Novel Golden Lipid Nanoparticles with Small Interference Ribonucleic Acid for Substrate Reduction Therapy in Fabry Disease
Abstract
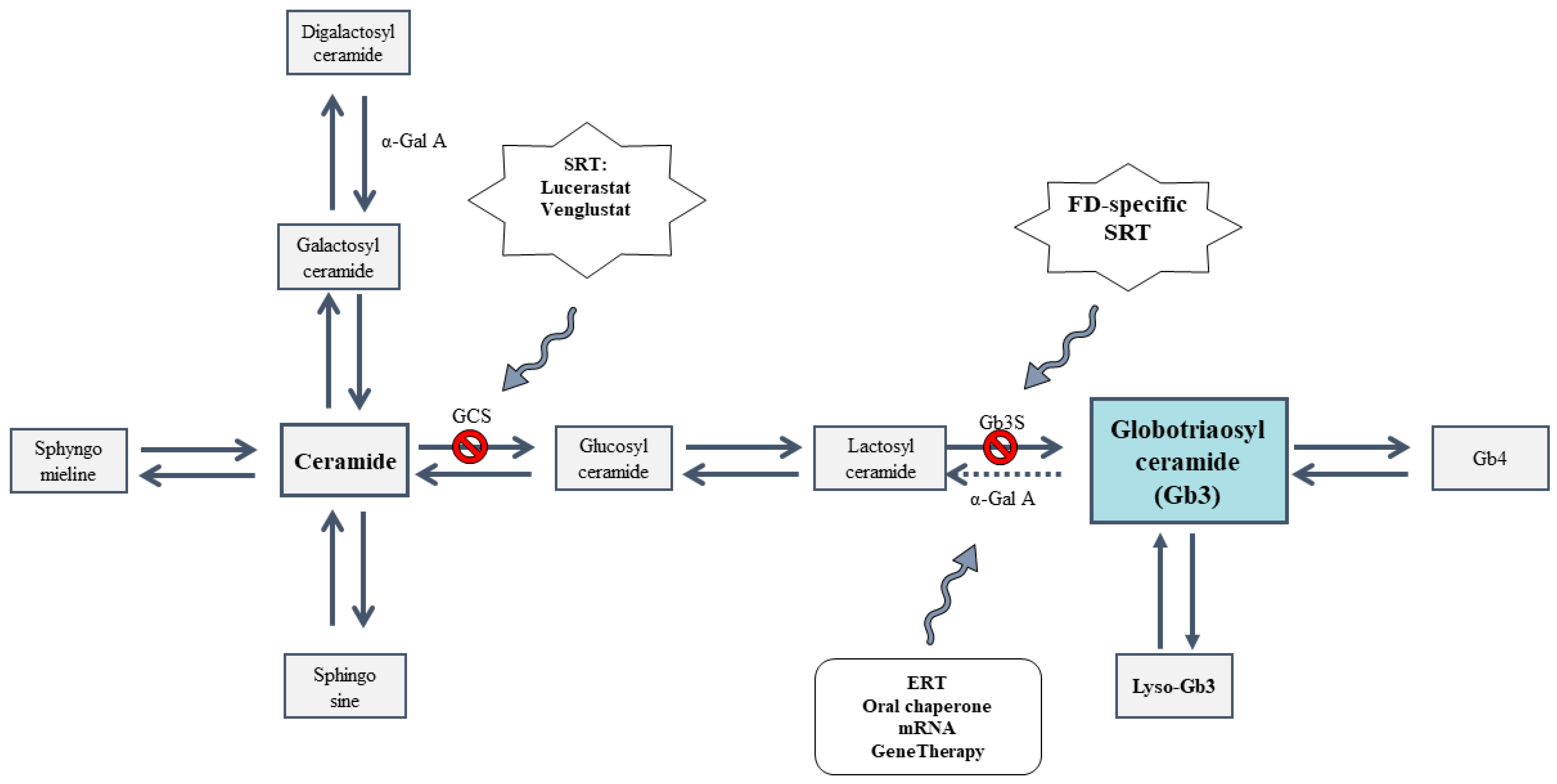
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Preparation of SLNs and Vectors
2.3. Size, Polidispersity Index and ζ-Potential Measurement
2.4. Stability Study of siRNA-Based Vectors: Size, PDI and ζ-Potential
2.5. Morphology Studies of siRNA Vectors
2.5.1. Transmission Electron Microscopy (TEM) Images
2.5.2. Cryo-Transmission Electron Microscopy (Cryo-TEM) Images
2.6. Agarose Gel Electrophoresis Assay
2.7. In Vitro Studies in IMFE-1 Cells
2.7.1. Cellular Uptake
2.7.2. Intracellular Disposition of the siRNA and GNs
2.7.3. Cell Viability
2.7.4. Silencing Efficacy
2.7.5. Gb3S Enzyme Expression
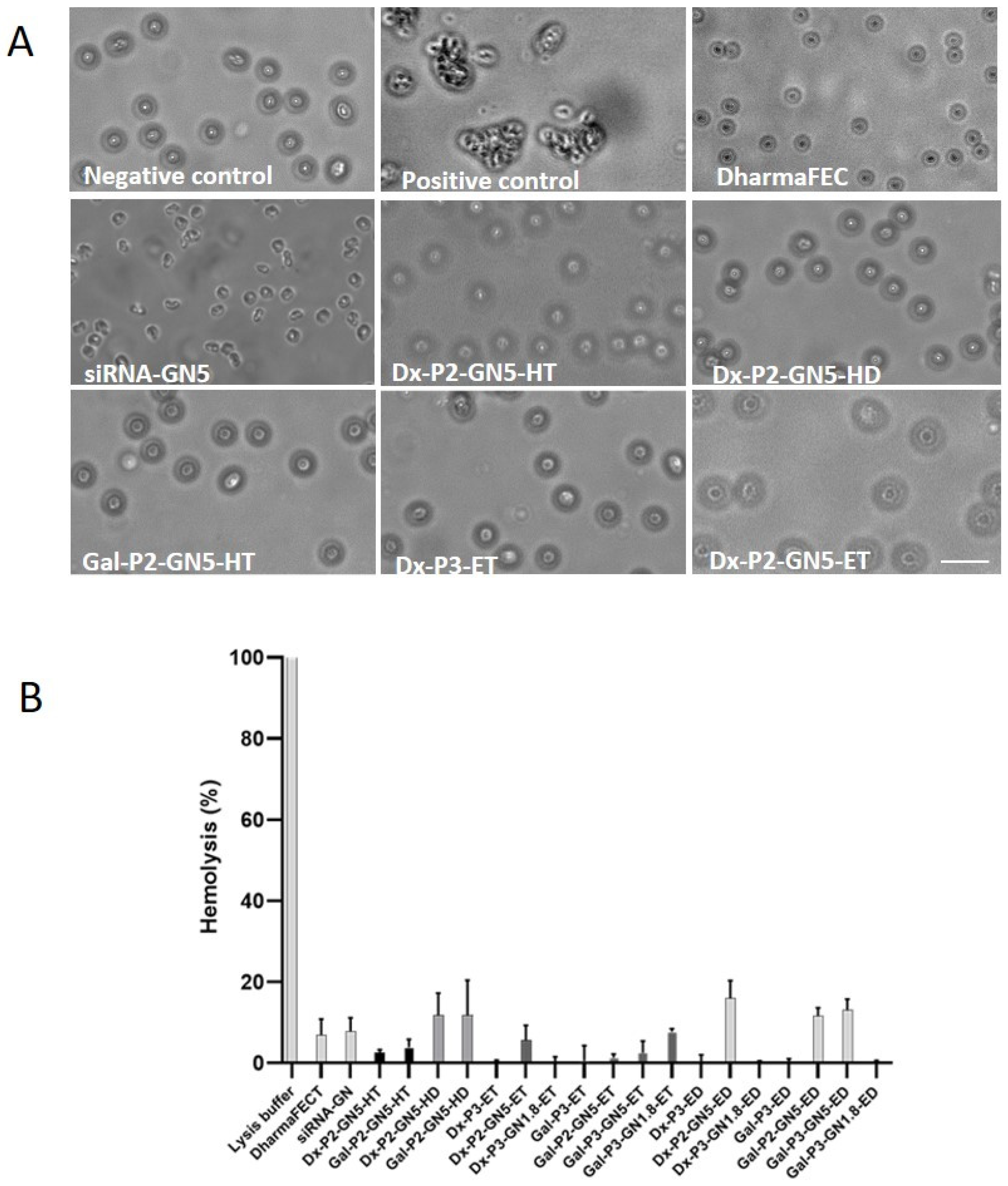
2.8. Interaction with Erythrocytes: Hemolysis and Hemagglutination
2.9. Data Analysis and Statics
3. Results and Discussion
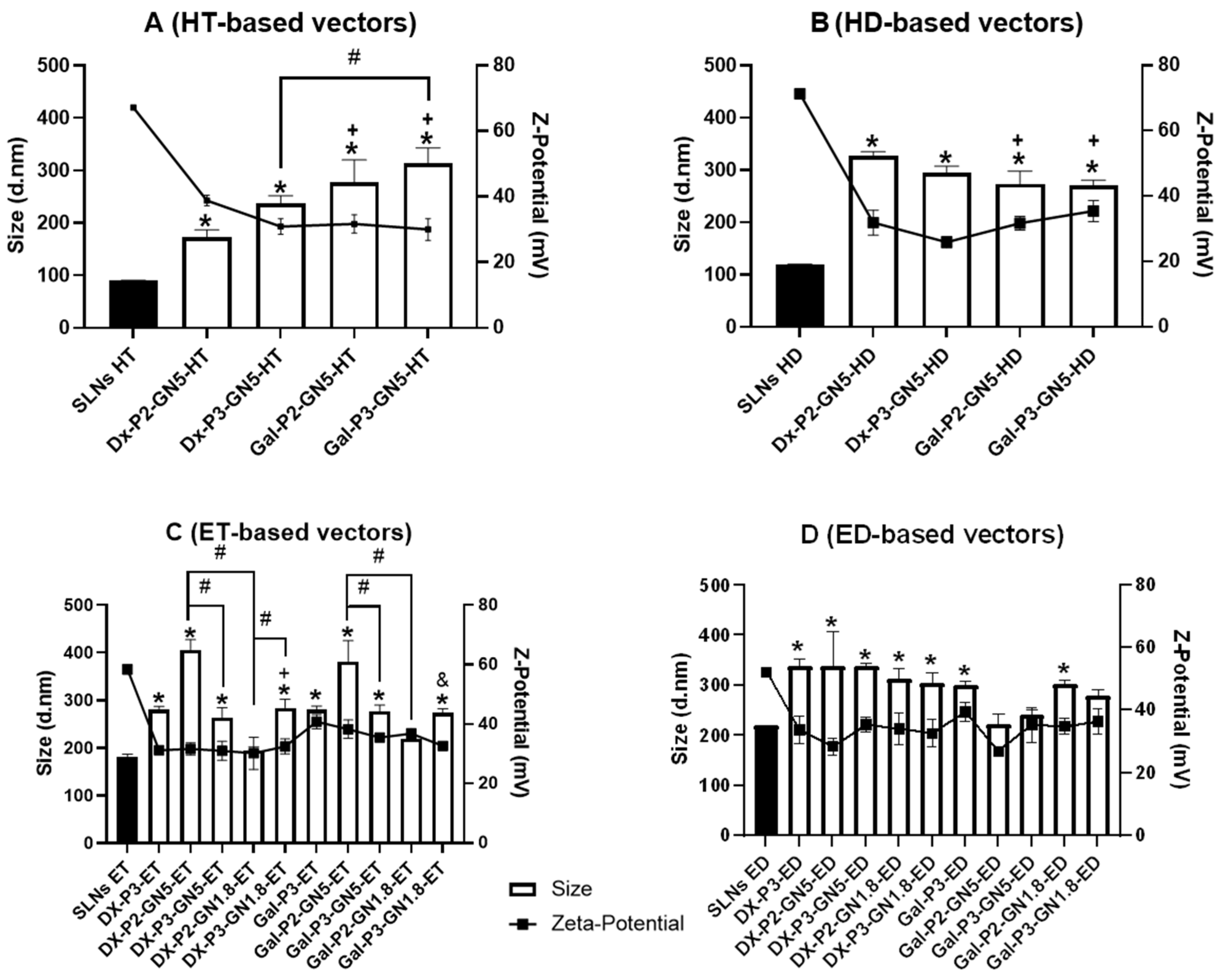
3.1. Size, PDI and ζ-Potential of the SLNs and Vectors
3.2. Stability Study of siRNA-Based Vectors: Size, PDI and ζ-Potential
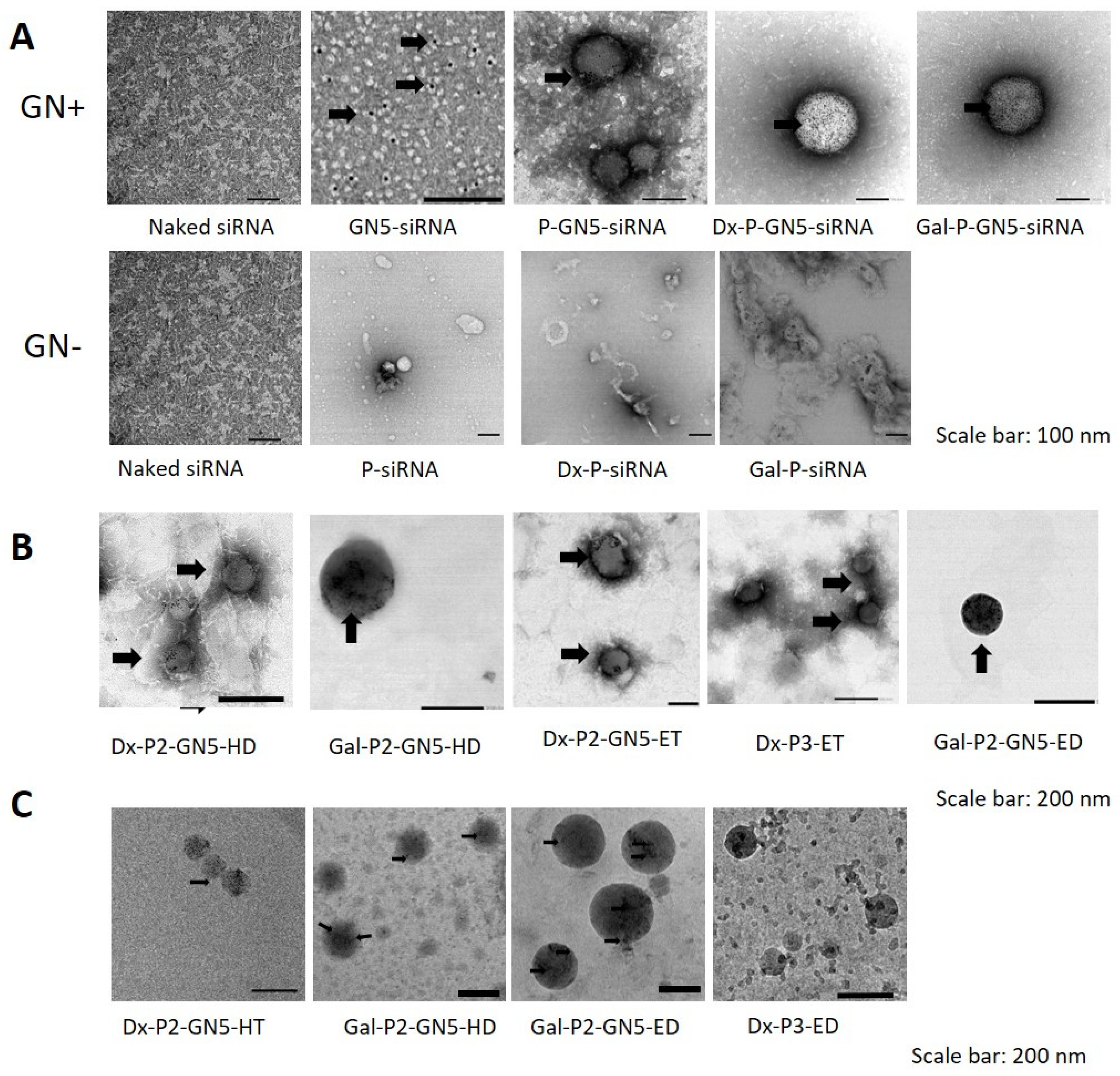
3.3. Morphology Studies of siRNA Vectors
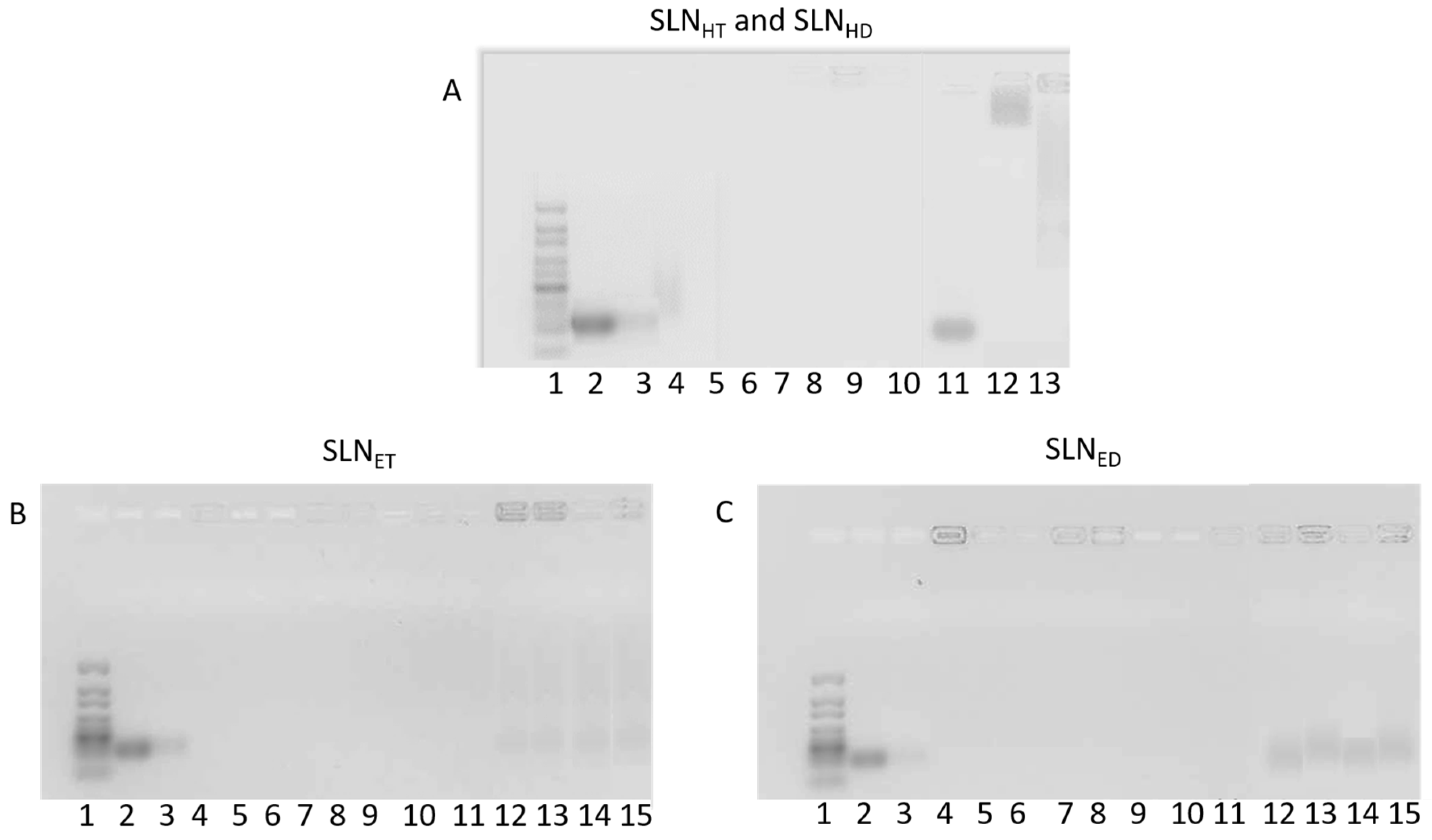
3.4. Agarose Gel Electrophoresis Assay
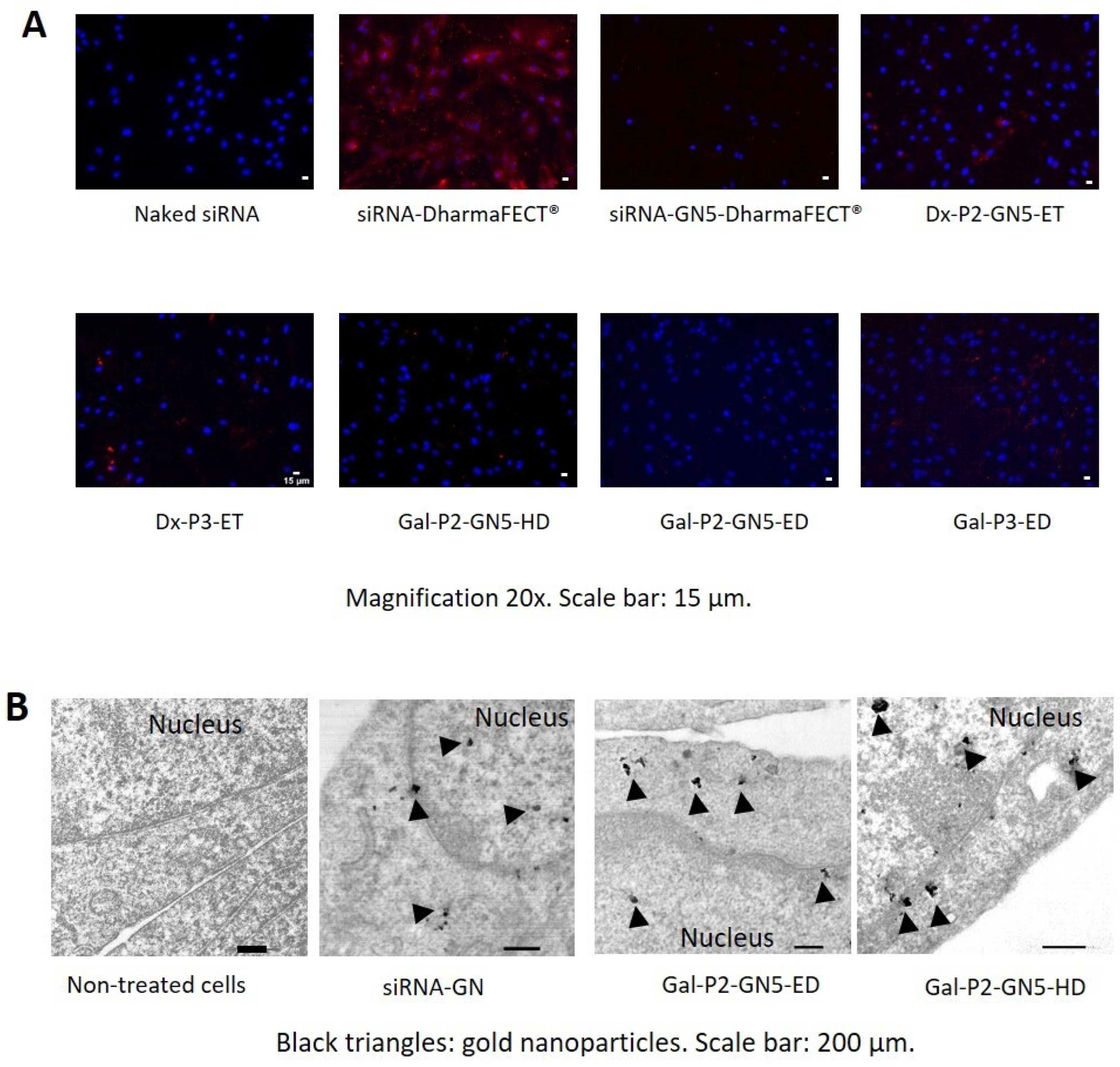
3.5. Cellular Uptake
3.6. Intracellular Disposition of the siRNA and GNs
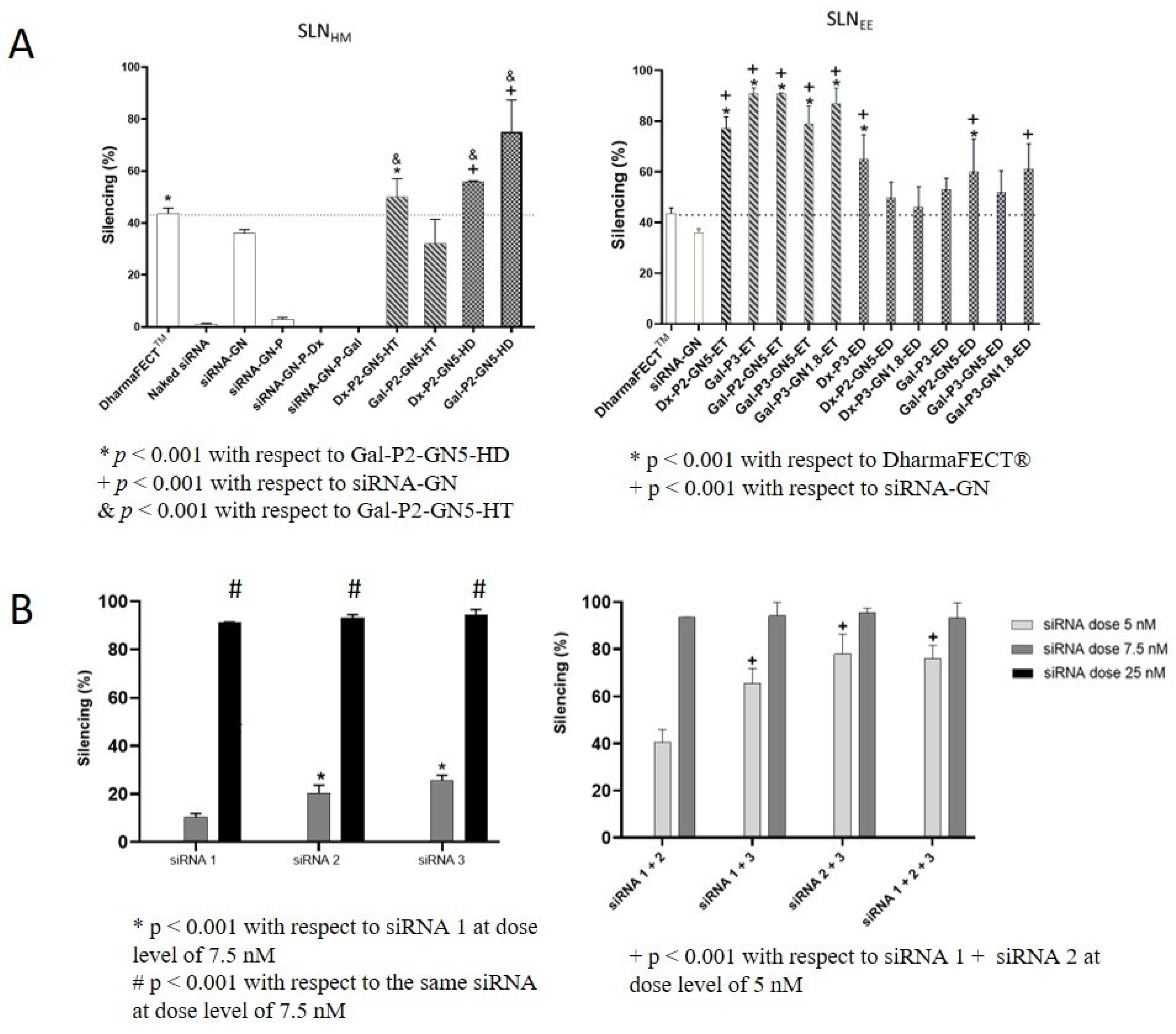
3.7. Silencing Efficacy
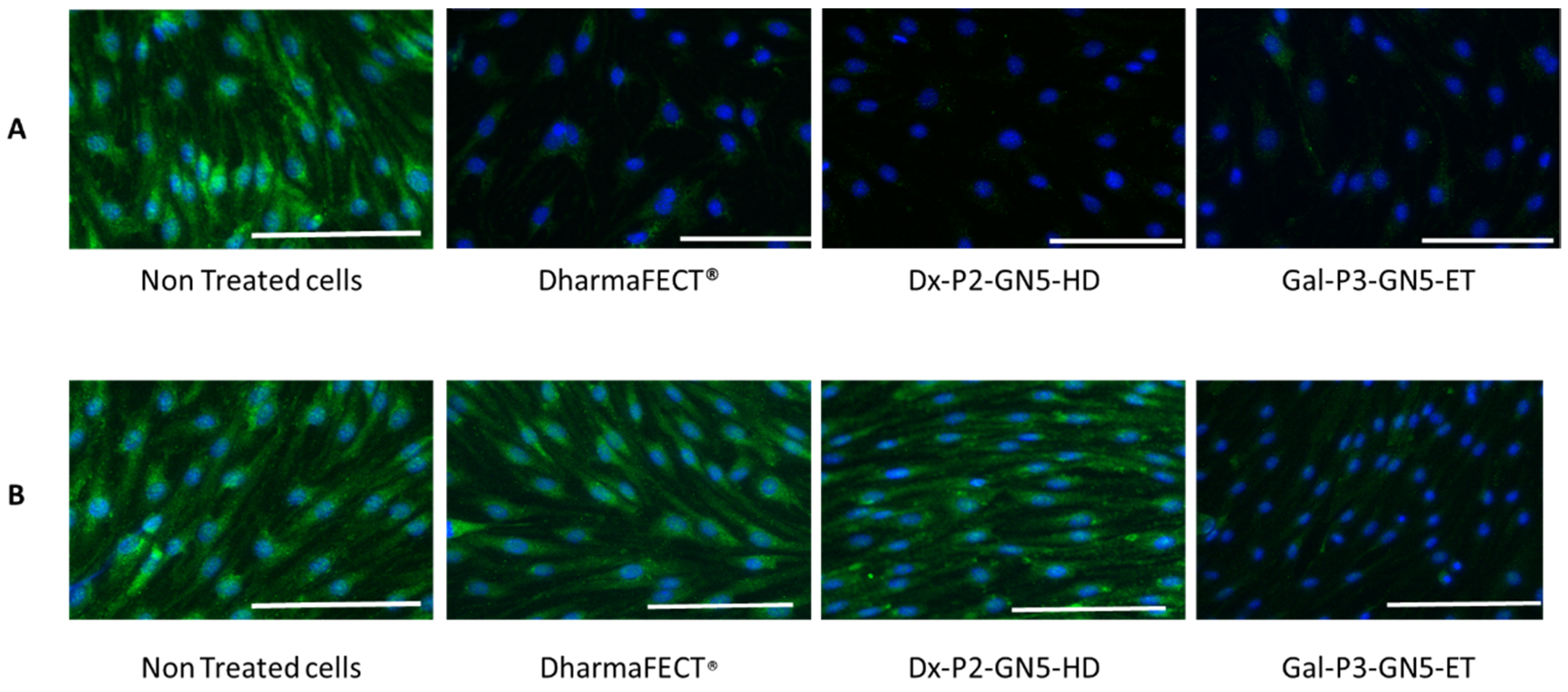
3.8. Gb3S Enzyme Expression
3.9. Cell Viability
3.10. Interaction with Erythrocytes: Hemolysis and Hemagglutination
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Arends, M.; Wanner, C.; Hughes, D.; Mehta, A.; Oder, D.; Watkinson, O.T.; Elliott, P.M.; Linthorst, G.E.; Wijburg, F.A.; Biegstraaten, M.; et al. Characterization of Classical and Nonclassical Fabry Disease: A Multicenter Study. J. Am. Soc. Nephrol. 2017, 28, 1631–1641. [Google Scholar] [CrossRef]
- Tuttolomondo, A.; Simonetta, I.; Riolo, R.; Todaro, F.; Di Chiara, T.; Miceli, S.; Pinto, A. Pathogenesis and Molecular Mechanisms of Anderson–Fabry Disease and Possible New Molecular Addressed Therapeutic Strategies. Int. J. Mol. Sci. 2021, 22, 88. [Google Scholar] [CrossRef]
- Azevedo, O.; Gago, M.F.; Miltenberger-Miltenyi, G.; Sousa, N.; Cunha, D. Fabry Disease Therapy: State-of-the-Art and Current Challenges. Int. J. Mol. Sci. 2021, 22, 206. [Google Scholar] [CrossRef]
