Anti-Cariogenic Effects of S. cerevisiae and S. boulardii in S. mutans–C. albicans Cross-Kingdom In Vitro Models
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Starter Preparation
2.2. Planktonic Model
2.3. PCR and Real-Time Quantitative PCR (Real-Time qPCR)
2.4. Statistical Analysis
3. Results
3.1. Growth Profile of Saccharomyces Species
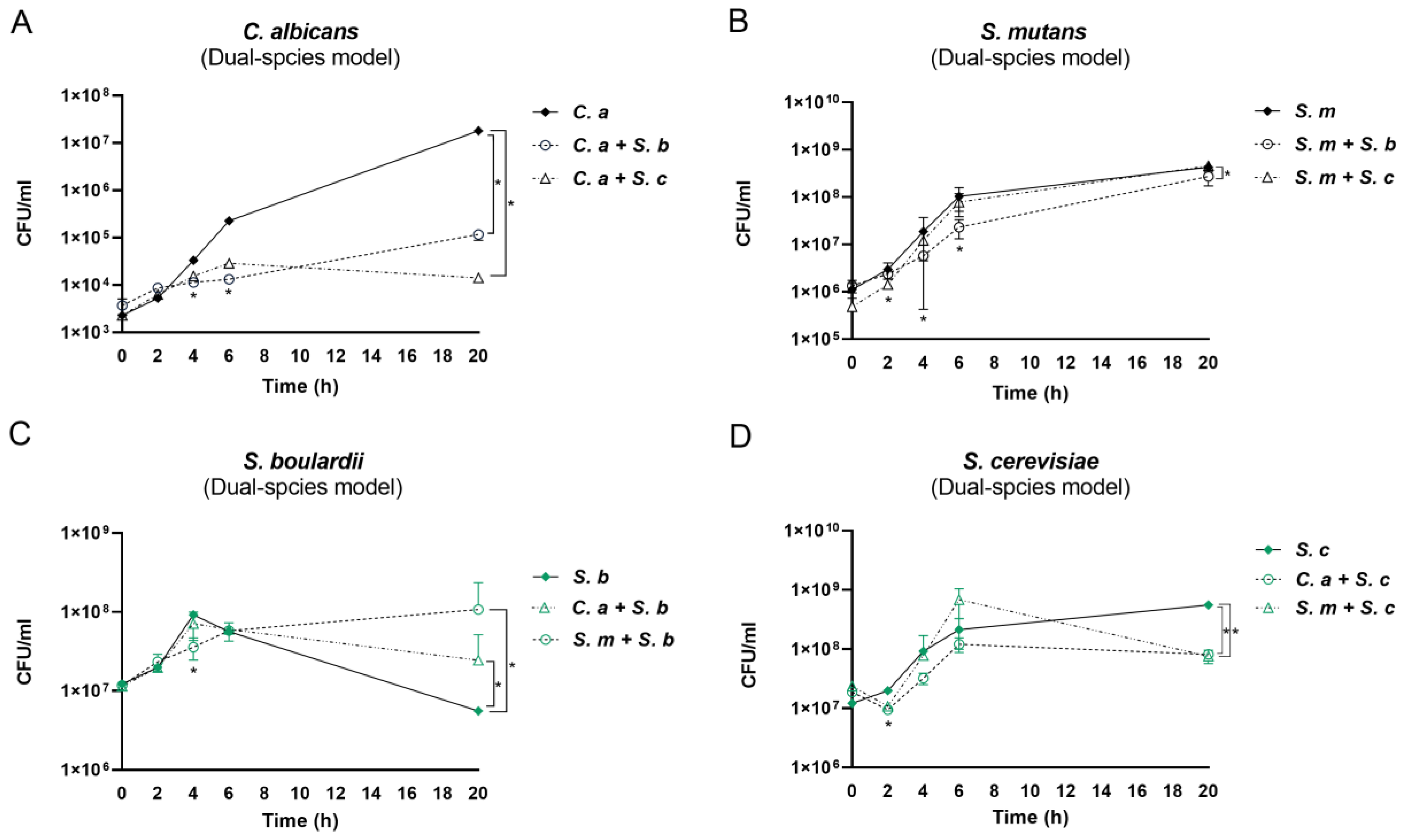
3.2. The Impact of Saccharomyces Species on C. albicans and S. mutans in the Dual-Species Conditions
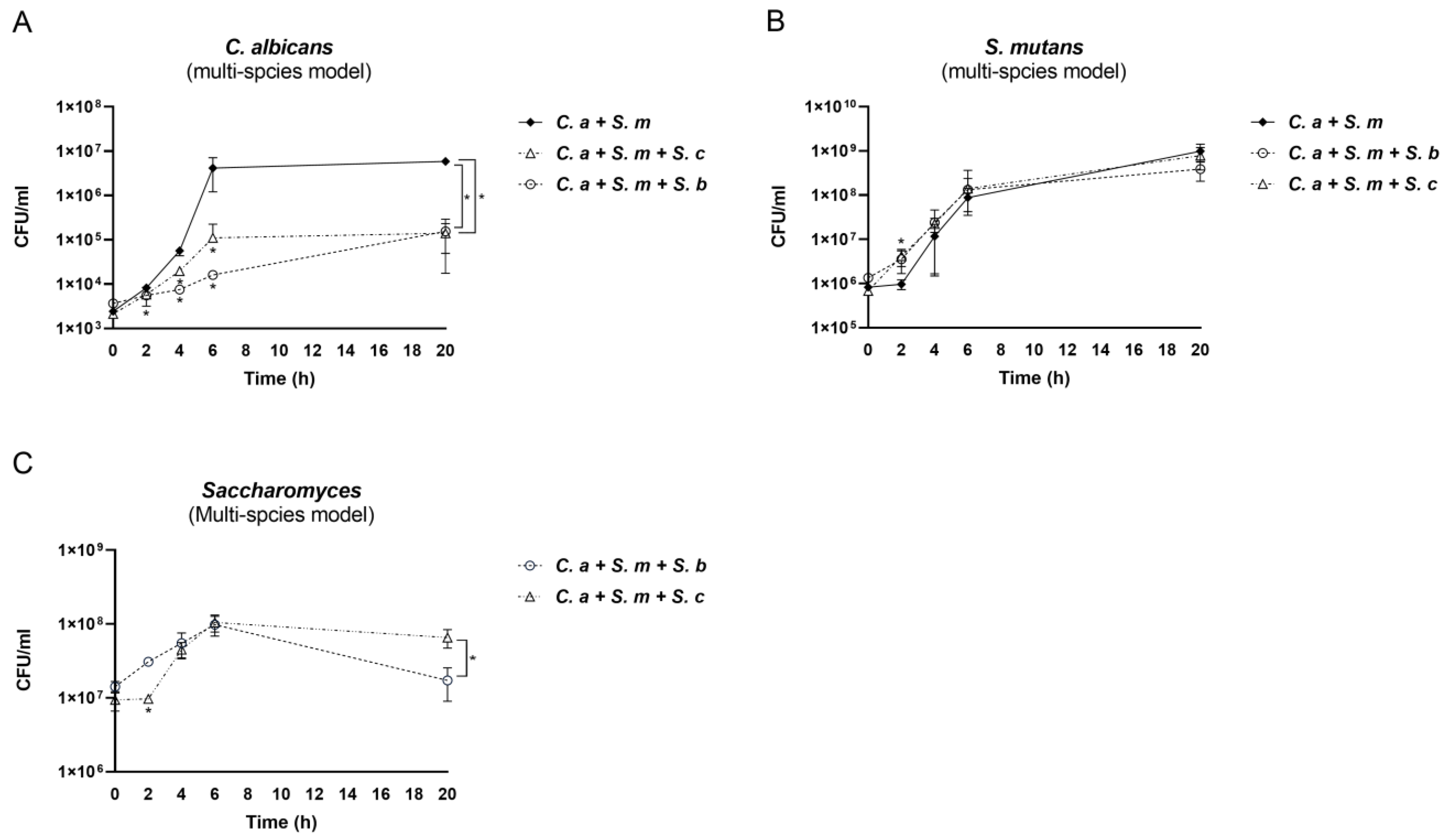
3.3. The Impact of Saccharomyces Species on C. albicans and S. mutans in the Multi-Species Conditions
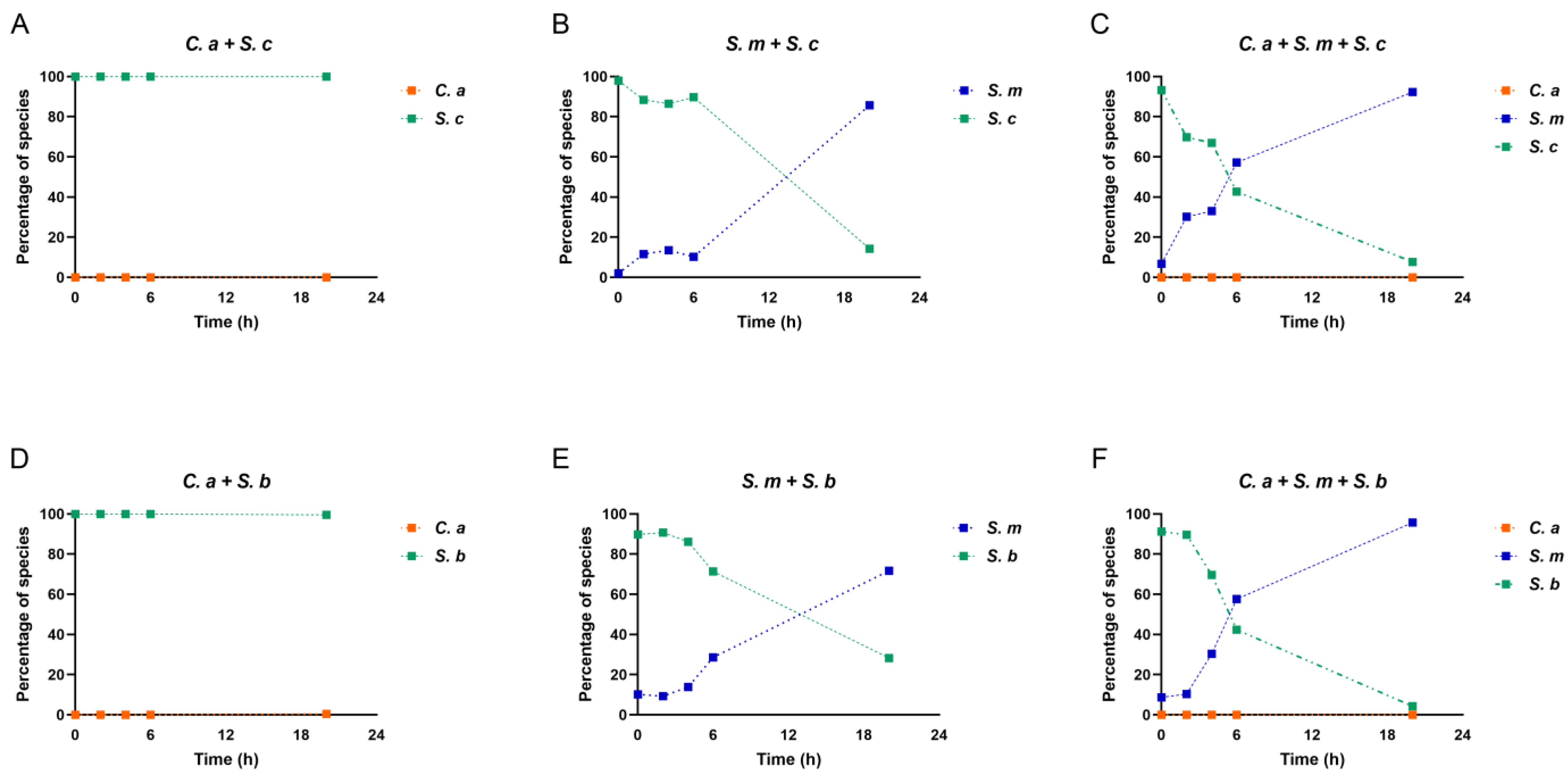
3.4. Compositional Changes in Saccharomyces Species, C. albicans, and S. mutans in the Multi-Species Model
3.5. Dynamic Changes in Culture pH in the Mono-, Dual-, and Multi-Species Conditions
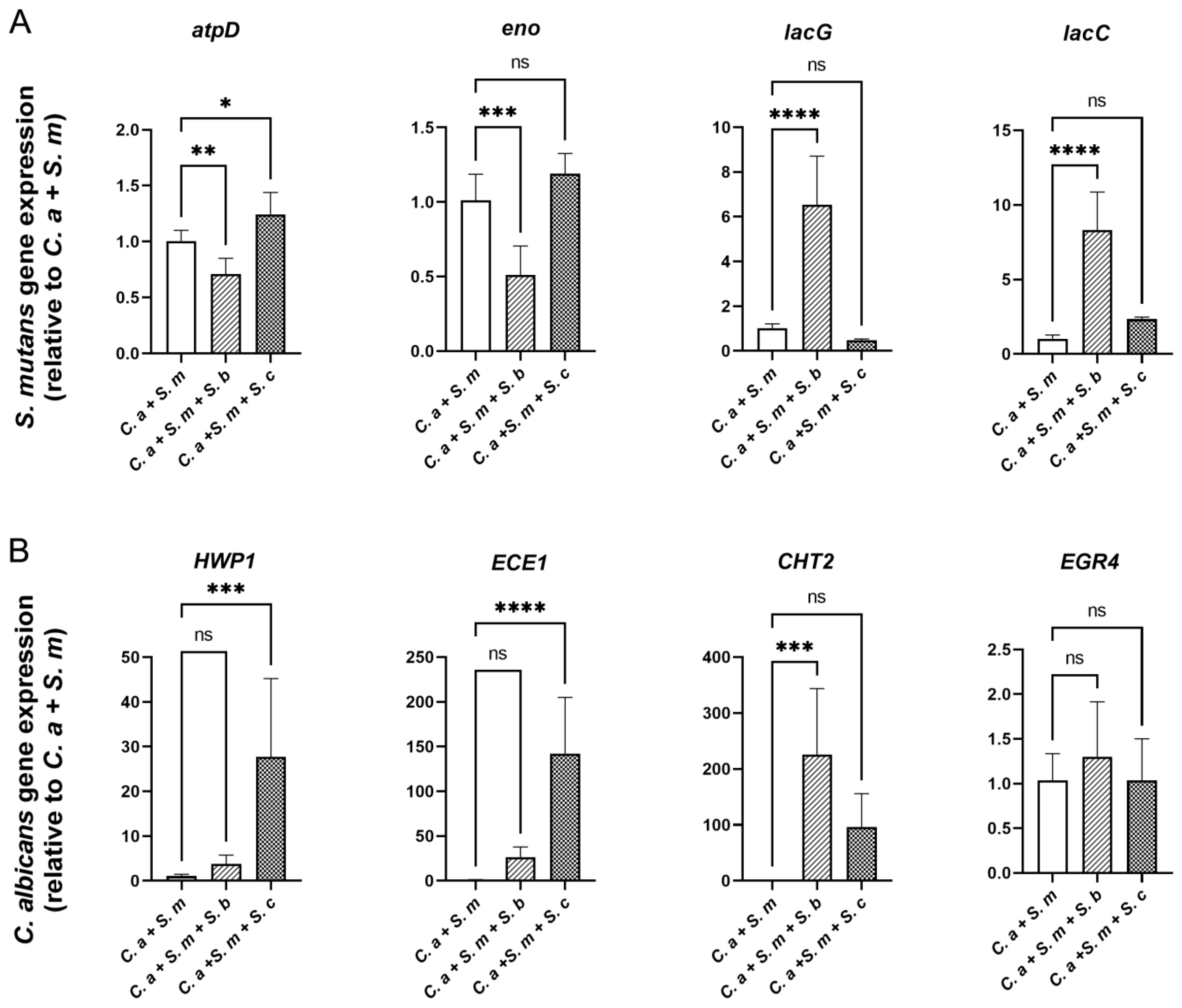
3.6. Regulation of S. mutans and C. albicans Virulence Genes by Saccharomyces Species
3.7. Inhibition of C. albicans Hyphae/Pseudohyphae Formation by Saccharomyces Species
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Meyer, F.; Enax, J. Early Childhood Caries: Epidemiology, Aetiology, and Prevention. Int. J. Dent. 2018, 2018, e1415873. [Google Scholar] [CrossRef]
- Alanzi, A.; Husain, F.; Husain, H.; Hanif, A.; Baskaradoss, J.K. Does the severity of untreated dental caries of preschool children influence the oral health-related quality of life? BMC Oral. Health 2023, 23, 552. [Google Scholar] [CrossRef]
- Anil, S.; Anand, P.S. Early Childhood Caries: Prevalence, Risk Factors, and Prevention. Front. Pediatr. 2017, 5, 157. [Google Scholar] [CrossRef]
- Zeng, Y.; Nikikova, A.; Abdelsalam, H.; Li, J.; Xiao, J. Activity of Quercetin and Kaemferol against Streptococcus mutans Biofilm. Arch. Oral. Biol. 2019, 98, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Ellepola, K.; Truong, T.; Liu, Y.; Lin, Q.; Lim, T.K.; Lee, Y.M.; Cao, T.; Koo, H.; Seneviratne, C.J. Multi-omics Analyses Reveal Synergistic Carbohydrate Metabolism in Streptococcus mutans-Candida albicans Mixed-Species Biofilms. Infect. Immun. 2019, 87, e00339-19. [Google Scholar] [CrossRef] [PubMed]
