Physiological Barriers to Nucleic Acid Therapeutics and Engineering Strategies for Lipid Nanoparticle Design, Optimization, and Clinical Translation
Abstract
1. Introduction
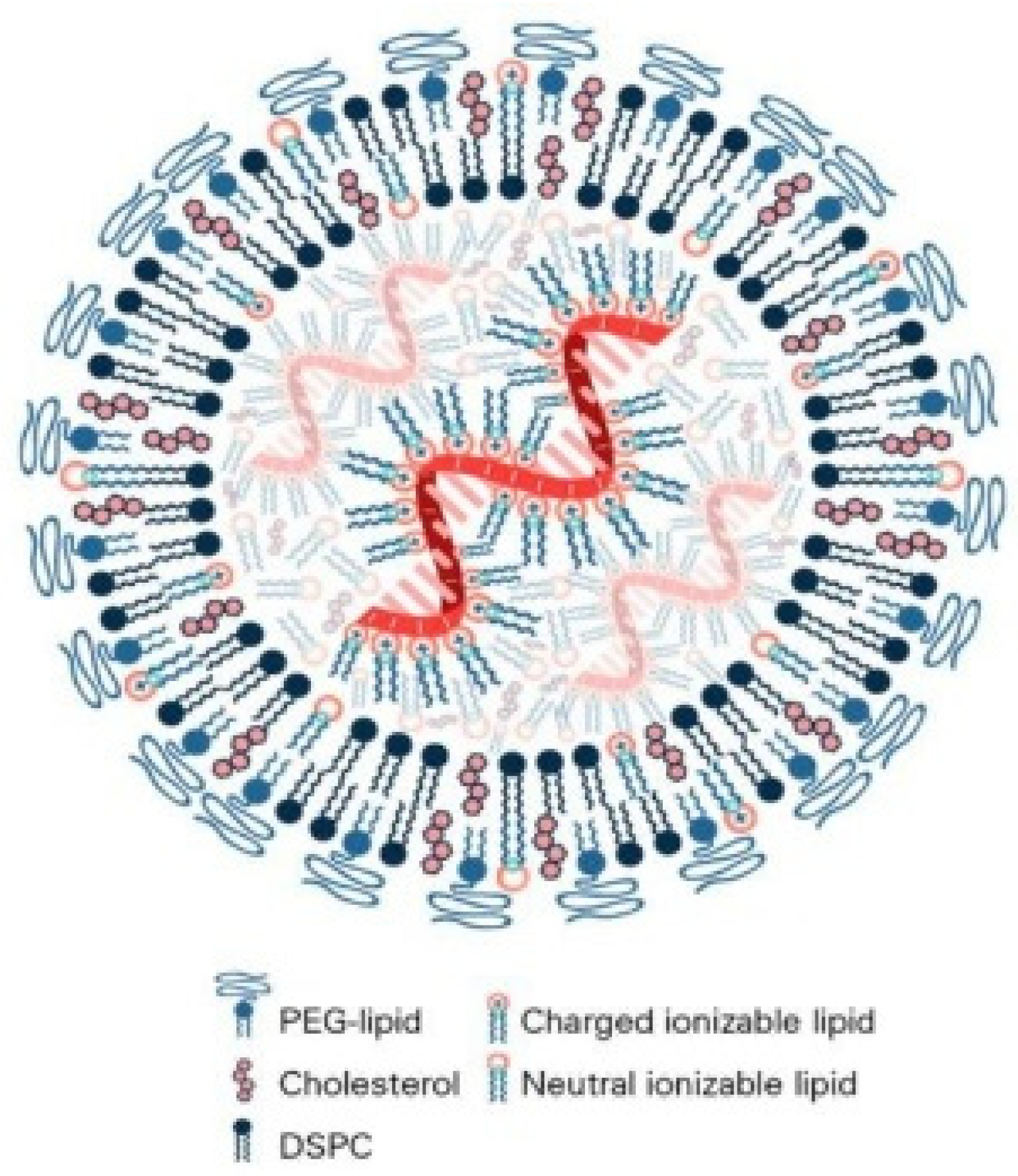
2. Composition, Physicochemical Properties, and Encapsulation of Lipid-Based Nanoparticles
2.1. Core Components and Functional Roles of Lipid-Based Nanoparticles
2.1.1. Ionizable Lipids
2.1.2. Helper Phospholipids
2.1.3. Cholesterol
2.1.4. PEG-Lipids
2.2. Physicochemical Properties of Lipid-Based Nanoparticles
2.2.1. Particle Size and In Vivo Fate
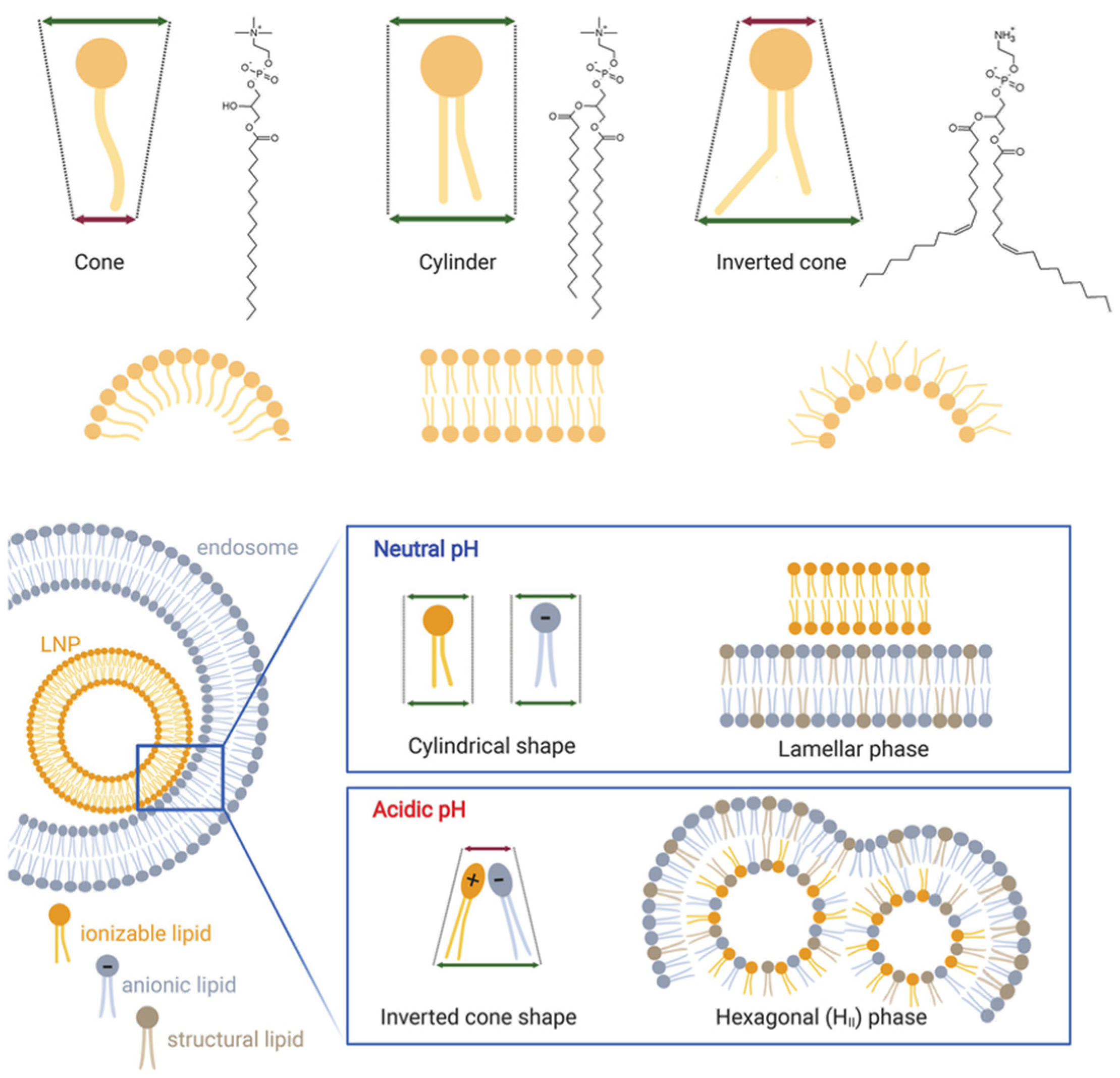
2.2.2. Morphological Architecture
2.2.3. Surface Charge
2.3. Nucleic Acid Encapsulation Mechanisms
3. Systemic Fate and Biological Barriers of Lipid-Based Nanoparticles
3.1. Hemodynamic Shear and Dilution in the Bloodstream
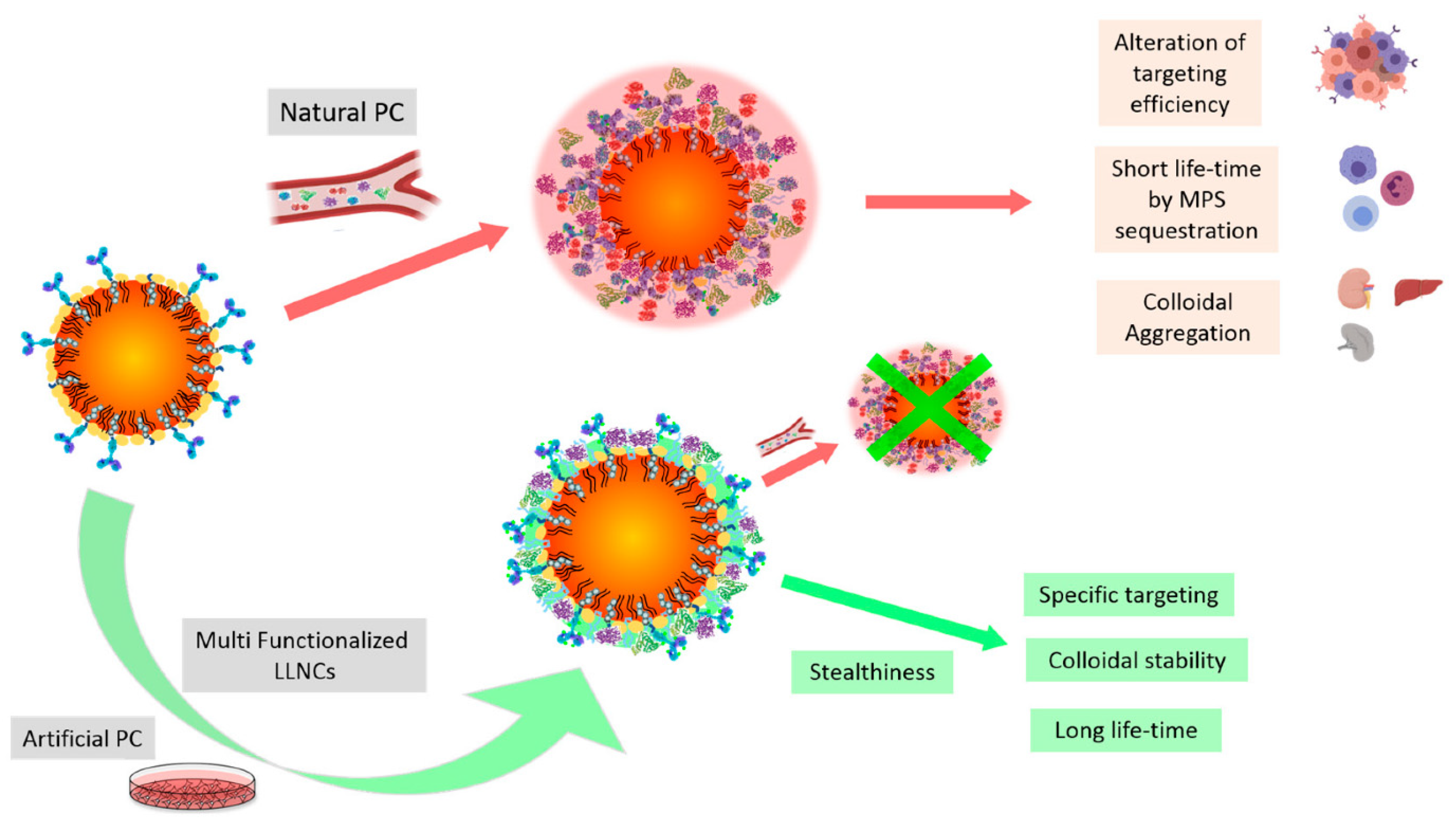
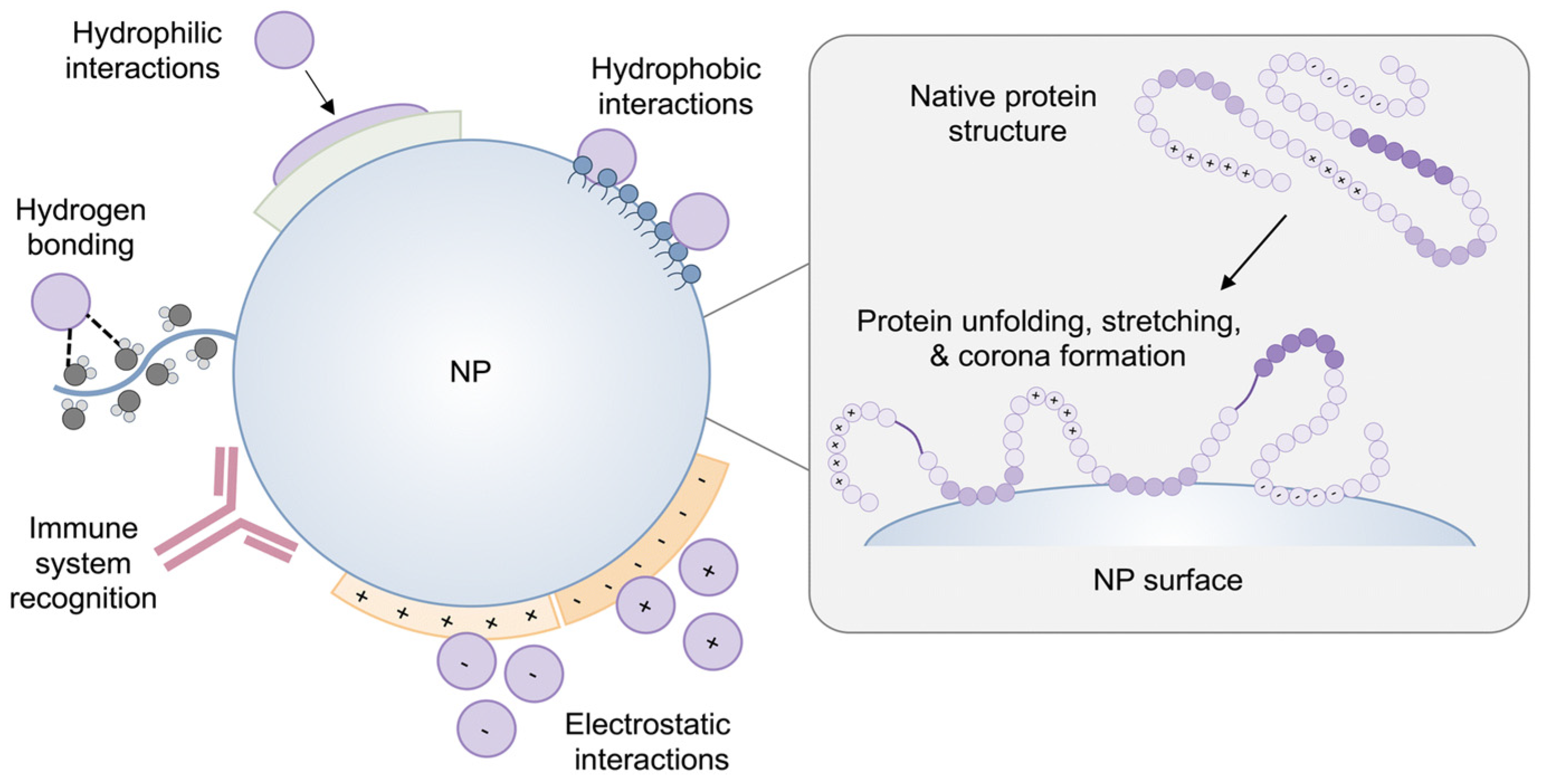
3.2. Protein Corona and Opsonization
Challenges and Opportunities of Protein Corona
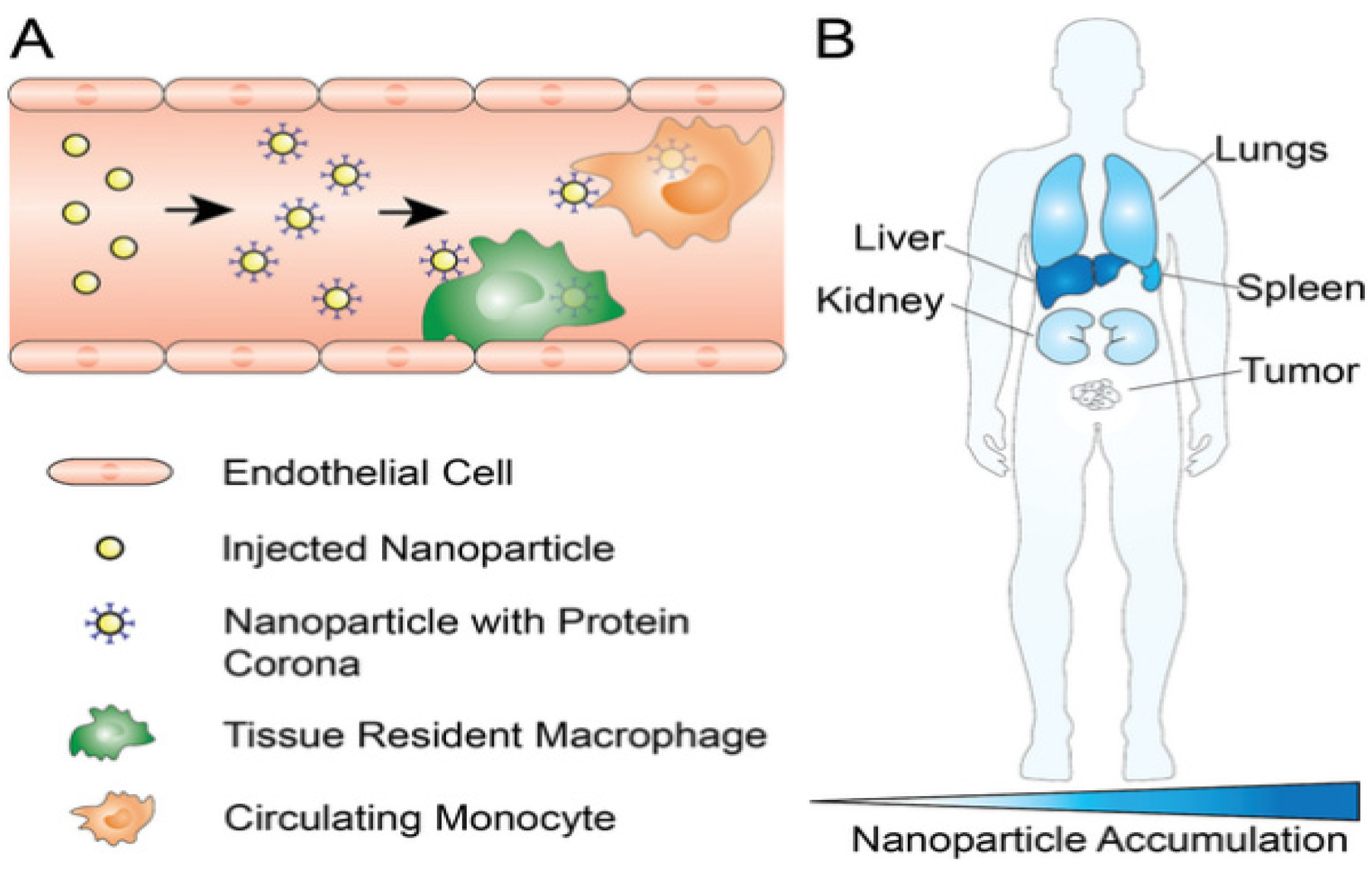
3.3. Reticuloendothelial System (RES) Clearance
PEG Stealth Effect and Emerging Immunological Concerns
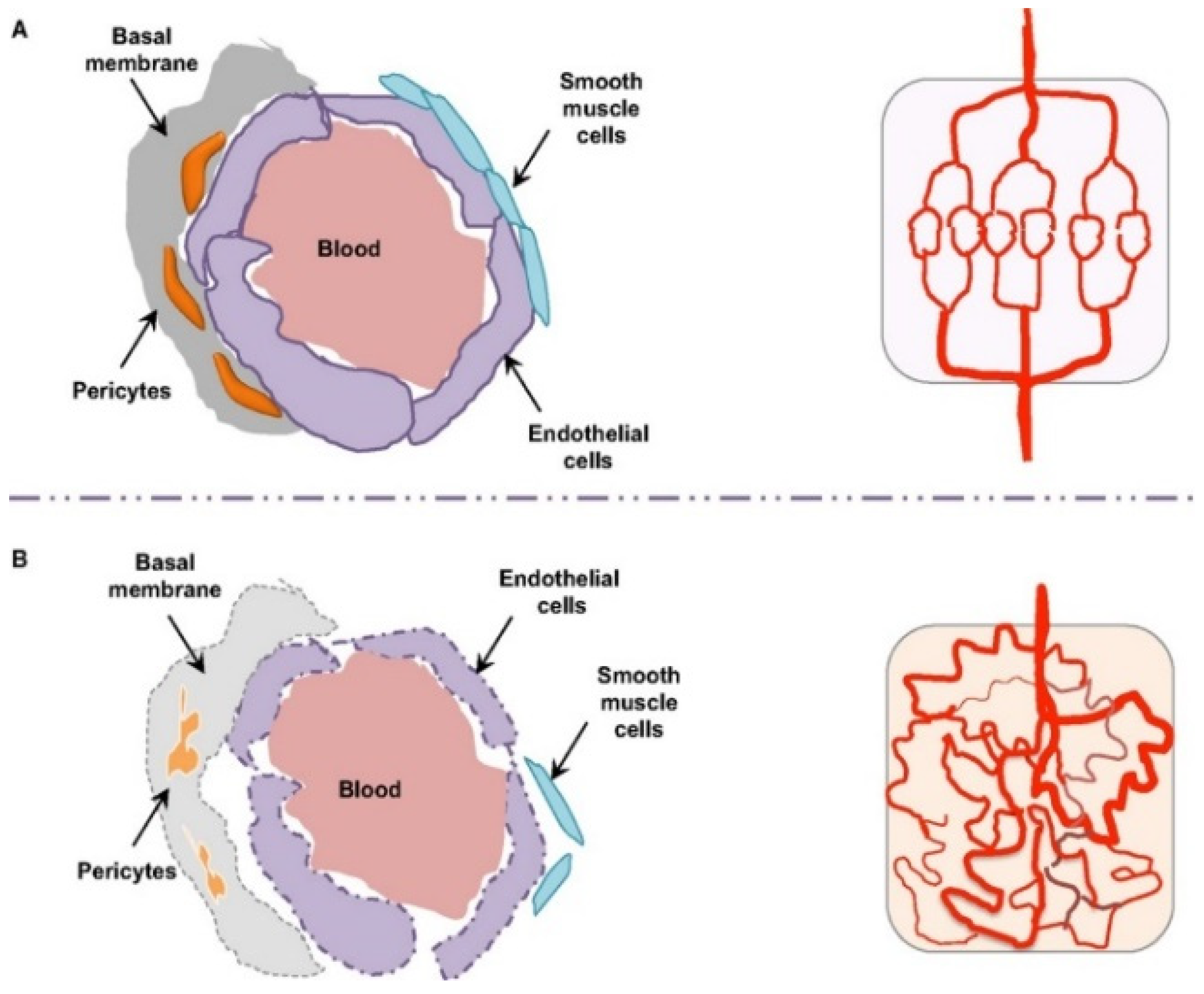
3.4. Tissue-Specific Vascular Endothelial Barriers
4. Microenvironmental and Cellular Barriers to Lipid Nanoparticle Delivery
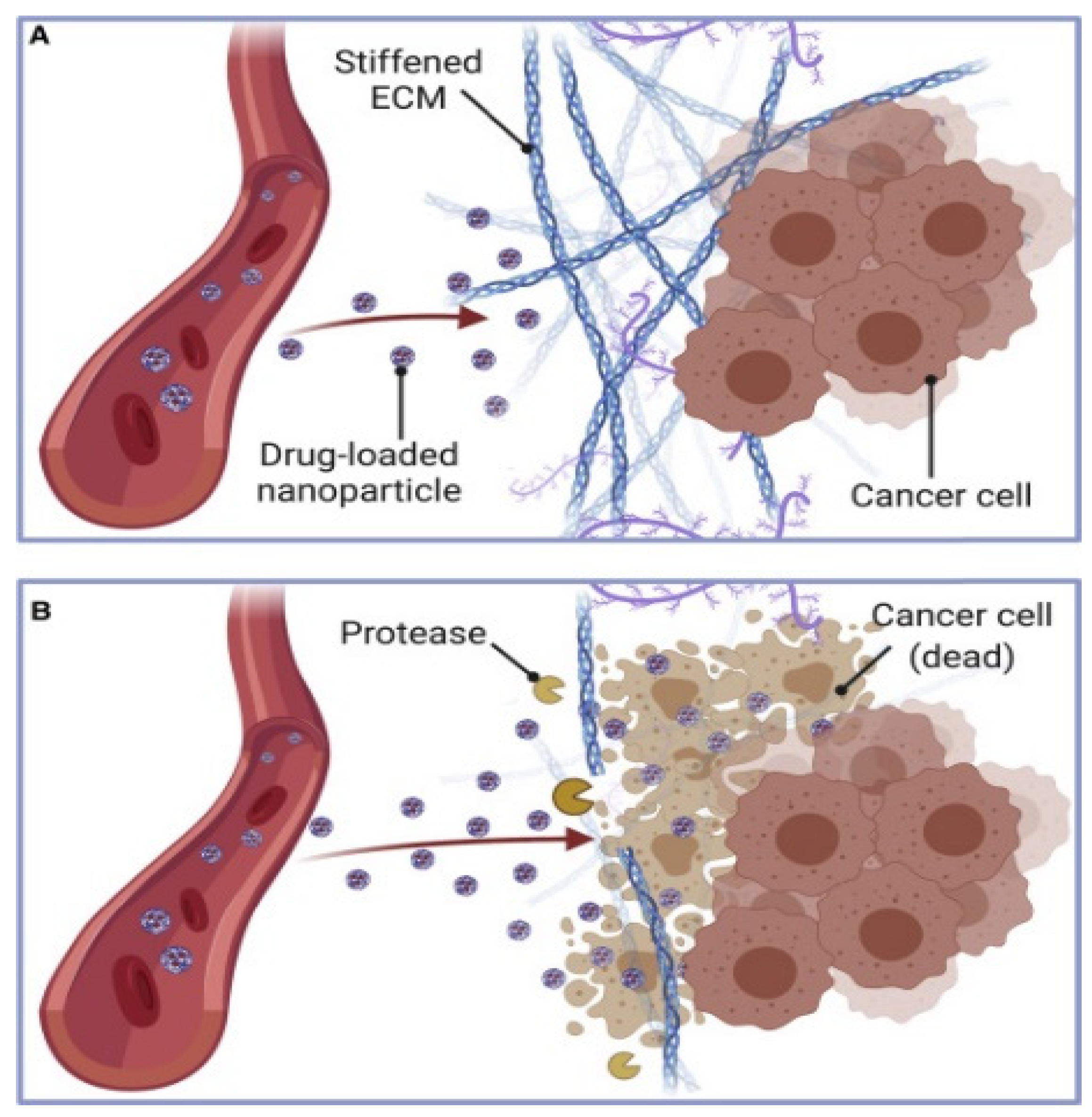
4.1. Extracellular Matrix Constraints on Nanoparticle Penetration
ECM-Associated Stability and Storage
4.2. Interstitial Fluid Pressure and Transport Barriers
4.3. Cellular Internalization Mechanisms
5. Intracellular Delivery Determinants of Lipid Nanoparticles
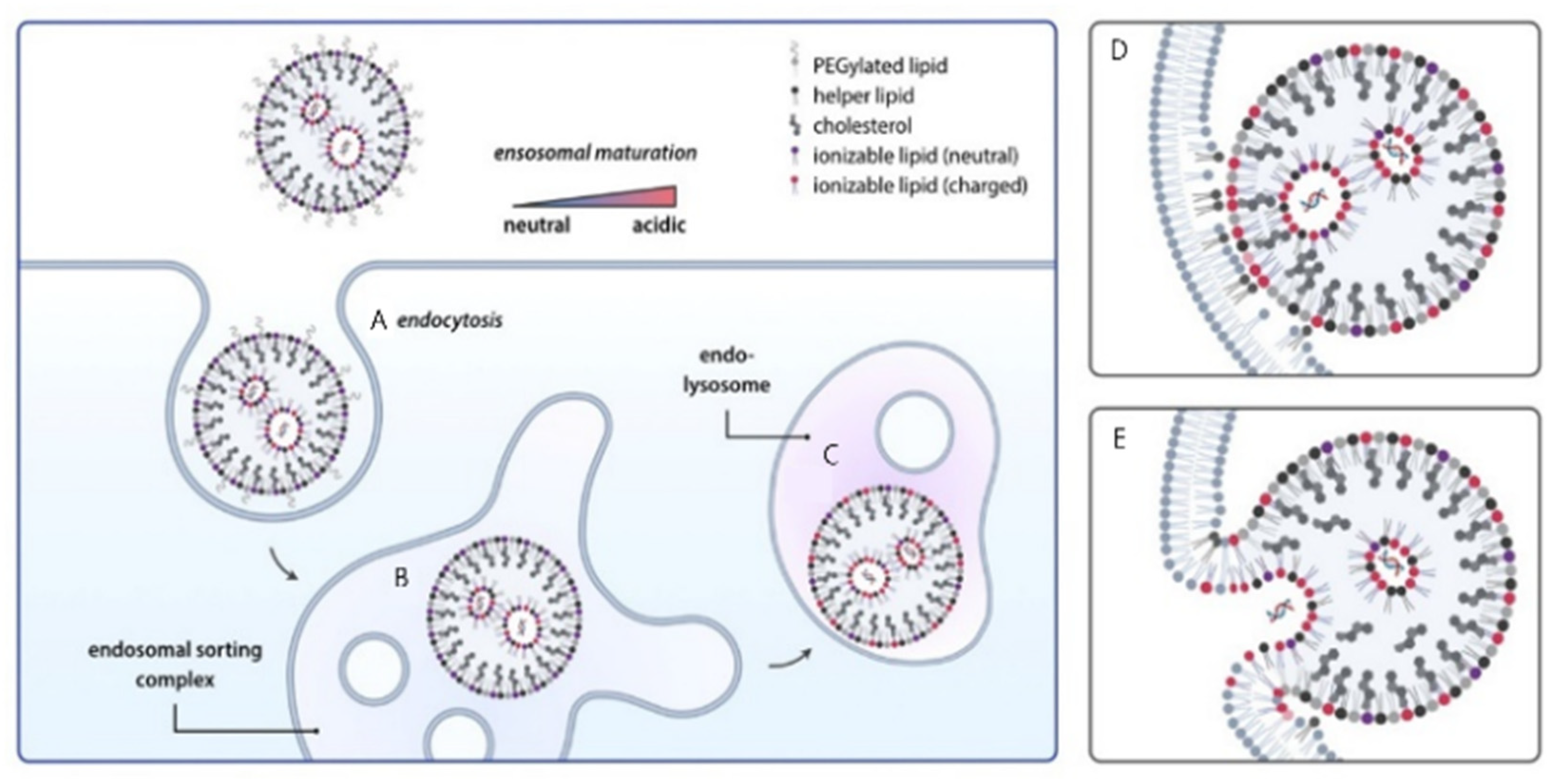
5.1. Endocytic Pathways and Endosomal Sequestration
5.2. Endosomal Escape Mechanisms
5.3. Cytoplasmic Stability and Nucleic Acid Protection
5.4. Nuclear Transport Requirements for Plasmid DNA
6. Integrated Engineering and Translation of Lipid Nanoparticles for Nucleic Acid Delivery
6.1. Lipid Composition Engineering for LNP Delivery
6.1.1. Ionizable Lipid Optimization
6.1.2. Helper Lipid and Cholesterol Optimization
6.1.3. PEG-Lipid Density and Shedding Control
6.1.4. Anti-PEG Immunity and Accelerated Blood Clearance
6.2. Tissue and Cell Targeting Strategies for Lipid Nanoparticles
6.2.1. Hepatocyte-Directed Targeting
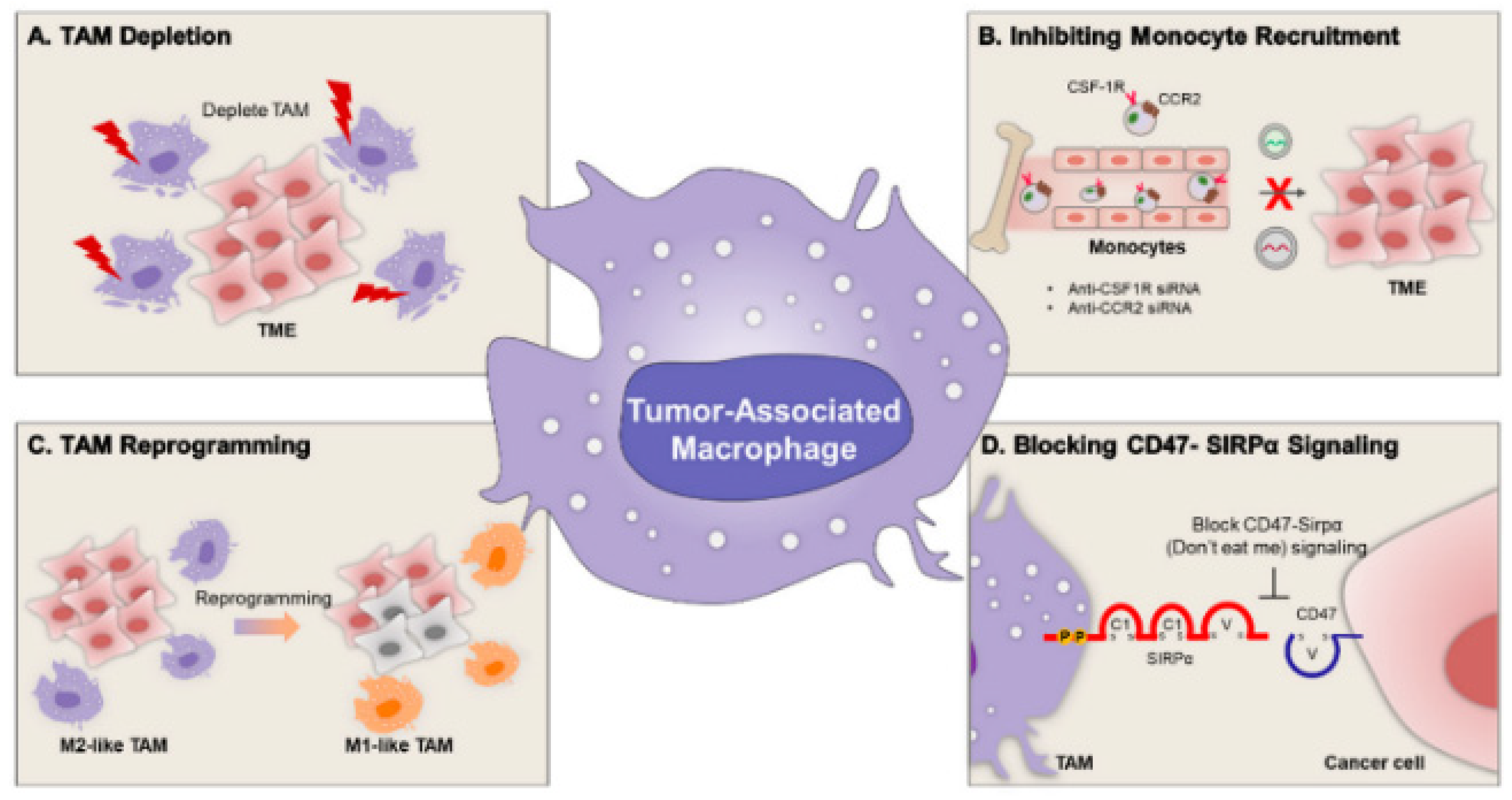
6.2.2. Immune-Cell or Tumor-Specific Targeting
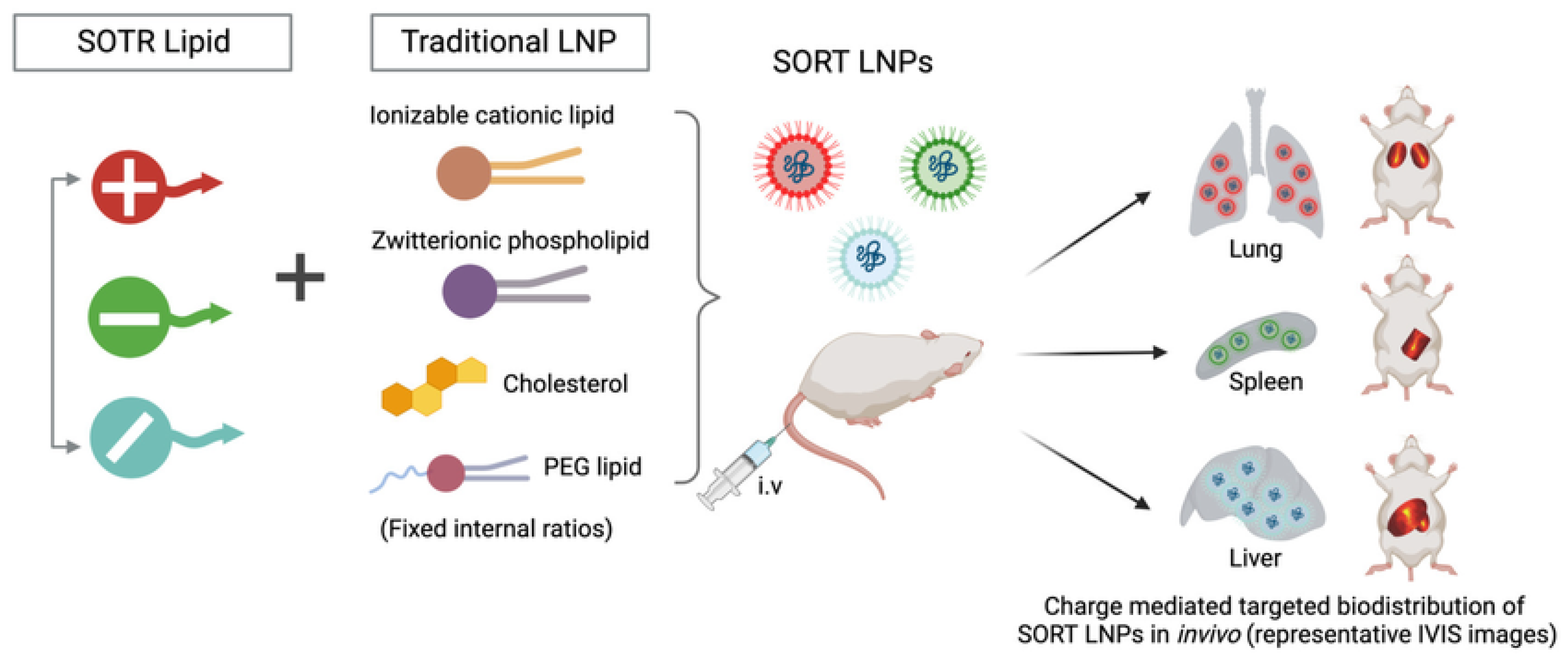
6.2.3. Organ-Selective Lipid Nanoparticles (SORT)
6.3. Endosomal Escape Engineering
6.3.1. pH-Responsive Lipid Engineering
6.3.2. Membrane-Fusion Peptides and Polymers
6.4. Immunosuppression Strategies and Administration Pathways for Lipid Nanoparticles
6.4.1. Incorporation of Immunosuppressive or Tolerogenic Agents
6.4.2. Administration Routes and Dosing Strategies
6.5. Manufacturing and Stability Engineering of Lipid Nanoparticles
6.5.1. Microfluidic Mixing Parameters
6.5.2. Lyophilization, Storage, and Long-Term Stability
7. Next-Generation LNP Systems and Precision Therapeutic Design
7.1. Hybrid and Multifunctional Nanocarriers Integrating Lipid Nanoparticles
7.2. Personalized and Precision Nanomedicine Approaches
7.3. Marketed Lipid Nanocarrier-Based Delivery Platforms
8. Conclusions
9. Patents
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| EPR | Enhanced Permeability and Retention |
| FRR | Flow Rate Ratio |
| IM | Intramuscular |
| IV | Intravenous |
| LNP | Lipid Nanoparticle |
| MPS | Mononuclear Phagocyte System |
| NLS | Nuclear Localization Signal |
| PEG | Polyethylene Glycol |
| RES | Reticuloendothelial System |
| SC | Subcutaneous |
| SORT | Selective Organ Targeting |
| TAM | Tumor-Associated Macrophage |
| TFR | Total Flow Rate |
| TME | Tumor Microenvironment |
References
- Xu, S.; Hu, Z.; Song, F.; Xu, Y.; Han, X. Lipid Nanoparticles: Composition, Formulation and Application. Mol. Ther. Methods Clin. Dev. 2025, 33, 101463. [Google Scholar] [CrossRef] [PubMed]
- Vasileva, O.; Zaborova, O.; Shmykov, B.; Ivanov, R.; Reshetnikov, V. Composition of lipid nanoparticles for targeted delivery: Application to mRNA therapeutics. Front. Pharmacol. 2024, 15, 1466337. [Google Scholar] [CrossRef]
- Tang, X.; Zhang, Y.; Han, X. Ionizable lipid nanoparticles for mRNA delivery. Adv. NanoBiomed Res. 2023, 3, 2300006. [Google Scholar] [CrossRef]
- Wei, P.-S.; Thota, N.; John, G.; Chang, E.; Lee, S.; Wang, Y.; Ma, Z.; Tsai, Y.-H.; Mei, K.-C. Enhancing RNA-lipid nanoparticle delivery: Organ-and cell-specificity and barcoding strategies. J. Control. Release 2024, 375, 366–388. [Google Scholar] [CrossRef]
- Amici, A.; Pozzi, D.; Marchini, C.; Caracciolo, G. The transformative potential of lipid nanoparticle–protein corona for next-generation vaccines and therapeutics. Mol. Pharm. 2023, 20, 5247–5253. [Google Scholar] [CrossRef]
- Younis, M.A.; Sato, Y.; Kimura, S.; Harashima, H. A new strategy for the extrahepatic delivery of lipid-based nanomedicines: A protein corona-mediated selective targeting system based on an ionizable cationic lipid library. RSC Pharm. 2025, 2, 982–1002. [Google Scholar] [CrossRef]
- Heredero, J.; Peña, Á.; Broset, E.; Blandín, B.; de Miguel, D.; Alejo, T.; Toro, A.; Mata, E.; López-Gavín, A.; Gallego-Lleyda, A. Predictive Lung-and Spleen-Targeted mRNA Delivery with Biodegradable Ionizable Lipids in Four-Component LNPs. Pharmaceutics 2025, 17, 459. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Wang, X.; Li, L.; Li, F.; Zhang, J.; Liang, X.J. Lipid nanoparticles optimized for targeting and release of nucleic acid. Adv. Mater. 2024, 36, 2305300. [Google Scholar] [CrossRef]
- Pinto, I.S.; Cordeiro, R.A.; Faneca, H. Polymer-and lipid-based gene delivery technology for CAR T cell therapy. J. Control. Release 2023, 353, 196–215. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Carter, J.; Santos, L.; Webster, C.; Van Der Walle, C.F.; Li, P.; Rogers, S.E.; Lu, J.R. Acidification-induced structure evolution of lipid nanoparticles correlates with their in vitro gene transfections. ACS Nano 2023, 17, 979–990. [Google Scholar] [CrossRef]
- Karmaker, S.; Rosales, P.D.; Tirumuruhan, B.; Viravalli, A.; Boehnke, N. More than a delivery system: The evolving role of lipid-based nanoparticles. Nanoscale 2025, 17, 11864–11893. [Google Scholar] [CrossRef]
- Amoako, K.; Mokhammad, A.; Malik, A.; Yesudasan, S.; Wheba, A.; Olagunju, O.; Gu, S.X.; Yarovinsky, T.; Faustino, E.V.S.; Nguyen, J. Enhancing nucleic acid delivery by the integration of artificial intelligence into lipid nanoparticle formulation. Front. Med. Technol. 2025, 7, 1591119. [Google Scholar] [CrossRef]