- Hendriksz, C.J.; Harmatz, P.; Giugliani, R.; Roberts, J.; Arul, G.S. Risks of Long-Term Port Use in Enzyme Replacement Therapy for Lysosomal Storage Disorders. Mol. Genet. Metab. Rep. 2018, 15, 71–73. [Google Scholar] [CrossRef] [PubMed]
- Müntze, J.; Lau, K.; Cybulla, M.; Brand, E.; Cairns, T.; Lorenz, L.; Üçeyler, N.; Sommer, C.; Wanner, C.; Nordbeck, P. Patient Reported Quality of Life and Medication Adherence in Fabry Disease Patients Treated with Migalastat: A Prospective, Multicenter Study. Mol. Genet. Metab. 2023, 138, 106981. [Google Scholar] [CrossRef]
- Germain, D.P.; Hughes, D.A.; Nicholls, K.; Bichet, D.G.; Giugliani, R.; Wilcox, W.R.; Feliciani, C.; Shankar, S.P.; Ezgu, F.; Amartino, H.; et al. Treatment of Fabry’s Disease with the Pharmacologic Chaperone Migalastat. N. Engl. J. Med. 2016, 375, 545–555. [Google Scholar] [CrossRef] [PubMed]
- Kant, S.; Atta, M.G. Therapeutic Advances in Fabry Disease: The Future Awaits. Biomed. Pharmacother. 2020, 131, 110779. [Google Scholar] [CrossRef]
- Felis, A.; Whitlow, M.; Kraus, A.; Warnock, D.G.; Wallace, E. Current and Investigational Therapeutics for Fabry Disease. Kidney Int. Rep. 2020, 5, 407–413. [Google Scholar] [CrossRef]
- Wanner, C.; Kimonis, V.; Politei, J.; Warnock, D.G.; Üçeyler, N.; Frey, A.; Cornelisse, P.; Hughes, D. Understanding and Modifying Fabry Disease: Rationale and Design of a Pivotal Phase 3 Study and Results from a Patient-Reported Outcome Validation Study. Mol. Genet. Metab. Rep. 2022, 31, 100862. [Google Scholar] [CrossRef] [PubMed]
- Deegan, P.B.; Goker-Alpan, O.; Geberhiwot, T.; Hopkin, R.J.; Lukina, E.; Tylki-Szymanska, A.; Zaher, A.; Sensinger, C.; Gaemers, S.J.M.; Modur, V.; et al. Venglustat, an Orally Administered Glucosylceramide Synthase Inhibitor: Assessment over 3 Years in Adult Males with Classic Fabry Disease in an Open-Label Phase 2 Study and Its Extension Study. Mol. Genet. Metab. 2022, 138, 106963. [Google Scholar] [CrossRef]
- Miller, J.J.; Kanack, A.J.; Dahms, N.M. Progress in the Understanding and Treatment of Fabry Disease. Biochim. Biophys. Acta—Gen. Subj. 2020, 1864, 129437. [Google Scholar] [CrossRef] [PubMed]
- Friedrich, M.; Aigner, A. Therapeutic SiRNA: State-of-the-Art and Future Perspectives. BioDrugs 2022, 36, 549–571. [Google Scholar] [CrossRef] [PubMed]
- Urits, I.; Swanson, D.; Swett, M.C.; Patel, A.; Berardino, K.; Amgalan, A.; Berger, A.A.; Kassem, H.; Kaye, A.; Viswanath, O. A Review of Patisiran (ONPATTRO®) for the Treatment of Polyneuropathy in People with Hereditary Transthyretin Amyloidosis. Neurol. Ther. 2020, 9, 301–315. [Google Scholar] [CrossRef] [PubMed]
