Remote Activation of Mechanotransduction via Integrin Alpha-5 via Aptamer-Conjugated Magnetic Nanoparticles Promotes Osteogenesis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. MNPs: Ligand Labeling
2.3. Loading Efficiency
2.4. Cell MNP Labeling
2.5. Magnetic Field Gradients
2.6. qRT-PCR
2.7. Flow Cytometry
2.8. ALP Activity
2.9. Western Blotting
2.10. Alizarin Red Staining
2.11. Statistical Analysis
3. Results and Discussion
3.1. Cell Tagging with MNP
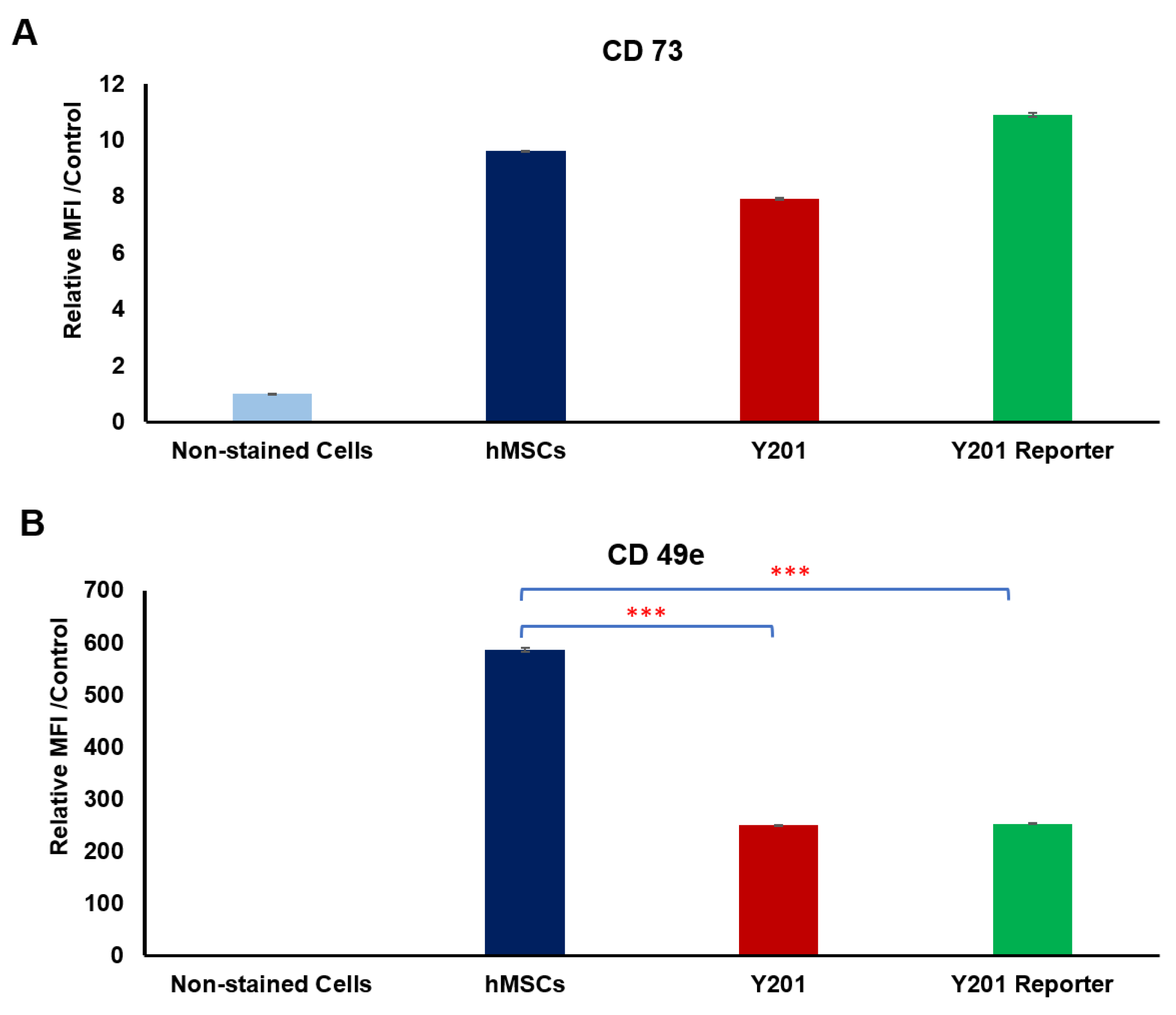
3.2. Receptor Selection for Osteogenesis
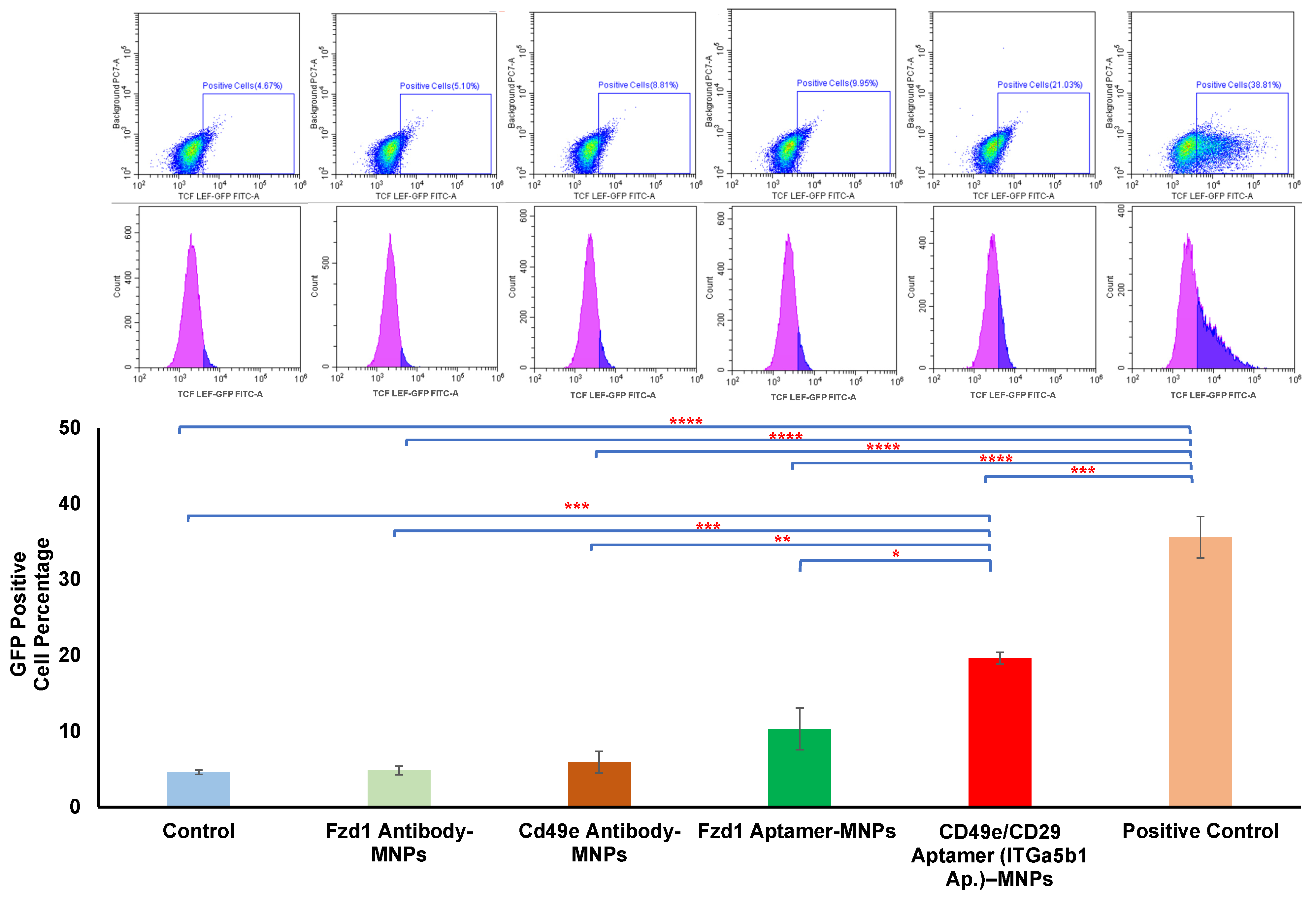
3.3. Comparison between Aptamer and Antibody
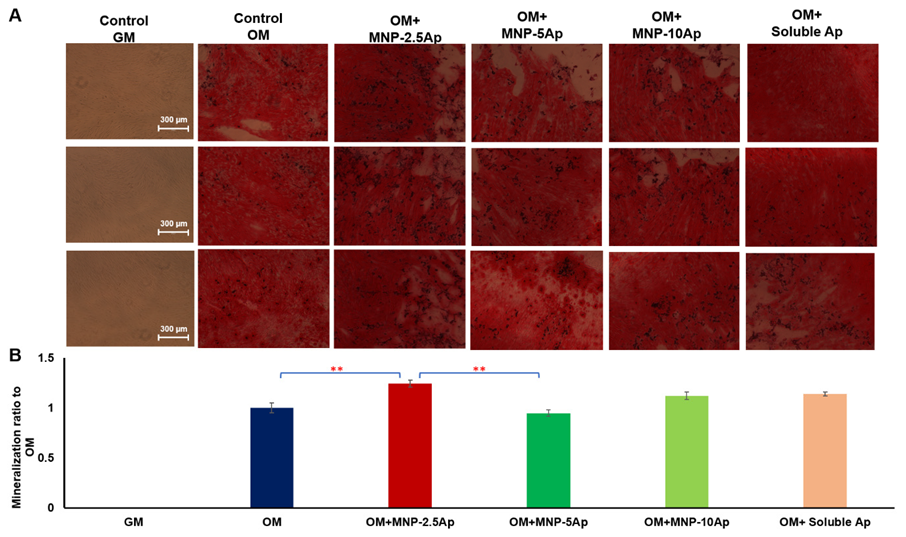
3.4. Optimization of Aptamer Concentration
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hutchings, G.; Moncrieff, L.; Dompe, C.; Janowicz, K.; Sibiak, R.; Bryja, A.; Jankowski, M.; Mozdziak, P.; Bukowska, D.; Antosik, P. Bone regeneration, reconstruction and use of osteogenic cells; from basic knowledge, animal models to clinical trials. J. Clin. Med. 2020, 9, 139. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Wu, X.; Lei, W.; Pang, L.; Wan, C.; Shi, Z.; Zhao, L.; Nagy, T.R.; Peng, X.; Hu, J. TGF-β1–induced migration of bone mesenchymal stem cells couples bone resorption with formation. Nat. Med. 2009, 15, 757–765. [Google Scholar] [CrossRef] [PubMed]
- Paspaliaris, V.; Kolios, G. Stem cells in osteoporosis: From biology to new therapeutic approaches. Stem Cells Int. 2019, 2019, 1730978. [Google Scholar] [CrossRef] [PubMed]
- Perez, J.R.; Kouroupis, D.; Li, D.J.; Best, T.M.; Kaplan, L.; Correa, D. Tissue engineering and cell-based therapies for fractures and bone defects. Front. Bioeng. Biotechnol. 2018, 6, 105. [Google Scholar] [CrossRef] [PubMed]
- Maciel, G.B.M.; Maciel, R.M.; Danesi, C.C. Bone cells and their role in physiological remodeling. Mol. Biol. Rep. 2023, 50, 2857–2863. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, C.A.; Komori, H.; Maruyama, Z.; Miyazaki, T.; Kawasaki, K.; Furuichi, T.; Fukuyama, R.; Mori, M.; Yamana, K.; Nakamura, K. SP7 inhibits osteoblast differentiation at a late stage in mice. PLoS ONE 2012, 7, e32364. [Google Scholar] [CrossRef]