- Falsetta, M.L.; Klein, M.I.; Colonne, P.M.; Scott-Anne, K.; Gregoire, S.; Pai, C.-H.; Gonzalez-Begne, M.; Watson, G.; Krysan, D.J.; Bowen, W.H.; et al. Symbiotic relationship between Streptococcus mutans and Candida albicans synergizes virulence of plaque biofilms in vivo. Infect. Immun. 2014, 82, 1968–1981. [Google Scholar] [CrossRef] [PubMed]
- Hwang, G.; Liu, Y.; Kim, D.; Li, Y.; Krysan, D.J.; Koo, H. Candida albicans mannans mediate Streptococcus mutans exoenzyme GtfB binding to modulate cross-kingdom biofilm development in vivo. PLoS Pathog. 2017, 13, e1006407. [Google Scholar] [CrossRef] [PubMed]
- Alkhars, N.; Zeng, Y.; Alomeir, N.; Al Jallad, N.; Wu, T.T.; Aboelmagd, S.; Youssef, M.; Jang, H.; Fogarty, C.; Xiao, J. Oral Candida Predicts Streptococcus mutans Emergence in Underserved US Infants. J. Dent. Res. 2022, 101, 54–62. [Google Scholar] [CrossRef]
- Raja, M.; Hannan, A.; Ali, K. Association of oral candidal carriage with dental caries in children. Caries Res. 2010, 44, 272–276. [Google Scholar] [CrossRef] [PubMed]
- Signoretto, C.; Burlacchini, G.; Faccioni, F.; Zanderigo, M.; Bozzola, N.; Canepari, P. Support for the role of Candida spp. in extensive caries lesions of children. New Microbiol. 2009, 32, 101–107. [Google Scholar]
- Colak, H.; Dulgergil, C.T.; Dalli, M.; Hamidi, M.M. Early childhood caries update: A review of causes, diagnoses, and treatments. J. Nat. Sci. Biol. Med. 2013, 4, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Tinanoff, N.; Baez, R.J.; Diaz Guillory, C.; Donly, K.J.; Feldens, C.A.; McGrath, C.; Phantumvanit, P.; Pitts, N.B.; Seow, W.K.; Sharkov, N.; et al. Early childhood caries epidemiology, aetiology, risk assessment, societal burden, management, education, and policy: Global perspective. Int. J. Paediatr. Dent. 2019, 29, 238–248. [Google Scholar] [CrossRef] [PubMed]
- Razeghi, S.; Amiri, P.; Mohebbi, S.Z.; Kharazifard, M.J. Impact of Health Promotion Interventions on Early Childhood Caries Prevention in Children Aged 2-5 Years Receiving Dental Treatment Under General Anesthesia. Front. Public. Health 2020, 8, 6. [Google Scholar] [CrossRef] [PubMed]
