Antibacterial Activity of AXOTL-13, a Novel Peptide Identified from the Transcriptome of the Salamander Ambystoma mexicanum
Abstract
1. Introduction
2. Materials and Methods
2.1. In Silico Identification of the Potential Antimicrobial Peptides to Be Evaluated
2.1.1. Acquisition of Hypothetical Proteins
2.1.2. Detection of Candidate Peptides in A. mexicanum
2.1.3. Selection of Candidate Peptides
2.1.4. Prediction of the Antimicrobial Activity of the Candidate Peptides
2.1.5. Molecular Confirmation of the Candidate Peptides
2.1.6. Peptide Synthesis
2.2. In Vitro Assays
2.2.1. Evaluation of the Antimicrobial Activity
2.2.2. Evaluation of Hemolytic Activity
2.3. Characterization of the Peptide AXOTL-13
2.3.1. Determination of Physicochemical Properties
2.3.2. Secondary Structure Modeling
2.3.3. Circular Dichroism Spectroscopy
2.3.4. Scanning Electron Microscopy (SEM)
2.4. Bioethical Considerations
2.5. Statistical Analysis
3. Results
3.1. Peptides Identified In Silico in the Transcriptome of Ambystoma mexicanum
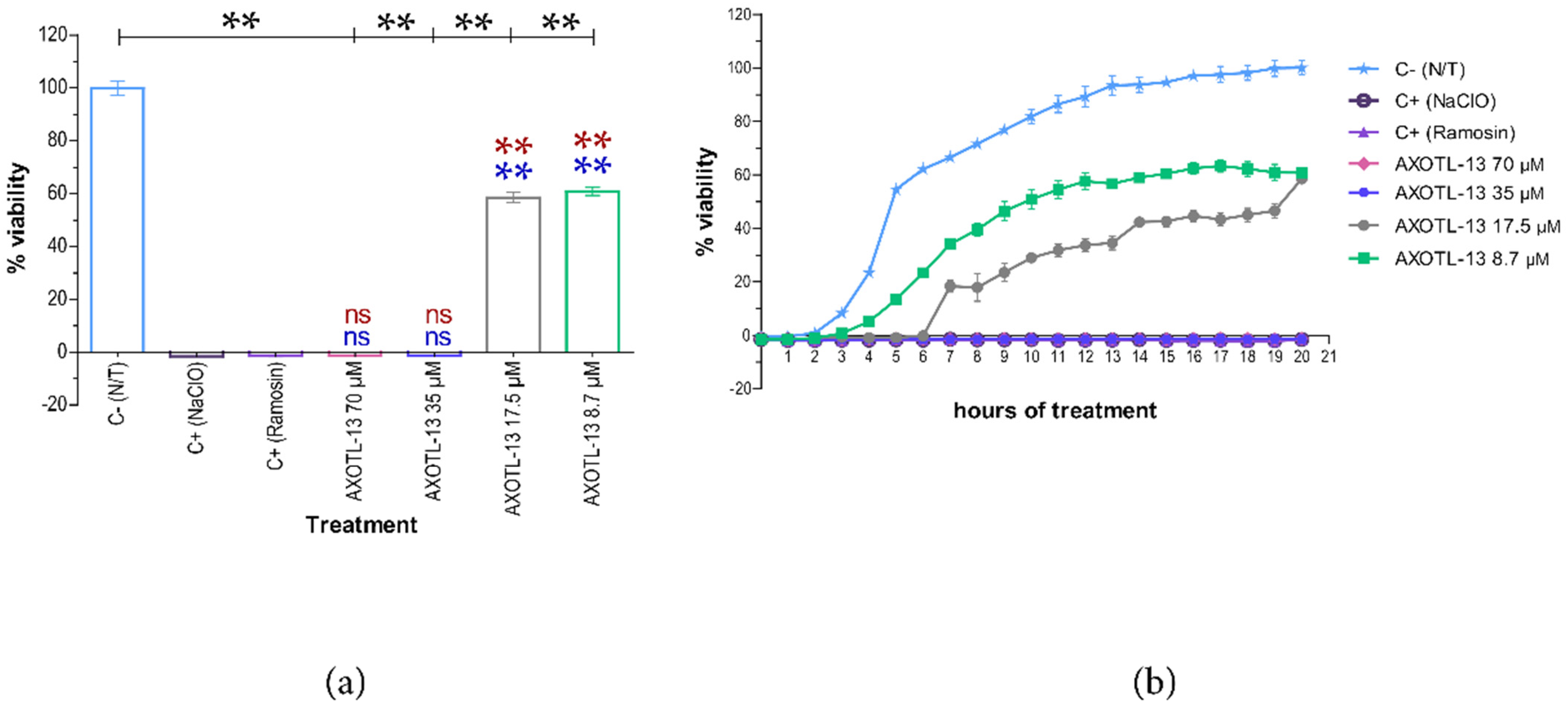
3.2. Antimicrobial Activity
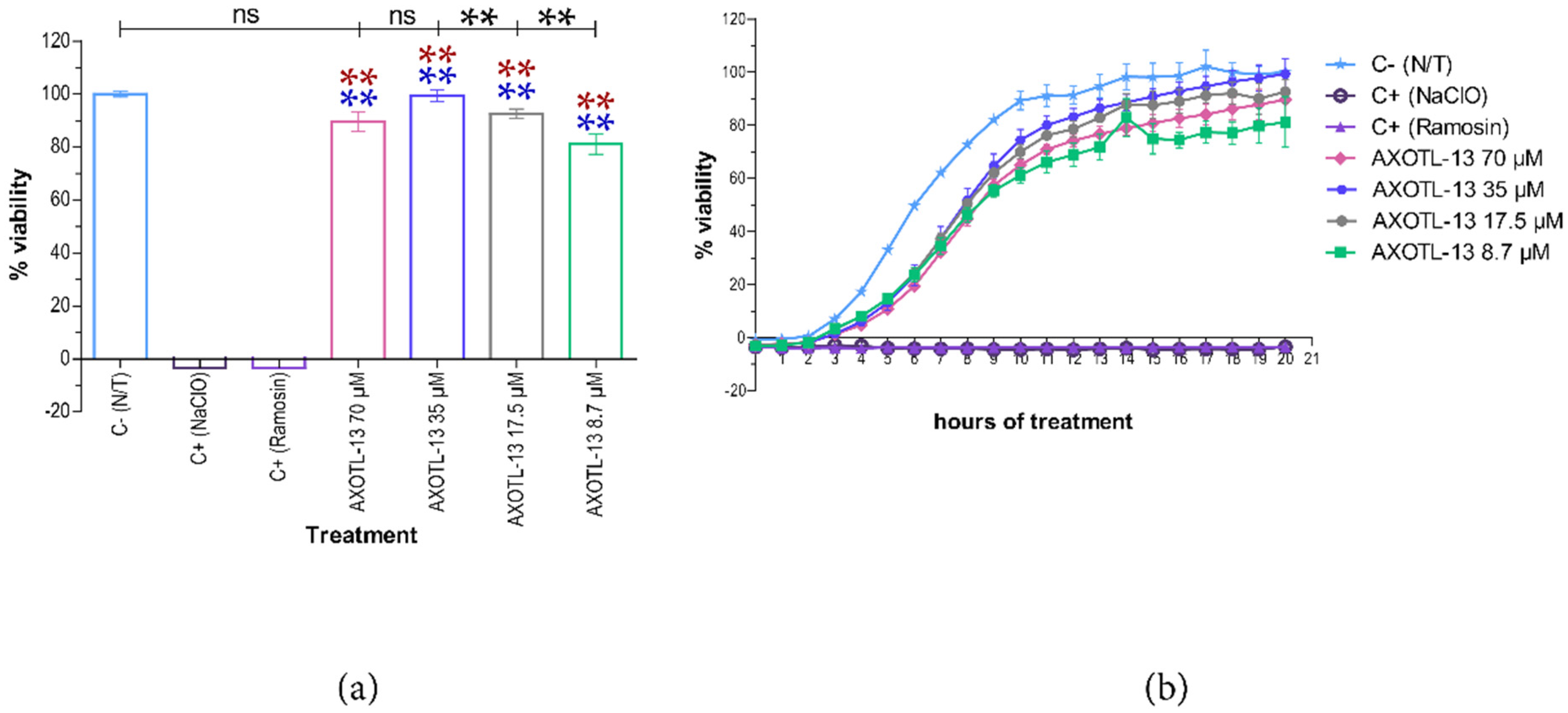
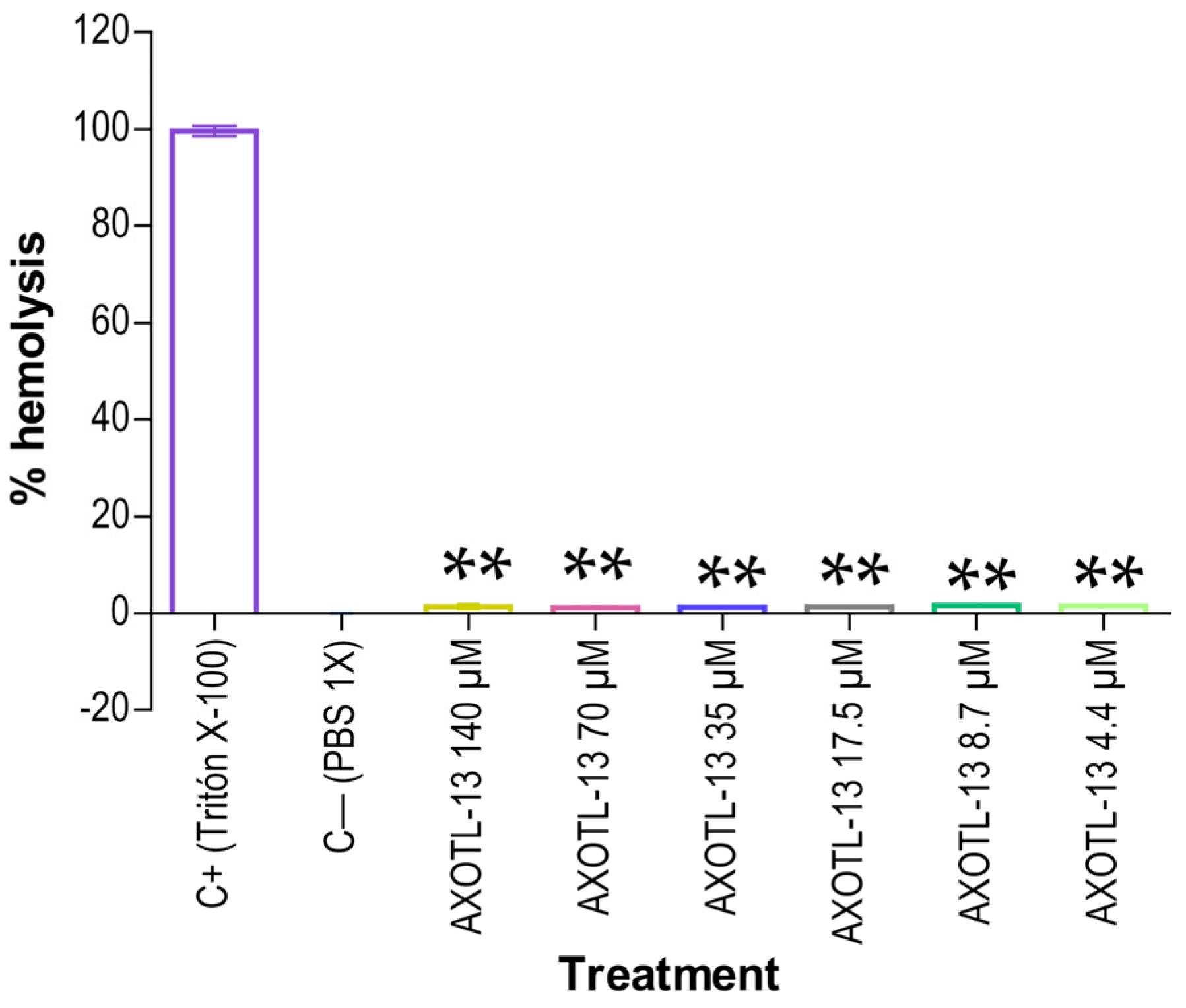
3.3. Hemolytic Activity of Peptide AXOTL-13
3.4. Physicochemical Properties of Peptide AXOTL-13
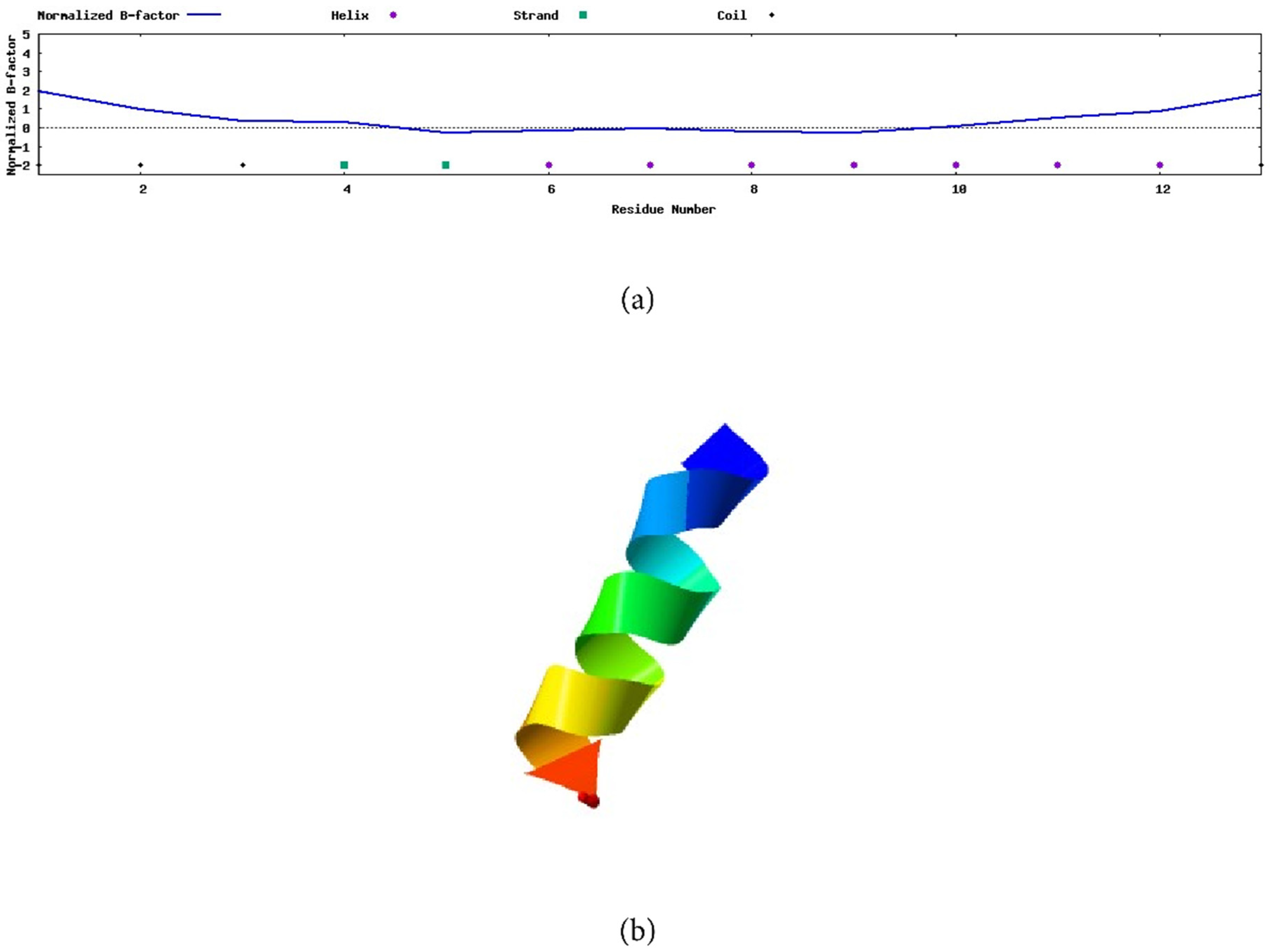
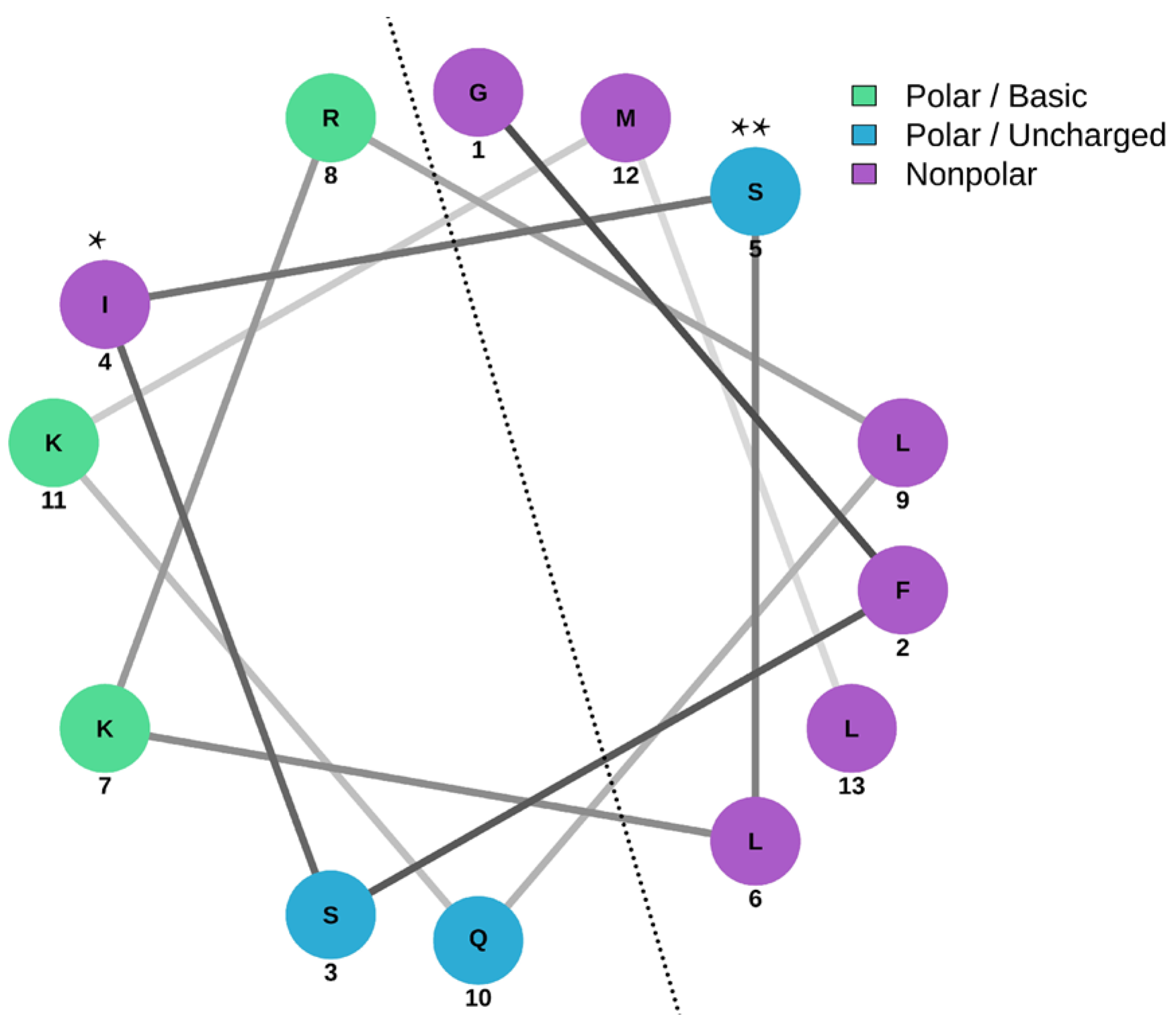
3.5. Secondary Structure of Peptide AXOTL-13
3.6. Circular Dichroism (CD)
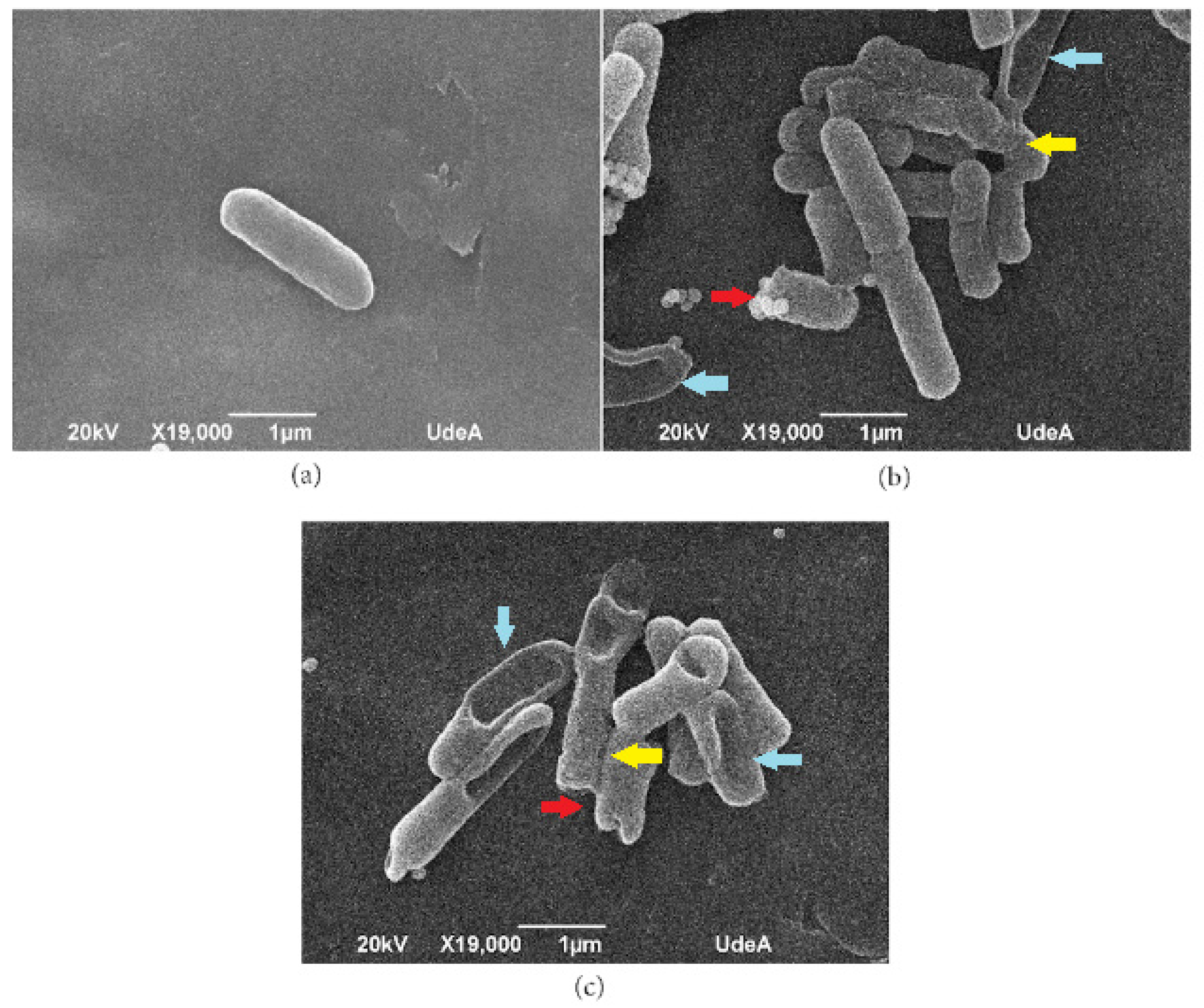
3.7. Scanning Electron Microscopy (SEM)
