, O.M.; Beyerle, A.; Librizzi, D.; Pfestroff, A.; Behr, T.M.; Sproat, B.; Barth, P.J.; Kissel, T. Nonviral SiRNA Delivery to the Lung: Investigation of PEG−PEI Polyplexes and Their In Vivo Performance. Mol. Pharm. 2009, 6, 1246–1260. [Google Scholar] [CrossRef]
- Zhupanyn, P.; Ewe, A.; Büch, T.; Malek, A.; Rademacher, P.; Müller, C.; Reinert, A.; Jaimes, Y.; Aigner, A. Extracellular Vesicle (ECV)-Modified Polyethylenimine (PEI) Complexes for Enhanced SiRNA Delivery in Vitro and in Vivo. J. Control. Release 2020, 319, 63–76. [Google Scholar] [CrossRef] [PubMed]
- Chuan, D.; Jin, T.; Fan, R.; Zhou, L.; Guo, G. Chitosan for Gene Delivery: Methods for Improvement and Applications. Adv. Colloid. Interface Sci. 2019, 268, 25–38. [Google Scholar] [CrossRef] [PubMed]
- Al-Absi, M.Y.; Caprifico, A.E.; Calabrese, G. Chitosan and Its Structural Modifications for SiRNA Delivery. Adv. Pharm. Bull. 2023, 13, 275–282. [Google Scholar] [CrossRef]
- Karayianni, M.; Sentoukas, T.; Skandalis, A.; Pippa, N.; Pispas, S. Chitosan-Based Nanoparticles for Nucleic Acid Delivery: Technological Aspects, Applications, and Future Perspectives. Pharmaceutics 2023, 15, 1849. [Google Scholar] [CrossRef]
- Şalva, E.; Özkan, N.; Kabasakal, L.; Turan, S.Ö.; Akbuğa, J. The Effect of Chitosan Complexes on Biodistribution of SiRNA. Clin. Exp. Health Sci. 2011, 1, 1. [Google Scholar]
- Holzerny, P.; Ajdini, B.; Heusermann, W.; Bruno, K.; Schuleit, M.; Meinel, L.; Keller, M. Biophysical Properties of Chitosan/SiRNA Polyplexes: Profiling the Polymer/SiRNA Interactions and Bioactivity. J. Control. Release 2012, 157, 297–304. [Google Scholar] [CrossRef]
- Cao, Y.; Tan, Y.F.; Wong, Y.S.; Aminuddin, M.; Ramya, B.; Liew, M.W.J.; Liu, J.; Venkatraman, S.S. Designing SiRNA/Chitosan-Methacrylate Complex Nanolipogel for Prolonged Gene Silencing Effects. Sci. Rep. 2022, 12, 5335. [Google Scholar] [CrossRef]
- Liu, X.; Ma, L.; Qin, W.; Gao, C. Effect of N/P Ratios on Physicochemical Stability, Cellular Association, and Gene Silencing Efficiency for Trimethyl Chitosan/Small Interfering RNA Complexes. J. Bioact. Compat. Polym. 2013, 28, 590–604. [Google Scholar] [CrossRef]
- Veilleux, D.; Gopalakrishna Panicker, R.K.; Chevrier, A.; Biniecki, K.; Lavertu, M.; Buschmann, M.D. Lyophilisation and Concentration of Chitosan/SiRNA Polyplexes: Influence of Buffer Composition, Oligonucleotide Sequence, and Hyaluronic Acid Coating. J. Colloid. Interface Sci. 2018, 512, 335–345. [Google Scholar] [CrossRef]
- Grząbka-Zasadzińska, A.; Amietszajew, T.; Borysiak, S. Thermal and Mechanical Properties of Chitosan Nanocomposites with Cellulose Modified in Ionic Liquids. J. Therm. Anal. Calorim. 2017, 130, 143–154. [Google Scholar] [CrossRef]
- Prakash, S.; Tomaro-Duchesneau, C.; Saha, S.; Kahouli; Malhotra, M. Development and Characterization of Chitosan-PEG-TAT Nanoparticles for the Intracellular Delivery of SiRNA. Int. J. Nanomed. 2013, 8, 2041. [Google Scholar] [CrossRef] [PubMed]
- Sun, P.; Huang, W.; Kang, L.; Jin, M.; Fan, B.; Jin, H.; Wang, Q.; Gao, Z. SiRNA-Loaded Poly(Histidine-Arginine)6-Modified Chitosan Nanoparticle with Enhanced Cell-Penetrating and Endosomal Escape Capacities for Suppressing Breast Tumor Metastasis. Int. J. Nanomed. 2017, 12, 3221–3234. [Google Scholar] [CrossRef] [PubMed]
- Muddineti, O.S.; Shah, A.; Rompicharla, S.V.K.; Ghosh, B.; Biswas, S. Cholesterol-Grafted Chitosan Micelles as a Nanocarrier System for Drug-SiRNA Co-Delivery to the Lung Cancer Cells. Int. J. Biol. Macromol. 2018, 118, 857–863. [Google Scholar] [CrossRef] [PubMed]
- Paredes-Hernández, U.; Aguilar-Peña, L.V.; Isaac-Olivé, K.; Ocampo-García, B.; Contreras, I.; Estrada, J.A.; Izquierdo, G.; Morales-Avila, E.; Aranda-Lara, L. Enhancing Photodynamic and Radionuclide Therapy by Small Interfering RNA (SiRNA)-RAD51 Transfection via Self-Emulsifying Delivery Systems (SNEDDS). Cytotherapy 2025, 27, 66–77. [Google Scholar] [CrossRef]
- Green, J.J.; Shi, J.; Chiu, E.; Leshchiner, E.S.; Langer, R.; Anderson, D.G. Biodegradable Polymeric Vectors for Gene Delivery to Human Endothelial Cells. Bioconjug. Chem. 2006, 17, 1162–1169. [Google Scholar] [CrossRef]
- Kozielski, K.L.; Tzeng, S.Y.; Green, J.J. A Bioreducible Linear Poly(β-Amino Ester) for SiRNA Delivery. Chem. Commun. 2013, 49, 5319–5321. [Google Scholar] [CrossRef]
- Zhang, J.; Cai, X.; Dou, R.; Guo, C.; Tang, J.; Hu, Y.; Chen, H.; Chen, J. Poly(β-Amino Ester)s-Based Nanovehicles: Structural Regulation and Gene Delivery. Mol. Ther. Nucleic. Acids. 2023, 32, 568–581. [Google Scholar] [CrossRef]
- Chou, J.J.; Berger, A.G.; Jalili-Firoozinezhad, S.; Hammond, P.T. A Design Approach for Layer-by-Layer Surface-Mediated SiRNA Delivery. Acta Biomater. 2021, 135, 331–341. [Google Scholar] [CrossRef]
- Rui, Y.; Wilson, D.R.; Choi, J.; Varanasi, M.; Sanders, K.; Karlsson, J.; Lim, M.; Green, J.J. Carboxylated Branched Poly(β-Amino Ester) Nanoparticles Enable Robust Cytosolic Protein Delivery and CRISPR-Cas9 Gene Editing. Sci. Adv. 2019, 5, 3255. [Google Scholar] [CrossRef] [PubMed]
- Berger, A.G.; DeLorenzo, C.; Vo, C.; Kaskow, J.A.; Nabar, N.; Hammond, P.T. Poly(β-Aminoester) Physicochemical Properties Govern the Delivery of SiRNA from Electrostatically Assembled Coatings. Biomacromolecules 2024, 25, 2934–2952. [Google Scholar] [CrossRef]
- Mohan, T.; Kleinschek, K.S.; Kargl, R. Polysaccharide Peptide Conjugates: Chemistry, Properties and Applications. Carbohydr. Polym. 2022, 280, 118875. [Google Scholar] [CrossRef]
- Zhou, J.; Patel, T.R.; Fu, M.; Bertram, J.P.; Saltzman, W.M. Octa-Functional PLGA Nanoparticles for Targeted and Efficient SiRNA Delivery to Tumors. Biomaterials 2012, 33, 583–591. [Google Scholar] [CrossRef]
- Abedi-Gaballu, F.; Dehghan, G.; Ghaffari, M.; Yekta, R.; Abbaspour-Ravasjani, S.; Baradaran, B.; Ezzati Nazhad Dolatabadi, J.; Hamblin, M.R. PAMAM Dendrimers as Efficient Drug and Gene Delivery Nanosystems for Cancer Therapy. Appl. Mater. Today 2018, 12, 177–190. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, J.A.; Cullis, P.R.; Van Der Meel, R. Lipid Nanoparticles Enabling Gene Therapies: From Concepts to Clinical Utility. Nucleic. Acid. Ther. 2018, 28, 146–157. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Zhang, H.; Li, G.; Su, J.; Wei, Y.; Xu, C. Lipid Nanovehicles Overcome Barriers to Systemic RNA Delivery: Lipid Components, Fabrication Methods, and Rational Design. Acta. Pharm. Sin. B 2024, 14, 579–601. [Google Scholar] [CrossRef]
- Kotelianski, V.; Zatsepin, T.; Kotelevtsev, Y. Lipid Nanoparticles for Targeted SiRNA Delivery—Going from Bench to Bedside. Int. J. Nanomed. 2016, 11, 3077–3086. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Jiang, Y.; He, Y.; Boucetta, H.; Wu, J.; Chen, Z.; He, W. Lipid Carriers for MRNA Delivery. Acta Pharm. Sin. B 2023, 13, 4105–4126. [Google Scholar] [CrossRef]
- Ponti, F.; Campolungo, M.; Melchiori, C.; Bono, N.; Candiani, G. Cationic Lipids for Gene Delivery: Many Players, One Goal. Chem. Phys. Lipids 2021, 235, 105032. [Google Scholar] [CrossRef]
- Tenchov, R.; Bird, R.; Curtze, A.E.; Zhou, Q. Lipid Nanoparticles─From Liposomes to MRNA Vaccine Delivery, a Landscape of Research Diversity and Advancement. ACS Nano 2021, 15, 16982–17015. [Google Scholar] [CrossRef]
- Kang, M.; Kim, H.; Leal, C. Self-Organization of Nucleic Acids in Lipid Constructs. Curr. Opin. Colloid Interface Sci. 2016, 26, 58–65. [Google Scholar] [CrossRef]
- Han, X.; Mitchell, M.J.; Nie, G. Nanomaterials for Therapeutic RNA Delivery. Matter 2020, 3, 1948–1975. [Google Scholar] [CrossRef]
- Zheng, L.I.; Bandara, S.R.; Tan, Z.I.; Leal, C. Lipid Nanoparticle Topology Regulates Endosomal Escape and Delivery of RNA to the Cytoplasm. Proc. Natl. Acad. Sci. USA 2023, 120, e2303567120. [Google Scholar] [CrossRef]
- Hattori, Y.; Arai, S.; Kikuchi, T.; Hamada, M.; Okamoto, R.; Machida, Y.; Kawano, K. Optimization of SiRNA Delivery Method into the Liver by Sequential Injection of Polyglutamic Acid and Cationic Lipoplex. Pharmacol. Pharm. 2015, 6, 302–310. [Google Scholar] [CrossRef]
- Mendonça, M.C.P.; Kont, A.; Kowalski, P.S.; O’Driscoll, C.M. Design of Lipid-Based Nanoparticles for Delivery of Therapeutic Nucleic Acids. Drug Discov. Today 2023, 28, 103505. [Google Scholar] [CrossRef] [PubMed]
- Mihailescu, M.; Worcester, D.L.; Carroll, C.L.; Chamberlin, A.R.; White, S.H. DOTAP: Structure, Hydration, and the Counterion Effect. Biophys. J. 2023, 122, 1086–1093. [Google Scholar] [CrossRef]
- Lechanteur, A.; Sanna, V.; Duchemin, A.; Evrard, B.; Mottet, D.; Piel, G. Cationic Liposomes Carrying SiRNA: Impact of Lipid Composition on Physicochemical Properties, Cytotoxicity and Endosomal Escape. Nanomaterials 2018, 8, 270. [Google Scholar] [CrossRef] [PubMed]
- Ni, R.; Feng, R.; Chau, Y. Synthetic Approaches for Nucleic Acid Delivery: Choosing the Right Carriers. Life 2019, 9, 59. [Google Scholar] [CrossRef] [PubMed]
- Sharma, D.; Arora, S.; Singh, J.; Layek, B. A Review of the Tortuous Path of Nonviral Gene Delivery and Recent Progress. Int. J. Biol. Macromol. 2021, 183, 2055–2073. [Google Scholar] [CrossRef]
- Kurakula, H.; Vaishnavi, S.; Sharif, M.Y.; Ellipilli, S. Emergence of Small Interfering RNA-Based Gene Drugs for Various Diseases. ACS Omega 2023, 8, 20234–20250. [Google Scholar] [CrossRef]
- Mai, Y.; Guo, J.; Zhao, Y.; Ma, S.; Hou, Y.; Yang, J. Intranasal Delivery of Cationic Liposome-Protamine Complex MRNA Vaccine Elicits Effective Anti-Tumor Immunity. Cell Immunol. 2020, 354, 104143. [Google Scholar] [CrossRef]
