In Vitro Characterization of Reversine-Treated Gingival Fibroblasts and Their Safety Evaluation after In Vivo Transplantation
Abstract
1. Introduction
2. Materials and Methods
2.1. Primary Culture of Gingival Fibroblasts
2.2. Cell Doubling Time
2.3. Reversine Administration
2.4. MTT Assay
2.5. SRB Assay
2.6. Crystal Violet Assay
2.7. Cell Cycle
2.8. Cell Morphology
2.9. Viability and Types of Cell Death
2.10. Comet Assay
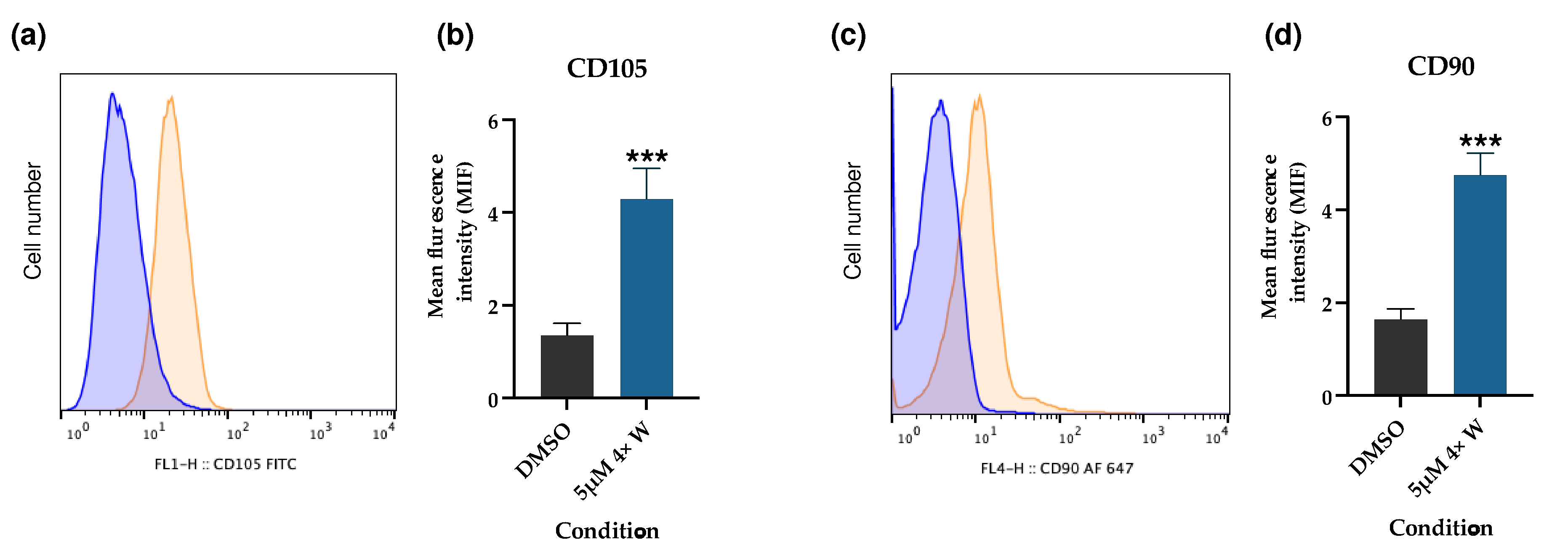
2.11. Mesenchymal Stem Cell (MSC) Markers
2.12. Cloning Efficiency
2.13. DNA Methylation
2.14. Cellular Senescence
2.15. Alkaline Phosphatase Gene Expression and Enzyme Activity
2.16. Western Blot
2.17. Trilineage Differentiation
2.18. Teratoma Formation
2.19. Statistical Analysis
3. Results
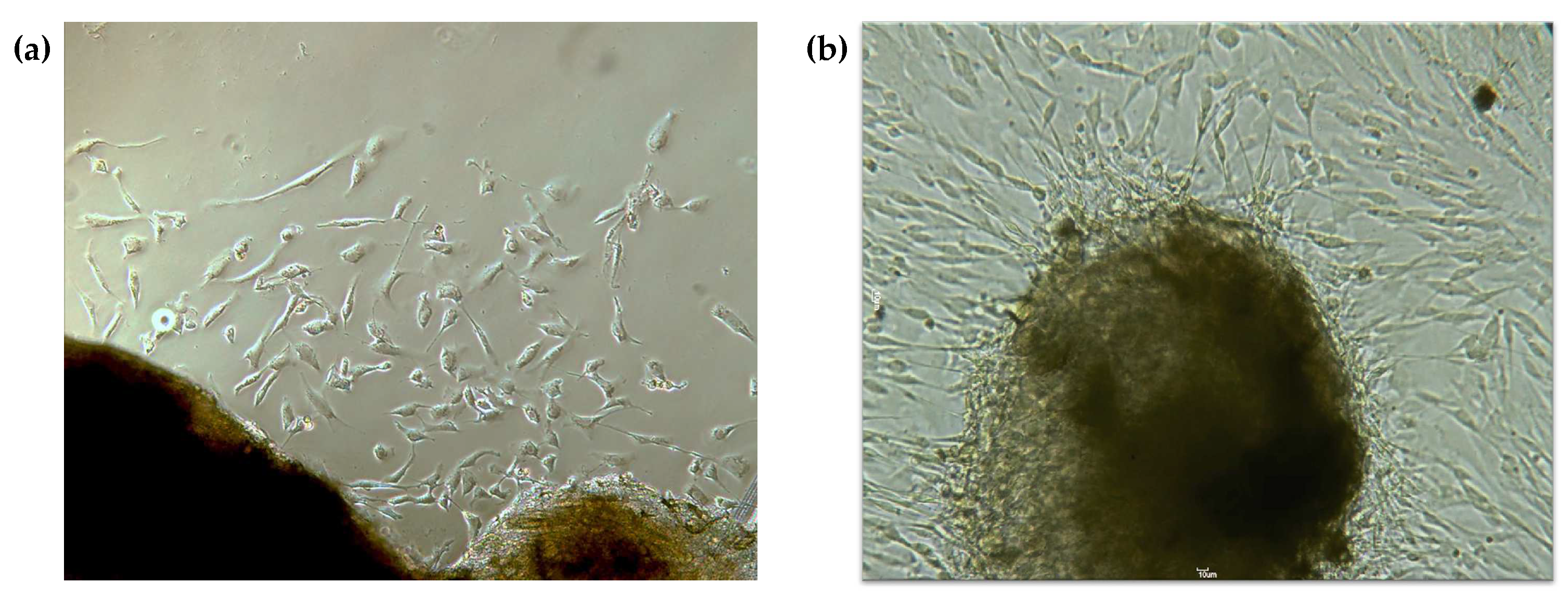
3.1. Gingival Fibroblasts Were Successfully Obtained from the Explants
3.2. Reversine Induces a Dose-Dependent Response in Metabolic Activity, Protein, and DNA Contents
3.3. Reversine Induces Changes in Cell Cycle and Cell Morphology
3.4. Reversine Induces Cell Death by Apoptosis and Genotoxicity at Higher Concentrations
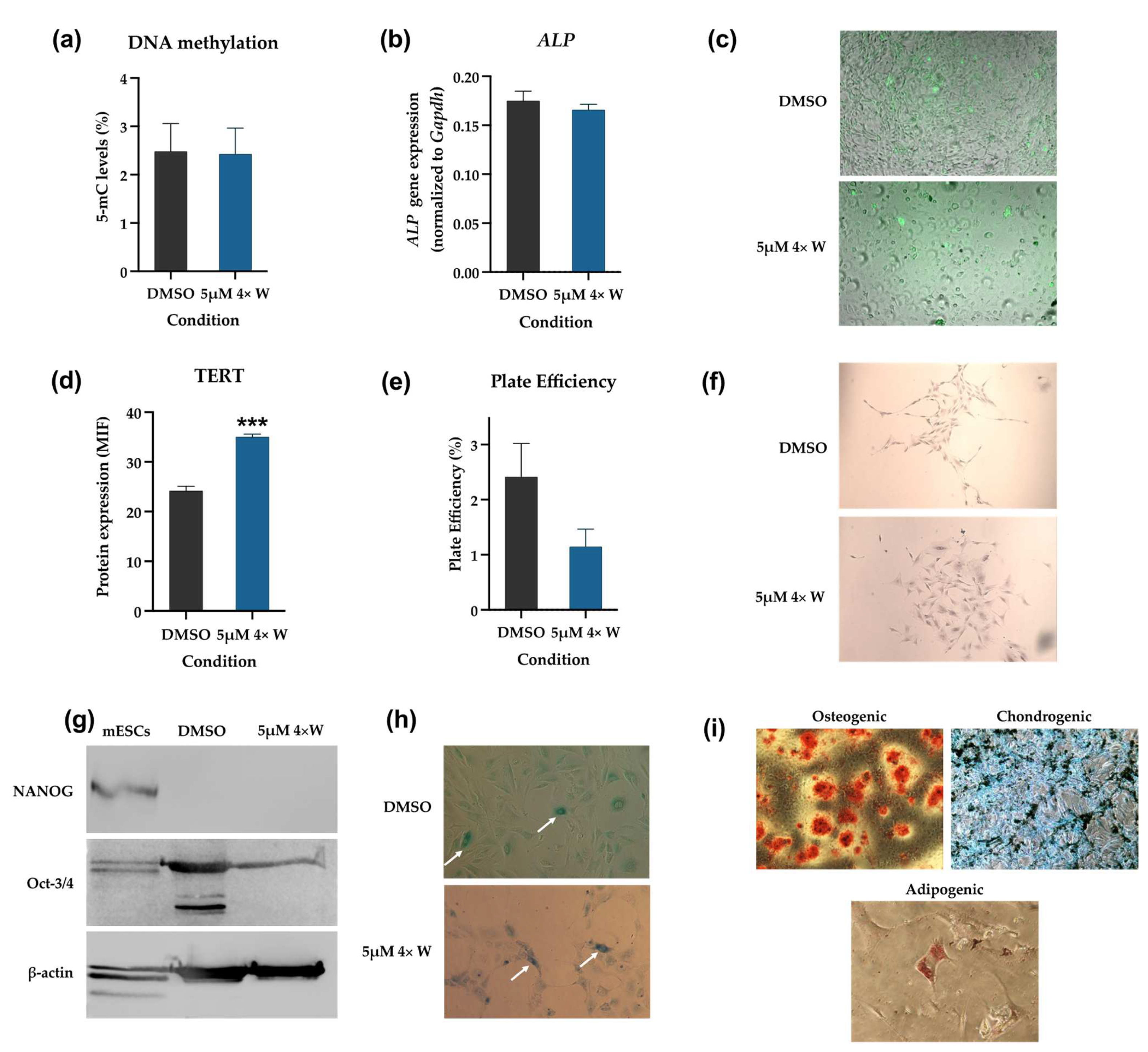
3.5. Reversine Induced Some Cells to Dedifferentiate and Acquire Stem-like Characteristics
3.6. Dedifferentiated Cells Do Not Induce Teratoma Formation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Conforti, E.; Arrigoni, E.; Piccoli, M.; Lopa, S.; de Girolamo, L.; Ibatici, A.; Di Matteo, A.; Tettamanti, G.; Brini, A.T.; Anastasia, L. Reversine Increases Multipotent Human Mesenchymal Cells Differentiation Potential. J. Biol. Regul. Homeost. Agents 2011, 25, S25–S33. [Google Scholar]
- Kim, Y.K.; Choi, H.Y.; Kim, N.H.; Lee, W.; Seo, D.W.; Kang, D.W.; Lee, H.Y.; Han, J.W.; Park, S.W.; Kim, S.N. Reversine Stimulates Adipocyte Differentiation and Downregulates Akt and P70s6k Signaling Pathways in 3T3-L1 Cells. Biochem. Biophys. Res. Commun. 2007, 358, 553–558. [Google Scholar] [CrossRef]
- Anastasia, L.; Sampaolesi, M.; Papini, N.; Oleari, D.; Lamorte, G.; Tringali, C.; Monti, E.; Galli, D.; Tettamanti, G.; Cossu, G.; et al. Reversine-Treated Fibroblasts Acquire Myogenic Competence in Vitro and in Regenerating Skeletal Muscle. Cell Death Differ. 2006, 13, 2042–2051. [Google Scholar] [CrossRef]
- Chen, S.; Zhang, Q.; Wu, X.; Schultz, P.G.; Ding, S. Dedifferentiation of Lineage-Committed Cells by a Small Molecule. J. Am. Chem. Soc. 2004, 126, 410–411. [Google Scholar] [CrossRef]
- Chen, S.; Takanashi, S.; Zhang, Q.; Xiong, W.; Zhu, S.; Peters, E.C.; Ding, S.; Schultz, P.G. Reversine Increases the Plasticity of Lineage-Committed Mammalian Cells. Proc. Natl. Acad. Sci. USA 2007, 104, 10482–10487. [Google Scholar] [CrossRef]
- Qu, G.; von Schroeder, H.P. Preliminary Evidence for the Dedifferentiation of RAW 264.7 Cells into Mesenchymal Progenitor-like Cells by a Purine Analog. Tissue Eng. Part A 2012, 18, 1890–1901. [Google Scholar] [CrossRef] [PubMed]
- Li, X.C.; Guo, Y.; Yao, Y.; Hua, J.; Ma, Y.H.; Liu, C.Q.; Guan, W.J. Reversine Increases the Plasticity of Long-Term Cryopreserved Fibroblasts to Multipotent Progenitor Cells through Activation of Oct4. Int. J. Biol. Sci. 2016, 12, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Saraiya, M.; Nasser, R.; Zeng, Y.; Addya, S.; Ponnappan, R.K.; Fortina, P.; Anderson, D.G.; Albert, T.J.; Shapiro, I.M.; Risbud, M.V. Reversine Enhances Generation of Progenitor-like Cells by Dedifferentiation of Annulus Fibrosus Cells. Tissue Eng. Part A 2010, 16, 1443–1455. [Google Scholar] [CrossRef] [PubMed]