- Yadav, D.N.; Ali, M.S.; Thanekar, A.M.; Pogu, S.V.; Rengan, A.K. Recent Advancements in the Design of Nanodelivery Systems of SiRNA for Cancer Therapy. Mol. Pharm. 2022, 19, 4506–4526. [Google Scholar] [CrossRef] [PubMed]
- Del Pozo-Rodríguez, A.; Solinís, M.Á.; Rodríguez-Gascón, A. Applications of Lipid Nanoparticles in Gene Therapy. Eur. J. Pharm. Biopharm. 2016, 109, 184–193. [Google Scholar] [CrossRef]
- Rodríguez-Castejón, J.; Alarcia-Lacalle, A.; Gómez-Aguado, I.; Vicente-Pascual, M.; Aspiazu, M.Á.S.; Del Pozo-Rodríguez, A.; Rodríguez-Gascón, A. α-Galactosidase A Augmentation by Non-Viral Gene Therapy: Evaluation in Fabry Disease Mice. Pharmaceutics 2021, 13, 771. [Google Scholar] [CrossRef]
- Del Pozo-Rodríguez, A.; Rodríguez-Gascón, A.; Gómez-Aguado, I.; Vicente-Pascual, M.; Solinís, M.Á. Golden Lipid Nanoparticles for Gene Therapy. Patent PCT/EP2020/087608 (A1), 22 December 2020. [Google Scholar]
- Rodríquez-Gascón, A.; Solinís, M.Á.; del Pozo-Rodríguez, A.; Delgado, D.; Pedraz, J.L. Lipid Nanoparticles for Gene Therapy. U.S. Patent 20120183589, 19 July 2012. [Google Scholar]
- Rodríguez-Gascón, A.; Solinís, M.Á.; del Pozo-Rodríguez, A.; Delgado, D.; Fernández, E. Lipid Nanoparticles for Treating Ocular Diseases. Patent P201031897, 21 December 2010. [Google Scholar]
- Kanu, G.A.; Parambath, J.B.M.; Odeh, R.O.A.; Mohamed, A.A. Gold Nanoparticle-Mediated Gene Therapy. Cancers 2022, 14, 5366. [Google Scholar] [CrossRef]
- Gómez-Aguado, I.; Rodríguez-Castejón, J.; Beraza-Millor, M.; Vicente-Pascual, M.; Rodríguez-Gascón, A.; Garelli, S.; Battaglia, L.; Del Pozo-Rodríguez, A.; Solinís, M.Á. Mrna-Based Nanomedicinal Products to Address Corneal Inflammation by Interleukin-10 Supplementation. Pharmaceutics 2021, 13, 1472. [Google Scholar] [CrossRef]
- Del Pozo-Rodríguez, A.; Delgado, D.; Solinís, M.A.; Gascón, A.R.; Pedraz, J.L. Solid Lipid Nanoparticles: Formulation Factors Affecting Cell Transfection Capacity. Int. J. Pharm. 2007, 339, 261–268. [Google Scholar] [CrossRef]
- Shen, J.S.; Meng, X.L.; Schiffmann, R.; Brady, R.O.; Kaneski, C.R. Establishment and Characterization of Fabry Disease Endothelial Cells with an Extended Lifespan. Mol. Genet. Metab. 2007, 92, 137–144. [Google Scholar] [CrossRef]
- Zumbrun, S.D.; Hanson, L.; Sinclair, J.F.; Freedy, J.; Melton-Celsa, A.R.; Rodriguez-Canales, J.; Hanson, J.C.; O’Brien, A.D. Human Intestinal Tissue and Cultured Colonic Cells Contain Globotriaosylceramide Synthase MRNA and the Alternate Shiga Toxin Receptor Globotetraosylceramide. Infect. Immun. 2010, 78, 4488–4499. [Google Scholar] [CrossRef]