- Alkhars, N.; Gaca, A.; Zeng, Y.; Al-Jallad, N.; Rustchenko, E.; Wu, T.T.; Eliav, E.; Xiao, J. Antifungal Susceptibility of Oral Candida Isolates from Mother-Infant Dyads to Nystatin, Fluconazole, and Caspofungin. J. Fungi 2023, 9, 580. [Google Scholar] [CrossRef] [PubMed]
- Badri, P.; Saltaji, H.; Flores-Mir, C.; Amin, M. Factors affecting children’s adherence to regular dental attendance: A systematic review. J. Am. Dent. Assoc. 2014, 145, 817–828. [Google Scholar] [CrossRef] [PubMed]
- Sharif, M.O.; Newton, T.; Cunningham, S.J. A systematic review to assess interventions delivered by mobile phones in improving adherence to oral hygiene advice for children and adolescents. Br. Dent. J. 2019, 227, 375–382. [Google Scholar] [CrossRef] [PubMed]
- Gendreau, L.; Loewy, Z.G. Epidemiology and etiology of denture stomatitis. J. Prosthodont. Implant. Esthet. Reconstr. Dent. 2011, 20, 251–260. [Google Scholar] [CrossRef] [PubMed]
- Mousa, M.; Lynch, E.; Kielbassa, A. Denture-related stomatitis in new complete denture wearers and its association with Candida species colonization: A prospective case-series. Quintessence Int. 2020, 51, 554–565. [Google Scholar] [CrossRef]
- Baena-Monroy, T.; Moreno-Maldonado, V.; Franco-Martínez, F.; Aldape-Barrios, B.; Quindós, G.; Sánchez-Vargas, L.O. Candida albicans, Staphylococcus aureus and Streptococcus mutans colonization in patients wearing dental prosthesis. Med. Oral Patol. Oral Cir. Bucal 2005, 10, E27–E39. [Google Scholar]
- Anilkumar, K.; Monisha, A.L.S. Role of friendly bacteria in oral health—A short review. Oral Health Prev. Dent. 2012, 10, 3–8. [Google Scholar]
- Bao, J.; Huang, X.; Zeng, Y.; Wu, T.; Lu, X.; Meng, G.; Ren, Y.; Xiao, J. Dose-Dependent Inhibitory Effect of Probiotic Lactobacillus plantarum on Streptococcus mutans-Candida albicans Cross-Kingdom Microorganisms. Pathogens 2023, 12, 848. [Google Scholar] [CrossRef]
- Zeng, Y.; Fadaak, A.; Alomeir, N.; Wu, Y.; Wu, T.T.; Qing, S.; Xiao, J. Effect of Probiotic Lactobacillus plantarum on Streptococcus mutans and Candida albicans Clinical Isolates from Children with Early Childhood Caries. Int. J. Mol. Sci. 2023, 24, 2991. [Google Scholar] [CrossRef] [PubMed]