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bankova, L.; Barrett, N. Innate Immunity. In Middleton’s Allergy: Principles and Practice, 9th ed.; Wesley, A., Ed.; Elsevier: Amsterdam, The Netherlands, 2020; Volume 2, pp. 1–19. [Google Scholar]
- Netea, M.G.; Schlitzer, A.; Placek, K.; Joosten, L.A.B.; Schultze, J.L. Innate and Adaptive Immune Memory: An Evolutionary Continuum in the Host’s Response to Pathogens. Cell Host Microbe 2019, 25, 13–26. [Google Scholar] [CrossRef] [PubMed]
- Lazzaro, B.P.; Zasloff, M.; Rolff, J. Antimicrobial peptides: Application informed by evolution. Science 2020, 368, eaau5480. [Google Scholar] [CrossRef] [PubMed]
- Tennessen, J.A. Molecular evolution of animal antimicrobial peptides: Widespread moderate positive selection. J. Evol. Biol. 2005, 18, 1387–1394. [Google Scholar] [CrossRef] [PubMed]
- Bakare, O.; Gokul, A.; Fadaka, A.O.; Wu, R.; Niekerk, L.A.; Barker, A.M.; Keyster, M.; Klein, A. Plant Antimicrobial Peptides (PAMPs): Features, Applications, Production, Expression, and Challenges. Molecules 2022, 27, 3703. [Google Scholar] [CrossRef] [PubMed]
- Tellez, G.; Castaño, J. Péptidos antimicrobianos. Infectio 2010, 14, 55–67. Available online: www.scielo.org.co/pdf/inf/v14n1/v14n1a07.pdf (accessed on 18 April 2020). [CrossRef]
- Wang, G. Tool developments for structure-function studies of host defense peptides. Protein Pept. Lett. 2007, 14, 57–69. [Google Scholar] [CrossRef]
- Moretta, A.; Scieuzo, C.; Petrone, A.M.; Salvia, R.; Manniello, M.; Franco, A.; Lucchetti, D.; Vassallo, A.; Vogel, H.; Sgambato, A.; et al. Antimicrobial Peptides: A New Hope in Biomedical and Pharmaceutical Fields. Front. Microbiol. 2021, 11, 668632. [Google Scholar] [CrossRef]
- Leite, M.L.; da Cunha, N.B.; Costa, F.F. Antimicrobial peptides, nanotechnology, and natural metabolites as novel approaches for cancer treatment. Pharmacol. Ther. 2018, 183, 160–176. [Google Scholar] [CrossRef]
- Kumar, P.; Kizhakkedathu, J.N.; Straus, S. Antimicrobial Peptides: Diversity, Mechanism of Action and Strategies to Improve the Activity and Biocompatibility In Vivo. Biomolecules 2018, 8, 4. [Google Scholar] [CrossRef]
- Bin, H.; Jiang, X.; Bergen, P.; Zhu, Y. Antimicrobial Peptides: An Update on Classifications Databases. Int. J. Mol. Sci. 2021, 22, 11691. [Google Scholar] [CrossRef]
- World Health Organization. Antimicrobial Resistance. 2021. Available online: www.who.int/es/news-room/fact-sheets/detail/antimicrobial-resistance (accessed on 31 March 2023).