- Wang, J.; Chen, R.; Xie, Y.; Qin, X.; Zhou, Y.; Xu, C. Endo/Lysosomal-Escapable Lipid Nanoparticle Platforms for Enhancing mRNA Delivery in Cancer Therapy. Pharmaceutics 2025, 17, 803. [Google Scholar] [CrossRef]
- Hou, X.; Zaks, T.; Langer, R.; Dong, Y. Lipid nanoparticles for mRNA delivery. Nat. Rev. Mater. 2021, 6, 1078–1094. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Ding, Y.; Chong, K.; Cui, M.; Cao, Z.; Tang, C.; Tian, Z.; Hu, Y.; Zhao, Y.; Jiang, S. Recent advances in lipid nanoparticles and their safety concerns for mRNA delivery. Vaccines 2024, 12, 1148. [Google Scholar] [CrossRef] [PubMed]
- Young, R.E.; Nelson, K.M.; Hofbauer, S.I.; Vijayakumar, T.; Alameh, M.-G.; Weissman, D.; Papachristou, C.; Gleghorn, J.P.; Riley, R.S. Systematic development of ionizable lipid nanoparticles for placental mRNA delivery using a design of experiments approach. Bioact. Mater. 2024, 34, 125–137. [Google Scholar] [CrossRef]
- Schober, G.B.; Story, S.; Arya, D.P. A careful look at lipid nanoparticle characterization: Analysis of benchmark formulations for encapsulation of RNA cargo size gradient. Sci. Rep. 2024, 14, 2403. [Google Scholar] [CrossRef] [PubMed]
- Haghighi, E.; Abolmaali, S.S.; Dehshahri, A.; Mousavi Shaegh, S.A.; Azarpira, N.; Tamaddon, A.M. Navigating the intricate in-vivo journey of lipid nanoparticles tailored for the targeted delivery of RNA therapeutics: A quality-by-design approach. J. Nanobiotechnol. 2024, 22, 710. [Google Scholar] [CrossRef]
- Biswas, B.; Shah, D.; Cox-Vázquez, S.J.; Vázquez, R.J. Sensing cholesterol-induced rigidity in model membranes with time-resolved fluorescence spectroscopy and microscopy. J. Mater. Chem. B 2024, 12, 6570–6576. [Google Scholar] [CrossRef]
- Zheng, L.; Bandara, S.R.; Tan, Z.; Leal, C. Lipid nanoparticle topology regulates endosomal escape and delivery of RNA to the cytoplasm. Proc. Natl. Acad. Sci. USA 2023, 120, e2301067120. [Google Scholar] [CrossRef]
- Mahmoudi, N.; Mahmoudi, M. Effects of cholesterol on biomolecular corona. Nat. Nanotechnol. 2023, 18, 974–976. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Zhang, H.; Li, G.; Su, J.; Wei, Y.; Xu, C. Lipid nanovehicles overcome barriers to systemic RNA delivery: Lipid components, fabrication methods, and rational design. Acta Pharm. Sin. B 2024, 14, 579–601. [Google Scholar] [CrossRef]
- Waggoner, L.E.; Miyasaki, K.F.; Kwon, E.J. Analysis of PEG-lipid anchor length on lipid nanoparticle pharmacokinetics and activity in a mouse model of traumatic brain injury. Biomater. Sci. 2023, 11, 4238–4253. [Google Scholar] [CrossRef]
- Zhang, H.; Barz, M. Investigating the stability of RNA-lipid nanoparticles in biological fluids: Unveiling its crucial role for understanding LNP performance. J. Control. Release 2025, 381, 113559. [Google Scholar] [CrossRef]
- Liu, M.; Chu, Y.; Liu, H.; Su, Y.; Zhang, Q.; Jiao, J.; Liu, M.; Ding, J.; Liu, M.; Hu, Y. Accelerated blood clearance of nanoemulsions modified with PEG-cholesterol and PEG-phospholipid derivatives in rats: The effect of PEG-lipid linkages and PEG molecular weights. Mol. Pharm. 2019, 17, 1059–1070. [Google Scholar] [CrossRef]
- Ishida, T.; Kiwada, H. Accelerated blood clearance (ABC) phenomenon upon repeated injection of PEGylated liposomes. Int. J. Pharm. 2008, 354, 56–62. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.-M.; Su, Y.-C.; Chang, C.-J.; Burnouf, P.-A.; Chuang, K.-H.; Chen, C.-H.; Cheng, T.-L.; Chen, Y.-T.; Wu, J.-Y.; Roffler, S.R. Measurement of pre-existing IgG and IgM antibodies against polyethylene glycol in healthy individuals. Anal. Chem. 2016, 88, 10661–10666. [Google Scholar] [CrossRef]
- Knop, K.; Hoogenboom, R.; Fischer, D.; Schubert, U.S. Poly (ethylene glycol) in drug delivery: Pros and cons as well as potential alternatives. Angew. Chem. Int. Ed. 2010, 49, 6288–6308. [Google Scholar] [CrossRef]
- Adhipandito, C.F.; Cheung, S.-H.; Lin, Y.-H.; Wu, S.-H. Atypical renal clearance of nanoparticles larger than the kidney filtration threshold. Int. J. Mol. Sci. 2021, 22, 11182. [Google Scholar] [CrossRef]
- Voke, E.; Arral, M.; Squire, H.J.; Lin, T.-J.; Coreas, R.; Lui, A.; Iavarone, A.T.; Pinals, R.L.; Whitehead, K.A.; Landry, M. Protein corona formed on lipid nanoparticles compromises delivery efficiency of mRNA cargo. bioRxiv 2025. [Google Scholar] [CrossRef] [PubMed]
- Wen, W.; Qiong, W.; Qi-Hui, L.; Yuan-Yuan, L.; Ping, R.; Yuan-Yu, W.; Fang-Fang, C. Identification and characterization of soft protein corona absorbed on iron oxide nanoparticles. Chin. J. Anal. Chem. 2023, 51, 100246. [Google Scholar] [CrossRef]
- Pattipeiluhu, R.; Zeng, Y.; Hendrix, M.M.; Voets, I.K.; Kros, A.; Sharp, T.H. Liquid crystalline inverted lipid phases encapsulating siRNA enhance lipid nanoparticle mediated transfection. Nat. Commun. 2024, 15, 1303. [Google Scholar] [CrossRef]
- Udepurkar, A.; Devos, C.; Sagmeister, P.; Destro, F.; Inguva, P.; Ahmadi, S.; Boulais, E.; Quan, Y.; Braatz, R.D.; Myerson, A.S. Structure and Morphology of Lipid Nanoparticles for Nucleic Acid Drug Delivery: A Review. ACS Nano 2025, 19, 21206–21242. [Google Scholar] [CrossRef] [PubMed]
- Mehta, M.; Bui, T.A.; Yang, X.; Aksoy, Y.; Goldys, E.M.; Deng, W. Lipid-based nanoparticles for drug/gene delivery: An overview of the production techniques and difficulties encountered in their industrial development. ACS Mater. Au 2023, 3, 600–619. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Wen, Y.; Shan, X.; Ma, X.; Yang, C.; Cheng, X.; Zhao, Y.; Li, J.; Mi, S.; Huo, H. Charge-assisted stabilization of lipid nanoparticles enables inhaled mRNA delivery for mucosal vaccination. Nat. Commun. 2024, 15, 9471. [Google Scholar] [CrossRef]
- Németh, Z.; Csóka, I.; Semnani Jazani, R.; Sipos, B.; Haspel, H.; Kozma, G.; Kónya, Z.; Dobó, D.G. Quality by design-driven zeta potential optimisation study of liposomes with charge imparting membrane additives. Pharmaceutics 2022, 14, 1798. [Google Scholar] [CrossRef]
- Borah, A.; Giacobbo, V.; Binici, B.; Baillie, R.; Perrie, Y. From in vitro to in vivo: The Dominant role of PEG-Lipids in LNP performance. Eur. J. Pharm. Biopharm. 2025, 212, 114726. [Google Scholar] [CrossRef]
- Liu, W.; Zhang, M.; Lv, H.; Yang, C. Formulation-Driven Optimization of PEG-Lipid Content in Lipid Nanoparticles for Enhanced mRNA Delivery In Vitro and In Vivo. Pharmaceutics 2025, 17, 950. [Google Scholar] [CrossRef]
- Haripriyaa, M.; Suthindhiran, K. Pharmacokinetics of nanoparticles: Current knowledge, future directions and its implications in drug delivery. Future J. Pharm. Sci. 2023, 9, 113. [Google Scholar] [CrossRef]