- Edwards-Ingram, L.; Gitsham, P.; Burton, N.; Warhurst, G.; Clarke, I.; Hoyle, D.; Oliver, S.G.; Stateva, L. Genotypic and physiological characterization of Saccharomyces boulardii, the probiotic strain of Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2007, 73, 2458–2467. [Google Scholar] [CrossRef] [PubMed]
- Moré, M.I.; Swidsinski, A. Saccharomyces boulardii CNCM I-745 supports regeneration of the intestinal microbiota after diarrheic dysbiosis–a review. Clin. Exp. Gastroenterol. 2015, 8, 237–255. [Google Scholar] [CrossRef] [PubMed]
- Neut, C.; Mahieux, S.; Dubreuil, L.J. Antibiotic susceptibility of probiotic strains: Is it reasonable to combine probiotics with antibiotics? Med. Mal. Infect. 2017, 47, 477–483. [Google Scholar] [CrossRef] [PubMed]
- Micklefield, G. Saccharomyces boulardii in the treatment and prevention of antibiotic-associated diarrhea. MMW-Fortschritte Med. 2014, 156, 18–22. [Google Scholar] [CrossRef]
- Na, X.; Kelly, C. Probiotics in Clostridium difficile infection. J. Clin. Gastroenterol. 2011, 45, S154. [Google Scholar] [CrossRef] [PubMed]
- Tung, J.M.; Dolovich, L.R.; Lee, C.H. Prevention of Clostridium difficile infection with Saccharomyces boulardii: A systematic review. Can. J. Gastroenterol. Hepatol. 2009, 23, 817–821. [Google Scholar] [CrossRef]
- Palma, M.L.; Zamith-Miranda, D.; Martins, F.S.; Bozza, F.A.; Nimrichter, L.; Montero-Lomeli, M.; Marques, E.T.A.; Douradinha, B. Probiotic Saccharomyces cerevisiae strains as biotherapeutic tools: Is there room for improvement? Appl. Microbiol. Biotechnol. 2015, 99, 6563–6570. [Google Scholar] [CrossRef]
- van der Aa Kühle, A.; Skovgaard, K.; Jespersen, L. In vitro screening of probiotic properties of Saccharomyces cerevisiae var. boulardii and food-borne Saccharomyces cerevisiae strains. Int. J. Food Microbiol. 2005, 101, 29–39. [Google Scholar] [CrossRef]
- Zanello, G.; Meurens, F.; Berri, M.; Salmon, H. Saccharomyces boulardii effects on gastrointestinal diseases. Curr. Issues Mol. Biol. 2009, 11, 47–58. [Google Scholar] [PubMed]
- Chandra, R.V.; Swathi, T.; Reddy, A.A.; Chakravarthy, Y.; Nagarajan, S.; Naveen, A. Effect of a locally delivered probiotic-prebiotic mixture as an adjunct to scaling and root planing in the management of chronic periodontitis. J. Int. Acad. Periodontol. 2016, 18, 67–75. [Google Scholar] [PubMed]