- Habrant, D.; Peuziat, P.; Colombani, T.; Dallet, L.; Gehin, J.; Goudeau, E.; Evrard, B.; Lambert, O.; Haudebourg, T.; Pitard, B. Design of Ionizable Lipids To Overcome the Limiting Step of Endosomal Escape: Application in the Intracellular Delivery of MRNA, DNA, and SiRNA. J. Med. Chem. 2016, 59, 3046–3062. [Google Scholar] [CrossRef]
- Kulkarni, J.A.; Darjuan, M.M.; Mercer, J.E.; Chen, S.; Van Der Meel, R.; Thewalt, J.L.; Tam, Y.Y.C.; Cullis, P.R. On the Formation and Morphology of Lipid Nanoparticles Containing Ionizable Cationic Lipids and SiRNA. ACS Nano 2018, 12, 4787–4795. [Google Scholar] [CrossRef]
- Fedorovskiy, A.G.; Antropov, D.N.; Dome, A.S.; Puchkov, P.A.; Makarova, D.M.; Konopleva, M.V.; Matveeva, A.M.; Panova, E.A.; Shmendel, E.V.; Maslov, M.A.; et al. Novel Efficient Lipid-Based Delivery Systems Enable a Delayed Uptake and Sustained Expression of MRNA in Human Cells and Mouse Tissues. Pharmaceutics 2024, 16, 684. [Google Scholar] [CrossRef]
- Pozzi, D.; Marchini, C.; Cardarelli, F.; Amenitsch, H.; Garulli, C.; Bifone, A.; Caracciolo, G. Transfection Efficiency Boost of Cholesterol-Containing Lipoplexes. Biochim. Biophys. Acta. Biomembr. 2012, 1818, 2335–2343. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Ahn, H.J. PEGylated DC-Chol/DOPE Cationic Liposomes Containing KSP SiRNA as a Systemic SiRNA Delivery Carrier for Ovarian Cancer Therapy. Biochem. Biophys. Res. Commun. 2018, 503, 1716–1722. [Google Scholar] [CrossRef]
- Semple, S.C.; Klimuk, S.K.; Harasym, T.O.; Dos Santos, N.; Ansell, S.M.; Wong, K.F.; Maurer, N.; Stark, H.; Cullis, P.R.; Hope, M.J.; et al. Efficient Encapsulation of Antisense Oligonucleotides in Lipid Vesicles Using Ionizable Aminolipids: Formation of Novel Small Multilamellar Vesicle Structures. Biochim. Biophys. Acta (BBA) Biomembr. 2001, 1510, 152–166. [Google Scholar] [CrossRef]
- Lin, P.J.C.; Tam, Y.Y.C.; Hafez, I.; Sandhu, A.; Chen, S.; Ciufolini, M.A.; Nabi, I.R.; Cullis, P.R. Influence of Cationic Lipid Composition on Uptake and Intracellular Processing of Lipid Nanoparticle Formulations of SiRNA. Nanomedicine 2013, 9, 233–246. [Google Scholar] [CrossRef]
- Dong, Y.; Love, K.T.; Dorkin, J.R.; Sirirungruang, S.; Zhang, Y.; Chen, D.; Bogorad, R.L.; Yin, H.; Chen, Y.; Vegas, A.J.; et al. Lipopeptide Nanoparticles for Potent and Selective SiRNA Delivery in Rodents and Nonhuman Primates. Proc. Natl. Acad. Sci. USA 2014, 111, 3955–3960. [Google Scholar] [CrossRef]
- Maier, M.A.; Jayaraman, M.; Matsuda, S.; Liu, J.; Barros, S.; Querbes, W.; Tam, Y.K.; Ansell, S.M.; Kumar, V.; Qin, J.; et al. Biodegradable Lipids Enabling Rapidly Eliminated Lipid Nanoparticles for Systemic Delivery of RNAi Therapeutics. Mol. Ther. 2013, 21, 1570–1578. [Google Scholar] [CrossRef]
- Buyens, K.; De Smedt, S.C.; Braeckmans, K.; Demeester, J.; Peeters, L.; van Grunsven, L.A.; de Mollerat du Jeu, X.; Sawant, R.; Torchilin, V.; Farkasova, K.; et al. Liposome Based Systems for Systemic SiRNA Delivery: Stability in Blood Sets the Requirements for Optimal Carrier Design. J. Control. Release 2012, 158, 362–370. [Google Scholar] [CrossRef]
- Cox, A.; Lim, S.A.; Chung, E.J. Strategies to Deliver RNA by Nanoparticles for Therapeutic Potential. Mol. Aspects Med. 2022, 83, 100991. [Google Scholar] [CrossRef]
- John, R.; Monpara, J.; Swaminathan, S.; Kalhapure, R. Chemistry and Art of Developing Lipid Nanoparticles for Biologics Delivery: Focus on Development and Scale-Up. Pharmaceutics 2024, 16, 131. [Google Scholar] [CrossRef] [PubMed]
- Nsairat, H.; Khater, D.; Sayed, U.; Odeh, F.; Al Bawab, A.; Alshaer, W. Liposomes: Structure, Composition, Types, and Clinical Applications. Heliyon 2022, 8, e09394. [Google Scholar] [CrossRef] [PubMed]
- Ong, S.G.M.; Ming, L.C.; Lee, K.S.; Yuen, K.H. Influence of the Encapsulation Efficiency and Size of Liposome on the Oral Bioavailability of Griseofulvin-Loaded Liposomes. Pharmaceutics 2016, 8, 25. [Google Scholar] [CrossRef]
- Fan, Y.; Marioli, M.; Zhang, K. Analytical Characterization of Liposomes and Other Lipid Nanoparticles for Drug Delivery. J. Pharm. Biomed. Anal. 2021, 192, 113642. [Google Scholar] [CrossRef] [PubMed]
- Rengaswamy, V.; Zimmer, D.; Süss, R.; Rössler, J. RGD Liposome-Protamine-SiRNA (LPR) Nanoparticles Targeting PAX3-FOXO1 for Alveolar Rhabdomyosarcoma Therapy. J. Control. Release 2016, 235, 319–327. [Google Scholar] [CrossRef]
- Jovanović, A.A.; Balanč, B.D.; Ota, A.; Ahlin Grabnar, P.; Djordjević, V.B.; Šavikin, K.P.; Bugarski, B.M.; Nedović, V.A.; Poklar Ulrih, N. Comparative Effects of Cholesterol and β-Sitosterol on the Liposome Membrane Characteristics. Eur. J. Lipid Sci. Technol. 2018, 120, 1800039. [Google Scholar] [CrossRef]
- Nosova, A.S.; Koloskova, O.O.; Nikonova, A.A.; Simonova, V.A.; Smirnov, V.V.; Kudlay, D.; Khaitov, M.R. Diversity of PEGylation Methods of Liposomes and Their Influence on RNA Delivery. Medchemcomm 2019, 10, 369–377. [Google Scholar] [CrossRef]
- Kulkarni, J.A.; Witzigmann, D.; Chen, S.; Cullis, P.R.; Van Der Meel, R. Lipid Nanoparticle Technology for Clinical Translation of SiRNA Therapeutics. Acc. Chem. Res. 2019, 52, 2435–2444. [Google Scholar] [CrossRef]
- Noble, G.T.; Stefanick, J.F.; Ashley, J.D.; Kiziltepe, T.; Bilgicer, B. Ligand-Targeted Liposome Design: Challenges and Fundamental Considerations. Trends Biotechnol. 2014, 32, 32–45. [Google Scholar] [CrossRef]
- Aranda-Lara, L.; Morales-Avila, E.; Luna-Gutiérrez, M.A.; Olivé-Alvarez, E.; Isaac-Olivé, K. Radiolabeled Liposomes and Lipoproteins as Lipidic Nanoparticles for Imaging and Therapy. Chem. Phys. Lipids 2020, 230, 104934. [Google Scholar] [CrossRef]
- Liang, Z.; Du, L.; Zhang, E.; Zhao, Y.; Wang, W.; Ma, P.; Dai, M.; Zhao, Q.; Xu, H.; Zhang, S.; et al. Targeted-Delivery of SiRNA via a Polypeptide-Modified Liposome for the Treatment of Gp96 over-Expressed Breast Cancer. Mater. Sci. Eng. C 2021, 121, 111847. [Google Scholar] [CrossRef]
- Belletti, D.; Tonelli, M.; Forni, F.; Tosi, G.; Vandelli, M.A.; Ruozi, B. AFM and TEM Characterization of SiRNAs Lipoplexes: A Combinatory Tools to Predict the Efficacy of Complexation. Colloids Surf. A Physicochem. Eng. Asp. 2013, 436, 459–466. [Google Scholar] [CrossRef]
- Park, J.; Park, J.; Pei, Y.; Xu, J.; Yeo, Y. Pharmacokinetics and Biodistribution of Recently-Developed SiRNA Nanomedicines. Adv. Drug Deliv. Rev. 2016, 104, 93–109. [Google Scholar] [CrossRef] [PubMed]
- Chono, S.; Li, S.D.; Conwell, C.C.; Huang, L. An Efficient and Low Immunostimulatory Nanoparticle Formulation for Systemic SiRNA Delivery to the Tumor. J. Control. Release 2008, 131, 64–69. [Google Scholar] [CrossRef]
- Buyens, K.; Demeester, J.; De Smedt, S.S.; Sanders, N.N. Elucidating the Encapsulation of Short Interfering RNA in PEGylated Cationic Liposomes. Langmuir 2009, 25, 4886–4891. [Google Scholar] [CrossRef]
- Chen, S.; Tam, Y.Y.C.; Lin, P.J.C.; Sung, M.M.H.; Tam, Y.K.; Cullis, P.R. Influence of Particle Size on the in Vivo Potency of Lipid Nanoparticle Formulations of SiRNA. J. Control. Release 2016, 235, 236–244. [Google Scholar] [CrossRef] [PubMed]
- Mokhtarieh, A.A.; Lee, J.; Kim, S.; Lee, M.K. Preparation of SiRNA Encapsulated Nanoliposomes Suitable for SiRNA Delivery by Simply Discontinuous Mixing. Biochim. Biophys. Acta Biomembr. 2018, 1860, 1318–1325. [Google Scholar] [CrossRef]
- Kim, H.K.; Davaa, E.; Myung, C.S.; Park, J.S. Enhanced SiRNA Delivery Using Cationic Liposomes with New Polyarginine-Conjugated PEG-Lipid. Int. J. Pharm. 2010, 392, 141–147. [Google Scholar] [CrossRef]
- Vader, P.; Crielaard, B.J.; Van Dommelen, S.M.; Van Der Meel, R.; Storm, G.; Schiffelers, R.M. Targeted Delivery of Small Interfering RNA to Angiogenic Endothelial Cells with Liposome-Polycation-DNA Particles. J. Control. Release 2012, 160, 211–216. [Google Scholar] [CrossRef]
- Grijalvo, S.; Alagia, A.; Jorge, A.; Eritja, R. Covalent Strategies for Targeting Messenger and Non-Coding RNAs: An Updated Review on SiRNA, MiRNA and AntimiR Conjugates. Genes 2018, 9, 74. [Google Scholar] [CrossRef] [PubMed]
- Kalita, T.; Dezfouli, S.A.; Pandey, L.M.; Uludag, H. SiRNA Functionalized Lipid Nanoparticles (LNPs) in Management of Diseases. Pharmaceutics 2022, 14, 2520. [Google Scholar] [CrossRef]
- Yonezawa, S.; Koide, H.; Asai, T. Recent Advances in SiRNA Delivery Mediated by Lipid-Based Nanoparticles. Adv. Drug Deliv. Rev. 2020, 154–155, 64–78. [Google Scholar] [CrossRef]
- Xue, H.Y.; Wong, H.L. Tailoring Nanostructured Solid-Lipid Carriers for Time-Controlled Intracellular SiRNA Kinetics to Sustain RNAi-Mediated Chemosensitization. Biomaterials 2011, 32, 2662–2672. [Google Scholar] [CrossRef]
- Cortesi, R.; Campioni, M.; Ravani, L.; Drechsler, M.; Pinotti, M.; Esposito, E. Cationic Lipid Nanosystems as Carriers for Nucleic Acids. N. Biotechnol. 2014, 31, 44–54. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.H.; Kim, E.; Park, D.E.; Shim, G.; Lee, S.; Kim, Y.B.; Kim, C.-W.; Oh, Y.-K. Cationic Solid Lipid Nanoparticles for Co-Delivery of Paclitaxel and SiRNA. Eur. J. Pharm. Biopharm. 2012, 80, 268–273. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, M.C.; Carbone, C.; Souto, E.B. Beyond Liposomes: Recent Advances on Lipid Based Nanostructures for Poorly Soluble/Poorly Permeable Drug Delivery. Prog. Lipid Res. 2017, 68, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, T.S.; Lee, A.C.H.; Akinc, A.; Bramlage, B.; Bumcrot, D.; Fedoruk, M.N.; Harborth, J.; Heyes, J.A.; Jeffs, L.B.; John, M.; et al. RNAi-Mediated Gene Silencing in Non-Human Primates. Nature 2006, 441, 111–114. [Google Scholar] [CrossRef]
- Sarisozen, C.; Salzano, G.P.; Torchilin, V. Lipid-Based SiRNA Delivery Systems: Challenges, Promises and Solutions Along the Long Journey. Curr. Pharm. Biotechnol. 2016, 17, 728–740. [Google Scholar] [CrossRef]