- Guo, W.X.; Hu, L.F.; Feng, Y.H.; Liu, Y.; Jing, L.Y.; Chen, B.Z.; Guo, X.D. Evaluation of Nanoparticle Stability under Blood Flow Shear. Langmuir 2022, 38, 12731–12738. [Google Scholar] [CrossRef]
- Li, Y.; Saba, L.; Scheinman, R.I.; Banda, N.K.; Holers, M.; Monte, A.; Dylla, L.; Moghimi, S.M.; Simberg, D. Nanoparticle-binding immunoglobulins predict variable complement responses in healthy and diseased cohorts. ACS Nano 2024, 18, 28649–28658. [Google Scholar] [CrossRef]
- Lebreton, V.; Legeay, S.; Vasylaki, A.; Lagarce, F.; Saulnier, P. Protein corona formation on lipidic nanocapsules: Influence of the interfacial PEG repartition. Eur. J. Pharm. Sci. 2023, 189, 106537. [Google Scholar] [CrossRef]
- Liu, K.; Nilsson, R.; Lázaro-Ibáñez, E.; Duàn, H.; Miliotis, T.; Strimfors, M.; Lerche, M.; Salgado Ribeiro, A.R.; Ulander, J.; Lindén, D. Multiomics analysis of naturally efficacious lipid nanoparticle coronas reveals high-density lipoprotein is necessary for their function. Nat. Commun. 2023, 14, 4007. [Google Scholar] [CrossRef]
- Chou, W.-C.; Lin, Z. Impact of protein coronas on nanoparticle interactions with tissues and targeted delivery. Curr. Opin. Biotechnol. 2024, 85, 103046. [Google Scholar] [CrossRef]
- Sun, X.; Jiang, X. Collagen targeting liposomes, a promising strategy for idiopathic pulmonary fibrosis. J. Control. Release Off. J. Control. Release Soc. 2023, 361, 896–897. [Google Scholar] [CrossRef]
- Tenzer, S.; Docter, D.; Rosfa, S.; Wlodarski, A.; Kuharev, J.; Rekik, A.; Knauer, S.K.; Bantz, C.; Nawroth, T.; Bier, C. Nanoparticle size is a critical physicochemical determinant of the human blood plasma corona: A comprehensive quantitative proteomic analysis. ACS Nano 2011, 5, 7155–7167. [Google Scholar] [CrossRef] [PubMed]
- Treuel, L.; Docter, D.; Maskos, M.; Stauber, R.H. Protein corona–from molecular adsorption to physiological complexity. Beilstein J. Nanotechnol. 2015, 6, 857–873. [Google Scholar] [CrossRef] [PubMed]
- Hosseini-Kharat, M.; Bremmell, K.E.; Prestidge, C.A. Why do lipid nanoparticles target the liver? Understanding of biodistribution and liver-specific tropism. Mol. Ther. Methods Clin. Dev. 2025, 33, 101436. [Google Scholar] [CrossRef] [PubMed]
- Zelepukin, I.V.; Shevchenko, K.G.; Deyev, S.M. Rediscovery of mononuclear phagocyte system blockade for nanoparticle drug delivery. Nat. Commun. 2024, 15, 4366. [Google Scholar] [CrossRef]
- Pan, J.; Wang, Y.; Chen, Y.; Zhang, C.; Deng, H.; Lu, J.; Chen, W. Emerging strategies against accelerated blood clearance phenomenon of nanocarrier drug delivery systems. J. Nanobiotechnology 2025, 23, 138. [Google Scholar] [CrossRef]
- Wen, P.; Ke, W.; Dirisala, A.; Toh, K.; Tanaka, M.; Li, J. Stealth and pseudo-stealth nanocarriers. Adv. Drug Deliv. Rev. 2023, 198, 114895. [Google Scholar] [CrossRef] [PubMed]
- Dal Magro, R.; Ornaghi, F.; Cambianica, I.; Beretta, S.; Re, F.; Musicanti, C.; Rigolio, R.; Donzelli, E.; Canta, A.; Ballarini, E. ApoE-modified solid lipid nanoparticles: A feasible strategy to cross the blood-brain barrier. J. Control. Release 2017, 249, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Sheth, V.; Wang, L.; Bhattacharya, R.; Mukherjee, P.; Wilhelm, S. Strategies for delivering nanoparticles across tumor blood vessels. Adv. Funct. Mater. 2021, 31, 2007363. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Wang, Y.; Wang, L.; Jiang, W.; Wilhelm, S. Understanding nanoparticle-liver interactions in nanomedicine. Expert Opin. Drug Deliv. 2024, 21, 829–843. [Google Scholar] [CrossRef]
- Wang, L.; Quine, S.; Frickenstein, A.N.; Lee, M.; Yang, W.; Sheth, V.M.; Bourlon, M.D.; He, Y.; Lyu, S.; Garcia-Contreras, L. Exploring and analyzing the systemic delivery barriers for nanoparticles. Adv. Funct. Mater. 2024, 34, 2308446. [Google Scholar] [CrossRef]
- Belyaev, I.B.; Griaznova, O.Y.; Yaremenko, A.V.; Deyev, S.M.; Zelepukin, I.V. Beyond the EPR effect: Intravital microscopy analysis of nanoparticle drug delivery to tumors. Adv. Drug Deliv. Rev. 2025, 219, 115550. [Google Scholar] [CrossRef]
- Li, X.; Hu, Y.; Zhang, X.; Shi, X.; Parak, W.J.; Pich, A. Transvascular transport of nanocarriers for tumor delivery. Nat. Commun. 2024, 15, 8172. [Google Scholar] [CrossRef]
- Lu, B.; Wang, J.; Hendriks, A.J.; Nolte, T.M. Clearance of nanoparticles from blood: Effects of hydrodynamic size and surface coatings. Environ. Sci. Nano 2024, 11, 406–417. [Google Scholar] [CrossRef]
- Attia, M.S.; Kijanka, G.; Nguyen, N.-T.; Zhang, J.; An, H. Advances and prospects of RNA delivery nanoplatforms for cancer therapy. Acta Pharm. Sin. B 2025, 15, 52–96. [Google Scholar] [CrossRef]
- Gong, N.; Zhong, W.; Alameh, M.-G.; Han, X.; Xue, L.; El-Mayta, R.; Zhao, G.; Vaughan, A.E.; Qin, Z.; Xu, F. Tumour-derived small extracellular vesicles act as a barrier to therapeutic nanoparticle delivery. Nat. Mater. 2024, 23, 1736–1747. [Google Scholar] [CrossRef]
- Jain, R.K. Transport of molecules, particles, and cells in solid tumors. Annu. Rev. Biomed. Eng. 1999, 1, 241–263. [Google Scholar] [CrossRef] [PubMed]
- Lu, P.; Weaver, V.M.; Werb, Z. The extracellular matrix: A dynamic niche in cancer progression. J. Cell Biol. 2012, 196, 395–406. [Google Scholar] [CrossRef]
- Stylianopoulos, T.; Jain, R.K. Design considerations for nanotherapeutics in oncology. Nanomed. Nanotechnol. Biol. Med. 2015, 11, 1893–1907. [Google Scholar] [CrossRef]
- Cruz-Acuña, R.; Vunjak-Novakovic, G.; Burdick, J.A.; Rustgi, A.K. Emerging technologies provide insights on cancer extracellular matrix biology and therapeutics. Iscience 2021, 24, 102475. [Google Scholar] [CrossRef]
- Cassani, M.; Fernandes, S.; Pagliari, S.; Cavalieri, F.; Caruso, F.; Forte, G. Unraveling the role of the tumor extracellular matrix to inform nanoparticle design for nanomedicine. Adv. Sci. 2025, 12, 2409898. [Google Scholar] [CrossRef] [PubMed]
- Meaney, C.; Stapleton, S.; Kohandel, M. Predicting intratumoral fluid pressure and liposome accumulation using physics informed deep learning. Sci. Rep. 2023, 13, 20548. [Google Scholar] [CrossRef]
- Vagena, I.-A.; Malapani, C.; Gatou, M.-A.; Lagopati, N.; Pavlatou, E.A. Enhancement of EPR effect for passive tumor targeting: Current status and future perspectives. Appl. Sci. 2025, 15, 3189. [Google Scholar] [CrossRef]
- Aloss, K.; Hamar, P. Augmentation of the EPR effect by mild hyperthermia to improve nanoparticle delivery to the tumor. Biochim. Biophys. Acta (BBA)-Rev. Cancer 2024, 1879, 189109. [Google Scholar] [CrossRef]