- Ranjith, A.; Nazimudeen, N.B.; Baiju, K.V. Probiotic mouthwash as an adjunct to mechanical therapy in the treatment of stage II periodontitis: A randomized controlled clinical trial. Int. J. Dent. Hyg. 2022, 20, 415–421. [Google Scholar] [CrossRef]
- Deshmukh, M.A.; Dodamani, A.S.; Karibasappa, G.; Khairnar, M.R.; Naik, R.G.; Jadhav, H.C. Comparative evaluation of the efficacy of probiotic, herbal and chlorhexidine mouthwash on gingival health: A randomized clinical trial. J. Clin. Diagn. Res. JCDR 2017, 11, ZC13. [Google Scholar] [CrossRef] [PubMed]
- Parapouli, M.; Vasileiadis, A.; Afendra, A.-S.; Hatziloukas, E. Saccharomyces cerevisiae and its industrial applications. AIMS Microbiol. 2020, 6, 1–31. [Google Scholar] [CrossRef] [PubMed]
- Pineton de Chambrun, G.; Neut, C.; Chau, A.; Cazaubiel, M.; Pelerin, F.; Justen, P.; Desreumaux, P. A randomized clinical trial of Saccharomyces cerevisiae versus placebo in the irritable bowel syndrome. Dig. Liver Dis. 2015, 47, 119–124. [Google Scholar] [CrossRef] [PubMed]
- Spiller, R.; Pélerin, F.; Cayzeele Decherf, A.; Maudet, C.; Housez, B.; Cazaubiel, M.; Jüsten, P. Randomized double blind placebo-controlled trial of Saccharomyces cerevisiae CNCM I-3856 in irritable bowel syndrome: Improvement in abdominal pain and bloating in those with predominant constipation. United Eur. Gastroenterol. J. 2016, 4, 353–362. [Google Scholar] [CrossRef] [PubMed]
- Pericolini, E.; Gabrielli, E.; Ballet, N.; Sabbatini, S.; Roselletti, E.; Cayzeele Decherf, A.; Pélerin, F.; Luciano, E.; Perito, S.; Jüsten, P.; et al. Therapeutic activity of a Saccharomyces cerevisiae-based probiotic and inactivated whole yeast on vaginal candidiasis. Virulence 2017, 8, 74–90. [Google Scholar] [CrossRef]
- Roselletti, E.; Sabbatini, S.; Ballet, N.; Perito, S.; Pericolini, E.; Blasi, E.; Mosci, P.; Cayzeele Decherf, A.; Monari, C.; Vecchiarelli, A. Saccharomyces cerevisiae CNCM I-3856 as a New Therapeutic Agent Against Oropharyngeal Candidiasis. Front. Microbiol. 2019, 10, 1469. [Google Scholar] [CrossRef]