- Jing, Z.; Liang, Z.; Yang, L.; Du, W.; Yu, T.; Tang, H.; Li, C.; Wei, W. Bone formation and bone repair: The roles and crosstalk of osteoinductive signaling pathways. Process Biochem. 2022, 118, 252–262. [Google Scholar] [CrossRef]
- Schupbach, D.; Comeau-Gauthier, M.; Harvey, E.; Merle, G. Wnt modulation in bone healing. Bone 2020, 138, 115491. [Google Scholar] [CrossRef]
- Leucht, P.; Lee, S.; Yim, N. Wnt signaling and bone regeneration: Can’t have one without the other. Biomaterials 2019, 196, 46–50. [Google Scholar] [CrossRef]
- Maeda, K.; Kobayashi, Y.; Koide, M.; Uehara, S.; Okamoto, M.; Ishihara, A.; Kayama, T.; Saito, M.; Marumo, K. The regulation of bone metabolism and disorders by Wnt signaling. Int. J. Mol. Sci. 2019, 20, 5525. [Google Scholar] [CrossRef]
- Sugimoto, A.; Miyazaki, A.; Kawarabayashi, K.; Shono, M.; Akazawa, Y.; Hasegawa, T.; Ueda-Yamaguchi, K.; Kitamura, T.; Yoshizaki, K.; Fukumoto, S. Piezo type mechanosensitive ion channel component 1 functions as a regulator of the cell fate determination of mesenchymal stem cells. Sci. Rep. 2017, 7, 17696. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Pan, Y.; Guo, S.; Sun, L.; Zhang, C.; Wang, L. The roles of mechanosensitive ion channels and associated downstream MAPK signaling pathways in PDLC mechanotransduction. Mol. Med. Rep. 2020, 21, 2113–2122. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Chi, S.; Li, Y.; Ling, S.; Tan, Y.; Xu, Y.; Jiang, F.; Li, J.; Liu, C.; Zhong, G. The mechanosensitive Piezo1 channel is required for bone formation. eLife 2019, 8, e47454. [Google Scholar] [CrossRef] [PubMed]
- Unnithan, A.R.; Sasikala, A.R.K.; Shrestha, B.K.; Lincoln, A.; Thomson, T.; El Haj, A.J. Remotely Actuated Magnetic Nanocarpets for Bone Tissue Engineering: Non-Invasive Modulation of Mechanosensitive Ion Channels for Enhanced Osteogenesis. Adv. Funct. Mater. 2022, 32, 2201311. [Google Scholar] [CrossRef]
- Rotherham, M.; Nahar, T.; Broomhall, T.J.; Telling, N.D.; El Haj, A.J. Remote magnetic actuation of cell signalling for tissue engineering. Curr. Opin. Biomed. Eng. 2022, 2022, 100410. [Google Scholar] [CrossRef]
- Pankhurst, Q.A.; Thanh, N.T.K.; Jones, S.K.; Dobson, J. Progress in applications of magnetic nanoparticles in biomedicine. J. Phys. D Appl. Phys. 2009, 42, 224001. [Google Scholar] [CrossRef]