- Gu, Q.; Zhu, L. Heating Induced Nanoparticle Migration and Enhanced Delivery in Tumor Treatment Using Nanotechnology. Bioengineering 2024, 11, 900. [Google Scholar] [CrossRef]
- Liu, S.; Ren, Z.; Yan, M.; Ye, W.; Hu, Y. Strategies to enhance the penetration of nanomedicine in solid tumors. Biomaterials 2025, 321, 123315. [Google Scholar] [CrossRef]
- Fan, D.; Cao, Y.; Cao, M.; Wang, Y.; Cao, Y.; Gong, T. Nanomedicine in cancer therapy. Signal Transduct. Target. Ther. 2023, 8, 293. [Google Scholar] [CrossRef]
- Chatterjee, S.; Kon, E.; Sharma, P.; Peer, D. Endosomal escape: A bottleneck for LNP-mediated therapeutics. Proc. Natl. Acad. Sci. USA 2024, 121, e2307800120. [Google Scholar] [CrossRef] [PubMed]
- Hagedorn, L.; Jürgens, D.C.; Merkel, O.M.; Winkeljann, B. Endosomal escape mechanisms of extracellular vesicle-based drug carriers: Lessons for lipid nanoparticle design. Extracell. Vesicles Circ. Nucleic Acids 2024, 5, 344. [Google Scholar] [CrossRef]
- Sadaqa, E.; Kurniawan, F.; Mudhakir, D. Mechanistic insights into endosomal escape by sodium oleate-modified liposomes. Beilstein J. Nanotechnol. 2024, 15, 1667–1685. [Google Scholar] [CrossRef] [PubMed]
- Johansson, J.M.; Du Rietz, H.; Hedlund, H.; Eriksson, H.C.; Oude Blenke, E.; Pote, A.; Harun, S.; Nordenfelt, P.; Lindfors, L.; Wittrup, A. Cellular and biophysical barriers to lipid nanoparticle mediated delivery of RNA to the cytosol. Nat. Commun. 2025, 16, 5354. [Google Scholar] [CrossRef]
- Chowdhury, C.R.; Hoover, E.C.; Day, E.S. Membrane-modified lipid nanoparticles for RNA delivery. Mol. Ther. Methods Clin. Dev. 2025, 33, 101505. [Google Scholar] [CrossRef]
- Pozzi, D.; Caracciolo, G. Looking back, moving forward: Lipid nanoparticles as a promising frontier in gene delivery. ACS Pharmacol. Transl. Sci. 2023, 6, 1561–1573. [Google Scholar] [CrossRef]
- Ritter, A.T.; Shtengel, G.; Xu, C.S.; Weigel, A.; Hoffman, D.P.; Freeman, M.; Iyer, N.; Alivodej, N.; Ackerman, D.; Voskoboinik, I. ESCRT-mediated membrane repair protects tumor-derived cells against T cell attack. Science 2022, 376, 377–382. [Google Scholar] [CrossRef]
- Zhao, Y.; Wang, Z.M.; Song, D.; Chen, M.; Xu, Q. Rational design of lipid nanoparticles: Overcoming physiological barriers for selective intracellular mRNA delivery. Curr. Opin. Chem. Biol. 2024, 81, 102499. [Google Scholar] [CrossRef]
- Lu, Z.-R.; Sun, D. Mechanism of pH-sensitive Amphiphilic Endosomal Escape of Ionizable Lipid Nanoparticles for Cytosolic Nucleic Acid Delivery. Pharm. Res. 2025, 42, 1065–1077. [Google Scholar] [CrossRef] [PubMed]
- Grau, M.; Wagner, E. Strategies and mechanisms for endosomal escape of therapeutic nucleic acids. Curr. Opin. Chem. Biol. 2024, 81, 102506. [Google Scholar] [CrossRef]
- Philipp, J.; Dabkowska, A.; Reiser, A.; Frank, K.; Krzysztoń, R.; Brummer, C.; Nickel, B.; Blanchet, C.E.; Sudarsan, A.; Ibrahim, M. pH-dependent structural transitions in cationic ionizable lipid mesophases are critical for lipid nanoparticle function. Proc. Natl. Acad. Sci. USA 2023, 120, e2310491120. [Google Scholar] [CrossRef]
- Omo-Lamai, S.; Wang, Y.; Patel, M.N.; Essien, E.-O.; Shen, M.; Majumdar, A.; Espy, C.; Wu, J.; Channer, B.; Tobin, M. Lipid nanoparticle-associated inflammation is triggered by sensing of endosomal damage: Engineering endosomal escape without side effects. bioRxiv 2024. [Google Scholar] [CrossRef]
- Padilla, M.S.; Mrksich, K.; Wang, Y.; Haley, R.M.; Li, J.J.; Han, E.L.; El-Mayta, R.; Kim, E.H.; Dias, S.; Gong, N. Branched endosomal disruptor (BEND) lipids mediate delivery of mRNA and CRISPR-Cas9 ribonucleoprotein complex for hepatic gene editing and T cell engineering. Nat. Commun. 2025, 16, 996. [Google Scholar] [CrossRef] [PubMed]
- Ewii, U.E.; Attama, A.A.; Olorunsola, E.O.; Onugwu, A.L.; Nwakpa, F.U.; Anyiam, C.; Chijioke, C.; Ogbulie, T. Nanoparticles For Drug Delivery: Insight Into In Vitro And In Vivo Drug Release From Nanomedicines. Nano TransMed 2025, 4, 100083. [Google Scholar] [CrossRef]
- Carton, F.; Malatesta, M. In vitro models of biological barriers for nanomedical research. Int. J. Mol. Sci. 2022, 23, 8910. [Google Scholar] [CrossRef]
- Gimondi, S.; Ferreira, H.; Reis, R.L.; Neves, N.M. Size-dependent polymeric nanoparticle distribution in a static versus dynamic microfluidic blood vessel model: Implications for nanoparticle-based drug delivery. ACS Appl. Nano Mater. 2023, 6, 7364–7374. [Google Scholar] [CrossRef]
- Liu, Y.; Huang, Y.; He, G.; Guo, C.; Dong, J.; Wu, L. Development of mRNA lipid nanoparticles: Targeting and therapeutic aspects. Int. J. Mol. Sci. 2024, 25, 10166. [Google Scholar] [CrossRef] [PubMed]
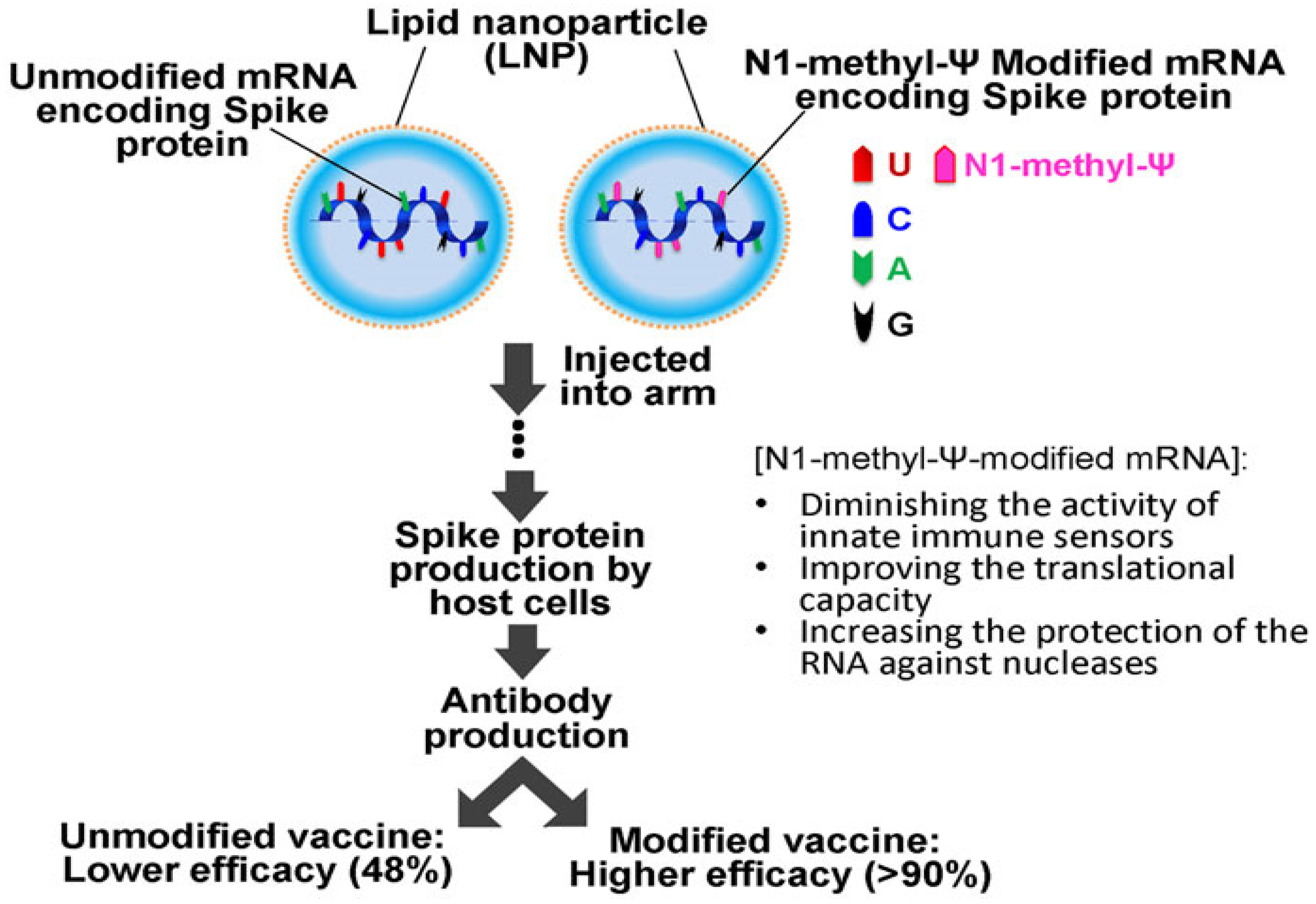
- Ho, L.L.; Schiess, G.H.; Miranda, P.; Weber, G.; Astakhova, K. Pseudouridine and N 1-methylpseudouridine as potent nucleotide analogues for RNA therapy and vaccine development. RSC Chem. Biol. 2024, 5, 418–425. [Google Scholar] [CrossRef] [PubMed]
- Mulroney, T.E.; Pöyry, T.; Yam-Puc, J.C.; Rust, M.; Harvey, R.F.; Kalmar, L.; Horner, E.; Booth, L.; Ferreira, A.P.; Stoneley, M. N 1-methylpseudouridylation of mRNA causes+ 1 ribosomal frameshifting. Nature 2024, 625, 189–194. [Google Scholar] [CrossRef]
- Lu, R.-M.; Hsu, H.-E.; Perez, S.J.L.P.; Kumari, M.; Chen, G.-H.; Hong, M.-H.; Lin, Y.-S.; Liu, C.-H.; Ko, S.-H.; Concio, C.A.P. Current landscape of mRNA technologies and delivery systems for new modality therapeutics. J. Biomed. Sci. 2024, 31, 89. [Google Scholar] [CrossRef]
- Qu, Y.; Liu, R.; Sun, D.; Dai, Z. Critical Considerations of mRNA-LNPs Technology for CAR-T Therapy: Components, Payloads and Emerging Horizons. Mater. Chem. Front. 2024, 8, 3106–3135. [Google Scholar] [CrossRef]
- Nance, K.D.; Meier, J.L. Modifications in an emergency: The role of N1-methylpseudouridine in COVID-19 vaccines. ACS Cent. Sci. 2021, 7, 748–756. [Google Scholar] [CrossRef]
- Hosseini-Kharat, M.; Bremmell, K.E.; Grubor-Bauk, B.; Prestidge, C.A. Enhancing non-viral DNA delivery systems: Recent advances in improving efficiency and target specificity. J. Control. Release 2025, 378, 170–194. [Google Scholar] [CrossRef]
- McCuskey, D.P.; Achiriloaie, R.E.; Benjamin, C.; Kushen, J.; Blacklow, I.; Mnfy, O.; Ross, J.L.; Robertson-Anderson, R.M.; Sheung, J.Y. Dna transport is topologically sculpted by active microtubule dynamics. PRX Life 2025, 3, 013015. [Google Scholar] [CrossRef]
- Bai, H.; Lester, G.M.S.; Petishnok, L.C.; Dean, D.A. Cytoplasmic transport and nuclear import of plasmid DNA. Biosci. Rep. 2017, 37, BSR20160616. [Google Scholar] [CrossRef] [PubMed]