- Jia, Z.; Gong, Y.; Pi, Y.; Liu, X.; Gao, L.; Kang, L.; Wang, J.; Yang, F.; Tang, J.; Lu, W.; et al. PPB Peptide-Mediated SiRNA-Loaded Stable Nucleic Acid Lipid Nanoparticles on Targeting Therapy of Hepatic Fibrosis. Mol. Pharm. 2018, 15, 53–62. [Google Scholar] [CrossRef]
- Ge, X.; Chen, L.; Zhao, B.; Yuan, W. Rationale and Application of PEGylated Lipid-Based System for Advanced Target Delivery of SiRNA. Front. Pharmacol. 2021, 11, 598175. [Google Scholar] [CrossRef]
- Chen, S.; Tam, Y.Y.C.; Lin, P.J.C.; Leung, A.K.K.; Tam, Y.K.; Cullis, P.R. Development of Lipid Nanoparticle Formulations of SiRNA for Hepatocyte Gene Silencing Following Subcutaneous Administration. J. Control. Release 2014, 196, 106–112. [Google Scholar] [CrossRef]
- Gao, L.Y.; Liu, X.Y.; Chen, C.J.; Wang, J.C.; Feng, Q.; Yu, M.Z.; Ma, X.F.; Pei, X.W.; Niu, Y.J.; Qiu, C.; et al. Core-Shell Type Lipid/RPAA-Chol Polymer Hybrid Nanoparticles for Invivo SiRNA Delivery. Biomaterials 2014, 35, 2066–2078. [Google Scholar] [CrossRef]
- Parveen, S.; Gupta, P.; Kumar, S.; Banerjee, M. Lipid Polymer Hybrid Nanoparticles as Potent Vehicles for Drug Delivery in Cancer Therapeutics. Med. Drug Discov. 2023, 20, 100165. [Google Scholar] [CrossRef]
- Yang, X.Z.; Dou, S.; Wang, Y.C.; Long, H.Y.; Xiong, M.H.; Mao, C.Q.; Yao, Y.D.; Wang, J. Single-Step Assembly of Cationic Lipid-Polymer Hybrid Nanoparticles for Systemic Delivery of SiRNA. ACS Nano 2012, 6, 4955–4965. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhao, P.; Su, W.; Wang, S.; Liao, Z.; Niu, R.; Chang, J. PLGA/Polymeric Liposome for Targeted Drug and Gene Co-Delivery. Biomaterials 2010, 31, 8741–8748. [Google Scholar] [CrossRef]
- Zhao, X.; Li, F.; Li, Y.; Wang, H.; Ren, H.; Chen, J.; Nie, G.; Hao, J. Co-Delivery of HIF1α SiRNA and Gemcitabine via Biocompatible Lipid-Polymer Hybrid Nanoparticles for Effective Treatment of Pancreatic Cancer. Biomaterials 2015, 46, 13–25. [Google Scholar] [CrossRef]
- Thanki, K.; Zeng, X.; Foged, C. Preparation, Characterization, and In Vitro Evaluation of Lipidoid–Polymer Hybrid Nanoparticles for SiRNA Delivery to the Cytosol. In Methods in Molecular Biology; Humana Press Inc.: Totowa, NJ, USA, 2019; Volume 1943, pp. 141–152. [Google Scholar]
- Li, J.; He, Y.Z.; Li, W.; Shen, Y.Z.; Li, Y.R.; Wang, Y.F. A Novel Polymer-Lipid Hybrid Nanoparticle for Efficient Nonviral Gene Delivery. Acta Pharmacol. Sin. 2010, 31, 509–514. [Google Scholar] [CrossRef]
- Hasan, W.; Chu, K.; Gullapalli, A.; Dunn, S.S.; Enlow, E.M.; Luft, J.C.; Tian, S.; Napier, M.E.; Pohlhaus, P.D.; Rolland, J.P.; et al. Delivery of Multiple SiRNAs Using Lipid-Coated PLGA Nanoparticles for Treatment of Prostate Cancer. Nano Lett. 2012, 12, 287–292. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, G.; Vila, M.; Portolés, M.T.; Vallet-Regi, M.; Gracio, J.; Marques, P.A.A.P. Nano-Graphene Oxide: A Potential Multifunctional Platform for Cancer Therapy. Adv. Healthc. Mater. 2013, 2, 1072–1090. [Google Scholar] [CrossRef] [PubMed]
- Bose, R.J.C.; Arai, Y.; Ahn, J.C.; Park, H.; Lee, S.H. Influence of Cationic Lipid Concentration on Properties of Lipid–Polymer Hybrid Nanospheres for Gene Delivery. Int. J. Nanomed. 2015, 10, 5367–5382. [Google Scholar] [CrossRef] [PubMed]
- Persano, F.; Gigli, G.; Leporatti, S. Lipid-Polymer Hybrid Nanoparticles in Cancer Therapy: Current Overview and Future Directions. Nano Express 2021, 2, 012006. [Google Scholar] [CrossRef]
- Wang, J.; Xie, L.; Wang, T.; Wu, F.; Meng, J.; Liu, J.; Xu, H. Visible Light-Switched Cytosol Release of SiRNA by Amphiphilic Fullerene Derivative to Enhance RNAi Efficacy in Vitro and in Vivo. Acta. Biomater. 2017, 59, 158–169. [Google Scholar] [CrossRef]
- Yang, Z.; Luo, X.; Zhang, X.; Liu, J.; Jiang, Q. Targeted Delivery of 10-Hydroxycamptothecin to Human Breast Cancers by Cyclic RGD-Modified Lipid-Polymer Hybrid Nanoparticles. Biomed. Mater. 2013, 8, 025012. [Google Scholar] [CrossRef]
- Krishnamurthy, S.; Vaiyapuri, R.; Zhang, L.; Chan, J.M. Lipid-Coated Polymeric Nanoparticles for Cancer Drug Delivery. Biomater. Sci. 2015, 3, 923–936. [Google Scholar] [CrossRef]
- Triantafyllopoulou, E.; Perinelli, D.R.; Forys, A.; Pantelis, P.; Gorgoulis, V.G.; Lagopati, N.; Trzebicka, B.; Bonacucina, G.; Valsami, G.; Pippa, N.; et al. Unveiling the Performance of Co-Assembled Hybrid Nanocarriers: Moving towards the Formation of a Multifunctional Lipid/Random Copolymer Nanoplatform. Pharmaceutics 2024, 16, 1204. [Google Scholar] [CrossRef]
- Rezaee, M.; Oskuee, R.K.; Nassirli, H.; Malaekeh-Nikouei, B. Progress in the Development of Lipopolyplexes as Efficient Non-Viral Gene Delivery Systems. J. Control. Release 2016, 236, 1–14. [Google Scholar] [CrossRef]
- Jain, S.; Kumar, M.; Kumar, P.; Verma, J.; Rosenholm, J.M.; Bansal, K.K.; Vaidya, A. Lipid–Polymer Hybrid Nanosystems: A Rational Fusion for Advanced Therapeutic Delivery. J. Funct. Biomater. 2023, 14, 437. [Google Scholar] [CrossRef]
- Morakul, B. Self-Nanoemulsifying Drug Delivery Systems (SNEDDS): An Advancement Technology for Oral Drug Delivery. Pharm. Sci. Asia 2020, 47, 205–220. [Google Scholar] [CrossRef]
- Kubackova, J.; Holas, O.; Zbytovska, J.; Vranikova, B.; Zeng, G.; Pavek, P.; Mullertz, A. Oligonucleotide Delivery across the Caco-2 Monolayer: The Design and Evaluation of Self-Emulsifying Drug Delivery Systems (Sedds). Pharmaceutics 2021, 13, 459. [Google Scholar] [CrossRef]
- Reyna-Lázaro, L.; Morales-Becerril, A.; Aranda-Lara, L.; Isaac-Olivé, K.; Ocampo-García, B.; Gibbens-Bandala, B.; Olea-Mejía, O.; Morales-Avila, E. Pharmaceutical Nanoplatforms Based on Self-Nanoemulsifying Drug Delivery Systems for Optimal Transport and Co-Delivery of SiRNAs and Anticancer Drugs. J. Pharm. Sci. 2024, 113, 1907–1918. [Google Scholar] [CrossRef] [PubMed]
- Attri, N.; Das, S.; Banerjee, J.; Shamsuddin, S.H.; Dash, S.K.; Pramanik, A. Liposomes to Cubosomes: The Evolution of Lipidic Nanocarriers and Their Cutting-Edge Biomedical Applications. ACS Appl. Bio. Mater. 2024, 7, 2677–2694. [Google Scholar] [CrossRef] [PubMed]
- Sivadasan, D.; Sultan, M.H.; Alqahtani, S.S.; Javed, S. Cubosomes in Drug Delivery—A Comprehensive Review on Its Structural Components, Preparation Techniques and Therapeutic Applications. Biomedicines 2023, 11, 1114. [Google Scholar] [CrossRef]
- Sarkar, S.; Tran, N.; Soni, S.K.; Nasa, Z.; Drummond, C.J.; Conn, C.E. Cuboplex-Mediated Nonviral Delivery of Functional SiRNA to Chinese Hamster Ovary (CHO) Cells. ACS Appl. Mater. Interfaces 2021, 13, 2336–2345. [Google Scholar] [CrossRef] [PubMed]
- Yaghmur, A.; Østergaard, J.; Mu, H. Lipid Nanoparticles for Targeted Delivery of Anticancer Therapeutics: Recent Advances in Development of SiRNA and Lipoprotein-Mimicking Nanocarriers. Adv. Drug Deliv. Rev. 2023, 203, 115136. [Google Scholar] [CrossRef]
- Busatto, S.; Walker, S.A.; Grayson, W.; Pham, A.; Tian, M.; Nesto, N.; Barklund, J.; Wolfram, J. Lipoprotein-Based Drug Delivery. Adv. Drug Deliv. Rev. 2020, 159, 377–390. [Google Scholar] [CrossRef]
- Dai, L.; Li, S.; Hao, Q.; Zhou, R.; Zhou, H.; Lei, W.; Kang, H.; Wu, H.; Li, Y.; Ma, X. Low-Density Lipoprotein: A Versatile Nanoscale Platform for Targeted Delivery. Nanoscale Adv. 2023, 5, 1011–1022. [Google Scholar] [CrossRef]
- McMahon, K.M.; Plebanek, M.P.; Thaxton, C.S. Properties of Native High-Density Lipoproteins Inspire Synthesis of Actively Targeted In Vivo SiRNA Delivery Vehicles. Adv. Funct. Mater. 2016, 26, 7824–7835. [Google Scholar] [CrossRef]
- McMahon, K.M.; Thaxton, C.S. High-Density Lipoproteins for the Systemic Delivery of Short Interfering RNA. Expert. Opin. Drug Deliv. 2014, 11, 231–247. [Google Scholar] [CrossRef] [PubMed]
- Lacko, A.G.; Sabnis, N.A.; Nagarajan, B.; McConathy, W.J. HDL as a Drug and Nucleic Acid Delivery Vehicle. Front. Pharmacol. 2015, 6, 247. [Google Scholar] [CrossRef] [PubMed]
- Lacko, A.G.; Nair, M.; Prokai, L.; McConathy, W.J. Prospects and Challenges of the Development of Lipoprotein-Based Formulations for Anti-Cancer Drugs. Expert. Opin. Drug Deliv. 2007, 4, 665–675. [Google Scholar] [CrossRef] [PubMed]
- Almer, G.; Mangge, H.; Zimmer, A.; Prassl, R. Lipoprotein-Related and Apolipoprotein-Mediated Delivery Systems for Drug Targeting and Imaging. Curr. Med. Chem. 2015, 22, 3631–3651. [Google Scholar] [CrossRef]
- Aranda-Lara, L.; Isaac-Olivé, K.; Ocampo-García, B.; Ferro-Flores, G.; González-Romero, C.; Mercado-López, A.; García-Marín, R.; Santos-Cuevas, C.; Estrada, J.A.; Morales-Avila, E. Engineered RHDL Nanoparticles as a Suitable Platform for Theranostic Applications. Molecules 2022, 27, 7046. [Google Scholar] [CrossRef]
- Ng, K.K.; Lovell, J.F.; Zheng, G. Lipoprotein-Inspired Nanoparticles for Cancer Theranostics. Acc. Chem. Res. 2011, 44, 1105–1113. [Google Scholar] [CrossRef]
- Yang, S.D.; Zhu, W.J.; Zhu, Q.L.; Chen, W.L.; Ren, Z.X.; Li, F.; Yuan, Z.Q.; Li, J.Z.; Liu, Y.; Zhou, X.F.; et al. Binary-Copolymer System Base on Low-Density Lipoprotein-Coupled N-Succinyl Chitosan Lipoic Acid Micelles for Co-Delivery MDR1 SiRNA and Paclitaxel, Enhances Antitumor Effects via Reducing Drug. J. Biomed. Mater. Res. B Appl. Biomater. 2017, 105, 1114–1125. [Google Scholar] [CrossRef]
- Quintos-Meneses, H.A.; Aranda-Lara, L.; Morales-Ávila, E.; Ocampo-García, B.; Contreras, I.; Ramírez-Nava, G.J.; Santos-Cuevas, C.L.; Estrada, J.A.; Luna-Gutiérrez, M.A.; Ferro-Flores, G.; et al. A Multimodal Theranostic System Prepared from High-Density Lipoprotein Carrier of Doxorubicin and 177 Lu. J. Biomed. Nanotechnol. 2021, 17, 2125–2141. [Google Scholar] [CrossRef]