- Hu, B.; Rotherham, M.; Farrow, N.; Roach, P.; Dobson, J.; El Haj, A.J. Immobilization of wnt fragment peptides on magnetic nanoparticles or synthetic surfaces regulate wnt signaling kinetics. Int. J. Mol. Sci. 2022, 23, 10164. [Google Scholar] [CrossRef]
- Bonnemay, L.; Hoffmann, C.; Gueroui, Z. Remote control of signaling pathways using magnetic nanoparticles, Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2015, 7, 342–354. [Google Scholar] [CrossRef]
- Henstock, J.R.; Rotherham, M.; El Haj, A.J. Magnetic ion channel activation of TREK1 in human mesenchymal stem cells using nanoparticles promotes osteogenesis in surrounding cells. J. Tissue Eng. 2018, 9, 2041731418808695. [Google Scholar] [CrossRef]
- Gonçalves, A.I.; Rotherham, M.; Markides, H.; Rodrigues, M.T.; Reis, R.L.; Gomes, M.E.; El Haj, A.J. Triggering the activation of Activin A type II receptor in human adipose stem cells towards tenogenic commitment using mechanomagnetic stimulation. Nanomed. Nanotechnol. Biol. Med. 2018, 14, 1149–1159. [Google Scholar] [CrossRef]
- Cartmell, S.H.; Dobson, J.; Verschueren, S.B.; El Haj, A.J. Development of magnetic particle techniques for long-term culture of bone cells with intermittent mechanical activation. IEEE Trans. Nanobiosci. 2002, 99, 92–97. [Google Scholar] [CrossRef] [PubMed]
- Rotherham, M.; Henstock, J.R.; Qutachi, O.; El Haj, A.J. Remote regulation of magnetic particle targeted Wnt signaling for bone tissue engineering, Nanomedicine Nanotechnology. Biol. Med. 2018, 14, 173–184. [Google Scholar]
- Byun, J. Recent progress and opportunities for nucleic acid aptamers. Life 2021, 11, 193. [Google Scholar] [CrossRef] [PubMed]
- Ramaswamy, V.; Monsalve, A.; Sautina, L.; Segal, M.S.; Dobson, J.; Allen, J.B. DNA aptamer assembly as a vascular endothelial growth factor receptor agonist. Nucleic Acid Ther. 2015, 25, 227–234. [Google Scholar] [CrossRef] [PubMed]
- Dobson, J.P.; Allen, J. Magnetic particle conjugates and methods of activating cell signaling. U.S. Patent 11,730,971, 8 September 2020. [Google Scholar]