- Skowicki, M.; Tarvirdipour, S.; Kraus, M.; Schoenenberger, C.-A.; Palivan, C.G. Nanoassemblies designed for efficient nuclear targeting. Adv. Drug Deliv. Rev. 2024, 211, 115354. [Google Scholar] [CrossRef] [PubMed]
- Vaughan, E.E.; DeGiulio, J.V.; Dean, D.A. Intracellular trafficking of plasmids for gene therapy: Mechanisms of cytoplasmic movement and nuclear import. Curr. Gene Ther. 2006, 6, 671–681. [Google Scholar] [CrossRef]
- Xu, Y.; Ma, S.; Cui, H.; Chen, J.; Xu, S.; Gong, F.; Golubovic, A.; Zhou, M.; Wang, K.C.; Varley, A. AGILE platform: A deep learning powered approach to accelerate LNP development for mRNA delivery. Nat. Commun. 2024, 15, 6305. [Google Scholar] [CrossRef]
- Tesei, G.; Hsiao, Y.W.; Dabkowska, A.; Gronberg, G.; Yanez Arteta, M.; Ulkoski, D.; Bray, D.J.; Trulsson, M.; Ulander, J.; Lund, M.; et al. Lipid shape and packing are key for optimal design of pH-sensitive mRNA lipid nanoparticles. Proc. Natl. Acad. Sci. USA 2024, 121, e2311700120. [Google Scholar] [CrossRef]
- Mrksich, K.; Padilla, M.S.; Mitchell, M.J. Breaking the final barrier: Evolution of cationic and ionizable lipid structure in lipid nanoparticles to escape the endosome. Adv. Drug Deliv. Rev. 2024, 214, 115446. [Google Scholar] [CrossRef]
- Gretskaya, N.; Akimov, M.; Andreev, D.; Zalygin, A.; Belitskaya, E.; Zinchenko, G.; Fomina-Ageeva, E.; Mikhalyov, I.; Vodovozova, E.; Bezuglov, V. Multicomponent lipid nanoparticles for RNA transfection. Pharmaceutics 2023, 15, 1289. [Google Scholar] [CrossRef]
- Khodadadi, E.; Khodadadi, E.; Chaturvedi, P.; Moradi, M. Comprehensive Insights into the Cholesterol-Mediated Modulation of Membrane Function Through Molecular Dynamics Simulations. Membranes 2025, 15, 173. [Google Scholar] [CrossRef] [PubMed]
- Garay, R.P.; El-Gewely, R.; Armstrong, J.K.; Garratty, G.; Richette, P. Antibodies against polyethylene glycol in healthy subjects and in patients treated with PEG-conjugated agents. Expert Opin. Drug Deliv. 2012, 9, 1319–1323. [Google Scholar] [CrossRef]
- Li, J.; Mooney, D.J. Designing hydrogels for controlled drug delivery. Nat. Rev. Mater. 2016, 1, 10671. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, J.K.; Hempel, G.; Koling, S.; Chan, L.; Fisher, T.; Meiselman, H.; Garratty, G. Antibody against poly (ethylene glycol) adversely affects PEG-asparaginase therapy in acute lymphoblastic leukemia patients. Cancer 2007, 110, 103–111. [Google Scholar] [CrossRef]
- Lila, A.S.A.; Shimizu, T.; Ishida, T. PEGylation and anti-PEG antibodies. In Engineering of Biomaterials for Drug Delivery Systems; Elsevier: Amsterdam, The Netherlands, 2018; pp. 51–68. [Google Scholar]
- Catenacci, L.; Rossi, R.; Sechi, F.; Buonocore, D.; Sorrenti, M.; Perteghella, S.; Peviani, M.; Bonferoni, M.C. Effect of lipid nanoparticle physico-chemical properties and composition on their interaction with the immune system. Pharmaceutics 2024, 16, 1521. [Google Scholar] [CrossRef] [PubMed]
- Suk, J.S.; Xu, Q.; Kim, N.; Hanes, J.; Ensign, L.M. PEGylation as a strategy for improving nanoparticle-based drug and gene delivery. Adv. Drug Deliv. Rev. 2016, 99, 28–51. [Google Scholar] [CrossRef]
- Verhoef, J.J.; Anchordoquy, T.J. Questioning the use of PEGylation for drug delivery. Drug Deliv. Transl. Res. 2013, 3, 499–503. [Google Scholar] [CrossRef]
- Ou, H.; Cheng, T.; Zhang, Y.; Liu, J.; Ding, Y.; Zhen, J.; Shen, W.; Xu, Y.; Yang, W.; Niu, P. Surface-adaptive zwitterionic nanoparticles for prolonged blood circulation time and enhanced cellular uptake in tumor cells. Acta Biomater. 2018, 65, 339–348. [Google Scholar] [CrossRef]
- Yang, Q.; Lai, S.K. Anti-PEG immunity: Emergence, characteristics, and unaddressed questions. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2015, 7, 655–677. [Google Scholar] [CrossRef] [PubMed]
- Steffens, R.C.; Wagner, E. Directing the Way-Receptor and Chemical Targeting Strategies for Nucleic Acid Delivery. Pharm. Res. 2023, 40, 47–76. [Google Scholar] [CrossRef]
- Zhang, T.; Yin, H.; Li, Y.; Yang, H.; Ge, K.; Zhang, J.; Yuan, Q.; Dai, X.; Naeem, A.; Weng, Y.; et al. Optimized lipid nanoparticles (LNPs) for organ-selective nucleic acids delivery in vivo. iScience 2024, 27, 109804. [Google Scholar] [CrossRef]
- Sabit, H.; Pawlik, T.M.; Radwan, F.; Abdel-Hakeem, M.; Abdel-Ghany, S.; Wadan, A.S.; Elzawahri, M.; El-Hashash, A.; Arneth, B. Precision nanomedicine: Navigating the tumor microenvironment for enhanced cancer immunotherapy and targeted drug delivery. Mol. Cancer 2025, 24, 160. [Google Scholar] [CrossRef]
- Parshad, B.; Arora, S.; Singh, B.; Pan, Y.; Tang, J.; Hu, Z.; Patra, H.K. Towards precision medicine using biochemically triggered cleavable conjugation. Commun. Chem. 2025, 8, 100. [Google Scholar] [CrossRef]
- Cheng, Q.; Wei, T.; Farbiak, L.; Johnson, L.T.; Dilliard, S.A.; Siegwart, D.J. Selective organ targeting (SORT) nanoparticles for tissue-specific mRNA delivery and CRISPR-Cas gene editing. Nat. Nanotechnol. 2020, 15, 313–320. [Google Scholar] [CrossRef]
- Wang, X.; Liu, S.; Sun, Y.; Yu, X.; Lee, S.M.; Cheng, Q.; Wei, T.; Gong, J.; Robinson, J.; Zhang, D. Preparation of selective organ-targeting (SORT) lipid nanoparticles (LNPs) using multiple technical methods for tissue-specific mRNA delivery. Nat. Protoc. 2023, 18, 265–291. [Google Scholar] [CrossRef]
- Żak, M.M.; Zangi, L. Lipid nanoparticles for organ-specific mRNA therapeutic delivery. Pharmaceutics 2021, 13, 1675. [Google Scholar] [CrossRef] [PubMed]
- Dilliard, S.A.; Sun, Y.; Brown, M.O.; Sung, Y.-C.; Chatterjee, S.; Farbiak, L.; Vaidya, A.; Lian, X.; Wang, X.; Lemoff, A. The interplay of quaternary ammonium lipid structure and protein corona on lung-specific mRNA delivery by selective organ targeting (SORT) nanoparticles. J. Control. Release 2023, 361, 361–372. [Google Scholar] [CrossRef] [PubMed]
- Ferraresso, F.; Strilchuk, A.W.; Juang, L.J.; Poole, L.G.; Luyendyk, J.P.; Kastrup, C.J. Comparison of DLin-MC3-DMA and ALC-0315 for siRNA delivery to hepatocytes and hepatic stellate cells. Mol. Pharm. 2022, 19, 2175–2182. [Google Scholar] [CrossRef] [PubMed]
- Larson, N.R.; Hu, G.; Wei, Y.; Tuesca, A.D.; Forrest, M.L.; Middaugh, C.R. pH-dependent phase behavior and stability of cationic lipid–mRNA nanoparticles. J. Pharm. Sci. 2022, 111, 690–698. [Google Scholar] [CrossRef]
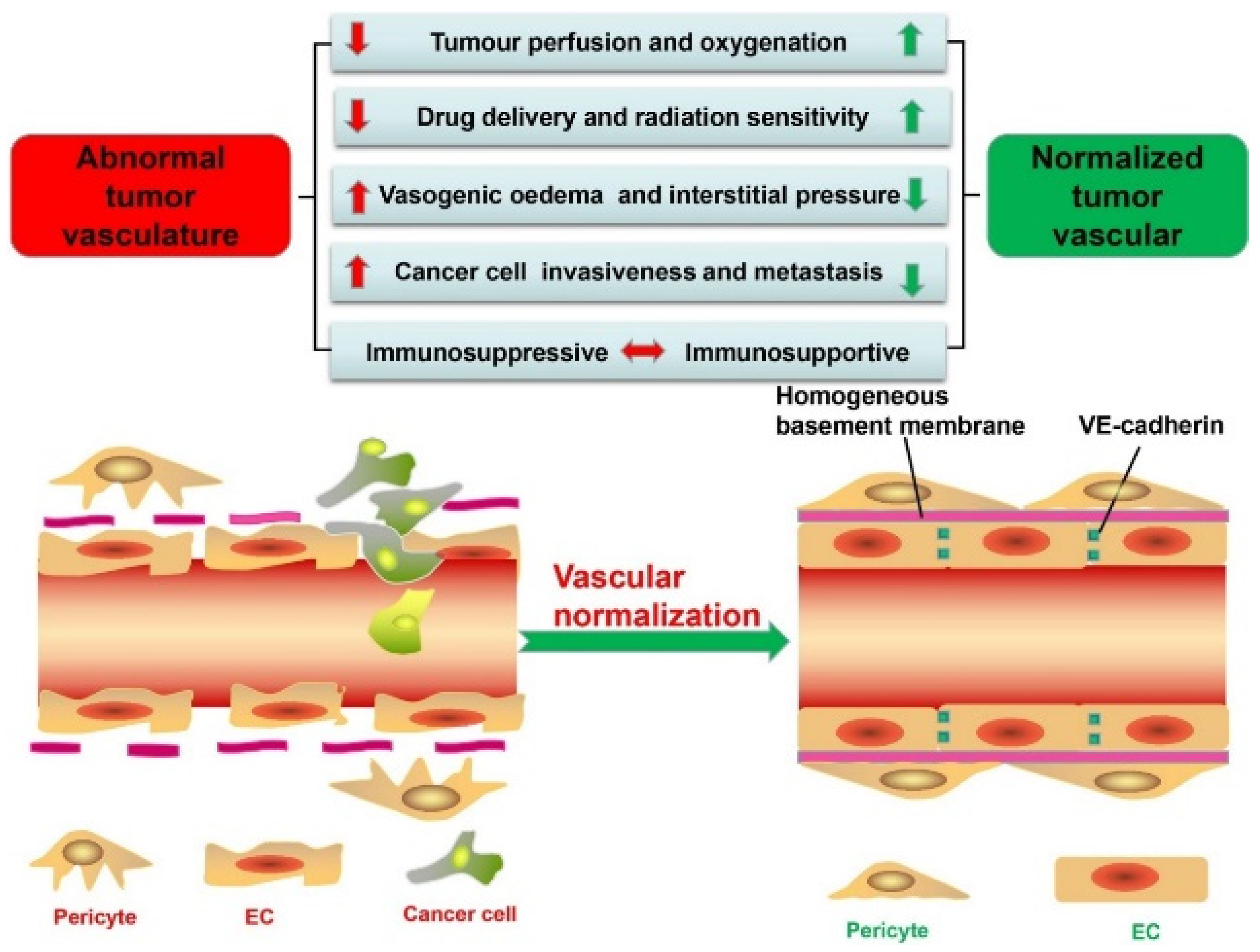
- Ha, H.; Choi, Y.; Kim, N.-H.; Kim, J.; Jang, J.; Niepa, T.H.; Tanaka, M.; Lee, H.-Y.; Choi, J. Lipid Nanoparticle Delivery System for Normalization of Tumor Microenvironment and Tumor Vascular Structure. Biomater. Res. 2025, 29, 0144. [Google Scholar] [CrossRef]