- He, B.; Yang, Q. Recent Development of LDL-Based Nanoparticles for Cancer Therapy. Pharmaceuticals 2022, 16, 18. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Li, Z.; Zheng, Q.; Yao, Q. Correlation Study of Serum Lipid Levels and Lipid Metabolism-Related Genes in Cervical Cancer. Front. Oncol. 2024, 14, 1384778. [Google Scholar] [CrossRef]
- Deng, C.-F.; Zhu, N.; Zhao, T.-J.; Li, H.-F.; Gu, J.; Liao, D.-F.; Qin, L. Involvement of LDL and Ox-LDL in Cancer Development and Its Therapeutical Potential. Front. Oncol. 2022, 12, 803473. [Google Scholar] [CrossRef]
- Jin, H.; Lovell, J.F.; Chen, J.; Lin, Q.; Ding, L.; Ng, K.K.; Pandey, R.K.; Manoharan, M.; Zhang, Z.; Zheng, G. Mechanistic Insights into LDL Nanoparticle-Mediated SiRNA Delivery. Bioconjug Chem. 2012, 23, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Duan, X.; Lv, Y.; Zhao, Y. Low Density Lipoprotein Receptor (LDLR)-Targeted Lipid Nanoparticles for the Delivery of Sorafenib and Dihydroartemisinin in Liver Cancers. Life Sci. 2019, 239, 117013. [Google Scholar] [CrossRef]
- Zhu, C.; Pradhan, P.; Huo, D.; Xue, J.; Shen, S.; Roy, K.; Xia, Y. Reconstitution of Low-Density Lipoproteins with Fatty Acids for the Targeted Delivery of Drugs into Cancer Cells. Angew. Chem. Int. Ed. 2017, 56, 10399–10402. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Wang, Y.; Huang, Y.; Zi, Y.; Yan, S.; Shi, X.; Cai, J.; Zhang, H.; Sang, J.; Zhang, W.; et al. Bioinspired Low-Density Lipoprotein Co-Delivery System for Targeting and Synergistic Cancer Therapy. Nanomedicine 2023, 48, 102641. [Google Scholar] [CrossRef]
- McMahon, K.M.; Foit, L.; Angeloni, N.L.; Giles, F.J.; Gordon, L.I.; Thaxton, C.S. Synthetic High-Density Lipoprotein-like Nanoparticles as Cancer Therapy. Cancer Treat. Res. 2015, 166, 129–150. [Google Scholar] [CrossRef]
- Wolfrum, C.; Shi, S.; Jayaprakash, K.N.; Jayaraman, M.; Wang, G.; Pandey, R.K.; Rajeev, K.G.; Nakayama, T.; Charrise, K.; Ndungo, E.M.; et al. Mechanisms and Optimization of in Vivo Delivery of Lipophilic SiRNAs. Nat. Biotechnol. 2007, 25, 1149–1157. [Google Scholar] [CrossRef]
- Kuwahara, H.; Nishina, K.; Yoshida, K.; Nishina, T.; Yamamoto, M.; Saito, Y.; Piao, W.; Yoshida, M.; Mizusawa, H.; Yokota, T. Efficient in Vivo Delivery of SiRNA into Brain Capillary Endothelial Cells along with Endogenous Lipoprotein. Mol. Ther. 2011, 19, 2213–2221. [Google Scholar] [CrossRef]
- Tanaka, M.; Hasegawa, M.; Yoshimoto, N.; Hoshikawa, K.; Mukai, T. Preparation of Lipid Nanodisks Containing Apolipoprotein E-Derived Synthetic Peptides for Biocompatible Delivery Vehicles Targeting Low-Density Lipoprotein Receptor. Biol. Pharm. Bull. 2019, 42, 1376–1383. [Google Scholar] [CrossRef]
- Zhu, Q.L.; Zhou, Y.; Guan, M.; Zhou, X.-F.; Yang, S.-D.; Liu, Y.; Chen, W.-L.; Zhang, C.-G.; Yuan, Z.-Q.; Liu, C.; et al. Low-Density Lipoprotein-Coupled N-Succinyl Chitosan Nanoparticles Co-Delivering SiRNA and Doxorubicin for Hepatocyte-Targeted Therapy. Biomaterials 2014, 35, 5965–5976. [Google Scholar] [CrossRef]
- Glickson, J.D.; Lund-Katz, S.; Zhou, R.; Choi, H.; Chen, I.-W.; Li, H.; Corbin, I.; Popov, A.V.; Cao, W.; Song, L.; et al. Lipoprotein Nanoplatform for Targeted Delivery of Diagnostic and Therapeutic Agents. In Oxygen Transport to Tissue XXX; Springer: Boston, MA, USA, 2009; Volume 645, pp. 227–239. [Google Scholar]
- Mei, Y.; Tang, L.; Xiao, Q.; Zhang, Z.; Zhang, Z.; Zang, J.; Zhou, J.; Wang, Y.; Wang, W.; Ren, M. Reconstituted High Density Lipoprotein (RHDL), a Versatile Drug Delivery Nanoplatform for Tumor Targeted Therapy. J. Mater. Chem. B 2021, 9, 612–633. [Google Scholar] [CrossRef]
- Shahzad, M.M.K.; Mangala, L.S.; Han, H.D.; Lu, C.; Bottsford-Miller, J.; Nishimura, M.; Mora, E.M.; Lee, J.W.; Stone, R.L.; Pecot, C.V.; et al. Targeted Delivery of Small Interfering RNA Using Reconstituted High-Density Lipoprotein Nanoparticles. Neoplasia 2011, 13, 309–319. [Google Scholar] [CrossRef] [PubMed]
- Pottash, A.E.; Kuffner, C.; Noonan-Shueh, M.; Jay, S.M. Protein-Based Vehicles for Biomimetic RNAi Delivery. J. Biol. Eng. 2019, 13, 30. [Google Scholar] [CrossRef] [PubMed]
- Zamanian-Daryoush, M.; Lindner, D.J.; Buffa, J.; Gopalan, B.; Na, J.; Hazen, S.L.; Didonato, J.A. Apolipoprotein A-I Anti-Tumor Activity Targets Cancer Cell Metabolism. Oncotarget 2020, 11, 1777–1796. [Google Scholar]
- Kannan Mutharasan, R.; Foit, L.; Shad Thaxton, C. High-Density Lipoproteins for Therapeutic Delivery Systems. J. Mater. Chem. B 2015, 4, 188–197. [Google Scholar] [CrossRef]
- Raut, S.; Mooberry, L.; Sabnis, N.; Garud, A.; Dossou, A.S.; Lacko, A. Reconstituted HDL: Drug Delivery Platform for Overcoming Biological Barriers to Cancer Therapy. Front. Pharmacol. 2018, 9, 1154. [Google Scholar] [CrossRef]
- Shen, J.; Zhang, W.; Qi, R.; Mao, Z.-W.; Shen, H. Engineering Functional Inorganic–Organic Hybrid Systems: Advances in SiRNA Therapeutics. Chem. Soc. Rev. 2018, 47, 1969–1995. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.; Chen, J.; Ng, K.K.; Cao, W.; Zhang, Z.; Zheng, G. Imaging the Cytosolic Drug Delivery Mechanism of HDL-like Nanoparticles. Pharm. Res. 2014, 31, 1438–1449. [Google Scholar] [CrossRef] [PubMed]
- Cruz, W.; Huang, H.; Barber, B.; Pasini, E.; Ding, L.; Zheng, G.; Chen, J.; Bhat, M. Lipoprotein-Like Nanoparticle Carrying Small Interfering RNA Against Spalt-Like Transcription Factor 4 Effectively Targets Hepatocellular Carcinoma Cells and Decreases Tumor Burden. Hepatol. Commun. 2020, 4, 2020. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Han, Y.; Wang, R.; Wang, Y.; Chi, C.; Zhao, Z.; Zhang, H.; Wang, W.; Yin, L.; Zhou, J. Rerouting Native HDL to Predetermined Receptors for Improved Tumor-Targeted Gene Silencing Therapy. ACS Appl. Mater. Interfaces 2017, 9, 30488–30501. [Google Scholar] [CrossRef] [PubMed]
- Uno, Y.; Piao, W.; Miyata, K.; Nishina, K.; Mizusawa, H.; Yokota, T. High-Density Lipoprotein Facilitates in Vivo Delivery of α-Tocopherol-Conjugated Short-Interfering RNA to the Brain. Hum. Gene Ther. 2011, 22, 711–719. [Google Scholar] [CrossRef]
- Rui, M.; Tang, H.; Li, Y.; Wei, X.; Xu, Y. Recombinant High Density Lipoprotein Nanoparticles for Target-Specific Delivery of SiRNA. Pharm. Res. 2013, 30, 1203–1214. [Google Scholar] [CrossRef]
- Yang, M.; Jin, H.; Chen, J.; Ding, L.; Ng, K.K.; Lin, Q.; Lovell, J.F.; Zhang, Z.; Zheng, G. Efficient Cytosolic Delivery of SiRNA Using HDL-Mimicking Nanoparticles. Small 2011, 7, 568–573. [Google Scholar] [CrossRef]
- Nakayama, T.; Butler, J.S.; Sehgal, A.; Severgnini, M.; Racie, T.; Sharman, J.; Ding, F.; Morskaya, S.S.; Brodsky, J.; Tchangov, L.; et al. Harnessing a Physiologic Mechanism for SiRNA Delivery with Mimetic Lipoprotein Particles. Mol. Ther. 2012, 20, 1582–1589. [Google Scholar] [CrossRef]
- Ding, Y.; Wang, Y.; Zhou, J.; Gu, X.; Wang, W.; Liu, C.; Bao, X.; Wang, C.; Li, Y.; Zhang, Q. Direct Cytosolic SiRNA Delivery by Reconstituted High Density Lipoprotein for Target-Specific Therapy of Tumor Angiogenesis. Biomaterials 2014, 35, 7214–7227. [Google Scholar] [CrossRef]
- Malajczuk, C.J.; Gandhi, N.S.; Mancera, R.L. Structure and Intermolecular Interactions in Spheroidal High-Density Lipoprotein Subpopulations. J. Struct. Biol. X 2021, 5, 100042. [Google Scholar] [CrossRef]
- Corbin, I.R. Ligand-Coupled Lipoprotein for Ovarian Cancer-Specific Drug Delivery. Methods Mol. Biol. 2013, 1049, 467–480. [Google Scholar] [CrossRef]
- Pérez-Medina, C.; Binderup, T.; Lobatto, M.E.; Tang, J.; Calcagno, C.; Giesen, L.; Wessel, C.H.; Witjes, J.; Ishino, S.; Baxter, S.; et al. In Vivo PET Imaging of HDL in Multiple Atherosclerosis Models. JACC Cardiovasc. Imaging 2016, 9, 950–961. [Google Scholar] [CrossRef] [PubMed]
- Kuai, R.; Li, D.; Chen, Y.E.; Moon, J.J.; Schwendeman, A. High-Density Lipoproteins: Nature’s Multifunctional Nanoparticles. ACS Nano 2016, 10, 3015–3041. [Google Scholar] [CrossRef]
- Ávila-Sánchez, M.A.; Isaac-Olivé, K.; Aranda-Lara, L.; Morales-Ávila, E.; Plata-Becerril, A.; Jiménez-Mancilla, N.P.; Ocampo-García, B.; Estrada, J.A.; Santos-Cuevas, C.L.; Torres-García, E.; et al. Targeted Photodynamic Therapy Using Reconstituted High-Density Lipoproteins as Rhodamine Transporters. Photodiagnosis Photodyn. Ther. 2022, 37, 102630. [Google Scholar] [CrossRef]
- Chuang, S.T.; Cruz, S.; Narayanaswami, V. Reconfiguring Nature’s Cholesterol Accepting Lipoproteins as Nanoparticle Platforms for Transport and Delivery of Therapeutic and Imaging Agents. Nanomaterials 2020, 10, 906. [Google Scholar] [CrossRef] [PubMed]
- Ubanako, P.; Mirza, S.; Ruff, P.; Penny, C. Exosome-Mediated Delivery of SiRNA Molecules in Cancer Therapy: Triumphs and Challenges. Front. Mol. Biosci. 2024, 11, 1447953. [Google Scholar] [CrossRef]
- Chen, J.; Fei, X.; Wang, J.; Cai, Z. Tumor-Derived Extracellular Vesicles: Regulators of Tumor Microenvironment and the Enlightenment in Tumor Therapy. Pharmacol. Res. 2020, 159, 105041. [Google Scholar] [CrossRef]
- Alvarez-Erviti, L.; Seow, Y.; Yin, H.; Betts, C.; Lakhal, S.; Wood, M.J.A. Delivery of SiRNA to the Mouse Brain by Systemic Injection of Targeted Exosomes. Nat. Biotechnol. 2011, 29, 341–345. [Google Scholar] [CrossRef]
- Kang, J.Y.; Mun, D.; Chun, Y.; Park, D.S.; Kim, H.; Yun, N.; Joung, B. Engineered Small Extracellular Vesicle-mediated NOX4 SiRNA Delivery for Targeted Therapy of Cardiac Hypertrophy. J. Extracell. Vesicles 2023, 12, e12371. [Google Scholar] [CrossRef] [PubMed]