- Kurosaki, T.; Kitahara, T.; Fumoto, S.; Nishida, K.; Yamamoto, K.; Nakagawa, H. Chondroitin Sulfate Capsule System for Efficient and Secure Gene Delivery. J. Pharm. Pharm. Sci. 2010, 13, 351–361. [Google Scholar] [CrossRef]
- Gómez-Aguado, I.; Rodríguez-Castejón, J.; Vicente-Pascual, M.; Rodríguez-Gascón, A.; Del Pozo-Rodríguez, A.; Solinís Aspiazu, M.Á. Nucleic Acid Delivery by Solid Lipid Nanoparticles Containing Switchable Lipids: Plasmid DNA vs. Messenger RNA. Molecules 2020, 25, 5995. [Google Scholar] [CrossRef]
- Torrecilla, J.; del Pozo-Rodríguez, A.; Solinís, M.Á.; Apaolaza, P.S.; Berzal-Herranz, B.; Romero-López, C.; Berzal-Herranz, A.; Rodríguez-Gascón, A. Silencing of Hepatitis C Virus Replication by a Non-Viral Vector Based on Solid Lipid Nanoparticles Containing a ShRNA Targeted to the Internal Ribosome Entry Site (IRES). Colloids Surf. B Biointerfaces 2016, 146, 808–817. [Google Scholar] [CrossRef]
- Graván, P.; Aguilera-Garrido, A.; Marchal, J.A.; Navarro-Marchal, S.A.; Galisteo-González, F. Lipid-Core Nanoparticles: Classification, Preparation Methods, Routes of Administration and Recent Advances in Cancer Treatment. Adv. Colloid Interface Sci. 2023, 314, 102871. [Google Scholar] [CrossRef]
- Delgado, D.; Gascón, A.R.; Del Pozo-Rodríguez, A.; Echevarría, E.; Ruiz De Garibay, A.P.; Rodríguez, J.M.; Solinís, M.Á. Dextran-Protamine-Solid Lipid Nanoparticles as a Non-Viral Vector for Gene Therapy: In Vitro Characterization and in Vivo Transfection after Intravenous Administration to Mice. Int. J. Pharm. 2012, 425, 35–43. [Google Scholar] [CrossRef]
- Apaolaza, P.S.; Delgado, D.; Del Pozo-Rodríguez, A.; Gascón, A.R.; Solinís, M.Á. A Novel Gene Therapy Vector Based on Hyaluronic Acid and Solid Lipid Nanoparticles for Ocular Diseases. Int. J. Pharm. 2014, 465, 413–426. [Google Scholar] [CrossRef]
- Koo, K.M.; Sina, A.A.I.; Carrascosa, L.G.; Shiddiky, M.J.A.; Trau, M. DNA-Bare Gold Affinity Interactions: Mechanism and Applications in Biosensing. Anal. Methods 2015, 7, 7042–7054. [Google Scholar] [CrossRef]
- Takeuchi, T.; Tagami, T.; Fukushige, K.; Ozeki, T. Useful Properties of SiRNA-Coated Gold Nanoparticles as a Mini-Nanocarrier Platform for Intraocular Administration. J. Drug Deliv. Sci. Technol. 2018, 47, 411–416. [Google Scholar] [CrossRef]
- Li, T.; Nowell, C.J.; Cipolla, D.; Rades, T.; Boyd, B.J. Direct Comparison of Standard Transmission Electron Microscopy and Cryogenic-TEM in Imaging Nanocrystals Inside Liposomes. Mol. Pharm. 2019, 16, 1775–1781. [Google Scholar] [CrossRef] [PubMed]