- Fania, C.; Anastasia, L.; Vasso, M.; Papini, N.; Capitanio, D.; Venerando, B.; Gelfi, C. Proteomic Signature of Reversine-Treated Murine Fibroblasts by 2-D Difference Gel Electrophoresis and MS: Possible Associations with Cell Signalling Networks. Electrophoresis 2009, 30, 2193–2206. [Google Scholar] [CrossRef] [PubMed]
- Anastasia, L.; Pelissero, G.; Venerando, B.; Tettamanti, G. Cell Reprogramming: Expectations and Challenges for Chemistry in Stem Cell Biology and Regenerative Medicine. Cell Death Differ. 2010, 17, 1230–1237. [Google Scholar] [CrossRef]
- Song, H.K.; Noh, E.M.; Kim, J.M.; You, Y.O.; Kwon, K.B.; Lee, Y.R. Reversine Inhibits MMP-3, IL-6 and IL-8 Expression through Suppression of ROS and JNK/AP-1 Activation in Interleukin-1β-Stimulated Human Gingival Fibroblasts. Arch. Oral. Biol. 2019, 108, 104530. [Google Scholar] [CrossRef]
- Huang, Y.; Huang, D.; Weng, J.; Zhang, S.; Zhang, Q.; Mai, Z.; Gu, W. Effect of Reversine on Cell Cycle, Apoptosis, and Activation of Hepatic Stellate Cells. Mol. Cell Biochem. 2016, 423, 9–20. [Google Scholar] [CrossRef]
- Lu, C.-H.; Liu, Y.-W.; Hua, S.-C.; Yu, H.-I.; Chang, Y.-P.; Lee, Y.-R. Autophagy Induction of Reversine on Human Follicular Thyroid Cancer Cells. Biomed. Pharmacother. 2012, 66, 642–647. [Google Scholar] [CrossRef] [PubMed]
- Bijian, K.; Lougheed, C.; Su, J.; Xu, B.; Yu, H.; Wu, J.H.; Riccio, K.; Alaoui-Jamali, M.A. Targeting Focal Adhesion Turnover in Invasive Breast Cancer Cells by the Purine Derivative Reversine. Br. J. Cancer 2013, 109, 2810–2818. [Google Scholar] [CrossRef] [PubMed]
- Righolt, A.J.; Jevdjevic, M.; Marcenes, W.; Listl, S. Global-, Regional-, and Country-Level Economic Impacts of Dental Diseases in 2015. J. Dent. Res. 2018, 97, 501–507. [Google Scholar] [CrossRef] [PubMed]
- Azzolino, D.; Passarelli, P.C.; De Angelis, P.; Piccirillo, G.B.; D’Addona, A.; Cesari, M. Poor Oral Health as a Determinant of Malnutrition and Sarcopenia. Nutrients 2019, 11, 2898. [Google Scholar] [CrossRef]
- Musacchio, E.; Binotto, P.; Perissinotto, E.; Sergi, G.; Zambon, S.; Corti, M.C.; Frigo, A.C.; Sartori, L. Tooth Retention Predicts Good Physical Performance in Older Adults. PLoS ONE 2021, 16, e0255741. [Google Scholar] [CrossRef]
- Muhammad, T.; Srivastava, S. Tooth Loss and Associated Self-Rated Health and Psychological and Subjective Wellbeing among Community-Dwelling Older Adults: A Cross-Sectional Study in India. BMC Public Health 2022, 22, 7. [Google Scholar] [CrossRef]
- Casagrande, L.; Cordeiro, M.M.; Nör, S.A.; Nör, J.E. Dental Pulp Stem Cells in Regenerative Dentistry. Odontology 2011, 99, 1–7. [Google Scholar] [CrossRef]
- Angelova Volponi, A.; Zaugg, L.K.; Neves, V.; Liu, Y.; Sharpe, P.T. Tooth Repair and Regeneration. Curr. Oral. Health Rep. 2018, 5, 295–303. [Google Scholar] [CrossRef]
- Yan, M.; Yu, Y.; Zhang, G.; Tang, C.; Yu, J. A Journey from Dental Pulp Stem Cells to a Bio-Tooth. Stem Cell Rev. Rep. 2011, 7, 161–171. [Google Scholar] [CrossRef]