- Ahmad, A.; Khan, J.M. pH-sensitive endosomolytic peptides in gene and drug delivery: Endosomal escape and current challenges. J. Drug Deliv. Sci. Technol. 2022, 76, 103786. [Google Scholar] [CrossRef]
- Klipp, A.; Burger, M.; Leroux, J.-C. Get out or die trying: Peptide-and protein-based endosomal escape of RNA therapeutics. Adv. Drug Deliv. Rev. 2023, 200, 115047. [Google Scholar] [CrossRef] [PubMed]
- Teo, S.L.; Rennick, J.J.; Yuen, D.; Al-Wassiti, H.; Johnston, A.P.; Pouton, C.W. Unravelling cytosolic delivery of cell penetrating peptides with a quantitative endosomal escape assay. Nat. Commun. 2021, 12, 3721. [Google Scholar] [CrossRef] [PubMed]
- Nayanathara, U.; Yang, F.; Zhang, C.; Wang, Y.; Herling, B.R.; Smith, S.A.; Beach, M.A.; Johnston, A.P.; Such, G.K. Unravelling the endosomal escape of pH-responsive nanoparticles using the split luciferase endosomal escape quantification assay. Biomater. Sci. 2025, 13, 1335–1346. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Ukwattage, V.; Xiong, Y.; Such, G.K. Advancing endosomal escape of polymeric nanoparticles: Towards improved intracellular delivery. Mater. Horiz. 2025, 12, 3622–3632. [Google Scholar] [CrossRef]
- Yim, E.Y.; Zhou, A.C.; Yim, Y.C.; Wang, X.; Xia, T. Antigen-specific mRNA lipid nanoparticle platforms for the prevention and treatment of allergy and autoimmune diseases. BMEMat 2024, 2, e12060. [Google Scholar] [CrossRef]
- Kishimoto, T.K.; Fournier, M.; Michaud, A.; Rizzo, G.; Roy, C.; Capela, T.; Nukolova, N.; Li, N.; Doyle, L.; Fu, F.-n. Rapamycin nanoparticles increase the therapeutic window of engineered interleukin-2 and drive expansion of antigen-specific regulatory T cells for protection against autoimmune disease. J. Autoimmun. 2023, 140, 103125. [Google Scholar] [CrossRef]
- Ramadan, E.; Ahmed, A.; Naguib, Y.W. Advances in mRNA LNP-based cancer vaccines: Mechanisms, formulation aspects, challenges, and future directions. J. Pers. Med. 2024, 14, 1092. [Google Scholar] [CrossRef]
- Jallow, M.B.; Huang, K.; Qiu, M. Versatility of LNPs across different administration routes for targeted RNA delivery. J. Mater. Chem. B 2025, 13, 7637–7652. [Google Scholar] [CrossRef]
- Ren, Y.; Lin, L.; Abdallah, M.; Zhu, X.; Liu, H.; Fabb, S.A.; Payne, T.J.; Pouton, C.W.; Johnston, A.P.; Trevaskis, N.L. Impact of ionizable lipid type on the pharmacokinetics and biodistribution of mRNA-lipid nanoparticles after intravenous and subcutaneous injection. J. Control. Release 2025, 384, 113945. [Google Scholar] [CrossRef]
- Münter, R.; Christensen, E.; Andresen, T.L.; Larsen, J.B. Studying how administration route and dose regulates antibody generation against LNPs for mRNA delivery with single-particle resolution. Mol. Ther. Methods Clin. Dev. 2023, 29, 450–459. [Google Scholar] [CrossRef]
- Tayyar, Y.; Idris, A.; Yassine, H. Intranasal and Pulmonary Lipid Nanoparticles for Gene Delivery: Turning Challenges into Opportunities. Int. J. Nanomed. 2025, 20, 8085–8099. [Google Scholar] [CrossRef]
- Lee, Y.; Jeong, M.; Park, J.; Jung, H.; Lee, H. Immunogenicity of lipid nanoparticles and its impact on the efficacy of mRNA vaccines and therapeutics. Exp. Mol. Med. 2023, 55, 2085–2096. [Google Scholar] [CrossRef] [PubMed]
- Witten, J.; Hu, Y.; Langer, R.; Anderson, D.G. Recent advances in nanoparticulate RNA delivery systems. Proc. Natl. Acad. Sci. USA 2024, 121, e2307798120. [Google Scholar] [CrossRef] [PubMed]
- Prakash, G.; Shokr, A.; Willemen, N.; Bashir, S.M.; Shin, S.R.; Hassan, S. Microfluidic fabrication of lipid nanoparticles for the delivery of nucleic acids. Adv. Drug Deliv. Rev. 2022, 184, 114197. [Google Scholar] [CrossRef]
- Petersen, D.M.S.; Chaudhary, N.; Arral, M.L.; Weiss, R.M.; Whitehead, K.A. The mixing method used to formulate lipid nanoparticles affects mRNA delivery efficacy and organ tropism. Eur. J. Pharm. Biopharm. 2023, 192, 126–135. [Google Scholar] [CrossRef]
- O’Brien Laramy, M.N.; Costa, A.P.; Cebrero, Y.M.; Joseph, J.; Sarode, A.; Zang, N.; Kim, L.J.; Hofmann, K.; Wang, S.; Goyon, A. Process robustness in lipid nanoparticle production: A comparison of microfluidic and turbulent jet mixing. Mol. Pharm. 2023, 20, 4285–4296. [Google Scholar] [CrossRef] [PubMed]
- Ripoll, M.; Martin, E.; Enot, M.; Robbe, O.; Rapisarda, C.; Nicolai, M.-C.; Deliot, A.; Tabeling, P.; Authelin, J.-R.; Nakach, M. Optimal self-assembly of lipid nanoparticles (LNP) in a ring micromixer. Sci. Rep. 2022, 12, 9483. [Google Scholar] [CrossRef]
- Shin, S.; Devos, C.; Udepurkar, A.P.; Inguva, P.K.; Myerson, A.S.; Braatz, R.D. Mechanistic Modeling of Lipid Nanoparticle (LNP) Precipitation via Population Balance Equations (PBEs). arXiv 2025, arXiv:2504.10533. [Google Scholar] [CrossRef]
- Young, R.E.; Hofbauer, S.I.; Riley, R.S. Overcoming the challenge of long-term storage of mRNA-lipid nanoparticle vaccines. Mol. Ther. 2022, 30, 1792–1793. [Google Scholar] [CrossRef] [PubMed]
- Muramatsu, H.; Lam, K.; Bajusz, C.; Laczkó, D.; Karikó, K.; Schreiner, P.; Martin, A.; Lutwyche, P.; Heyes, J.; Pardi, N. Lyophilization provides long-term stability for a lipid nanoparticle-formulated, nucleoside-modified mRNA vaccine. Mol. Ther. 2022, 30, 1941–1951. [Google Scholar] [CrossRef]
- Li, M.; Jia, L.; Xie, Y.; Ma, W.; Yan, Z.; Liu, F.; Deng, J.; Zhu, A.; Siwei, X.; Su, W. Lyophilization process optimization and molecular dynamics simulation of mRNA-LNPs for SARS-CoV-2 vaccine. npj Vaccines 2023, 8, 153. [Google Scholar] [CrossRef]
- Fan, Y.; Rigas, D.; Kim, L.J.; Chang, F.-P.; Zang, N.; McKee, K.; Kemball, C.C.; Yu, Z.; Winkler, P.; Su, W.-C. Physicochemical and structural insights into lyophilized mRNA-LNP from lyoprotectant and buffer screenings. J. Control. Release 2024, 373, 727–737. [Google Scholar] [CrossRef]
- Ruppl, A.; Kiesewetter, D.; Koell-Weber, M.; Lemazurier, T.; Süss, R.; Allmendinger, A. Formulation screening of lyophilized mRNA-lipid nanoparticles. Int. J. Pharm. 2025, 671, 125272. [Google Scholar] [CrossRef]
- Arte, K.S.; Chen, M.; Patil, C.D.; Huang, Y.; Qu, L.; Zhou, Q. Recent advances in drying and development of solid formulations for stable mRNA and siRNA lipid nanoparticles. J. Pharm. Sci. 2025, 114, 805–815. [Google Scholar] [CrossRef]
- Hidayat, A.F.; Wardhana, Y.W.; Suwendar, S.; Mohammed, A.F.A.; Mahmoud, S.A.; Elamin, K.M.; Wathoni, N. A Review on QbD-Driven Optimization of Lipid Nanoparticles for Oral Drug Delivery: From Framework to Formulation. Int. J. Nanomed. 2025, 20, 8611–8651. [Google Scholar] [CrossRef] [PubMed]