- Buyens, K.; De Smedt, S.C.; Braeckmans, K.; Demeester, J.; Peeters, L.; Van Grunsven, L.A.; De Mollerat Du Jeu, X.; Sawant, R.; Torchilin, V.; Farkasova, K.; et al. Liposome Based Systems for Systemic SiRNA Delivery: Stability in Blood Sets the Requirements for Optimal Carrier Design. J. Control. Release 2012, 158, 362–370. [Google Scholar] [CrossRef]
- Franken, L.E.; Boekema, E.J.; Stuart, M.C.A. Transmission Electron Microscopy as a Tool for the Characterization of Soft Materials: Application and Interpretation. Adv. Sci. 2017, 4, 1600476. [Google Scholar] [CrossRef]
- Thompson, R.F.; Walker, M.; Siebert, C.A.; Muench, S.P.; Ranson, N.A. An Introduction to Sample Preparation and Imaging by Cryo-Electron Microscopy for Structural Biology. Methods 2016, 100, 3–15. [Google Scholar] [CrossRef]
- Patterson, J.P.; Xu, Y.; Moradi, M.A.; Sommerdijk, N.A.J.M.; Friedrich, H. CryoTEM as an Advanced Analytical Tool for Materials Chemists. Acc. Chem. Res. 2017, 50, 1495–1501. [Google Scholar] [CrossRef]
- Kuntsche, J.; Horst, J.C.; Bunjes, H. Cryogenic Transmission Electron Microscopy (Cryo-TEM) for Studying the Morphology of Colloidal Drug Delivery Systems. Int. J. Pharm. 2011, 417, 120–137. [Google Scholar] [CrossRef]
- Lee, S.H.; Kang, Y.Y.; Jang, H.E.; Mok, H. Current Preclinical Small Interfering RNA (SiRNA)-Based Conjugate Systems for RNA Therapeutics. Adv. Drug Deliv. Rev. 2016, 104, 78–92. [Google Scholar] [CrossRef]
- Delgado, D.; Del Pozo-Rodríguez, A.; Solinís, M.Á.; Rodríguez-Gascón, A. Understanding the Mechanism of Protamine in Solid Lipid Nanoparticle-Based Lipofection: The Importance of the Entry Pathway. Eur. J. Pharm. Biopharm. 2011, 79, 495–502. [Google Scholar] [CrossRef] [PubMed]
- Huo, S.; Jin, S.; Ma, X.; Xue, X.; Yang, K.; Kumar, A.; Wang, P.C.; Zhang, J.; Hu, Z.; Liang, X.J. Ultrasmall Gold Nanoparticles as Carriers for Nucleus-Based Gene Therapy Due to Size-Dependent Nuclear Entry. ACS Nano 2014, 8, 5852–5862. [Google Scholar] [CrossRef] [PubMed]
- Van Lehn, R.C.; Atukorale, P.U.; Carney, R.P.; Yang, Y.S.; Stellacci, F.; Irvine, D.J.; Alexander-Katz, A. Effect of Particle Diameter and Surface Composition on the Spontaneous Fusion of Monolayer-Protected Gold Nanoparticles with Lipid Bilayers. Nano Lett. 2013, 13, 4060–4067. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Yang, H.; Yu, Z.; Li, Y.; Pan, W.; Wang, H.; Tang, B. Nuclear-Targeted SiRNA Delivery for Long-Term Gene Silencing. Chem. Sci. 2017, 8, 2816–2822. [Google Scholar] [CrossRef] [PubMed]