- Sartaj, R.; Sharpe, P. Biological Tooth Replacement. J. Anat. 2006, 209, 503. [Google Scholar] [CrossRef]
- Olaru, M.; Sachelarie, L.; Calin, G. Hard Dental Tissues Regeneration—Approaches and Challenges. Materials 2021, 14, 2558. [Google Scholar] [CrossRef]
- Liang, C.; Liao, L.; Tian, W. Stem Cell-based Dental Pulp Regeneration: Insights from Signaling Pathways. Stem Cell Rev. Rep. 2021, 17, 1251–1263. [Google Scholar] [CrossRef]
- Zhang, W.; Yelick, P.C. Tooth Repair and Regeneration: Potential of Dental Stem Cells. Trends Mol. Med. 2021, 27, 501–511. [Google Scholar] [CrossRef]
- Mosaddad, S.A.; Rasoolzade, B.; Namanloo, R.A.; Azarpira, N.; Dortaj, H. Stem Cells and Common Biomaterials in Dentistry: A Review Study. J. Mater. Sci. Mater. Med. 2022, 33, 55. [Google Scholar] [CrossRef]
- Baena, A.R.Y.; Casasco, A.; Monti, M. Hypes and Hopes of Stem Cell Therapies in Dentistry: A Review. Stem Cell Rev. Rep. 2022, 18, 1294–1308. [Google Scholar] [CrossRef]
- Chalisserry, E.P.; Nam, S.Y.; Park, S.H.; Anil, S. Therapeutic Potential of Dental Stem Cells. J. Tissue Eng. 2017, 8, 2041731417702531. [Google Scholar] [CrossRef] [PubMed]
- Paz, A.G.; Maghaireh, H.; Mangano, F.G. Stem Cells in Dentistry: Types of Intra- and Extraoral Tissue-Derived Stem Cells and Clinical Applications. Stem Cells Int. 2018, 2018, 4313610. [Google Scholar] [CrossRef] [PubMed]
- Ibarretxe, G.; Alvarez, A.; Cañavate, M.-L.; Hilario, E.; Aurrekoetxea, M.; Unda, F. Cell Reprogramming, IPS Limitations, and Overcoming Strategies in Dental Bioengineering. Stem Cells Int. 2012, 2012, 365932. [Google Scholar] [CrossRef] [PubMed]
- Otsu, K.; Kishigami, R.; Oikawa-Sasaki, A.; Fukumoto, S.; Yamada, A.; Fujiwara, N.; Ishizeki, K.; Harada, H. Differentiation of Induced Pluripotent Stem Cells into Dental Mesenchymal Cells. Stem Cells Dev. 2012, 21, 1156–1164. [Google Scholar] [CrossRef]
- Lee, E.K.; Bae, G.-U.; You, J.S.; Lee, J.C.; Jeon, Y.J.; Park, J.W.; Park, J.H.; Ahn, S.H.; Kim, Y.K.; Choi, W.S.; et al. Reversine Increases the Plasticity of Lineage-Committed Cells toward Neuroectodermal Lineage. J. Biol. Chem. 2009, 284, 2891–2901. [Google Scholar] [CrossRef]
- Ferreira, M.J.S.; Mancini, F.E.; Humphreys, P.A.; Ogene, L.; Buckley, M.; Domingos, M.A.N.; Kimber, S.J. Pluripotent Stem Cells for Skeletal Tissue Engineering. Crit. Rev. Biotechnol. 2022, 42, 774–793. [Google Scholar] [CrossRef]
- Kimbrel, E.A.; Lanza, R. Next-Generation Stem Cells—Ushering in a New Era of Cell-Based Therapies. Nat. Rev. Drug Discov. 2020, 19, 463–479. [Google Scholar] [CrossRef]
- Aly, R.M.; Mohamed, R. Current State of Stem Cell-Based Therapies: An Overview. Stem Cell Investig. 2020, 7, 8. [Google Scholar] [CrossRef]
- Yang, X.; Chen, B.; Liu, T.; Chen, X. Reversal of Myofibroblast Differentiation: A Review. Eur. J. Pharmacol. 2014, 734, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, H.; Kazama, T.; Hagikura, K.; Yamamoto, C.; Kazama, M.; Nagaoka, Y.; Matsumoto, T. An Efficient Method to Obtain Dedifferentiated Fat Cells. J. Vis. Exp. 2016, 113, e54177. [Google Scholar] [CrossRef]
- Duckmanton, A.; Kumar, A.; Chang, Y.T.; Brockes, J.P. A Single-Cell Analysis of Myogenic Dedifferentiation Induced by Small Molecules. Chem. Biol. 2005, 12, 1117–1126. [Google Scholar] [CrossRef] [PubMed]