- Ahmadi-Kashani, M.; Dehghani, H.; Zarrabi, A. A Biocompatible Nanoplatform Formed by MgAl-Layered Double Hydroxide Modified Mn3O4/N-Graphene Quantum Dot Conjugated-Polyaniline for PH-Triggered Release of Doxorubicin. Mater. Sci. Eng. C 2020, 114, 111055. [Google Scholar] [CrossRef]
- Jiang, Y.; Huo, S.; Hardie, J.; Liang, X.-J.; Rotello, V.M. Progress and Perspective of Inorganic Nanoparticle-Based SiRNA Delivery Systems. Expert. Opin. Drug Deliv. 2016, 13, 547–559. [Google Scholar] [CrossRef]
- Mobarakeh, V.; Modarressi, M.H.; Rahimi, P.; Bolhassani, A.; Arefian, E. Modification of SPION Nanocarriers for SiRNA Delivery: A Therapeutic Strategy against HIV Infection. Vaccine Res. 2019, 6, 43–49. [Google Scholar] [CrossRef]
- Chhour, P.; Naha, P.C.; Cheheltani, R.; Benardo, B.; Mian, S.; Cormode, D.P. Gold Nanoparticles for Biomedical Applications: Synthesis and In Vitro Evaluation. In Methods in Pharmacology and Toxicology; Humana Press Inc.: Totowa, NJ, USA, 2016; Volume 39, pp. 87–111. [Google Scholar]
- Wu, J.; Liu, B.; Wu, H.; Wu, Y.; Zhang, W.; Zhao, S.; Zhang, L.; Pan, X.; Gao, W.; Wang, X.; et al. A Gold Nanoparticle Platform for the Delivery of Functional TGF-Β1 SiRNA into Cancer Cells. J. Biomed. Nanotechnol. 2016, 12, 800–810. [Google Scholar] [CrossRef]
- Liu, X.Y.; Wang, J.Q.; Ashby, C.R.; Zeng, L.; Fan, Y.F.; Chen, Z.S. Gold Nanoparticles: Synthesis, Physiochemical Properties and Therapeutic Applications in Cancer. Drug Discov. Today 2021, 26, 1284–1292. [Google Scholar] [CrossRef]
- Cruz-Rodríguez, J.C.; Camacho-López, M.A.; Torres-García, E.; Aranda-Lara, L.; Morales-Avila, E.; Díaz-Sánchez, L.E.; Jiménez-Mancilla, N.P.; Isaac-Olivé, K. Characterization of the Absorption Properties of 5 Nm Spherical Gold Nanoparticles Functionalized with Dodecanothiol and without Functionalization with Potential Therapeutic Applications. Phys. Scr. 2023, 98, 065005. [Google Scholar] [CrossRef]
- DeLong, R.K.; Akhtar, U.; Sallee, M.; Parker, B.; Barber, S.; Zhang, J.; Craig, M.; Garrad, R.; Hickey, A.J.; Engstrom, E. Characterization and Performance of Nucleic Acid Nanoparticles Combined with Protamine and Gold. Biomaterials 2009, 30, 6451–6459. [Google Scholar] [CrossRef]
- Kong, L.; Wu, Y.; Alves, C.S.; Shi, X. Efficient Delivery of Therapeutic SiRNA into Glioblastoma Cells Using Multifunctional Dendrimer-Entrapped Gold Nanoparticles. Nanomedicine 2016, 12, 3103–3115. [Google Scholar] [CrossRef]
- Melamed, J.R.; Riley, R.S.; Valcourt, D.M.; Billingsley, M.M.; Kreuzberger, N.L.; Day, E.S. Quantification of SiRNA Duplexes Bound to Gold Nanoparticle Surfaces. In Methods in Molecular Biology; Humana Press Inc.: Totowa, NJ, USA, 2017; Volume 1570, pp. 1–15. [Google Scholar]
- Bloise, N.; Strada, S.; Dacarro, G.; Visai, L. Gold Nanoparticles Contact with Cancer Cell: A Brief Update. Int. J. Mol. Sci. 2022, 23, 7683. [Google Scholar] [CrossRef]
- Artiga, Á.; Serrano-Sevilla, I.; De Matteis, L.; Mitchell, S.G.; de la Fuente, J.M. Current Status and Future Perspectives of Gold Nanoparticle Vectors for SiRNA Delivery. J. Mater. Chem. B 2019, 7, 876–896. [Google Scholar] [CrossRef]
- Ekin, A.; Karatas, O.F.; Culha, M.; Ozen, M. Designing a Gold Nanoparticle-Based Nanocarrier for MicroRNA Transfection into the Prostate and Breast Cancer Cells. J. Gene Med. 2014, 16, 331–335. [Google Scholar] [CrossRef]
- Cho, T.J.; Gorham, J.M.; Pettibone, J.M.; Liu, J.; Tan, J.; Hackley, V.A. Parallel Multi-Parameter Study of PEI-Functionalized Gold Nanoparticle Synthesis for Bio-Medical Applications: Part 1—A Critical Assessment of Methodology, Properties, and Stability. J. Nanoparticle Res. 2019, 21, 148. [Google Scholar] [CrossRef]
- Elbakry, A.; Zaky, A.; Liebl, R.; Rachel, R.; Goepferich, A.; Breunig, M. Layer-by-Layer Assembled Gold Nanoparticles for Sirna Delivery. Nano Lett. 2009, 9, 2059–2064. [Google Scholar] [CrossRef]
- Kim, S.T.; Chompoosor, A.; Yeh, Y.; Agasti, S.S.; Solfiell, D.J.; Rotello, V.M. Dendronized Gold Nanoparticles for SiRNA Delivery. Small 2012, 8, 3253–3256. [Google Scholar] [CrossRef]
- Labala, S.; Jose, A.; Venuganti, V.V.K. Transcutaneous Iontophoretic Delivery of STAT3 SiRNA Using Layer-by-Layer Chitosan Coated Gold Nanoparticles to Treat Melanoma. Colloids Surf. B Biointerfaces 2016, 146, 188–197. [Google Scholar] [CrossRef]
- Shaabani, E.; Sharifiaghdam, M.; De Keersmaecker, H.; De Rycke, R.; De Smedt, S.; Faridi-Majidi, R.; Braeckmans, K.; Fraire, J.C. Layer by Layer Assembled Chitosan-Coated Gold Nanoparticles for Enhanced SiRNA Delivery and Silencing. Int. J. Mol. Sci. 2021, 22, 831. [Google Scholar] [CrossRef]
- Baghani, L.; Noroozi Heris, N.; Khonsari, F.; Dinarvand, S.; Dinarvand, M.; Atyabi, F. Trimethyl-Chitosan Coated Gold Nanoparticles Enhance Delivery, Cellular Uptake and Gene Silencing Effect of EGFR-SiRNA in Breast Cancer Cells. Front. Mol. Biosci. 2022, 9, 871541. [Google Scholar] [CrossRef]
- Elizarova, T.N.; Antopolsky, M.L.; Novichikhin, D.O.; Skirda, A.M.; Orlov, A.V.; Bragina, V.A.; Nikitin, P.I. A Straightforward Method for the Development of Positively Charged Gold Nanoparticle-Based Vectors for Effective SiRNA Delivery. Molecules 2023, 28, 3318. [Google Scholar] [CrossRef]
- Wu, L.; Zhou, W.; Lin, L.; Chen, A.; Feng, J.; Qu, X.; Zhang, H.; Yue, J. Delivery of Therapeutic Oligonucleotides in Nanoscale. Bioact. Mater. 2022, 7, 292–323. [Google Scholar] [CrossRef]
- Pędziwiatr-Werbicka, E.; Gorzkiewicz, M.; Michlewska, S.; Ionov, M.; Shcharbin, D.; Klajnert-Maculewicz, B.; Peña-González, C.E.; Sánchez-Nieves, J.; Gómez, R.; de la Mata, F.J.; et al. Evaluation of Dendronized Gold Nanoparticles as SiRNAs Carriers into Cancer Cells. J. Mol. Liq. 2021, 324, 114726. [Google Scholar] [CrossRef]
- Huang, N.; Liu, Y.; Fang, Y.; Zheng, S.; Wu, J.; Wang, M.; Zhong, W.; Shi, M.; Xing, M.; Liao, W. Gold Nanoparticles Induce Tumor Vessel Normalization and Impair Metastasis by Inhibiting Endothelial Smad2/3 Signaling. ACS Nano 2020, 14, 7940–7958. [Google Scholar] [CrossRef]
- Issa, B.; Obaidat, I.; Albiss, B.; Haik, Y. Magnetic Nanoparticles: Surface Effects and Properties Related to Biomedicine Applications. Int. J. Mol. Sci. 2013, 14, 21266–21305. [Google Scholar] [CrossRef]
- Abdelrahman, M.; Douziech Eyrolles, L.; Alkarib, S.Y.; Hervé-Aubert, K.; Ben Djemaa, S.; Marchais, H.; Chourpa, I.; David, S. SiRNA Delivery System Based on Magnetic Nanovectors: Characterization and Stability Evaluation. Eur. J. Pharm. Sci. 2017, 106, 287–293. [Google Scholar] [CrossRef]
- Gozuacik, D.; Yagci-Acar, H.F.; Akkoc, Y.; Kosar, A.; Dogan-Ekici, A.I.; Ekici, S. Anticancer Use of Nanoparticles as Nucleic Acid Carriers. J. Biomed. Nanotechnol. 2014, 10, 1751–1783. [Google Scholar] [CrossRef]
- Dalmina, M.; Pittella, F.; Sierra, J.A.; Souza, G.R.R.; Silva, A.H.; Pasa, A.A.; Creczynski-Pasa, T.B. Magnetically Responsive Hybrid Nanoparticles for in Vitro SiRNA Delivery to Breast Cancer Cells. Mater. Sci. Eng. C 2019, 99, 1182–1190. [Google Scholar] [CrossRef]
- Arango, D.; Cifuentes, J.; Puentes, P.R.; Beltran, T.; Bittar, A.; Ocasión, C.; Muñoz-Camargo, C.; Bloch, N.I.; Reyes, L.H.; Cruz, J.C. Tailoring Magnetite-Nanoparticle-Based Nanocarriers for Gene Delivery: Exploiting CRISPRa Potential in Reducing Conditions. Nanomaterials 2023, 13, 1782. [Google Scholar] [CrossRef]
- Dobson Magnetic Nanoparticles for Gene and Drug Delivery. Int. J. Nanomed. 2008, 3, 169. [CrossRef]
- Bruniaux, J.; Allard-Vannier, E.; Aubrey, N.; Lakhrif, Z.; Ben Djemaa, S.; Eljack, S.; Marchais, H.; Hervé-Aubert, K.; Chourpa, I.; David, S. Magnetic Nanocarriers for the Specific Delivery of SiRNA: Contribution of Breast Cancer Cells Active Targeting for down-Regulation Efficiency. Int. J. Pharm. 2019, 569, 118572. [Google Scholar] [CrossRef]
- Mahajan, U.M.; Teller, S.; Sendler, M.; Palankar, R.; van den Brandt, C.; Schwaiger, T.; Kühn, J.-P.; Ribback, S.; Glöckl, G.; Evert, M.; et al. Tumour-Specific Delivery of SiRNA-Coupled Superparamagnetic Iron Oxide Nanoparticles, Targeted against PLK1, Stops Progression of Pancreatic Cancer. Gut 2016, 65, 1838–1849. [Google Scholar] [CrossRef]
- Panday, R.; Abdalla, A.M.E.; Yu, M.; Li, X.; Ouyang, C.; Yang, G. Functionally Modified Magnetic Nanoparticles for Effective SiRNA Delivery to Prostate Cancer Cells in Vitro. J. Biomater. Appl. 2020, 34, 952–964. [Google Scholar] [CrossRef]
- Yang, Z.; Duan, J.; Wang, J.; Liu, Q.; Shang, R.; Yang, X.; Lu, P.; Xia, C.; Wang, L.; Dou, K. Superparamagnetic Iron Oxide Nanoparticles Modified with Polyethylenimine and Galactose for SiRNA Targeted Delivery in Hepatocellular Carcinoma Therapy. Int. J. Nanomed. 2018, 13, 1851–1865. [Google Scholar] [CrossRef]
- Lin, G.; Huang, J.; Zhang, M.; Chen, S.; Zhang, M. Chitosan-Crosslinked Low Molecular Weight PEI-Conjugated Iron Oxide Nanoparticle for Safe and Effective DNA Delivery to Breast Cancer Cells. Nanomaterials 2022, 12, 584. [Google Scholar] [CrossRef]
- Licciardi, M.; Li Volsi, A.; Sardo, C.; Mauro, N.; Cavallaro, G.; Giammona, G. Inulin-Ethylenediamine Coated SPIONs Magnetoplexes: A Promising Tool for Improving SiRNA Delivery. Pharm. Res. 2015, 32, 3674–3687. [Google Scholar] [CrossRef]
- Bi, Q.; Song, X.; Hu, A.; Luo, T.; Jin, R.; Ai, H.; Nie, Y. Magnetofection: Magic Magnetic Nanoparticles for Efficient Gene Delivery. Chin. Chem. Lett. 2020, 31, 3041–3046. [Google Scholar] [CrossRef]
- Mondal, S.; Raut, J.; Sahoo, P. Gene Silencing and Gene Delivery in Therapeutics: Insights Using Quantum Dots. Front. Biosci. 2023, 28, 364. [Google Scholar] [CrossRef]
- Alizadeh-Ghodsi, M.; Pourhassan-Moghaddam, M.; Zavari-Nematabad, A.; Walker, B.; Annabi, N.; Akbarzadeh, A. State-of-the-Art and Trends in Synthesis, Properties, and Application of Quantum Dots-Based Nanomaterials. Part. Part. Syst. Charact. 2019, 36, 1900197. [Google Scholar] [CrossRef]