- Castel, S.E.; Martienssen, R.A. RNA Interference in the Nucleus: Roles for Small RNAs in Transcription, Epigenetics and Beyond. Nat. Rev. Genet. 2013, 14, 100–112. [Google Scholar] [CrossRef]
- Chery, J. RNA Therapeutics: RNAi and Antisense Mechanisms and Clinical Applications. Postdoc J. J. Postdr. Res. Postdr. Aff. 2016, 4, 35–50. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, J.A.; Witzigmann, D.; Thomson, S.B.; Chen, S.; Leavitt, B.R.; Cullis, P.R.; van der Meel, R. The Current Landscape of Nucleic Acid Therapeutics. Nat. Nanotechnol. 2021, 16, 630–643. [Google Scholar] [CrossRef]
- Ruiz De Garibay, A.P.; Solinís, M.A.; Del Pozo-Rodríguez, A.; Apaolaza, P.S.; Shen, J.S.; Rodríguez-Gascón, A. Solid Lipid Nanoparticles as Non-Viral Vectors for Gene Transfection in a Cell Model of Fabry Disease. J. Biomed. Nanotechnol. 2015, 11, 500–511. [Google Scholar] [CrossRef]
- Charbe, N.B.; Amnerkar, N.D.; Ramesh, B.; Tambuwala, M.M.; Bakshi, H.A.; Aljabali, A.A.A.; Khadse, S.C.; Satheeshkumar, R.; Satija, S.; Metha, M.; et al. Small Interfering RNA for Cancer Treatment: Overcoming Hurdles in Delivery. Acta Pharm. Sin. B 2020, 10, 2075–2109. [Google Scholar] [CrossRef] [PubMed]
- Shaabani, E.; Sharifiaghdam, M.; Lammens, J.; De Keersmaecker, H.; Vervaet, C.; De Beer, T.; Motevaseli, E.; Ghahremani, M.H.; Mansouri, P.; De Smedt, S.; et al. Increasing Angiogenesis Factors in Hypoxic Diabetic Wound Conditions by Sirna Delivery: Additive Effect of Lbl-Gold Nanocarriers and Desloratadine-Induced Lysosomal Escape. Int. J. Mol. Sci. 2021, 22, 9216. [Google Scholar] [CrossRef] [PubMed]
- Sahay, G.; Querbes, W.; Alabi, C.; Eltoukhy, A.; Sarkar, S.; Zurenko, C.; Karagiannis, E.; Love, K.; Chen, D.; Zoncu, R.; et al. Efficiency of SiRNA Delivery by Lipid Nanoparticles Is Limited by Endocytic Recycling. Physiol. Behav. 2017, 176, 139–148. [Google Scholar] [CrossRef]
- Berger, M.; Lechanteur, A.; Evrard, B.; Piel, G. Innovative Lipoplexes Formulations with Enhanced SiRNA Efficacy for Cancer Treatment: Where Are We Now? Int. J. Pharm. 2021, 605, 120851. [Google Scholar] [CrossRef]
- Kulkarni, J.A.; Witzigmann, D.; Chen, S.; Cullis, P.R.; Van Der Meel, R. Lipid Nanoparticle Technology for Clinical Translation of SiRNA Therapeutics. Acc. Chem. Res. 2019, 52, 2435–2444. [Google Scholar] [CrossRef]
- Degors, I.M.S.; Wang, C.; Rehman, Z.U.; Zuhorn, I.S. Carriers Break Barriers in Drug Delivery: Endocytosis and Endosomal Escape of Gene Delivery Vectors. Acc. Chem. Res. 2019, 52, 1750–1760. [Google Scholar] [CrossRef]