- James, S.; Fox, J.; Afsari, F.; Lee, J.; Clough, S.; Knight, C.; Ashmore, J.; Ashton, P.; Preham, O.; Hoogduijn, M. Multiparameter analysis of human bone marrow stromal cells identifies distinct immunomodulatory and differentiation-competent subtypes. Stem Cell Rep. 2015, 4, 1004–1015. [Google Scholar] [CrossRef] [PubMed]
- Saleh, F.; Carstairs, A.; Etheridge, S.L.; Genever, P. Real-time analysis of endogenous Wnt signalling in 3D mesenchymal stromal cells. Stem Cells Int. 2016, 2016, 7132529. [Google Scholar] [CrossRef]
- Rotherham, M.; El Haj, A.J. Remote activation of the Wnt/β-catenin signalling pathway using functionalised magnetic particles. PLoS ONE 2015, 10, e0121761. [Google Scholar] [CrossRef]
- Vlashi, R.; Zhang, X.; Wu, M.; Chen, G. Wnt signaling: Essential roles in osteoblast differentiation, bone metabolism and therapeutic implications for bone and skeletal disorders. Genes Dis. 2023, 10, 1291–1317. [Google Scholar] [CrossRef]
- Etheridge, S.L.; Spencer, G.J.; Heath, D.J.; Genever, P.G. Expression profiling and functional analysis of wnt signaling mechanisms in mesenchymal stem cells. Stem Cells 2004, 22, 849–860. [Google Scholar] [CrossRef]
- Lojk, J.; Marc, J. Roles of non-canonical Wnt signalling pathways in bone biology. Int. J. Mol. Sci. 2021, 22, 10840. [Google Scholar] [CrossRef]
- Saal, H.M.; Prows, C.A.; Guerreiro, I.; Donlin, M.; Knudson, L.; Sund, K.L.; Chang, C.-F.; Brugmann, S.A.; Stottmann, R.W. A mutation in FRIZZLED2 impairs Wnt signaling and causes autosomal dominant omodysplasia. Hum. Mol. Genet. 2015, 24, 3399–3409. [Google Scholar] [CrossRef] [PubMed]
- Liang, D.; Wang, X.; Mittal, A.; Dhiman, S.; Hou, S.-Y.; Degenhardt, K.; Astrof, S. Mesodermal expression of integrin α5β1 regulates neural crest development and cardiovascular morphogenesis. Dev. Biol. 2014, 395, 232–244. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zheng, F.; Song, R.; Zhuang, L.; Yang, M.; Suo, J.; Li, L. Integrins in the regulation of mesenchymal stem cell differentiation by mechanical signals. Stem Cell Rev. Rep. 2022, 18, 126–141. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Kaneko, S.; Soma, K. Expression of integrinα5β1, focal adhesion kinase and integrin-linked kinase in rat condylar cartilage during mandibular lateral displacement. Arch. Oral Biol. 2008, 53, 701–708. [Google Scholar] [CrossRef] [PubMed]