- Premanathan, M.; Shakurfow, F.A.A.; Ismail, A.A.; Berfad, M.A.; Ebrahim, A.T.; Awaj, M.M. Treatment of oral candidiasis (thrush) by Saccharomyces cerevisiae. Int. J. Med. Med. Sci. 2011, 3, 83–86. [Google Scholar]
- Collins, J.H.; Kunyeit, L.; Weintraub, S.; Sharma, N.; White, C.; Haq, N.; Anu-Appaiah, K.A.; Rao, R.P.; Young, E.M. Genetic basis for probiotic yeast phenotypes revealed by nanopore sequencing. G3 Genes|Genomes|Genet. 2023, 13, jkad093. [Google Scholar] [CrossRef]
- Chen, H.; Fink, G.R. Feedback control of morphogenesis in fungi by aromatic alcohols. Genes Dev. 2006, 20, 1150–1161. [Google Scholar] [CrossRef] [PubMed]
- Kunyeit, L.; Kurrey, N.K.; Anu-Appaiah, K.A.; Rao, R.P. Secondary Metabolites from Food-Derived Yeasts Inhibit Virulence of Candida albicans. mBio 2021, 12, e01891-21. [Google Scholar] [CrossRef]
- Krasowska, A.; Murzyn, A.; Dyjankiewicz, A.; Łukaszewicz, M.; Dziadkowiec, D. The antagonistic effect of Saccharomyces boulardii on Candida albicans filamentation, adhesion and biofilm formation. FEMS Yeast Res. 2009, 9, 1312–1321. [Google Scholar] [CrossRef]
- Murzyn, A.; Krasowska, A.; Stefanowicz, P.; Dziadkowiec, D.; Łukaszewicz, M. Capric acid secreted by S. boulardii inhibits C. albicans filamentous growth, adhesion and biofilm formation. PLoS ONE 2010, 5, e12050. [Google Scholar] [CrossRef]
- Gabris, K.; Nagy, G.; Madlena, M.; Denes, Z.; Marton, S.; Keszthelyi, G.; Banoczy, J. Associations between microbiological and salivary caries activity tests and caries experience in Hungarian adolescents. Caries Res. 1999, 33, 191–195. [Google Scholar] [CrossRef]
- Epstein, J.B. Oral and pharyngeal candidiasis. Topical agents for management and prevention. Postgrad. Med. 1989, 85, 257–269. [Google Scholar] [CrossRef] [PubMed]
- Wasfi, R.; Abd El-Rahman, O.A.; Zafer, M.M.; Ashour, H.M. Probiotic Lactobacillus sp. inhibit growth, biofilm formation and gene expression of caries-inducing Streptococcus mutans. J. Cell Mol. Med. 2018, 22, 1972–1983. [Google Scholar] [CrossRef] [PubMed]
- Kellis, M.; Birren, B.W.; Lander, E.S. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature 2004, 428, 617–624. [Google Scholar] [CrossRef]