- Xia, Y.; Tian, J.; Chen, X. Effect of Surface Properties on Liposomal SiRNA Delivery. Biomaterials 2016, 79, 56–68. [Google Scholar] [CrossRef] [PubMed]
- Kimura, S.; Khalil, I.A.; Elewa, Y.H.A.; Harashima, H. Novel Lipid Combination for Delivery of Plasmid DNA to Immune Cells in the Spleen. J. Control. Release 2021, 330, 753–764. [Google Scholar] [CrossRef] [PubMed]
- Yonezawa, S.; Koide, H.; Asai, T. Recent Advances in SiRNA Delivery Mediated by Lipid-Based Nanoparticles. Adv. Drug Deliv. Rev. 2020, 154, 64–78. [Google Scholar] [CrossRef]
- He, F.; Han, Y.; Gong, J.; Song, J.; Wang, H.; Li, Y. Predicting SiRNA Efficacy Based on Multiple Selective SiRNA Representations and Their Combination at Score Level. Sci. Rep. 2017, 7, 44836. [Google Scholar] [CrossRef]
- Li, X.; Ren, X.; Zhang, Y.; Ding, L.; Huo, M.; Li, Q. Fabry Disease: Mechanism and Therapeutics Strategies. Front. Pharmacol. 2022, 13, 1025740. [Google Scholar] [CrossRef] [PubMed]
- Kok, K.; Zwiers, K.C.; Boot, R.G.; Overkleeft, H.S.; Aerts, J.M.F.G.; Artola, M. Fabry Disease: Molecular Basis, Pathophysiology, Diagnostics and Potential Therapeutic Directions. Biomolecules 2021, 11, 271. [Google Scholar] [CrossRef]
- Kim, I.G.; Jung, W.H.; You, G.; Lee, H.; Shin, Y.J.; Lim, S.W.; Chung, B.H.; Mok, H. Efficient Delivery of Globotriaosylceramide Synthase SiRNA Using Polyhistidine-Incorporated Lipid Nanoparticles. Macromol. Biosci. 2023, 23, 2200423. [Google Scholar] [CrossRef] [PubMed]
- Scioli Montoto, S.; Muraca, G.; Ruiz, M.E. Solid Lipid Nanoparticles for Drug Delivery: Pharmacological and Biopharmaceutical Aspects. Front. Mol. Biosci. 2020, 7, 319. [Google Scholar] [CrossRef]
- Sani, A.; Cao, C.; Cui, D. Toxicity of Gold Nanoparticles (AuNPs): A Review. Biochem. Biophys. Rep. 2021, 26, 100991. [Google Scholar] [CrossRef]
- Mikhailova, E.O. Gold Nanoparticles: Biosynthesis and Potential of Biomedical Application. J. Funct. Biomater. 2021, 12, 70. [Google Scholar] [CrossRef]
- Mellor, R.D.; Uchegbu, I.F. Ultrasmall-in-Nano: Why Size Matters. Nanomaterials 2022, 12, 2476. [Google Scholar] [CrossRef]
- Elsaesser, A.; Howard, C.V. Toxicology of Nanoparticles. Adv. Drug Deliv. Rev. 2012, 64, 129–137. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Wang, L.; Mettenbrink, E.M.; Deangelis, P.L.; Wilhelm, S. Nanoparticle Toxicology. Annu. Rev. Pharmacol. Toxicol. 2021, 61, 269–289. [Google Scholar] [CrossRef]