- McIlhenny, S.E.; Hager, E.S.; Grabo, D.J.; DiMatteo, C.; Shapiro, I.M.; Tulenko, T.N.; DiMuzio, P.J. Linear shear conditioning improves vascular graft retention of adipose-derived stem cells by upregulation of the α5β1 integrin. Tissue Eng. Part A 2010, 16, 245–255. [Google Scholar] [CrossRef] [PubMed]
- Wang, N. Instant integrin mechanosensing. Nat. Mater. 2017, 16, 1173–1174. [Google Scholar] [CrossRef]
- Kong, F.; García, A.J.; Mould, A.P.; Humphries, M.J.; Zhu, C. Demonstration of catch bonds between an integrin and its ligand. J. Cell Biol. 2009, 185, 1275–1284. [Google Scholar] [CrossRef]
- Fromigué, O.; Brun, J.; Marty, C.; Da Nascimento, S.; Sonnet, P.; Marie, P.J. Peptide-based activation of alpha5 integrin for promoting osteogenesis. J. Cell. Biochem. 2012, 113, 3029–3038. [Google Scholar] [CrossRef]
- Sonowal, H.; Kumar, A.; Bhattacharyya, J.; Gogoi, P.K.; Jaganathan, B.G. Inhibition of actin polymerization decreases osteogeneic differentiation of mesenchymal stem cells through p38 MAPK pathway. J. Biomed. Sci. 2013, 20, 71. [Google Scholar] [CrossRef]
- Du, J.; Zu, Y.; Li, J.; Du, S.; Xu, Y.; Zhang, L.; Jiang, L.; Wang, Z.; Chien, S.; Yang, C. Extracellular matrix stiffness dictates Wnt expression through integrin pathway. Sci. Rep. 2016, 6, 20395. [Google Scholar] [CrossRef]
- Subjakova, V.; Oravczova, V.; Hianik, T. Polymer nanoparticles and nanomotors modified by DNA/RNA aptamers and antibodies in targeted therapy of cancer. Polymers 2021, 13, 341. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Lai, B.S.; Juhas, M. Recent advances in aptamer discovery and applications. Molecules 2019, 24, 941. [Google Scholar] [CrossRef]
- Li, K.; Liu, S.; Li, J.; Yi, D.; Shao, D.; Hu, T.; Zheng, X. Manganese supplementation of orthopedic implants: A new strategy for enhancing integrin-mediated cellular responses. Biomater. Sci. 2023, 11, 3893–3905. [Google Scholar] [CrossRef] [PubMed]
- Giudice, V.; Mensitieri, F.; Izzo, V.; Filippelli, A.; Selleri, C. Aptamers and antisense oligonucleotides for diagnosis and treatment of hematological diseases. Int. J. Mol. Sci. 2020, 21, 3252. [Google Scholar] [CrossRef]