- Banerjee, A.; Pons, T.; Lequeux, N.; Dubertret, B. Quantum Dots–DNA Bioconjugates: Synthesis to Applications. Interface Focus. 2016, 6, 20160064. [Google Scholar] [CrossRef]
- Li, J.-M.; Zhao, M.-X.; Su, H.; Wang, Y.-Y.; Tan, C.-P.; Ji, L.-N.; Mao, Z.-W. Multifunctional Quantum-Dot-Based SiRNA Delivery for HPV18 E6 Gene Silence and Intracellular Imaging. Biomaterials 2011, 32, 7978–7987. [Google Scholar] [CrossRef]
- Li, J.M.; Wang, Y.Y.; Zhao, M.X.; Tan, C.P.; Li, Y.Q.; Le, X.Y.; Ji, L.N.; Mao, Z.W. Multifunctional QD-Based Co-Delivery of SiRNA and Doxorubicin to HeLa Cells for Reversal of Multidrug Resistance and Real-Time Tracking. Biomaterials 2012, 33, 2780–2790. [Google Scholar] [CrossRef]
- Bae, K.H.; Lee, J.Y.; Lee, S.H.; Park, T.G.; Nam, Y.S. Optically Traceable Solid Lipid Nanoparticles Loaded with Sirna and Paclitaxel for Synergistic Chemotherapy with in Situ Imaging. Adv. Healthc. Mater. 2013, 2, 576–584. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Dai, W.; Ju, H.; Lu, H.; Wang, S.; Xu, L.; Zhou, S.F.; Zhang, Y.; Zhang, X. Multifunctional Poly(l-Lactide)-Polyethylene Glycol-Grafted Graphene Quantum Dots for Intracellular MicroRNA Imaging and Combined Specific-Gene-Targeting Agents Delivery for Improved Therapeutics. ACS Appl. Mater. Interfaces 2015, 7, 11015–11023. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.; Solanki, A.; Memoli, K.A.; Kamei, K.; Kim, H.; Drahl, M.A.; Williams, L.J.; Tseng, H.; Lee, K. Selective Inhibition of Human Brain Tumor Cells through Multifunctional Quantum-Dot-Based SiRNA Delivery. Angew. Chem. Int. Ed. 2010, 49, 103–107. [Google Scholar] [CrossRef] [PubMed]
- Endres, T.; Zheng, M.; Kiliç, A.; Turowska, A.; Beck-Broichsitter, M.; Renz, H.; Merkel, O.M.; Kissel, T. Amphiphilic Biodegradable PEG-PCL-PEI Triblock Copolymers for FRET-Capable in Vitro and in Vivo Delivery of SiRNA and Quantum Dots. Mol. Pharm. 2014, 11, 1273–1281. [Google Scholar] [CrossRef]
- Zhu, H.; Zhang, S.; Ling, Y.; Meng, G.; Yang, Y.; Zhang, W. PH-Responsive Hybrid Quantum Dots for Targeting Hypoxic Tumor SiRNA Delivery. J. Control. Release 2015, 220, 529–544. [Google Scholar] [CrossRef]
- Lee, H.; Kim, I.-K.; Park, T.G. Intracellular Trafficking and Unpacking of SiRNA/Quantum Dot-PEI Complexes Modified with and without Cell Penetrating Peptide: Confocal and Flow Cytometric FRET Analysis. Bioconjug Chem. 2010, 21, 289–295. [Google Scholar] [CrossRef] [PubMed]
- Lin, G.; Chen, T.; Zou, J.; Wang, Y.; Wang, X.; Li, J.; Huang, Q.; Fu, Z.; Zhao, Y.; Lin, M.C.M.; et al. Quantum Dots-SiRNA Nanoplexes for Gene Silencing in Central Nervous System Tumor Cells. Front. Pharmacol. 2017, 8, 182. [Google Scholar] [CrossRef]
- Derfus, A.M.; Chen, A.A.; Min, D.H.; Ruoslahti, E.; Bhatia, S.N. Targeted Quantum Dot Conjugates for SiRNA Delivery. Bioconjug Chem. 2007, 18, 1391–1396. [Google Scholar] [CrossRef]
- Bagalkot, V.; Gao, X. SiRNA-Aptamer Chimeras on Nanoparticles: Preserving Targeting Functionality for Effective Gene Silencing. ACS Nano 2011, 5, 8131–8139. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yang, C.; Hu, R.; Toh, H.T.; Liu, X.; Lin, G.; Yin, F.; Yoon, H.S.; Yong, K.T. Assembling Mn:ZnSe Quantum Dots-SiRNA Nanoplexes for Gene Silencing in Tumor Cells. Biomater. Sci. 2015, 3, 192–202. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, D.; Sil, M.; Goswami, A.; Lahiri, D.; Nag, M. Antibiofilm Activities of Carbon-Based Nanoparticles and Nanocomposites: A Comparative Review. J. Inorg. Organomet. Polym. Mater. 2023, 33, 3961–3983. [Google Scholar] [CrossRef]
- Yazdani, S.; Mozaffarian, M.; Pazuki, G.; Hadidi, N.; Villate-Beitia, I.; Zárate, J.; Puras, G.; Pedraz, J.L. Carbon-Based Nanostructures as Emerging Materials for Gene Delivery Applications. Pharmaceutics 2024, 16, 288. [Google Scholar] [CrossRef]
- Riley, P.R.; Narayan, R.J. Recent Advances in Carbon Nanomaterials for Biomedical Applications: A Review. Curr. Opin. Biomed. Eng. 2021, 17, 100262. [Google Scholar] [CrossRef]
- Siu, K.S.; Zheng, X.; Liu, Y.; Zhang, Y.; Zhang, X.; Chen, D.; Yuan, K.; Gillies, E.R.; Koropatnick, J.; Min, W.-P. Single-Walled Carbon Nanotubes Noncovalently Functionalized with Lipid Modified Polyethylenimine for SiRNA Delivery in Vitro and in Vivo. Bioconjug Chem. 2014, 25, 1744–1751. [Google Scholar] [CrossRef]
- Siu, K.S.; Zhang, Y.; Zheng, X.; Koropatnick, J.; Min, W.P. Non-Covalently Functionalized of Single-Walled Carbon Nanotubes by DSPE-PEG-PEI for SiRNA Delivery. In Methods in Molecular Biology; Humana Press Inc.: Totowa, NJ, USA, 2016; Volume 1364, pp. 151–163. [Google Scholar]
- Varkouhi, A.K.; Foillard, S.; Lammers, T.; Schiffelers, R.M.; Doris, E.; Hennink, W.E.; Storm, G. SiRNA Delivery with Functionalized Carbon Nanotubes. Int. J. Pharm. 2011, 416, 419–425. [Google Scholar] [CrossRef]
- Zhang, L.; Zheng, W.; Tang, R.; Wang, N.; Zhang, W.; Jiang, X. Gene Regulation with Carbon-Based SiRNA Conjugates for Cancer Therapy. Biomaterials 2016, 104, 269–278. [Google Scholar] [CrossRef]
- Meran, M.; Akkus, P.D.; Kurkcuoglu, O.; Baysak, E.; Hizal, G.; Haciosmanoglu, E.; Unlu, A.; Karatepe, N.; Güner, F.S. Noncovalent Pyrene-Polyethylene Glycol Coatings of Carbon Nanotubes Achieve in Vitro Biocompatibility. Langmuir 2018, 34, 12071–12082. [Google Scholar] [CrossRef]
- Imani, R.; Shao, W.; Taherkhani, S.; Emami, S.H.; Prakash, S.; Faghihi, S. Dual-Functionalized Graphene Oxide for Enhanced SiRNA Delivery to Breast Cancer Cells. Colloids Surf. B Biointerfaces 2016, 147, 315–325. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Liu, Z.; Welsher, K.; Robinson, J.T.; Goodwin, A.; Zaric, S.; Dai, H. Nano-Graphene Oxide for Cellular Imaging and Drug Delivery. Nano Res. 2008, 1, 203–212. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Guo, Z.; Huang, D.; Liu, Z.; Guo, X.; Zhong, H. Synergistic Effect of Chemo-Photothermal Therapy Using PEGylated Graphene Oxide. Biomaterials 2011, 32, 8555–8561. [Google Scholar] [CrossRef]
- Yin, F.; Hu, K.; Chen, Y.; Yu, M.; Wang, D.; Wang, Q.; Yong, K.T.; Lu, F.; Liang, Y.; Li, Z. SiRNA Delivery with PEGylated Graphene Oxide Nan Osheets for Combined Photothermal and Genetherapy for Pancreatic Cancer. Theranostics 2017, 7, 1133–1148. [Google Scholar] [CrossRef]
- Liu, J.; Cui, L.; Losic, D. Graphene and Graphene Oxide as New Nanocarriers for Drug Delivery Applications. Acta Biomater. 2013, 9, 9243–9257. [Google Scholar] [CrossRef]
- Feng, L.; Yang, X.; Shi, X.; Tan, X.; Peng, R.; Wang, J.; Liu, Z. Polyethylene Glycol and Polyethylenimine Dual-Functionalized Nano-Graphene Oxide for Photothermally Enhanced Gene Delivery. Small 2013, 9, 1989–1997. [Google Scholar] [CrossRef] [PubMed]
- Georgakilas, V.; Tiwari, J.N.; Kemp, K.C.; Perman, J.A.; Bourlinos, A.B.; Kim, K.S.; Zboril, R. Noncovalent Functionalization of Graphene and Graphene Oxide for Energy Materials, Biosensing, Catalytic, and Biomedical Applications. Chem. Rev. 2016, 116, 5464–5519. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Ding, R.; Zhao, X.; Li, Y.; Qu, L.; Pei, H.; Yildirimer, L.; Wu, Z.; Zhang, W. Graphene-Based Nanomaterials for Drug and/or Gene Delivery, Bioimaging, and Tissue Engineering. Drug Discov. Today 2017, 22, 1302–1317. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.Y.; Zhang, W.; Liu, H.; Jiang, J.J.; Wang, W.J.; Jia, Z.Y. Cell-Penetrating Peptide-Modified Graphene Oxide Nanoparticles Loaded with Rictor SiRNA for the Treatment of Triple-Negative Breast Cancer. Drug Devel. Ther. 2021, 15, 4961–4972. [Google Scholar] [CrossRef]
- Montellano, A.; Da Ros, T.; Bianco, A.; Prato, M. Fullerene C60 as a Multifunctional System for Drug and Gene Delivery. Nanoscale 2011, 3, 4035–4041. [Google Scholar] [CrossRef] [PubMed]
- Minami, K.; Okamoto, K.; Doi, K.; Harano, K.; Noiri, E.; Nakamura, E. SiRNA Delivery Targeting to the Lung via Agglutination-Induced Accumulation and Clearance of Cationic Tetraamino Fullerene. Sci. Rep. 2014, 4, 4916. [Google Scholar] [CrossRef]
- Korzuch, J.; Rak, M.; Balin, K.; Zubko, M.; Głowacka, O.; Dulski, M.; Musioł, R.; Madeja, Z.; Serda, M. Towards Water-Soluble [60]Fullerenes for the Delivery of SiRNA in a Prostate Cancer Model. Sci. Rep. 2021, 11, 10565. [Google Scholar] [CrossRef]
- Minami, K.; Okamoto, K.; Harano, K.; Noiri, E.; Nakamura, E. Hierarchical Assembly of SiRNA with Tetraamino Fullerene in Physiological Conditions for Efficient Internalization into Cells and Knockdown. ACS Appl. Mater. Interfaces 2018, 10, 19347–19354. [Google Scholar] [CrossRef]
- Liu, S.; Sun, X.; Lu, H.; Chen, D.; Li, X.; Li, L.; Su, S.; Zhao, Z.; Cao, X.; Liu, L.; et al. Fullerene-Based Nanocomplex Assists Pulmonary Delivery of SiRNA for Treating Metastatic Lung Cancer. Nano Today 2023, 50, 101878. [Google Scholar] [CrossRef]
- Chen, F.; Gao, W.; Qiu, X.; Zhang, H.; Liu, L.; Liao, P.; Fu, W.; Luo, Y. Graphene Quantum Dots in Biomedical Applications: Recent Advances and Future Challenges. Front. Lab. Med. 2017, 1, 192–199. [Google Scholar] [CrossRef]
- Das, S.; Mondal, S.; Ghosh, D. Carbon Quantum Dots in Bioimaging and Biomedicines. Front. Bioeng. Biotechnol. 2024, 11, 1333752. [Google Scholar] [CrossRef]
- Mansuriya, B.D.; Altintas, Z. Carbon Dots: Classification, Properties, Synthesis, Characterization, and Applications in Health Care-An Updated Review (2018–2021). Nanomaterials 2021, 11, 2525. [Google Scholar] [CrossRef]
- Yang, H.-L.; Bai, L.-F.; Geng, Z.-R.; Chen, H.; Xu, L.-T.; Xie, Y.-C.; Wang, D.-J.; Gu, H.-W.; Wang, X.-M. Carbon Quantum Dots: Preparation, Optical Properties, and Biomedical Applications. Mater. Today Adv. 2023, 18, 100376. [Google Scholar] [CrossRef]