- Rima, M.; Rima, M.; Fajloun, Z.; Sabatier, J.M.; Bechinger, B.; Naas, T. Antimicrobial Peptides: A Potent Alternative to Antibiotics. Antibiotics 2021, 10, 1095. [Google Scholar] [CrossRef] [PubMed]
- Waghu, F.H.; Idicula-Thomas, S. Collection of antimicrobial peptides database and its derivatives: Applications and beyond. Protein Sci. 2020, 29, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Huan, Y.; Kong, Q.; Mou, H.; Yi, H. Antimicrobial Peptides: Classification, Design, Application and Research Progress in Multiple Fields. Front. Microbiol. 2020, 11, 582779. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Li, X.; Wang, Z. APD3: The antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 2016, 44, D1087–D1093. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Li, J.; Anboukaria, H. Comparative transcriptome analyses of seven anurans reveal functions and adaptations of amphibian skin. Sci. Rep. 2016, 6, 24069. [Google Scholar] [CrossRef]
- Cunliffe, R.; Mahida, Y. Expression regulation of antimicrobial peptides in the gastrointestinal tract. J. Leukoc. Biol. 2004, 75, 49–58. [Google Scholar] [CrossRef]
- Chen, J.; Zhang, C.Y.; Ma, L.; Xin, R.; Ding, G.H. Host defense peptide LEAP2 contributes to antimicrobial activity in a mustache toad (Leptobrachium liui). Res Sq. 2022. [Google Scholar] [CrossRef]
- Amiche, M.; Seon, A.A.; Wroblewski, H.; Nicolas, P. Isolation of dermatoxin from frog skin, an antibacterial peptide encoded by a novel member of the dermaseptin genes family. Eur. J. Biochem. 2000, 267, 4583–4592. [Google Scholar] [CrossRef]
- Hao, X.; Yang, H.; Wei, L.; Yang, S.; Zhu, W.; Ma, D.; Yu, H.; Lai, R. Amphibian cathelicidin fills the evolutionary gap of cathelicidin in vertebrate. Amino Acids 2012, 43, 677–685. [Google Scholar] [CrossRef]
- Chiu, K.W.; Lee, Y.C. Cardiac activity of some peptide hormones in the frog, Rana tigrina. Comp. Biochem. Physiol. C Comp. Pharmacol. Toxicol. 1992, 103, 483–487. [Google Scholar] [CrossRef]
- Sheafor, B.; Davidson, E.; Parr, L.; Rollins-Smith, L. Antimicrobial peptide defenses in the salamander, Ambystoma tigrinum, against emerging amphibian pathogens. J. Wildl. Dis. 2008, 44, 226–236. [Google Scholar] [CrossRef] [PubMed]
- Cera-Domínguez, E.; Arenas-Ballesteros, G.; Varela-Rodríguez, L.; Camarillo-Cisneros, J.; Guzman-Pando, A.; Varela-Rodríguez, H. Transcriptional Expression of Bioactive Antimicrobial Peptides with Biomedical Potential in Diverse Organs of the Mexican Axolotl. In XLVI Mexican Conference on Biomedical Engineering; de Jesús Agustín Flores Cuautle, J., Benítez-Mata, B., Salido-Ruiz, R.A., Alonso-Silverio, G.A., Dorantes-Méndez, G., Zúñiga-Aguilar, E., Vélez-Pérez, H.A., Del Hierro-Gutiérrez, E., Mejía-Rodríguez, A.R., Eds.; CNIB 2023; IFMBE Proceedings; Springer: Cham, Switzerland, 2024; Volume 96. [Google Scholar] [CrossRef]
- Helbing, C.C.; Hammond, S.A.; Jackman, S.H.; Houston, S.; Warren, R.L.; Cameron, C.E.; Birol, I. Antimicrobial peptides from Rana [Lithobates] catesbeiana: Gene structure and bioinformatic identification of novel forms from tadpoles. Sci. Rep. 2019, 9, 1529. [Google Scholar] [CrossRef] [PubMed]
- Rollins-Smith, L.A. The role of amphibian antimicrobial peptides in protection of amphibians from pathogens linked to global amphibian declines. Biochim. Biophys. Acta 2009, 1788, 1593–1599. [Google Scholar] [CrossRef]
- Seifert, A.W.; Monaghan, J.R.; Voss, S.R.; Maden, M. Skin Regeneration in Adult Axolotls: A Blueprint for ScarFree Healing in Vertebrates. PLoS ONE 2012, 7, e32875. [Google Scholar] [CrossRef]
- Dwaraka, V.B.; Smith, J.J.; Woodcock, M.R.; Voss, S.R. Comparative transcriptomics of limb regeneration: Identification of conserved expression changes among three species of Ambystoma. Genomics 2019, 111, 1216–1225. [Google Scholar] [CrossRef]
- TransDecoder. Find Coding Regions Within Transcripts. 2018. Available online: https://Github.com/TransDecoder/TransDecoder (accessed on 18 April 2020).
- Pearson, W.R. Finding Protein Nucleotide Similarities with FASTA. Curr. Protoc. Bioinform. 2016, 53, 3.9.1–3.9.25. [Google Scholar] [CrossRef]
- Medina, L.; Guzmán, F.; Álvarez, C.; Delgado, J.P.; Carbonell, M.B. Ramosin: The First Antibacterial Peptide Identified on Bolitoglossa ramosi Colombian Salamander. Pharmaceutics 2022, 14, 2579. [Google Scholar] [CrossRef] [PubMed]
- Waghu, F.H.; Barai, R.S.; Gurung, P.; Idicula-Thomas, S. CAMPR3: A database on sequences, structures and signatures of antimicrobial peptides. Nucleic Acids Res. 2016, 4, 44. [Google Scholar] [CrossRef]
- Carbonell, M.B.; Zapata, J.; Delgado, J.P. Post-amputation reactive oxygen species production is necessary for axolotls limb regeneration. Front. Cell Dev. Biol. 2022, 10, 921520. [Google Scholar] [CrossRef]
- Guzmán, F.; Gauna, A.; Luna, O.; Román, T.; Álvarez, C.; Albericio, F.; Cárdenas, C. The tea-bag protocol for comparison of Fmoc removal reagents in solid-phase peptide synthesis. Amino Acids 2020, 52, 1201–1205. [Google Scholar] [CrossRef]
- Wiegand, I.; Hilpert, K.; Hancock, R.E.W. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 2008, 3, 163–175. [Google Scholar] [CrossRef] [PubMed]
- Methods for Determining Bactericidal Activity of Antimicrobial Agents; Approved Guideline; CLSI Document M26-A; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 1999; Available online: https://clsi.org/standards/products/microbiology/documents/m26/ (accessed on 15 July 2020).