- Jopling, C.; Boue, S.; Belmonte, J.C.I. Dedifferentiation, Transdifferentiation and Reprogramming: Three Routes to Regeneration. Nat. Rev. Mol. Cell Biol. 2011, 12, 79–89. [Google Scholar] [CrossRef]
- Odelberg, S.J.; Kollhoff, A.; Keating, M.T. Dedifferentiation of Mammalian Myotubes Induced by Msx1. Cell 2000, 103, 1099–1109. [Google Scholar] [CrossRef] [PubMed]
- Guan, J.; Wang, G.; Wang, J.; Zhang, Z.; Fu, Y.; Cheng, L.; Meng, G.; Lyu, Y.; Zhu, J.; Li, Y.; et al. Chemical Reprogramming of Human Somatic Cells to Pluripotent Stem Cells. Nature 2022, 605, 325–331. [Google Scholar] [CrossRef]
- Kim, M.; Yi, S.A.; Lee, H.; Bang, S.Y.; Park, E.K.; Lee, M.G.; Nam, K.H.; Yoo, J.H.; Lee, D.H.; Ryu, H.-W.; et al. Reversine Induces Multipotency of Lineage-Committed Cells through Epigenetic Silencing of MiR-133a. Biochem. Biophys. Res. Commun. 2014, 445, 255–262. [Google Scholar] [CrossRef]
- Serrano-Lopez, R.; Morandini, A.C. Fibroblasts at the Curtain Call: From Ensemble to Principal Dancers in Immunometabolism and Inflammaging. J. Appl. Oral. Sci. 2023, 31, e20230050. [Google Scholar] [CrossRef]
- Alfonso García, S.L.; Parada-Sanchez, M.T.; Arboleda Toro, D. The Phenotype of Gingival Fibroblasts and Their Potential Use in Advanced Therapies. Eur. J. Cell Biol. 2020, 99, 151123. [Google Scholar] [CrossRef]
- Häkkinen, L.; Larjava, H.; Fournier, B.P.J. Distinct Phenotype and Therapeutic Potential of Gingival Fibroblasts. Cytotherapy 2014, 16, 1171–1186. [Google Scholar] [CrossRef]
- Fadl, A.; Leask, A. Hiding in Plain Sight: Human Gingival Fibroblasts as an Essential, Yet Overlooked, Tool in Regenerative Medicine. Cells 2023, 12, 2021. [Google Scholar] [CrossRef]
- Mah, W.; Jiang, G.; Olver, D.; Cheung, G.; Kim, B.; Larjava, H.; Häkkinen, L. Human Gingival Fibroblasts Display a Non-Fibrotic Phenotype Distinct from Skin Fibroblasts in Three-Dimensional Cultures. PLoS ONE 2014, 9, e90715. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gölz, L.; Vestewig, E.; Blankart, M.M.; Kraus, D.; Appel, T.; Frede, S.; Jäger, A. Differences in Human Gingival and Dermal Fibroblasts May Contribute to Oral-Induced Tolerance against Nickel. J. Allergy Clin. Immunol. 2016, 138, 1202–1205.e3. [Google Scholar] [CrossRef] [PubMed]
- Paulo, S.; Laranjo, M.; Abrantes, A.M.; Casalta-Lopes, J.; Santos, K.; Gonçalves, A.C.; Paula, A.B.; Marto, C.M.; Sarmento-Ribeiro, A.B.; Carrilho, E.; et al. Synthetic Calcium Phosphate Ceramics as a Potential Treatment for Bisphosphonate-Related Osteonecrosis of the Jaw. Materials 2019, 12, 1840. [Google Scholar] [CrossRef]
- Strober, W. Trypan Blue Exclusion Test of Cell Viability. In Current Protocols in Immunology; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2001; Volume Appendix 3, pp. A.3B.1–A.3B.2. [Google Scholar]
- Laranjo, M.; Serra, A.C.; Abrantes, M.; Piñeiro, M.; Gonçalves, A.C.; Casalta-Lopes, J.; Carvalho, L.; Sarmento-Ribeiro, A.B.; Rocha-Gonsalves, A.; Botelho, F. 2-Bromo-5-Hydroxyphenylporphyrins for Photodynamic Therapy: Photosensitization Efficiency, Subcellular Localization and in Vivo Studies. Photodiagnosis Photodyn. Ther. 2013, 10, 51–61. [Google Scholar] [CrossRef]
- Vichai, V.; Kirtikara, K. Sulforhodamine B Colorimetric Assay for Cytotoxicity Screening. Nat. Protoc. 2006, 1, 1112–1116. [Google Scholar] [CrossRef] [PubMed]