- Carnovale, C.; Bryant, G.; Shukla, R.; Bansal, V. Identifying Trends in Gold Nanoparticle Toxicity and Uptake: Size, Shape, Capping Ligand, and Biological Corona. ACS Omega 2019, 4, 242–256. [Google Scholar] [CrossRef]
- Nguyen, N.H.A.; Falagan-Lotsch, P. Mechanistic Insights into the Biological Effects of Engineered Nanomaterials: A Focus on Gold Nanoparticles. Int. J. Mol. Sci. 2023, 24, 4109. [Google Scholar] [CrossRef] [PubMed]
- Balfourier, A.; Luciani, N.; Wang, G.; Lelong, G.; Ersen, O.; Khelfa, A.; Alloyeau, D.; Gazeau, F.; Carn, F. Unexpected Intracellular Biodegradation and Recrystallization of Gold Nanoparticles. Proc. Natl. Acad. Sci. USA 2020, 117, 103–113. [Google Scholar] [CrossRef] [PubMed]
| Type of SLN | Preparation Method | Lipid (%) | Tween 80 (%) | Size (d.nm) | PDI | ζ-Potential (mV) | |
|---|---|---|---|---|---|---|---|
| DOTAP | DODAP | ||||||
| SLNHT | HM | 0.4 | - | 0.1 | 90.0 ± 0.8 | 0.27 ± 0.01 | +67.2 ± 0.7 |
| SLNHD | HM | 0.2 | 0.2 | 0.1 | 119.3 ± 0.6 | 0.23 ± 0.01 | +71.3 ± 0.9 |
| SLNET | EE | 0.4 | - | 0.1 | 181.2 ± 5.5 | 0.26 ± 0.03 | +58.4 ± 1.6 |
| SLNED | EE | 0.2 | 0.2 | 0.1 | 218.8 ± 1.2 | 0.26 ± 0.05 | +52.2 ± 1.6 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Beraza-Millor, M.; Rodríguez-Castejón, J.; Miranda, J.; del Pozo-Rodríguez, A.; Rodríguez-Gascón, A.; Solinís, M.Á. Novel Golden Lipid Nanoparticles with Small Interference Ribonucleic Acid for Substrate Reduction Therapy in Fabry Disease. Pharmaceutics 2023, 15, 1936. https://doi.org/10.3390/pharmaceutics15071936
Beraza-Millor M, Rodríguez-Castejón J, Miranda J, del Pozo-Rodríguez A, Rodríguez-Gascón A, Solinís MÁ. Novel Golden Lipid Nanoparticles with Small Interference Ribonucleic Acid for Substrate Reduction Therapy in Fabry Disease. Pharmaceutics. 2023; 15(7):1936. https://doi.org/10.3390/pharmaceutics15071936
Chicago/Turabian StyleBeraza-Millor, Marina, Julen Rodríguez-Castejón, Jonatan Miranda, Ana del Pozo-Rodríguez, Alicia Rodríguez-Gascón, and María Ángeles Solinís. 2023. "Novel Golden Lipid Nanoparticles with Small Interference Ribonucleic Acid for Substrate Reduction Therapy in Fabry Disease" Pharmaceutics 15, no. 7: 1936. https://doi.org/10.3390/pharmaceutics15071936
APA StyleBeraza-Millor, M., Rodríguez-Castejón, J., Miranda, J., del Pozo-Rodríguez, A., Rodríguez-Gascón, A., & Solinís, M. Á. (2023). Novel Golden Lipid Nanoparticles with Small Interference Ribonucleic Acid for Substrate Reduction Therapy in Fabry Disease. Pharmaceutics, 15(7), 1936. https://doi.org/10.3390/pharmaceutics15071936