- Jones, E.; McGonagle, D. Human bone marrow mesenchymal stem cells in vivo. Rheumatology 2008, 47, 126–131. [Google Scholar] [CrossRef] [PubMed]
- Mildmay-White, A.; Khan, W. Cell surface markers on adipose-derived stem cells: A systematic review. Curr. Stem Cell Res. Ther. 2017, 12, 484–492. [Google Scholar] [CrossRef]
- Huang, W.; Yang, S.; Shao, J.; Li, Y.-P. Signaling and transcriptional regulation in osteoblast commitment and differentiation. Front. Biosci. J. Virtual Libr. 2007, 12, 3068. [Google Scholar] [CrossRef] [PubMed]
- Marupanthorn, K.; Tantrawatpan, C.; Kheolamai, P.; Tantikanlayaporn, D.; Manochantr, S. MicroRNA treatment modulates osteogenic differentiation potential of mesenchymal stem cells derived from human chorion and placenta. Sci. Rep. 2021, 11, 7670. [Google Scholar] [CrossRef]
- Wülfing, C.; Dovedi, S.J. For optimal antibody effectiveness, sometimes less is more. Nature 2023, 2, 416–418. [Google Scholar] [CrossRef]
- Hajiali, H.; Shahgasempour, S.; Naimi-Jamal, M.R.; Peirovi, H. Electrospun PGA/gelatin nanofibrous scaffolds and their potential application in vascular tissue engineering. Int. J. Nanomed. 2011, 9, 2133–2141. [Google Scholar] [CrossRef]
- Kilian, K.A.; Mrksich, M. Directing stem cell fate by controlling the affinity and density of ligand–receptor interactions at the biomaterials interface. Angew. Chem. Int. Ed. 2012, 51, 4891–4895. [Google Scholar] [CrossRef] [PubMed]
- Mann, B.K.; Tsai, A.T.; Scott-Burden, T.; West, J.L. Modification of surfaces with cell adhesion peptides alters extracellular matrix deposition. Biomaterials 1999, 20, 2281–2286. [Google Scholar] [CrossRef] [PubMed]
- Neff, J.A.; Tresco, P.A.; Caldwell, K.D. Surface modification for controlled studies of cell–ligand interactions. Biomaterials 1999, 20, 2377–2393. [Google Scholar] [CrossRef] [PubMed]
- Palecek, S.P.; Loftus, J.C.; Ginsberg, M.H.; Lauffenburger, D.A.; Horwitz, A.F. Integrin-ligand binding properties govern cell migration speed through cell-substratum adhesiveness. Nature 1997, 385, 537–540. [Google Scholar] [CrossRef] [PubMed]
- Hlavacek, W.S.; Posner, R.G.; Perelson, A.S. Steric effects on multivalent ligand-receptor binding: Exclusion of ligand sites by bound cell surface receptors. Biophys. J. 1999, 76, 3031–3043. [Google Scholar] [CrossRef] [PubMed]