- Feoktistova, M.; Geserick, P.; Leverkus, M. Crystal Violet Assay for Determining Viability of Cultured Cells. Cold Spring Harb. Protoc. 2016, 2016, 343–346. [Google Scholar] [CrossRef] [PubMed]
- Jemaà, M.; Kifagi, C.; Serrano, S.S.; Massoumi, R. Preferential Killing of Tetraploid Colon Cancer Cells by Targeting the Mitotic Kinase Plk1. Cell. Physiol. Biochem. 2020, 54, 303–320. [Google Scholar] [CrossRef]
- Cardoso, M.; Coelho, A.; Marto, C.M.; Gonçalves, A.C.; Paula, A.; Ribeiro, A.B.S.; Ferreira, M.M.; Botelho, M.F.; Laranjo, M.; Carrilho, E. Effects of AdperTM ScotchbondTM 1 XT, ClearfilTM SE Bond 2 and ScotchbondTM Universal in Odontoblasts. Materials 2021, 14, 6435. [Google Scholar] [CrossRef]
- Almeida-Ferreira, C.; Silva-Teixeira, R.; Gonçalves, A.C.; Marto, C.M.; Sarmento-Ribeiro, A.B.; Caramelo, F.; Botelho, M.F.; Laranjo, M. Cold Atmospheric Plasma Apoptotic and Oxidative Effects on MCF7 and HCC1806 Human Breast Cancer Cells. Int. J. Mol. Sci. 2022, 23, 1698. [Google Scholar] [CrossRef]
- Santos, K.; Laranjo, M.; Abrantes, A.M.; Brito, A.F.; Gonçalves, C.; Sarmento Ribeiro, A.B.; Botelho, M.F.; Soares, M.I.L.; Oliveira, A.S.R.; Pinho e Melo, T.M.V.D. Targeting Triple-Negative Breast Cancer Cells with 6,7-Bis(Hydroxymethyl)-1H,3H-Pyrrolo [1,2-c]Thiazoles. Eur. J. Med. Chem. 2014, 79, 273–281. [Google Scholar] [CrossRef]
- Gyori, B.M.; Venkatachalam, G.; Thiagarajan, P.S.; Hsu, D.; Clement, M.V. OpenComet: An Automated Tool for Comet Assay Image Analysis. Redox Biol. 2014, 2, 457–465. [Google Scholar] [CrossRef]
- Franken, N.A.P.; Rodermond, H.M.; Stap, J.; Haveman, J.; van Bree, C. Clonogenic Assay of Cells in Vitro. Nat. Protoc. 2006, 1, 2315–2319. [Google Scholar] [CrossRef] [PubMed]
- Koh, C.M. Isolation of Genomic DNA from Mammalian Cells. In Methods in Enzymology; Academic Press Inc.: Cambridge, MA, USA, 2013; Volume 529, pp. 161–169. ISBN 9780124186873. [Google Scholar]
- Hernandez-Segura, A.; Brandenburg, S.; Demaria, M. Induction and Validation of Cellular Senescence in Primary Human Cells. J. Vis. Exp. 2018, 136, e57782. [Google Scholar] [CrossRef]
- Sousa, M.I.; Correia, B.; Branco, A.F.; Rodrigues, A.S.; Ramalho-Santos, J. Effects of DMSO on the Pluripotency of Cultured Mouse Embryonic Stem Cells (MESCs). Stem Cells Int. 2020, 2020, 8835353. [Google Scholar] [CrossRef]
- Qu, G.; Li, Y.; Chen, L.; Chen, Q.; Zou, D.; Yang, C.; Zhou, Q. Comparison of Osteogenic Differentiation Potential of Human Dental-Derived Stem Cells Isolated from Dental Pulp, Periodontal Ligament, Dental Follicle, and Alveolar Bone. Stem Cells Int. 2021, 2021, 6631905. [Google Scholar] [CrossRef]
- Baptista Paula, A.; Laranjo, M.; Coelho, A.S.; Abrantes, A.M.; Gonçalves, A.C.; Sarmento-Ribeiro, A.B.; Ferreira, M.M.; Botelho, M.F.; Marto, C.M.; Carrilho, E. Accessing the Cytotoxicity and Cell Response to Biomaterials. J. Vis. Exp. 2021, 173, e61512. [Google Scholar] [CrossRef]
- Quesada, M.P.; García-Bernal, D.; Pastor, D.; Estirado, A.; Blanquer, M.; García-Hernández, A.M.; Moraleda, J.M.; Martínez, S. Safety and Biodistribution of Human Bone Marrow-Derived Mesenchymal Stromal Cells Injected Intrathecally in Non-Obese Diabetic Severe Combined Immunodeficiency Mice: Preclinical Study. Tissue Eng. Regen. Med. 2019, 16, 525–538. [Google Scholar] [CrossRef] [PubMed]