- Wolfe, K.H. Comparative genomics and genome evolution in yeasts. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2006, 361, 403–412. [Google Scholar] [CrossRef][Green Version]
- Goffeau, A.; Barrell, B.G.; Bussey, H.; Davis, R.W.; Dujon, B.; Feldmann, H.; Galibert, F.; Hoheisel, J.D.; Jacq, C.; Johnston, M.; et al. Life with 6000 genes. Science 1996, 274, 546–567. [Google Scholar] [CrossRef] [PubMed]
- McFarland, L.V. Systematic review and meta-analysis of Saccharomyces boulardii in adult patients. World J. Gastroenterol. 2010, 16, 2202–2222. [Google Scholar] [CrossRef] [PubMed]
- Murzyn, A.; Krasowska, A.; Augustyniak, D.; Majkowska-Skrobek, G.; Lukaszewicz, M.; Dziadkowiec, D. The effect of Saccharomyces boulardii on Candida albicans-infected human intestinal cell lines Caco-2 and Intestin 407. FEMS Microbiol. Lett. 2010, 310, 17–23. [Google Scholar] [CrossRef]
- Berg, R.; Bernasconi, P.; Fowler, D.; Gautreaux, M. Inhibition of Candida albicans translocation from the gastrointestinal tract of mice by oral administration of Saccharomyces boulardii. J. Infect. Dis. 1993, 168, 1314–1318. [Google Scholar] [CrossRef]
- Laurian, R.; Ravent, J.; Dementhon, K.; Lemaire, M.; Soulard, A.; Cotton, P. Candida albicans Hexokinase 2 Challenges the Saccharomyces cerevisiae Moonlight Protein Model. Microorganisms 2021, 9, 848. [Google Scholar] [CrossRef]
- Mendes-Ferreira, A.; Cosme, F.; Barbosa, C.; Falco, V.; Ines, A.; Mendes-Faia, A. Optimization of honey-must preparation and alcoholic fermentation by Saccharomyces cerevisiae for mead production. Int. J. Food Microbiol. 2010, 144, 193–198. [Google Scholar] [CrossRef]
- Morton, C.O.; Hayes, A.; Wilson, M.; Rash, B.M.; Oliver, S.G.; Coote, P. Global phenotype screening and transcript analysis outlines the inhibitory mode(s) of action of two amphibian-derived, alpha-helical, cationic peptides on Saccharomyces cerevisiae. Antimicrob. Agents Chemother. 2007, 51, 3948–3959. [Google Scholar] [CrossRef] [PubMed]
- Abranches, J.; Chen, Y.Y.; Burne, R.A. Galactose metabolism by Streptococcus mutans. Appl. Environ. Microbiol. 2004, 70, 6047–6052. [Google Scholar] [CrossRef]