- Hernández, Y.; Mollineda, N.; González, D.M. Estudio de la Actividad Hemolítica de los Posibles Taninos Extraídos a Partir de la Boldoa purpurascens. Cav REDVET 2006, 7, 1–5. Available online: www.redalyc.org/articulo.oa?id=63617167024 (accessed on 15 May 2020).
- NovoPro Bioscience Inc. 2014. Available online: www.novoprolabs.com (accessed on 8 May 2023).
- Yang, J.; Yan, R.; Roy, A.; Xu, D.; Poisson, J.; Zhang, Y. The I-TASSER Suite: Protein structure and function prediction. Nat. Methods 2015, 12, 7–8. [Google Scholar] [CrossRef] [PubMed]
- Roy, A.; Kucukural, A.; Zhang, Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Protoc. 2010, 5, 725–738. [Google Scholar] [CrossRef]
- Zhang, Y. I-TASSER server for protein 3D structure prediction. BMC Bioinform. 2008, 9, 40. [Google Scholar] [CrossRef]
- Mól, A.R.; Castro, M.S.; Fontes, W. NetWheels: A web application to create high quality peptide helical wheel and net projections. bioRxiv 2018. [Google Scholar] [CrossRef]
- Santana, P.A.; Álvarez, C.A.; Guzmán, F.; Mercado, L. Development of a Sandwich ELISA for Quantifying Hepcidin in Rainbow Trout. Fish. Shellfish. Immunol. 2013, 35, 748–755. [Google Scholar] [CrossRef]
- Santana, P.; Salinas, N.; Álvarez, C.; Mercado, L.; Guzmán, F. Alpha-Helical Domain from IL-8 of Salmonids: Mechanism of Action and Identification of a Novel Antimicrobial Function. Biochem. Biophys. Res. Commun. 2018, 498, 803–809. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2023; Available online: www.R-project.org/ (accessed on 25 January 2023).
- GraphPad Prism Versión 5.0.0 for Windows; GraphPad Software: San Diego, CA, USA, 2007; Available online: www.graphpad.com (accessed on 23 March 2023).
- Pei, J.; Jiang, L. Antimicrobial peptide from mucus of Andrias davidianus: Screening and purification by magnetic cell membrane separation technique. Int. J. Antimicrob. Agents 2017, 50, 41–46. [Google Scholar] [CrossRef]
- Pei, J.; Feng, Z.; Ren, T.; Sun, H.; Han, H.; Jin, W.; Dang, J.; Tao, Y. Purification, characterization and application of a novel antimicrobial peptide from Andrias davidianus blood. Lett. Appl. Microbiol. 2018, 66, 38–43. [Google Scholar] [CrossRef]
- Yakovlev, I.A.; Lysøe, E.; Heldal, I.; Steen, H.; Hagen, S.B.; Clarke, J.L. Transcriptome profiling and in silico detection of the antimicrobial peptides of red king crab Paralithodes camtschaticus. Sci. Rep. 2020, 10, 12679. [Google Scholar] [CrossRef] [PubMed]
- Kim, I.W.; Lee, J.H.; Subramaniyam, S.; Yun, E.Y.; Kim, I.; Park, J.; Hwang, J.S. De Novo Transcriptome Analysis and Detection of Antimicrobial Peptides of the American Cockroach Periplaneta americana (Linnaeus). PLoS ONE 2016, 11, e0155304. [Google Scholar] [CrossRef] [PubMed]
- Rozek, T.; Wegener, K.L.; Bowie, J.H.; Olver, I.N.; Carver, J.A.; Wallace, J.C.; Tyler, M.J. The antibiotic and anticancer active aurein peptides from the Australian Bell Frogs Litoria aurea and Litoria raniformis. Eur. J. Biochem. 2000, 267, 5330–5341. [Google Scholar] [CrossRef] [PubMed]
- Rollins-Smith, L.A. The importance of antimicrobial peptides (AMPs) in amphibian skin defense. Dev. Comp. Immunol. 2023, 142, 104657. [Google Scholar] [CrossRef]
- Demircan, T.; İlhan, A.E.; Aytürk, N.; Yıldırım, B.; Öztürk, G.; Keskin, İ. A histological atlas of the tissues and organs of neotenic and metamorphosed axolotl. Acta Histochem. 2016, 118, 746–759. [Google Scholar] [CrossRef]
- Fellah, J.S.; Vaulot, D.; Tournefier, A.; Charlemagne, J. Ontogeny of immunoglobulin expression in the Mexican axolotl. Development 1989, 107, 253–263. [Google Scholar] [CrossRef]
- Risso, A. Leukocyte antimicrobial peptides: Multifunctional effector molecules of innate immunity. J. Leukoc. Biol. 2000, 68, 785–792. [Google Scholar] [CrossRef]
- Mozsolits, H.; Wirth, H.J.; Werkmeister, J.; Aguilar, M.I. Analysis of antimicrobial peptide interactions with hybrid bilayer membrane systems using surface plasmon resonance. Biochim. Biophys. Acta 2001, 1512, 64–76. [Google Scholar] [CrossRef]
- Dos Santos Cabrera, M.P.; Rangel, M.; Ruggiero Neto, J.; Konno, K. Chemical and Biological Characteristics of Antimicrobial α-Helical Peptides Found in Solitary Wasp Venoms and Their Interactions with Model Membranes. Toxins 2019, 11, 559. [Google Scholar] [CrossRef]