- Kraus, N.A.; Ehebauer, F.; Zapp, B.; Rudolphi, B.; Kraus, B.J.; Kraus, D. Quantitative Assessment of Adipocyte Differentiation in Cell Culture. Adipocyte 2016, 5, 351–358. [Google Scholar] [CrossRef]
- Zhang, L.; Su, P.; Xu, C.; Yang, J.; Yu, W.; Huang, D. Chondrogenic Differentiation of Human Mesenchymal Stem Cells: A Comparison between Micromass and Pellet Culture Systems. Biotechnol. Lett. 2010, 32, 1339–1346. [Google Scholar] [CrossRef] [PubMed]
- Zuliani, C.C.; Bombini, M.F.; de Andrade, K.C.; Mamoni, R.; Pereira, A.H.; Coimbra, I.B. Micromass Cultures Are Effective for Differentiation of Human Amniotic Fluid Stem Cells into Chondrocytes. Clinics 2018, 73, e268. [Google Scholar] [CrossRef]
- Lezmi, E.; Weissbein, U.; Golan-Lev, T.; Nissim-Rafinia, M.; Meshorer, E.; Benvenisty, N. The Chromatin Regulator ZMYM2 Restricts Human Pluripotent Stem Cell Growth and Is Essential for Teratoma Formation. Stem Cell Rep. 2020, 15, 1275. [Google Scholar] [CrossRef]
- Workman, P.; Aboagye, E.O.; Balkwill, F.; Balmain, A.; Bruder, G.; Chaplin, D.J.; Double, J.A.; Everitt, J.; Farningham, D.A.H.; Glennie, M.J.; et al. Guidelines for the Welfare and Use of Animals in Cancer Research. Br. J. Cancer 2010, 102, 1555–1577. [Google Scholar] [CrossRef] [PubMed]
- Demšar, J.; Erjavec, A.; Hočevar, T.; Milutinovič, M.; Možina, M.; Toplak, M.; Umek, L.; Zbontar, J.; Zupan, B. Orange: Data Mining Toolbox in Python Tomaž Curk Matija Polajnar Laň Zagar. J. Mach. Learn. Res. 2013, 14, 2349–2353. [Google Scholar]
- Fournier, B.P.J.; Larjava, H.; Häkkinen, L. Gingiva as a Source of Stem Cells with Therapeutic Potential. Stem Cells Dev. 2013, 22, 3157–3177. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Yang, P.; Ge, S. Isolation and Characterization of Human Gingiva-Derived Mesenchymal Stem Cells Using Limiting Dilution Method. J. Dent. Sci. 2016, 11, 304–314. [Google Scholar] [CrossRef]
- Kuo, C.-H.; Lu, Y.-C.; Tseng, Y.-S.; Shi, C.-S.; Chen, S.-H.; Chen, P.-T.; Wu, F.-L.; Chang, Y.-P.; Lee, Y.-R. Reversine Induces Cell Cycle Arrest, Polyploidy, and Apoptosis in Human Breast Cancer Cells. Breast Cancer 2014, 21, 358–369. [Google Scholar] [CrossRef] [PubMed]
- Hua, S.-C.; Chang, T.-C.; Chen, H.-R.; Lu, C.-H.; Liu, Y.-W.; Chen, S.-H.; Yu, H.-I.; Chang, Y.-P.; Lee, Y.-R. Reversine, a 2,6-Disubstituted Purine, as an Anti-Cancer Agent in Differentiated and Undifferentiated Thyroid Cancer Cells. Pharm. Res. 2012, 29, 1990–2005. [Google Scholar] [CrossRef] [PubMed]
- Park, J.G.; Lee, D.-H.; Moon, Y.S.; Kim, K.-H. Reversine Increases the Plasticity of Lineage-Committed Preadipocytes to Osteogenesis by Inhibiting Adipogenesis through Induction of TGF-β Pathway in Vitro. Biochem. Biophys. Res. Commun. 2014, 446, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.-R.; Wu, W.-C.; Ji, W.-T.; Chen, J.Y.-F.; Cheng, Y.-P.; Chiang, M.-K.; Chen, H.-R. Reversine Suppresses Oral Squamous Cell Carcinoma via Cell Cycle Arrest and Concomitantly Apoptosis and Autophagy. J. Biomed. Sci. 2012, 19, 9. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, T.-C.; Traganos, F.; Darzynkiewicz, Z.; Wu, J.M. The 2,6-Disubstituted Purine Reversine Induces Growth Arrest and Polyploidy in Human Cancer Cells. Int. J. Oncol. 2007, 31, 1293–1300. [Google Scholar] [CrossRef] [PubMed]