- O’Brien Laramy, M.; Foley, D.A.; Pak, R.H.; Lewis, J.A.; McKinney, E.; Egan, P.M.; Yerabolu, R.; Dane, E.; Dirat, O.; Saunders Gorka, L. Chemistry, manufacturing and controls strategies for using novel excipients in lipid nanoparticles. Nat. Nanotechnol. 2025, 20, 331–344. [Google Scholar] [CrossRef]
- Gajbhiye, K.R.; Salve, R.; Narwade, M.; Sheikh, A.; Kesharwani, P.; Gajbhiye, V. Lipid polymer hybrid nanoparticles: A custom-tailored next-generation approach for cancer therapeutics. Mol. Cancer 2023, 22, 160. [Google Scholar] [CrossRef] [PubMed]
- Hassan, A.A.; Ramadan, E.; Kristó, K.; Regdon, G., Jr.; Sovány, T. Lipid-Polymer Hybrid Nanoparticles as a Smart Drug Delivery System for Peptide/Protein Delivery. Pharmaceutics 2025, 17, 797. [Google Scholar] [CrossRef]
- Yadava, S.K.; Reddy, B.P.K.; Prausnitz, M.R.; Cicerone, M.T. Hybrid Lipid Nanocapsules: A Robust Platform for mRNA Delivery. ACS Appl. Mater. Interfaces 2024, 16, 15981–15992. [Google Scholar] [CrossRef]
- Manturthi, S.; El-Sahli, S.; Bo, Y.; Durocher, E.; Kirkby, M.; Popatia, A.; Mediratta, K.; Daniel, R.; Lee, S.-H.; Iqbal, U. Nanoparticles Codelivering mRNA and SiRNA for Simultaneous Restoration and Silencing of Gene/Protein Expression In Vitro and In Vivo. ACS Nanosci. Au 2024, 4, 416–425. [Google Scholar] [CrossRef]
- Maeki, M.; Uno, S.; Sugiura, K.; Sato, Y.; Fujioka, Y.; Ishida, A.; Ohba, Y.; Harashima, H.; Tokeshi, M. Development of polymer–lipid hybrid nanoparticles for large-sized plasmid DNA transfection. ACS Appl. Mater. Interfaces 2023, 16, 2110–2119. [Google Scholar] [CrossRef] [PubMed]
- Manda, A.; Alhazza, A.; Uludağ, H.; Montazeri Aliabadi, H. Engineering Lipid–Polymer Nanoparticles for siRNA Delivery to Cancer Cells. Pharmaceuticals 2025, 18, 864. [Google Scholar] [CrossRef]
- Yadav, M.; Sharma, A.; Patne, K.; Tabasum, S.; Suryavanshi, J.; Rawat, L.; Machaalani, M.; Eid, M.; Singh, R.P.; Choueiri, T.K. AXL signaling in cancer: From molecular insights to targeted therapies. Signal Transduct. Target. Ther. 2025, 10, 37. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Xu, L.; Dou, Y.; He, Y. AXL: Shapers of tumor progression and immunosuppressive microenvironments. Mol. Cancer 2025, 24, 11. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Cheng, X.; Shao, P.; Dong, Y.; Wu, Y.; Xiao, L.; Cui, Z.; Sun, X.; Gao, C.; Chen, J. Bacterial protoplast-derived nanovesicles carrying CRISPR-Cas9 tools re-educate tumor-associated macrophages for enhanced cancer immunotherapy. Nat. Commun. 2024, 15, 950. [Google Scholar] [CrossRef]
- Sun, L.; Liu, H.; Ye, Y.; Lei, Y.; Islam, R.; Tan, S.; Tong, R.; Miao, Y.-B.; Cai, L. Smart nanoparticles for cancer therapy. Signal Transduct. Target. Ther. 2023, 8, 418. [Google Scholar] [CrossRef]
- Teng, L.; Bi, Y.; Xing, X.; Yao, G. Nano-oncology revisited: Insights on precise therapeutic advances and challenges in tumor. Fundam. Res. 2025. [Google Scholar] [CrossRef]
- Navarro-Marchal, S.A.; Martín-Contreras, M.; Castro-Santiago, D.; del Castillo-Santaella, T.; Graván, P.; Jódar-Reyes, A.B.; Marchal, J.A.; Peula-García, J.M. Effect of the Protein Corona Formation on Antibody Functionalized Liquid Lipid Nanocarriers. Int. J. Mol. Sci. 2023, 24, 16759. [Google Scholar] [CrossRef] [PubMed]
- Lu, Q.; Kou, D.; Lou, S.; Ashrafizadeh, M.; Aref, A.R.; Canadas, I.; Tian, Y.; Niu, X.; Wang, Y.; Torabian, P. Nanoparticles in tumor microenvironment remodeling and cancer immunotherapy. J. Hematol. Oncol. 2024, 17, 16. [Google Scholar] [CrossRef] [PubMed]
- Thi, T.T.H.; Suys, E.J.; Lee, J.S.; Nguyen, D.H.; Park, K.D.; Truong, N.P. Lipid-based nanoparticles in the clinic and clinical trials: From cancer nanomedicine to COVID-19 vaccines. Vaccines 2021, 9, 359. [Google Scholar] [CrossRef] [PubMed]
- Akinc, A.; Maier, M.A.; Manoharan, M.; Fitzgerald, K.; Jayaraman, M.; Barros, S.; Ansell, S.; Du, X.; Hope, M.J.; Madden, T.D. The Onpattro story and the clinical translation of nanomedicines containing nucleic acid-based drugs. Nat. Nanotechnol. 2019, 14, 1084–1087. [Google Scholar] [CrossRef]
- Albertsen, C.H.; Kulkarni, J.A.; Witzigmann, D.; Lind, M.; Petersson, K.; Simonsen, J.B. The role of lipid components in lipid nanoparticles for vaccines and gene therapy. Adv. Drug Deliv. Rev. 2022, 188, 114416. [Google Scholar] [CrossRef]
- Xu, Y.; Golubovic, A.; Xu, S.; Pan, A.; Li, B. Rational design and combinatorial chemistry of ionizable lipids for RNA delivery. J. Mater. Chem. B 2023, 11, 6527–6539. [Google Scholar] [CrossRef]
- An, K.; Kurek, D.; Kulkarni, J.; Witzigmann, D. Lipid Nanoparticle Formulations with High Sterol Content for Nucleic Acid Delivery. U.S. Patent 11,951,177 B2, 9 April 2022. [Google Scholar]
- Berger, M.; Schaeffner, P. Multipurpose Lipid Nanoparticles for Delivery of Diverse Nucleic Acids Including CRISPR Cargo. U.S. Patent 2023/0097090 A1, 30 March 2023. [Google Scholar]
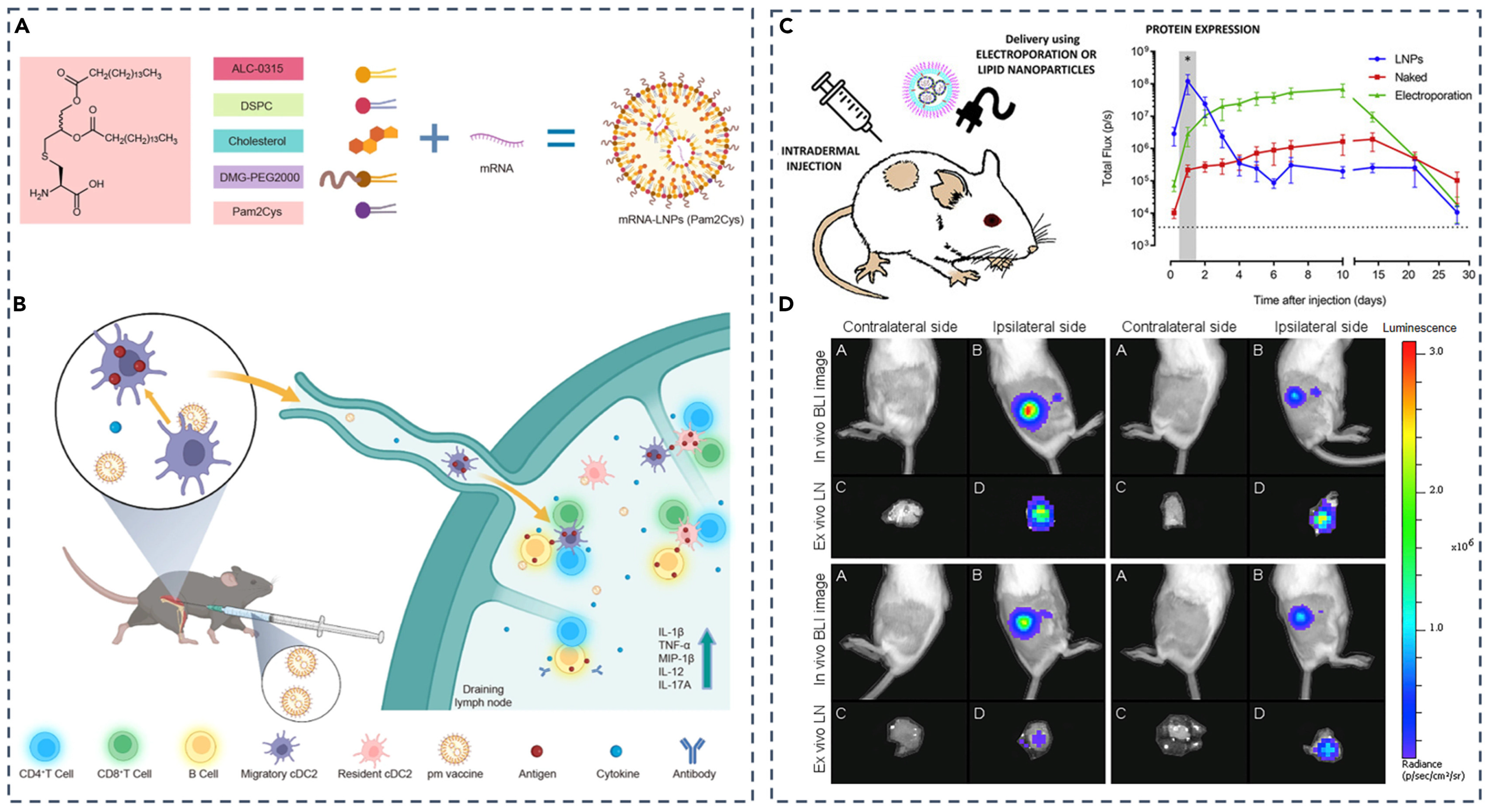
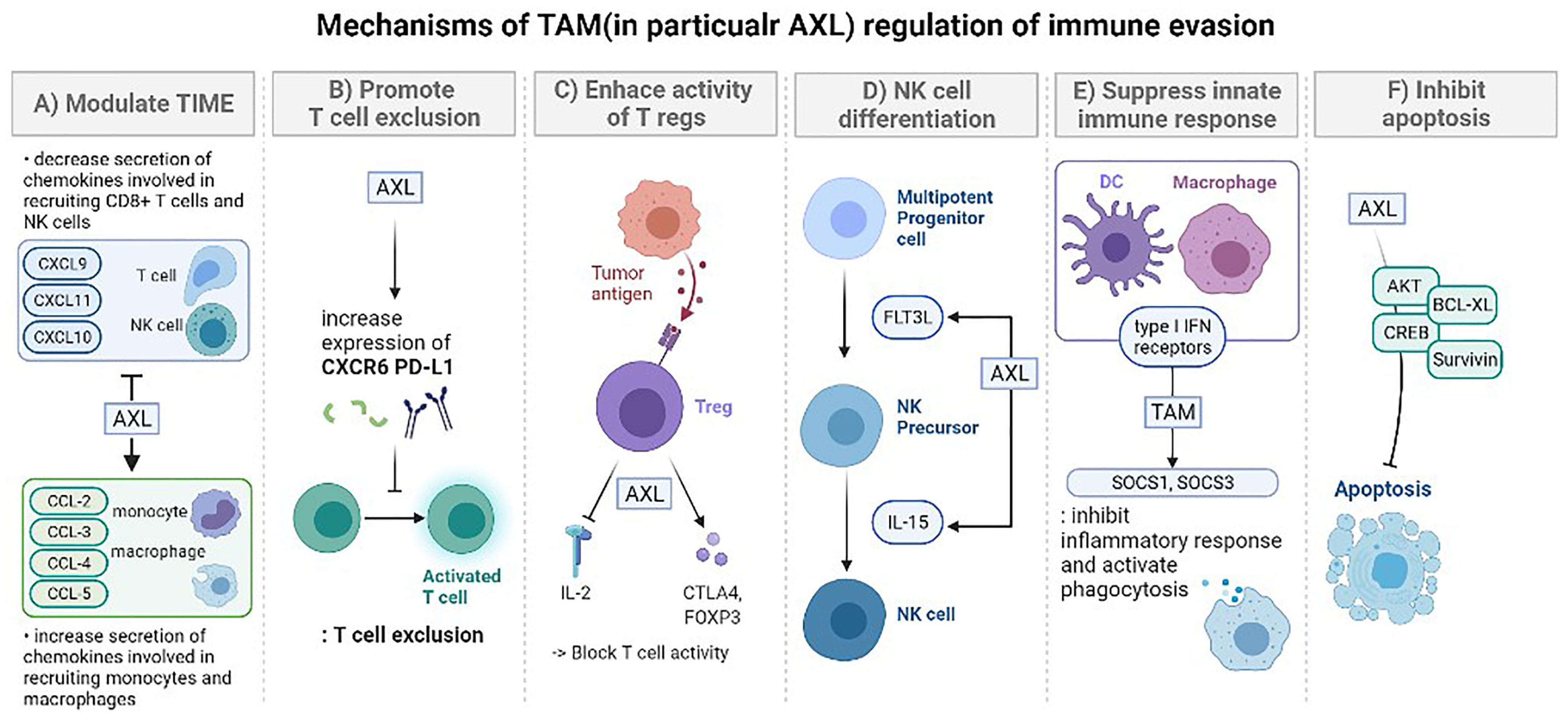
| Barrier | Key Challenge | Representative Strategies | Advantages | Limitations/Trade-offs |
|---|---|---|---|---|
| Circulation | Rapid clearance, opsonization | PEGylation, zwitterionic coatings | Prolonged circulation, stealth effect | Anti-PEG immunity, reduced uptake efficiency |
| Protein corona | Ligand masking, altered biodistribution | Pre-coating, corona engineering | Potential for controlled tropism | Variability across species/patient |
| Vasclar/ECM penetration | High interstitial pressure, dense matrix | Enzyme co-delivery, deformable/neutral particles | Improved tissue penetration | Risk of off-target toxicity, tissue remodeling |
| Cellular uptake | Heterogeneous endocytosis routes | Ligand conjugation, receptor targeting | Higher uptake in specific cells | Corona masking, receptor downregulation |
| Endosomal escape | <2% efficiency into cytoplasm | Ionizable lipids, fusion peptides, photosensitizers | Enhanced cytosolic release | Safety issues, poor scalability, limited reproducibility |
| Cytoplasmic stability | Nuclease degradation, innate immunity | Nucleoside modification, shielding polymers | Extended RNA stability | May alter translation efficiency |
| Nuclear access (DNA) | Nuclear envelope barrier | Nuclear localization signals, mitosis coupling | Enables plasmid/CRISPR delivery | Low efficiency, cell cycle dependence |
| Component | Function in LNP | Structural Role | Notes and Limitations |
|---|---|---|---|
| Ionizable lipids | Encapsulation and endosomal escape | Neutral at physiological pH, protonates in endosomes to disrupt membranes | Apparent pKa, tail architecture, and linker design require tuning for efficacy and safety |