- Xia, C.; Zhu, S.; Feng, T.; Yang, M.; Yang, B. Evolution and Synthesis of Carbon Dots: From Carbon Dots to Carbonized Polymer Dots. Adv. Sci. 2019, 6, 1901316. [Google Scholar] [CrossRef]
- Pierrat, P.; Wang, R.; Kereselidze, D.; Lux, M.; Didier, P.; Kichler, A.; Pons, F.; Lebeau, L. Efficient Invitro and Invivo Pulmonary Delivery of Nucleic Acid by Carbon Dot-Based Nanocarriers. Biomaterials 2015, 51, 290–302. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Zhang, C.; Shen, G.; Liu, H.; Fu, H.; Cui, D. Fluorescent Carbon Dots as an Efficient SiRNA Nanocarrier for Its Interference Therapy in Gastric Cancer Cells. J. Nanobiotechnol. 2014, 12, 58. [Google Scholar] [CrossRef]
- Wang, L.; Wang, X.; Bhirde, A.; Cao, J.; Zeng, Y.; Huang, X.; Sun, Y.; Liu, G.; Chen, X. Carbon-Dot-Based Two-Photon Visible Nanocarriers for Safe and Highly Efficient Delivery of SiRNA and DNA. Adv. Healthc. Mater. 2014, 3, 1203–1209. [Google Scholar] [CrossRef]
- Adrita, S.; Tasnim, K.; Ryu, J.; Sharker, S. Nanotheranostic Carbon Dots as an Emerging Platform for Cancer Therapy. J. Nanotheranostics 2020, 58–77. [Google Scholar] [CrossRef]
- Vallet-Regí, M.; Colilla, M.; Izquierdo-Barba, I.; Manzano, M. Mesoporous Silica Nanoparticles for Drug Delivery: Current Insights. Molecules 2018, 23, 47. [Google Scholar] [CrossRef]
- Jafari, S.; Derakhshankhah, H.; Alaei, L.; Fattahi, A.; Varnamkhasti, B.S.; Saboury, A.A. Mesoporous Silica Nanoparticles for Therapeutic/Diagnostic Applications. Biomed. Pharmacother. 2019, 109, 1100–1111. [Google Scholar] [CrossRef] [PubMed]
- Rastegari, E.; Hsiao, Y.-J.; Lai, W.-Y.; Lai, Y.-H.; Yang, T.-C.; Chen, S.-J.; Huang, P.-I.; Chiou, S.-H.; Mou, C.-Y.; Chien, Y. An Update on Mesoporous Silica Nanoparticle Applications in Nanomedicine. Pharmaceutics 2021, 13, 1067. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Shi, S.; Goel, S.; Shen, X.; Xie, X.; Chen, Z.; Zhang, H.; Li, S.; Qin, X.; Yang, H.; et al. Recent Advancements in Mesoporous Silica Nanoparticles towards Therapeutic Applications for Cancer. Acta Biomater. 2019, 89, 1–13. [Google Scholar] [CrossRef]
- Xie, X.; Yue, T.; Gu, W.; Cheng, W.; He, L.; Ren, W.; Li, F.; Piao, J.-G. Recent Advances in Mesoporous Silica Nanoparticles Delivering SiRNA for Cancer Treatment. Pharmaceutics 2023, 15, 2483. [Google Scholar] [CrossRef]
- Ma, X.; Zhao, Y.; Ng, K.W.; Zhao, Y. Integrated Hollow Mesoporous Silica Nanoparticles for Target Drug/SiRNA Co-Delivery. Chem. Eur. J. 2013, 19, 15593–15603. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, Q.; Han, N.; Bai, L.; Li, J.; Liu, J.; Che, E.; Hu, L.; Zhang, Q.; Jiang, T.; et al. Mesoporous Silica Nanoparticles in Drug Delivery and Biomedical Applications. Nanomedicine 2015, 11, 313–327. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, Y.; Tang, C.; Tian, C.; Sun, Q.; Su, Z.; Xue, L.; Yin, Y.; Ju, C.; Zhang, C. Co-Delivery of Paclitaxel and Anti-Survivin SiRNA via Redox-Sensitive Oligopeptide Liposomes for the Synergistic Treatment of Breast Cancer and Metastasis. Int. J. Pharm. 2017, 529, 102–115. [Google Scholar] [CrossRef]
- Xiao, D.; Hu, J.-J.; Zhu, J.-Y.; Wang, S.-B.; Zhuo, R.-X.; Zhang, X.-Z. A Redox-Responsive Mesoporous Silica Nanoparticle with a Therapeutic Peptide Shell for Tumor Targeting Synergistic Therapy. Nanoscale 2016, 8, 16702–16709. [Google Scholar] [CrossRef]
- Chan, M.-H.; Lin, H.-M. Preparation and Identification of Multifunctional Mesoporous Silica Nanoparticles for in Vitro and in Vivo Dual-Mode Imaging, Theranostics, and Targeted Tracking. Biomaterials 2015, 46, 149–158. [Google Scholar] [CrossRef] [PubMed]
- Girija, A.R.; Balasubramanian, S. Theragnostic Potentials of Core/Shell Mesoporous Silica Nanostructures. Nanotheranostics 2019, 3, 1–40. [Google Scholar] [CrossRef] [PubMed]
- Hanafi-Bojd, M.Y.; Ansari, L.; Malaekeh-Nikouei, B. Codelivery of Anticancer Drugs and SiRNA By Mesoporous Silica Nanoparticles. Ther. Deliv. 2016, 7, 649–655. [Google Scholar] [CrossRef]
- Meng, H.; Mai, W.X.; Zhang, H.; Xue, M.; Xia, T.; Lin, S.; Wang, X.; Zhao, Y.; Ji, Z.; Zink, J.I.; et al. Codelivery of an Optimal Drug/SiRNA Combination Using Mesoporous Silica Nanoparticles To Overcome Drug Resistance in Breast Cancer in Vitro and in Vivo. ACS Nano 2013, 7, 994–1005. [Google Scholar] [CrossRef] [PubMed]
- Paris, J.L.; Vallet-Regí, M. Mesoporous Silica Nanoparticles for Co-Delivery of Drugs and Nucleic Acids in Oncology: A Review. Pharmaceutics 2020, 12, 526. [Google Scholar] [CrossRef]
- Carvalho, A.M.; Cordeiro, R.A.; Faneca, H. Silica-Based Gene Delivery Systems: From Design to Therapeutic Applications. Pharmaceutics 2020, 12, 649. [Google Scholar] [CrossRef]
- Jin, M.; Hou, Y.; Quan, X.; Chen, L.; Gao, Z.; Huang, W. Smart Polymeric Nanoparticles with PH-Responsive and Peg-Detachable Properties (II): Co-Delivery of Paclitaxel and VEGF SiRNA for Synergistic Breast Cancer Therapy in Mice. Int. J. Nanomed. 2021, 16, 5479–5494. [Google Scholar] [CrossRef] [PubMed]
- Lv, W.; Xu, C.; Wu, H.; Zhu, Y.; Akakuru, O.U.; Du, H.; Nie, F.; Wu, A.; Li, J. Ultrasound-Visualized Nanocarriers with SiRNA for Targeted Inhibition of M2-like TAM Polarization to Enhance Photothermal Therapy in NSCLC. Nano Res. 2023, 16, 882–893. [Google Scholar] [CrossRef]
- Dang, J.; Ye, H.; Li, Y.; Liang, Q.; Li, X.; Yin, L. Multivalency-Assisted Membrane-Penetrating SiRNA Delivery Sensitizes Photothermal Ablation via Inhibition of Tumor Glycolysis Metabolism. Biomaterials 2019, 223, 119437. [Google Scholar] [CrossRef] [PubMed]
- Zheng, N.; Luo, X.; Zhang, Z.; Wang, A.; Song, W. Cationic Polyporphyrins as SiRNA Delivery Vectors for Photodynamic and Gene Synergistic Anticancer Therapy. ACS App.l Mater. Interfaces 2021, 13, 27513–27521. [Google Scholar] [CrossRef]
- Camorani, S.; Tortorella, S.; Agnello, L.; Spanu, C.; d’Argenio, A.; Nilo, R.; Zannetti, A.; Locatelli, E.; Fedele, M.; Comes Franchini, M.; et al. Aptamer-Functionalized Nanoparticles Mediate PD-L1 SiRNA Delivery for Effective Gene Silencing in Triple-Negative Breast Cancer Cells. Pharmaceutics 2022, 14, 2225. [Google Scholar] [CrossRef]
- Tan, S.; Chen, Z.; Mironchik, Y.; Mori, N.; Penet, M.F.; Si, G.; Krishnamachary, B.; Bhujwalla, Z.M. VEGF Overexpression Significantly Increases Nanoparticle-Mediated SiRNA Delivery and Target-Gene Downregulation. Pharmaceutics 2022, 14, 1260. [Google Scholar] [CrossRef]
- Ding, L.; Tang, S.; Yu, A.; Wang, A.; Tang, W.; Jia, H.; Oupický, D. Nanoemulsion-Assisted SiRNA Delivery to Modulate the Nervous Tumor Microenvironment in the Treatment of Pancreatic Cancer. ACS Appl. Mater. Interfaces 2022, 14, 10015–10029. [Google Scholar] [CrossRef]
- Vaidya, S.; Jeengar, M.K.; Wadaan, M.A.; Mahboob, S.; Kumar, P.; Reece, L.M.; Bathula, S.R.; Dutta, M. Design and In Vitro Evaluation of Novel Cationic Lipids for SiRNA Delivery in Breast Cancer Cell Lines. Evid. Based Complement. Altern. Med. 2022, 2022, 1–9. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, T.; Gao, J. Biocompatible Iron Oxide Nanoparticles for Targeted Cancer Gene Therapy: A Review. Nanomaterials 2022, 12, 3323. [Google Scholar] [CrossRef]
- Yu, S.; Bi, X.; Yang, L.; Wu, S.; Yu, Y.; Jiang, B.; Zhang, A.; Lan, K.; Duan, S. Co-Delivery of Paclitaxel and PLK1-Targeted SiRNA Using Aptamer-Functionalized Cationic Liposome for Synergistic Anti-Breast Cancer Effects in Vivo. J. Biomed. Nanotechnol. 2019, 15, 1135–1148. [Google Scholar] [CrossRef]
- Lee, J.; Cho, Y.J.; Lee, J.W.; Ahn, H.J. KSP SiRNA/Paclitaxel-Loaded PEGylated Cationic Liposomes for Overcoming Resistance to KSP Inhibitors: Synergistic Antitumor Effects in Drug-Resistant Ovarian Cancer. J. Control. Release 2020, 321, 184–197. [Google Scholar] [CrossRef]
- Xie, Y.; Lu, X.; Wang, Z.; Liu, M.; Liu, L.; Wang, R.; Yang, K.; Xiao, H.; Li, J.; Tang, X.; et al. A Hypoxia-Dissociable SiRNA Nanoplatform for Synergistically Enhanced Chemo-Radiotherapy of Glioblastoma. Biomater. Sci. 2022, 10, 6791–6803. [Google Scholar] [CrossRef]
- Çağdaş Tunalı, B.; Çelik, E.; Budak Yıldıran, F.A.; Türk, M. Delivery of SiRNA Using Hyaluronic Acid-Guided Nanoparticles for Downregulation of CXCR4. Biopolymers 2023, 114, e23535. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Kim, S.; Kim, T. Il Polyethylenimine-Conjugated Hydroxyethyl Cellulose for Doxorubicin/Bcl-2 SiRNA Co-Delivery Systems. Pharmaceutics 2023, 15, 708. [Google Scholar] [CrossRef] [PubMed]
- Aranda-Lara, L.; Trujillo-Nolasco, M.; López-Marmolejo, C.M.; Cárdenas-Rodríguez, K.; Gómez-Pulido, V.O.; Morales-Avila, E.; Ocampo-García, B.; Jiménez-Mancilla, N.P.; Estrada, J.A.; Otero, G.; et al. Chitosan/SiRNA Complex in Mimetic High-Density Lipoproteins as Specific/Hybrid Transfection Technology for SR-B1 Expressing Cancer Cells. J. Drug Deliv. Sci. Technol. 2024, 101, 106274. [Google Scholar] [CrossRef]
- Abbasi, A.; Lindqvist-Reis, P.; Eriksson, L.; Sandström, D.; Lidin, S.; Persson, I.; Sandström, M. Highly Hydrated Cations: Deficiency, Mobility, and Coordination of Water in Crystalline Nonahydrated Scandium(III), Yttrium(III), and Lanthanoid(III) Trifluoromethanesulfonates. Chem. Eur. J. 2005, 11, 4065–4077. [Google Scholar] [CrossRef]
- Hattab, D.; Gazzali, A.M.; Bakhtiar, A. Clinical Advances of Sirna-Based Nanotherapeutics for Cancer Treatment. Pharmaceutics 2021, 13, 1009. [Google Scholar] [CrossRef]
- Drago, S.E.; Utzeri, M.A.; Mauro, N.; Cavallaro, G. Polyamidoamine-Carbon Nanodot Conjugates with Bioreducible Building Blocks: Smart Theranostic Platforms for Targeted SiRNA Delivery. Biomacromolecules 2024, 25, 1191–1204. [Google Scholar] [CrossRef]