- D’Alise, A.M.; Amabile, G.; Iovino, M.; Di Giorgio, F.P.; Bartiromo, M.; Sessa, F.; Villa, F.; Musacchio, A.; Cortese, R. Reversine, a Novel Aurora Kinases Inhibitor, Inhibits Colony Formation of Human Acute Myeloid Leukemia Cells. Mol. Cancer Ther. 2008, 7, 1140–1149. [Google Scholar] [CrossRef] [PubMed]
- Hakura, A. Improved AMES Test for Genotoxicity Assessment of Drugs: Preincubation Assay Using a Low Concentration of Dimethyl Sulfoxide. In Optimization in Drug Discovery. Methods in Pharmacology and Toxicology; Humana Press: Totowa, NJ, USA, 2014; pp. 545–559. [Google Scholar] [CrossRef]
- Verheijen, M.; Lienhard, M.; Schrooders, Y.; Clayton, O.; Nudischer, R.; Boerno, S.; Timmermann, B.; Selevsek, N.; Schlapbach, R.; Gmuender, H.; et al. DMSO Induces Drastic Changes in Human Cellular Processes and Epigenetic Landscape in Vitro. Sci. Rep. 2019, 9, 4641. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, S.; Nguyen, S.T.; Nguyen, H.T.-L.; Truong, K.D. Comparative Cytotoxic Effects of Methanol, Ethanol and DMSO on Human Cancer Cell Lines. Biomed. Res. Ther. 2020, 7, 3855–3859. [Google Scholar] [CrossRef]
- Dominici, M.; Le Blanc, K.; Mueller, I.; Slaper-Cortenbach, I.; Marini, F.; Krause, D.; Deans, R.; Keating, A.; Prockop, D.; Horwitz, E. Minimal Criteria for Defining Multipotent Mesenchymal Stromal Cells. The International Society for Cellular Therapy Position Statement. Cytotherapy 2006, 8, 315–317. [Google Scholar] [CrossRef]
- Kong, D.-H.; Kim, Y.; Kim, M.; Jang, J.; Lee, S. Emerging Roles of Vascular Cell Adhesion Molecule-1 (VCAM-1) in Immunological Disorders and Cancer. Int. J. Mol. Sci. 2018, 19, 1057. [Google Scholar] [CrossRef]
- Halfon, S.; Abramov, N.; Grinblat, B.; Ginis, I. Markers Distinguishing Mesenchymal Stem Cells from Fibroblasts Are Downregulated with Passaging. Stem Cells Dev. 2011, 20, 53–66. [Google Scholar] [CrossRef] [PubMed]
- Kundrotas, G. Surface Markers Distinguishing Mesenchymal Stem Cells from Fibroblasts. Acta Med. Litu. 2012, 19, 75–79. [Google Scholar] [CrossRef]
- Denu, R.A.; Nemcek, S.; Bloom, D.D.; Goodrich, A.D.; Kim, J.; Mosher, D.F.; Hematti, P. Fibroblasts and Mesenchymal Stromal/Stem Cells Are Phenotypically Indistinguishable. Acta Haematol. 2016, 136, 85–97. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Costello, J. DNA Methylation: An Epigenetic Mark of Cellular Memory. Exp. Mol. Med. 2017, 49, e322. [Google Scholar] [CrossRef]
- Liao, Y.; Zeng, Z.; Lu, F.; Dong, Z.; Chang, Q.; Gao, J. In Vivo Dedifferentiation of Adult Adipose Cells. PLoS ONE 2015, 10, e0125254. [Google Scholar] [CrossRef]
- Ugurlu, B.; Karaoz, E. Comparison of Similar Cells: Mesenchymal Stromal Cells and Fibroblasts. Acta Histochem. 2020, 122, 151634. [Google Scholar] [CrossRef]
- Štefková, K.; Procházková, J.; Pacherník, J. Alkaline Phosphatase in Stem Cells. Stem Cells Int. 2015, 2015, 628368. [Google Scholar] [CrossRef]
- Yuan, X.; Xu, D. Telomerase Reverse Transcriptase (TERT) in Action: Cross-Talking with Epigenetics. Int. J. Mol. Sci. 2019, 20, 3338. [Google Scholar] [CrossRef]
- Jemaà, M.; Galluzzi, L.; Kepp, O.; Boilève, A.; Lissa, D.; Senovilla, L.; Harper, F.; Pierron, G.; Berardinelli, F.; Antoccia, A.; et al. Preferential Killing of P53-Deficient Cancer Cells by Reversine. Cell Cycle 2012, 11, 2149–2158. [Google Scholar] [CrossRef]