- Dathe, M.; Wieprecht, T. Structural features of helical antimicrobial peptides: Their potential to modulate activity on model membranes and biological cells. Biochim. Biophys. Acta 1999, 1462, 71–87. [Google Scholar] [CrossRef]
- Mathews, C.; Van Holde, K.; Appling, D.; Anthony-Cahill, S. Introducción a las proteínas: Nivel primario de la estructura proteica. In Bioquímica, 4th ed.; Martin, M., Ed.; Pearson Educación S.A.: Madrid, Spain, 2013; pp. 136–159. [Google Scholar]
- Ko, S.J.; Kim, M.K.; Bang, J.K.; Seo, C.H.; Luchian, T.; Park, Y. Macropis fulvipes Venom component Macropin Exerts its Antibacterial and Anti-Biofilm Properties by Damaging the Plasma Membranes of Drug Resistant Bacteria. Sci. Rep. 2017, 7, 16580. [Google Scholar] [CrossRef] [PubMed]
- Che, Q.; Zhou, Y.; Yang, H.; Li, J.; Xu, X.; Lai, R. A novel antimicrobial peptide from amphibian skin secretions of Odorrana grahami. Peptides 2008, 29, 529–535. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhang, L.; Ma, C.; Zhang, Y.; Xi, X.; Wang, L.; Zhou, M.; Burrows, J.; Chen, T. A novel antimicrobial peptide, Ranatuerin-2PLx, showing therapeutic potential in inhibiting proliferation of cancer cells. Biosci. Rep. 2018, 38, BSR20180710. [Google Scholar] [CrossRef] [PubMed]
- Zelezetsky, I.; Tossi, A. Alpha-helical antimicrobial peptides—Using a sequence template to guide structure–activity relationship studies. Biochim. Biophys. Acta 2006, 1758, 1436–1449. [Google Scholar] [CrossRef]

| Parameter | Inclusion Criteria | Justification |
|---|---|---|
| Identity | Hits with a percentage lower than 100% | A 100% identity affirms that it is the same peptide as in the database. |
| Similarity | Hits with a percentage higher than 80% | The higher the similarity between two sequences, the greater the likelihood that they have a shared biological function. |
| Length | 5–20 amino acids | Synthesizing short peptides can affect their stability due to the fewer peptide bonds. With a higher number of amino acids, more complications arise since a higher number of coupling steps is required. |
| Activity | Antimicrobial | Alignment with a peptide sequence whose antimicrobial activity has been previously tested. |
| Peptide | Sequence | Molecular Weight (g/mol) | Net Charge | (%) Hydrophobicity | Similar Peptide from APD3 | (%) Similarity with APD3 Peptide |
|---|---|---|---|---|---|---|
| 4766 | LIGGQLGGLIKAL | 1252.8 | 1 | 53 | AP02274 | 92.3 |
| 4767 | YITGLIAPILKSL | 1401.9 | 1 | 53 | AP02275 | 92.3 |
| 4768 | VLGSILGALKAI | 1124.6 | 1 | 66 | AP00521 | 91.7 |
| 4769 | LHPLIGRVIGGVI | 1343.9 | 1.25 | 53 | AP00094 | 92.3 |
| 4770 | GFSISLKRLQKML | 1521.1 | 3 | 46 | AP01956 | 92.3 |
| 4771 | LLDTIGKIFGSLL | 1389.9 | 0 | 53 | AP00875 | 100 |
| Features | Description |
|---|---|
| Length | 13 amino acid residues |
| Chemical formula | C69H121N19O17S |
| Extinction coefficient | 0 |
| Isoelectric point | 11.65 |
| Net charge | +3 * |
| Grand average of hydropathicity (GRAVY) | 0.22 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Córdoba, L.; López, D.; Mejía, M.; Guzmán, F.; Beltrán, D.; Carbonell, B.; Medina, L. Antibacterial Activity of AXOTL-13, a Novel Peptide Identified from the Transcriptome of the Salamander Ambystoma mexicanum. Pharmaceutics 2024, 16, 1445. https://doi.org/10.3390/pharmaceutics16111445
Córdoba L, López D, Mejía M, Guzmán F, Beltrán D, Carbonell B, Medina L. Antibacterial Activity of AXOTL-13, a Novel Peptide Identified from the Transcriptome of the Salamander Ambystoma mexicanum. Pharmaceutics. 2024; 16(11):1445. https://doi.org/10.3390/pharmaceutics16111445
Chicago/Turabian StyleCórdoba, Laura, Daniela López, Mariana Mejía, Fanny Guzmán, Dina Beltrán, Belfran Carbonell, and Laura Medina. 2024. "Antibacterial Activity of AXOTL-13, a Novel Peptide Identified from the Transcriptome of the Salamander Ambystoma mexicanum" Pharmaceutics 16, no. 11: 1445. https://doi.org/10.3390/pharmaceutics16111445
APA StyleCórdoba, L., López, D., Mejía, M., Guzmán, F., Beltrán, D., Carbonell, B., & Medina, L. (2024). Antibacterial Activity of AXOTL-13, a Novel Peptide Identified from the Transcriptome of the Salamander Ambystoma mexicanum. Pharmaceutics, 16(11), 1445. https://doi.org/10.3390/pharmaceutics16111445