- Ranjbar, S.; Zhong, X.; Manautou, J.; Lu, X. A Holistic Analysis of the Intrinsic and Delivery-Mediated Toxicity of SiRNA Therapeutics. Adv. Drug Deliv. Rev. 2023, 201, 115052. [Google Scholar] [CrossRef]
- Liu, Z.; Sun, Q.; Wang, X. PLK1, A Potential Target for Cancer Therapy. Transl. Oncol. 2017, 10, 22–32. [Google Scholar] [CrossRef]
- El Dika, I.; Lim, H.Y.; Yong, W.P.; Lin, C.-C.; Yoon, J.-H.; Modiano, M.; Freilich, B.; Choi, H.J.; Chao, T.-Y.; Kelley, R.K.; et al. An Open-Label, Multicenter, Phase I, Dose Escalation Study with Phase II Expansion Cohort to Determine the Safety, Pharmacokinetics, and Preliminary Antitumor Activity of Intravenous TKM-080301 in Subjects with Advanced Hepatocellular Carcinoma. Oncologist 2019, 24, 747-e218. [Google Scholar] [CrossRef] [PubMed]
- Chiappa, M.; Petrella, S.; Damia, G.; Broggini, M.; Guffanti, F.; Ricci, F. Present and Future Perspective on PLK1 Inhibition in Cancer Treatment. Front. Oncol. 2022, 12, 903016. [Google Scholar] [CrossRef]
- Mukai, H.; Muramatsu, A.; Mashud, R.; Kubouchi, K.; Tsujimoto, S.; Hongu, T.; Kanaho, Y.; Tsubaki, M.; Nishida, S.; Shioi, G.; et al. PKN3 Is the Major Regulator of Angiogenesis and Tumor Metastasis in Mice. Sci. Rep. 2016, 6, 18979. [Google Scholar] [CrossRef]
- Schultheis, B.; Strumberg, D.; Kuhlmann, J.; Wolf, M.; Link, K.; Seufferlein, T.; Kaufmann, J.; Feist, M.; Gebhardt, F.; Khan, M.; et al. Safety, Efficacy and Pharcacokinetics of Targeted Therapy with the Liposomal RNA Interference Therapeutic Atu027 Combined with Gemcitabine in Patients with Pancreatic Adenocarcinoma. A Randomized Phase Ib/IIa Study. Cancers 2020, 12, 3130. [Google Scholar] [CrossRef]
- Temme, L.; Laitinen, T.; Asquith, C.R.M. PKN3: A Target in Cancer Metastasis. Nat. Rev. Drug Discov. 2022, 21, 704. [Google Scholar] [CrossRef] [PubMed]
- Unsal-Kacmaz, K.; Ragunathan, S.; Rosfjord, E.; Dann, S.; Upeslacis, E.; Grillo, M.; Hernandez, R.; Mack, F.; Klippel, A. The Interaction of PKN3 with RhoC Promotes Malignant Growth. Mol. Oncol. 2012, 6, 284–298. [Google Scholar] [CrossRef]
- London, M.; Gallo, E. The EphA2 and Cancer Connection: Potential for Immune-Based Interventions. Mol. Biol. Rep. 2020, 47, 8037–8048. [Google Scholar] [CrossRef]
- Oner, E.; Kotmakci, M.; Baird, A.M.; Gray, S.G.; Debelec Butuner, B.; Bozkurt, E.; Kantarci, A.G.; Finn, S.P. Development of EphA2 SiRNA-Loaded Lipid Nanoparticles and Combination with a Small-molecule Histone Demethylase Inhibitor in Prostate Cancer Cells and Tumor Spheroids. J. Nanobiotechnol. 2021, 19, 40. [Google Scholar] [CrossRef] [PubMed]
- Wagner, M.J.; Mitra, R.; Mcarthur, M.J.; Baze, W.; Barnhart, K.; Wu, S.Y.; Rodriguez-Aguayo, C.; Zhang, X.; Coleman, R.L.; Lopez-Berestein, G.; et al. Preclinical Mammalian Safety Studies of EPHARNA (DOPC Nanoliposomal EphA2-Targeted SiRNA). Mol. Cancer Ther. 2017, 16, 1114–1123. [Google Scholar] [CrossRef]
- Golan, T.; Khvalevsky, E.Z.; Hubert, A.; Gabai, R.M.; Hen, N.; Segal, A.; Domb, A.; Harari, G.; David, E.B.; Raskin, S.; et al. RNAi Therapy Targeting KRAS in Combination with Chemotherapy for Locally Advanced Pancreatic Cancer Patients. Oncotarget 2015, 6, 24560–24570. [Google Scholar] [CrossRef] [PubMed]
- Hunter, J.C.; Manandhar, A.; Carrasco, M.A.; Gurbani, D.; Gondi, S.; Westover, K.D. Biochemical and Structural Analysis of Common Cancer-Associated KRAS Mutations. Mol. Cancer Res. 2015, 13, 1325–1335. [Google Scholar] [CrossRef]
- Huang, L.; Guo, Z.; Wang, F.; Fu, L. KRAS Mutation: From Undruggable to Druggable in Cancer. Signal Transduct. Target. Ther. 2021, 6, 386. [Google Scholar] [CrossRef] [PubMed]
- Jani, V.; Sonavane, U.; Joshi, R. Insight into Structural Dynamics Involved in Activation Mechanism of Full Length KRAS Wild Type and P-Loop Mutants. Heliyon 2024, 10, e36161. [Google Scholar] [CrossRef]
- Baba, A.B.; Rah, B.; Bhat, G.R.; Mushtaq, I.; Parveen, S.; Hassan, R.; Hameed Zargar, M.; Afroze, D. Transforming Growth Factor-Beta (TGF-β) Signaling in Cancer-A Betrayal Within. Front. Pharmacol. 2022, 13, 791272. [Google Scholar] [CrossRef] [PubMed]
- Derynck, R.; Turley, S.J.; Akhurst, R.J. TGFβ Biology in Cancer Progression and Immunotherapy. Nat. Rev. Clin. Oncol. 2021, 18, 9–34. [Google Scholar] [CrossRef]
- Hashemi Goradel, N.; Najafi, M.; Salehi, E.; Farhood, B.; Mortezaee, K. Cyclooxygenase-2 in Cancer: A Review. J. Cell Physiol. 2019, 234, 5683–5699. [Google Scholar] [CrossRef]
- Li, S.; Jiang, M.; Wang, L.; Yu, S. Combined Chemotherapy with Cyclooxygenase-2 (COX-2) Inhibitors in Treating Human Cancers: Recent Advancement. Biomed. Pharmacother. 2020, 129, 110389. [Google Scholar] [CrossRef]
- Liu, S.; Zhang, Y.; Chen, Y.; Su, Y.; Chen, B.; Wang, Y.; Xu, M.; Qiao, K.; Li, S.; Liu, Z. Isolation and Purification of Protamine from the Cultured Takifugu Flavidus and Its Physicochemical Properties. Molecules 2024, 29, 263. [Google Scholar] [CrossRef]
- Massagué, J. TGFβ in Cancer. Cell 2008, 134, 215–230. [Google Scholar] [CrossRef]
- Chien, S.T.; Yang, T.F.; Yang, M.C.; Hsu, C.M.; Hong, Y.R.; Lee, T.M. Differential Roles of Bcl2L12 and Its Short Variant in Breast Cancer Lymph Node Metastasis. Oncol. Rep. 2015, 34, 961–971. [Google Scholar] [CrossRef]
- Kouri, F.M.; Jensen, S.A.; Stegh, A.H. The Role of Bcl-2 Family Proteins in Therapy Responses of Malignant Astrocytic Gliomas: Bcl2L12 and Beyond. Sci. World J. 2012, 2012, 1–8. [Google Scholar] [CrossRef]
- Kumthekar, P.; Ko, C.H.; Paunesku, T.; Dixit, K.; Sonabend, A.M.; Bloch, O.; Tate, M.; Schwartz, M.; Zuckerman, L.; Lezon, R.; et al. A First-in-Human Phase 0 Clinical Study of RNA Interference-Based Spherical Nucleic Acids in Patients with Recurrent Glioblastoma. Sci. Transl. Med. 2021, 13, eabb3945. [Google Scholar] [CrossRef] [PubMed]
- Stegh, A.H.; Brennan, C.; Mahoney, J.A.; Forloney, K.L.; Jenq, H.T.; Luciano, J.P.; Protopopov, A.; Chin, L.; DePinho, R.A. Glioma Oncoprotein Bc12L12 Inhibits the P53 Tumor Suppressor. Genes. Dev. 2010, 24, 2194–2204. [Google Scholar] [CrossRef]
- Apte, R.S.; Chen, D.S.; Ferrara, N. VEGF in Signaling and Disease: Beyond Discovery and Development. Cell 2019, 176, 1248–1264. [Google Scholar] [CrossRef]
- Ahmad, A.; Nawaz, M.I. Molecular Mechanism of VEGF and Its Role in Pathological Angiogenesis. J. Cell Biochem. 2022, 123, 1938–1965. [Google Scholar] [CrossRef]
- Kang, Y.; Li, H.; Liu, Y.; Li, Z. Regulation of VEGF-A Expression and VEGF-A-Targeted Therapy in Malignant Tumors. J. Cancer Res. Clin. Oncol. 2024, 150, 221. [Google Scholar] [CrossRef] [PubMed]
- Elebiyo, T.C.; Rotimi, D.; Evbuomwan, I.O.; Maimako, R.F.; Iyobhebhe, M.; Ojo, O.A.; Oluba, O.M.; Adeyemi, O.S. Reassessing Vascular Endothelial Growth Factor (VEGF) in Anti-Angiogenic Cancer Therapy. Cancer Treat. Res. Commun. 2022, 32, 100620. [Google Scholar] [CrossRef]
- Tabernero, J.; Shapiro, G.I.; LoRusso, P.M.; Cervantes, A.; Schwartz, G.K.; Weiss, G.J.; Paz-Ares, L.; Cho, D.C.; Infante, J.R.; Alsina, M.; et al. First-in-Humans Trial of an RNA Interference Therapeutic Targeting VEGF and KSP in Cancer Patients with Liver Involvement. Cancer Discov. 2013, 3, 406–417. [Google Scholar] [CrossRef] [PubMed]
- Tolcher, A.W.; Spira, A.I.; Nemunaitis, J.J.; Vandross, L.; Mamdani, H.; O’brien, Z.; Huynh, A.; Albaugh, Z.; Gullbo, J.; Zabludoff, S. First-in-Human Dose-Escalation Study of NBF-006, a Novel Investigational SiRNA Targeting GSTP, in Patients with Non-Small Cell Lung, Pancreatic, or Colorectal Cancer. J. Clin. Oncol. 2023, 41, 3084. [Google Scholar] [CrossRef]
- Kennedy, L.; Sandhu, J.K.; Harper, M.-E.; Cuperlovic-Culf, M. Role of Glutathione in Cancer: From Mechanisms to Therapies. Biomolecules 2020, 10, 1429. [Google Scholar] [CrossRef]
- Singh, R.R.; Reindl, K.M. Glutathione S-Transferases in Cancer. Antioxidants 2021, 10, 701. [Google Scholar] [CrossRef]
- Belete, T.M. The Current Status of Gene Therapy for the Treatment of Cancer. Biologics 2021, 15, 67–77. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Kang, G.; Wang, T.; Huang, H. Tumor Angiogenesis and Anti-angiogenic Gene Therapy for Cancer (Review). Oncol. Lett. 2018, 16, 687–702. [Google Scholar] [CrossRef]
- Younis, M.; Harashima, H. Understanding Gene Involvement in Hepatocellular Carcinoma: Implications for Gene Therapy and Personalized Medicine. Pharmgenomics Pers. Med. 2024, 17, 193–213. [Google Scholar] [CrossRef] [PubMed]
- Nussinov, R.; Yavuz, B.R.; Jang, H. Molecular Principles Underlying Aggressive Cancers. Signal Transduct. Target. Ther. 2025, 10, 42. [Google Scholar] [CrossRef]
- Man, R.C.H.; Qiu, Y.; Leung, S.W.S.; Fruhwirth, G.O.; Lam, J.K.W. Co-Delivery of PD-L1- and EGFR-Targeting SiRNAs by Synthetic PEG12-KL4 Peptide to the Lungs as Potential Strategy against Non-Small Cell Lung Cancer. Eur. J. Pharm. Biopharm. 2024, 195, 114177. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.J.; Kim, H.; Gao, X.; Ryu, J.H.; Yang, Y.; Kwon, I.C.; Roberts, T.M.; Kim, S.H. Multi-Targeting SiRNA Nanoparticles for Simultaneous Inhibition of PI3K and Rac1 in PTEN-Deficient Prostate Cancer. J. Ind. Eng. Chem. 2021, 99, 196–203. [Google Scholar] [CrossRef]
- Jamalinia, M.; Weiskirchen, R. Advances in Personalized Medicine: Translating Genomic Insights into Targeted Therapies for Cancer Treatment. Ann. Transl. Med. 2025, 13, 18. [Google Scholar] [CrossRef]
- Sousa, C.; Videira, M. Dual Approaches in Oncology: The Promise of SiRNA and Chemotherapy Combinations in Cancer Therapies. Oncology 2025, 5, 2. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).