- Liu, J.J.; Zhang, G.C.; Kong, I.I.; Yun, E.J.; Zheng, J.Q.; Kweon, D.H.; Jin, Y.S. A Mutation in PGM2 Causing Inefficient Galactose Metabolism in the Probiotic Yeast Saccharomyces boulardii. Appl. Environ. Microbiol. 2018, 84, e02858-1. [Google Scholar] [CrossRef]
- Saghrouni, F.; Ben Abdeljelil, J.; Boukadida, J.; Ben Said, M. Molecular methods for strain typing of Candida albicans: A review. J. Appl. Microbiol. 2013, 114, 1559–1574. [Google Scholar] [CrossRef]
- Thomson, D.D.; Wehmeier, S.; Byfield, F.J.; Janmey, P.A.; Caballero-Lima, D.; Crossley, A.; Brand, A.C. Contact-induced apical asymmetry drives the thigmotropic responses of Candida albicans hyphae. Cell Microbiol. 2015, 17, 342–354. [Google Scholar] [CrossRef] [PubMed]
- Uwamahoro, N.; Verma-Gaur, J.; Shen, H.H.; Qu, Y.; Lewis, R.; Lu, J.; Bambery, K.; Masters, S.L.; Vince, J.E.; Naderer, T.; et al. The pathogen Candida albicans hijacks pyroptosis for escape from macrophages. mBio 2014, 5, e00003–e00014. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yousif, D.; Wu, Y.; Gonzales, A.A.; Mathieu, C.; Zeng, Y.; Sample, L.; Terando, S.; Li, T.; Xiao, J. Anti-Cariogenic Effects of S. cerevisiae and S. boulardii in S. mutans–C. albicans Cross-Kingdom In Vitro Models. Pharmaceutics 2024, 16, 215. https://doi.org/10.3390/pharmaceutics16020215
Yousif D, Wu Y, Gonzales AA, Mathieu C, Zeng Y, Sample L, Terando S, Li T, Xiao J. Anti-Cariogenic Effects of S. cerevisiae and S. boulardii in S. mutans–C. albicans Cross-Kingdom In Vitro Models. Pharmaceutics. 2024; 16(2):215. https://doi.org/10.3390/pharmaceutics16020215
Chicago/Turabian StyleYousif, Dina, Yan Wu, Alexandria Azul Gonzales, Christa Mathieu, Yan Zeng, Lee Sample, Sabrina Terando, Ting Li, and Jin Xiao. 2024. "Anti-Cariogenic Effects of S. cerevisiae and S. boulardii in S. mutans–C. albicans Cross-Kingdom In Vitro Models" Pharmaceutics 16, no. 2: 215. https://doi.org/10.3390/pharmaceutics16020215
APA StyleYousif, D., Wu, Y., Gonzales, A. A., Mathieu, C., Zeng, Y., Sample, L., Terando, S., Li, T., & Xiao, J. (2024). Anti-Cariogenic Effects of S. cerevisiae and S. boulardii in S. mutans–C. albicans Cross-Kingdom In Vitro Models. Pharmaceutics, 16(2), 215. https://doi.org/10.3390/pharmaceutics16020215