- Kilinc, D.; Dennis, C.L.; Lee, G.U. Bio-Nano-Magnetic Materials for Localized Mechanochemical Stimulation of Cell Growth and Death. Adv. Mater. 2016, 28, 5672–5680. [Google Scholar] [CrossRef]
- Evans, A.; Calderwood, D.A. Forces and bond dynamics in cell adhesion. Science 2007, 316, 1148–1153. [Google Scholar] [CrossRef]
- Na, S.; Collin, O.; Chowdhury, F.; Tay, B.; Ouyang, M.; Wang, Y.; Wang, N. Rapid signal transduction in living cells is a unique feature of mechanotransduction. Proc. Natl. Acad. Sci. USA 2008, 105, 6626–6631. [Google Scholar] [CrossRef]




| MNP-Uncoated | MNP-Piezo1 | MNP-Fzd1 | MNP-Cd49e | MNP-Fzd2 | |
|---|---|---|---|---|---|
| Size (nm) | 263.6 ± 17.6 | 253.5 ± 23.17 | 273.7 ± 28.6 | 265.1 ± 15.1 | 270.8 ± 27.4 |
| Zeta (mV) | −15.4 ± 0.6 | −11.1 ± 0.4 | −11.5 ± 0.6 | −12.3 ± 0.1 | −11.8 ± 0.8 |
| MNP-Uncoated | MNP-2.5 Ap | MNP-5 Ap | MNP-10 Ap | MNP-20 Ap | |
|---|---|---|---|---|---|
| Size (nm) | 263.6 ± 17.6 | 258.1 ± 4.6 | 265 ± 3.5 | 276.8 ± 12.5 | 277.7 ± 16.9 |
| Zeta (mV) | −15.4 ± 0.6 | −15.8 ± 0.2 | −15.7 ± 0.7 | −16.2 ± 0.3 | −16.0 ± 0.4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hajiali, H.; Rotherham, M.; El Haj, A.J. Remote Activation of Mechanotransduction via Integrin Alpha-5 via Aptamer-Conjugated Magnetic Nanoparticles Promotes Osteogenesis. Pharmaceutics 2024, 16, 21. https://doi.org/10.3390/pharmaceutics16010021
Hajiali H, Rotherham M, El Haj AJ. Remote Activation of Mechanotransduction via Integrin Alpha-5 via Aptamer-Conjugated Magnetic Nanoparticles Promotes Osteogenesis. Pharmaceutics. 2024; 16(1):21. https://doi.org/10.3390/pharmaceutics16010021
Chicago/Turabian StyleHajiali, Hadi, Michael Rotherham, and Alicia J. El Haj. 2024. "Remote Activation of Mechanotransduction via Integrin Alpha-5 via Aptamer-Conjugated Magnetic Nanoparticles Promotes Osteogenesis" Pharmaceutics 16, no. 1: 21. https://doi.org/10.3390/pharmaceutics16010021
APA StyleHajiali, H., Rotherham, M., & El Haj, A. J. (2024). Remote Activation of Mechanotransduction via Integrin Alpha-5 via Aptamer-Conjugated Magnetic Nanoparticles Promotes Osteogenesis. Pharmaceutics, 16(1), 21. https://doi.org/10.3390/pharmaceutics16010021