- Olariu, V.; Lövkvist, C.; Sneppen, K. Nanog, Oct4 and Tet1 Interplay in Establishing Pluripotency. Sci. Rep. 2016, 6, 25438. [Google Scholar] [CrossRef]
- Wang, X.; Dai, J. Concise Review: Isoforms of OCT4 Contribute to the Confusing Diversity in Stem Cell Biology. Stem Cells 2010, 28, 885–893. [Google Scholar] [CrossRef] [PubMed]
- Jez, M.; Ambady, S.; Kashpur, O.; Grella, A.; Malcuit, C.; Vilner, L.; Rozman, P.; Dominko, T. Expression and Differentiation between OCT4A and Its Pseudogenes in Human ESCs and Differentiated Adult Somatic Cells. PLoS ONE 2014, 9, e89546. [Google Scholar] [CrossRef] [PubMed]
- Furukawa, M.; Matsuda, K.; Aoki, Y.; Yamada, M.; Wang, J.; Watanabe, M.; Kurosawa, M.; Shikama, Y.; Matsushita, K. Analysis of Senescence in Gingival Tissues and Gingival Fibroblast Cultures. Clin. Exp. Dent. Res. 2022, 8, 939–949. [Google Scholar] [CrossRef]
- Tao, H.; Chen, X.; Wei, A.; Song, X.; Wang, W.; Liang, L.; Zhao, Q.; Han, Z.; Han, Z.; Wang, X.; et al. Comparison of Teratoma Formation between Embryonic Stem Cells and Parthenogenetic Embryonic Stem Cells by Molecular Imaging. Stem Cells Int. 2018, 2018, 7906531. [Google Scholar] [CrossRef] [PubMed]
- Soares, A.S.L.S.; Scelza, M.Z.; Spoladore, J.; Gallito, M.A.; Oliveira, F.; de Cássia Martins Moraes, R.; Alves, G.G. Comparison of Primary Human Gingival Fibroblasts from an Older and a Young Donor on the Evaluation of Cytotoxicity of Denture Adhesives. J. Appl. Oral. Sci. 2018, 26, e20160594. [Google Scholar] [CrossRef]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marto, C.M.; Laranjo, M.; Gonçalves, A.C.; Paula, A.; Jorge, J.; Caetano-Oliveira, R.; Sousa, M.I.; Oliveiros, B.; Ramalho-Santos, J.; Sarmento-Ribeiro, A.B.; et al. In Vitro Characterization of Reversine-Treated Gingival Fibroblasts and Their Safety Evaluation after In Vivo Transplantation. Pharmaceutics 2024, 16, 207. https://doi.org/10.3390/pharmaceutics16020207
Marto CM, Laranjo M, Gonçalves AC, Paula A, Jorge J, Caetano-Oliveira R, Sousa MI, Oliveiros B, Ramalho-Santos J, Sarmento-Ribeiro AB, et al. In Vitro Characterization of Reversine-Treated Gingival Fibroblasts and Their Safety Evaluation after In Vivo Transplantation. Pharmaceutics. 2024; 16(2):207. https://doi.org/10.3390/pharmaceutics16020207
Chicago/Turabian StyleMarto, Carlos Miguel, Mafalda Laranjo, Ana Cristina Gonçalves, Anabela Paula, Joana Jorge, Rui Caetano-Oliveira, Maria Inês Sousa, Bárbara Oliveiros, João Ramalho-Santos, Ana Bela Sarmento-Ribeiro, and et al. 2024. "In Vitro Characterization of Reversine-Treated Gingival Fibroblasts and Their Safety Evaluation after In Vivo Transplantation" Pharmaceutics 16, no. 2: 207. https://doi.org/10.3390/pharmaceutics16020207
APA StyleMarto, C. M., Laranjo, M., Gonçalves, A. C., Paula, A., Jorge, J., Caetano-Oliveira, R., Sousa, M. I., Oliveiros, B., Ramalho-Santos, J., Sarmento-Ribeiro, A. B., Marques-Ferreira, M., Cabrita, A., Botelho, M. F., & Carrilho, E. (2024). In Vitro Characterization of Reversine-Treated Gingival Fibroblasts and Their Safety Evaluation after In Vivo Transplantation. Pharmaceutics, 16(2), 207. https://doi.org/10.3390/pharmaceutics16020207