| Helper phospholipids | Bilayer stability and fusion support | Maintain lamellar structure and reduce leakage | Highly ordered high-Tm species can impede fusion and slow release |
| Cholesterol | Membrane packing and fluidity modulation | Adjusts rigidity and can promote nonlamellar phases | Excess may stiffen membranes and alter organ tropism |
| PEG-lipids | Stealth and prolonged circulation | Steric shielding that reduces opsonization | High surface density or slow deshielding can hinder uptake and endosomal escape |
| Barrier | Key Challenges | Major Influencing Factors | Representative Strategies |
|---|---|---|---|
| Hemodynamic shear and dilution | Shear-induced instability, dilution upon injection, reduced vascular residence | Particle size, surface charge, PEG-lipid density | Optimize PEG density, use near-neutral surface charge, size tuning |
| Protein corona formation and opsonization | Non-specific protein binding, rapid MPS clearance, loss of targeting ability | Surface chemistry, PEGylation, lipid composition | PEGylation, zwitterionic/bioinspired coatings, controlled corona engineering |
| RES (MPS) clearance | Phagocytic uptake by liver/spleen macrophages, reduced delivery to target tissues | Size > 150 nm, cationic charge, protein corona | Size optimization (<150 nm), PEG stealth layer, decoy particles, immune-inert coatings |
| Vascular endothelial barriers | Limited extravasation into target tissues, especially in non- fenestrated organs | Tissue-specific endothelium, particle deformability, ligand use | EPR effect exploitation (in tumors), receptor-mediated transcytosis, active targeting ligands |
| Tissue Type | Endothelial Type | Fenestration/ Permeability | LNP Extravasation Potential | Remarks |
|---|---|---|---|---|
| Liver (sinusoid) | Fenestrated, discontinuous | High | High | Rapid uptake by Kupffer cells |
| Spleen | Fenestrated, slit-like pores | Moderate | Moderate | Filtration-based selectivity |
| Tumor | Leaky, disorganized vasculature | Very high (EPR effect) | High (but heterogeneous) | Interstitial pressure hinders deep delivery |
| Normal tissue | Continuous, tight junction | Low | Low | Minimal passive permeability |
| Strategy | Mechanism of Action | Advantages |
|---|---|---|
| Novel ionizable lipids | Protonation at acidic pH → membrane destabilization | Tunable chemistry, high efficiency |
| Fusion peptides | Viral-mimetic membrane fusion and pore formation | Potent endosomal release, biomimetic mechanism |
| Photosensitization | Light-activated ROSs disrupt endosomal membranes | Spatiotemporal control, efficient release |
| Endocytosis Pathway | Size & Structural Features | Characteristics of mRNA-LNP Internalization |
|---|---|---|
| Clathrin-mediated (Receptor-mediated) | ~100 nm, clathrin-coated pits | Receptor-specific uptake; rapid and efficient internalization |
| Caveolae-mediated | 50–80 nm, caveolin-enriched flask-shaped invaginations | Cholesterol-rich; involved in certain cell-specific uptake |
| Macropinocytosis | Several hundred nm to micrometers | Non-selective bulk uptake; important in large-volume internalization |
| Aspect | In Vitro (Static Cell Culture) | In Vivo Environment |
|---|---|---|
| Fluid dynamics | Static media, no flow | Shear stress, blood/lymph flow, dynamic perfusion |
| Extracellular matrix | Minimal or absent | Dense, heterogeneous ECM impeding penetration |
| Cellular composition | Single or limited cell types | Multiple cell types incl. stromal, endothelial, immune |
| Protein corona | Often absent or serum- supplement artifact | Dynamic, biofluid-specific coronas altering uptake |
| Readouts | Uptake %, colocalization, reporter expression | Biodistribution, pharmacokinetics, immune clearance |
| Administration Route | Primary Tissue Distribution | mRNA Expression & Immune Activation Pattern |
|---|---|---|
| IM (Intramuscular) | Muscle, lymph nodes, liver, spleen | Induces systemic immune responses; strong CD8+ T-cell activation |
| IV (Intravenous) | Primarily liver, spleen (RES) | Dominant hepatic uptake; limited immune stimulation |
| SC (Subcutaneous) | Draining lymph nodes | Specialized for local lymph node immune activation |
| Product Name | Company/Developer | Nucleic Acid Cargo | Indication | Approval Year |
|---|---|---|---|---|
| Onpattro® (patisiran) | Alnylam Pharmaceuticals | siRNA | Hereditary transthyretin-mediated amyloidosis (hATTR) | 2018 |
| Comirnaty® (BNT162b2) | Pfizer/BioNTech | mRNA | COVID-19 vaccine | 2020 |
| Spikevax® (mRNA-1273) | Moderna Therapeutics | mRNA | COVID-19 vaccine | 2020 |
| Irdelisiran (ALN-TTRsc02, in late-stage trials) | Alnylam Pharmaceuticals | siRNA | Transthyretin amyloidosis | Ongoing (Phase III) |
| Patent Number | Jurisdiction | Year | Key Focus | REF |
|---|---|---|---|---|
| US 11951177 B2 | USA | 2024 | High-sterol LNP formulations for nucleic acid delivery | [168] |
| US 20230097090 A1 | USA | 2023 | Multipurpose LNPs for diverse nucleic acids, including CRISPR cargo | [169] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, Y.; Park, J.; Choi, J.; Kim, M.; Seo, G.; Kim, J.; Park, J.-A.; Lim, K.S.; Ha, S.-J.; Kim, H.-O. Physiological Barriers to Nucleic Acid Therapeutics and Engineering Strategies for Lipid Nanoparticle Design, Optimization, and Clinical Translation. Pharmaceutics 2025, 17, 1309. https://doi.org/10.3390/pharmaceutics17101309
Kim Y, Park J, Choi J, Kim M, Seo G, Kim J, Park J-A, Lim KS, Ha S-J, Kim H-O. Physiological Barriers to Nucleic Acid Therapeutics and Engineering Strategies for Lipid Nanoparticle Design, Optimization, and Clinical Translation. Pharmaceutics. 2025; 17(10):1309. https://doi.org/10.3390/pharmaceutics17101309
Chicago/Turabian StyleKim, Yerim, Jisu Park, Jaewon Choi, Minse Kim, Gyeongsu Seo, Jeongeun Kim, Jeong-Ann Park, Kwang Suk Lim, Suk-Jin Ha, and Hyun-Ouk Kim. 2025. "Physiological Barriers to Nucleic Acid Therapeutics and Engineering Strategies for Lipid Nanoparticle Design, Optimization, and Clinical Translation" Pharmaceutics 17, no. 10: 1309. https://doi.org/10.3390/pharmaceutics17101309
APA StyleKim, Y., Park, J., Choi, J., Kim, M., Seo, G., Kim, J., Park, J.-A., Lim, K. S., Ha, S.-J., & Kim, H.-O. (2025). Physiological Barriers to Nucleic Acid Therapeutics and Engineering Strategies for Lipid Nanoparticle Design, Optimization, and Clinical Translation. Pharmaceutics, 17(10), 1309. https://doi.org/10.3390/pharmaceutics17101309









