Catestatin: Antimicrobial Functions and Potential Therapeutics
Abstract
1. Introduction
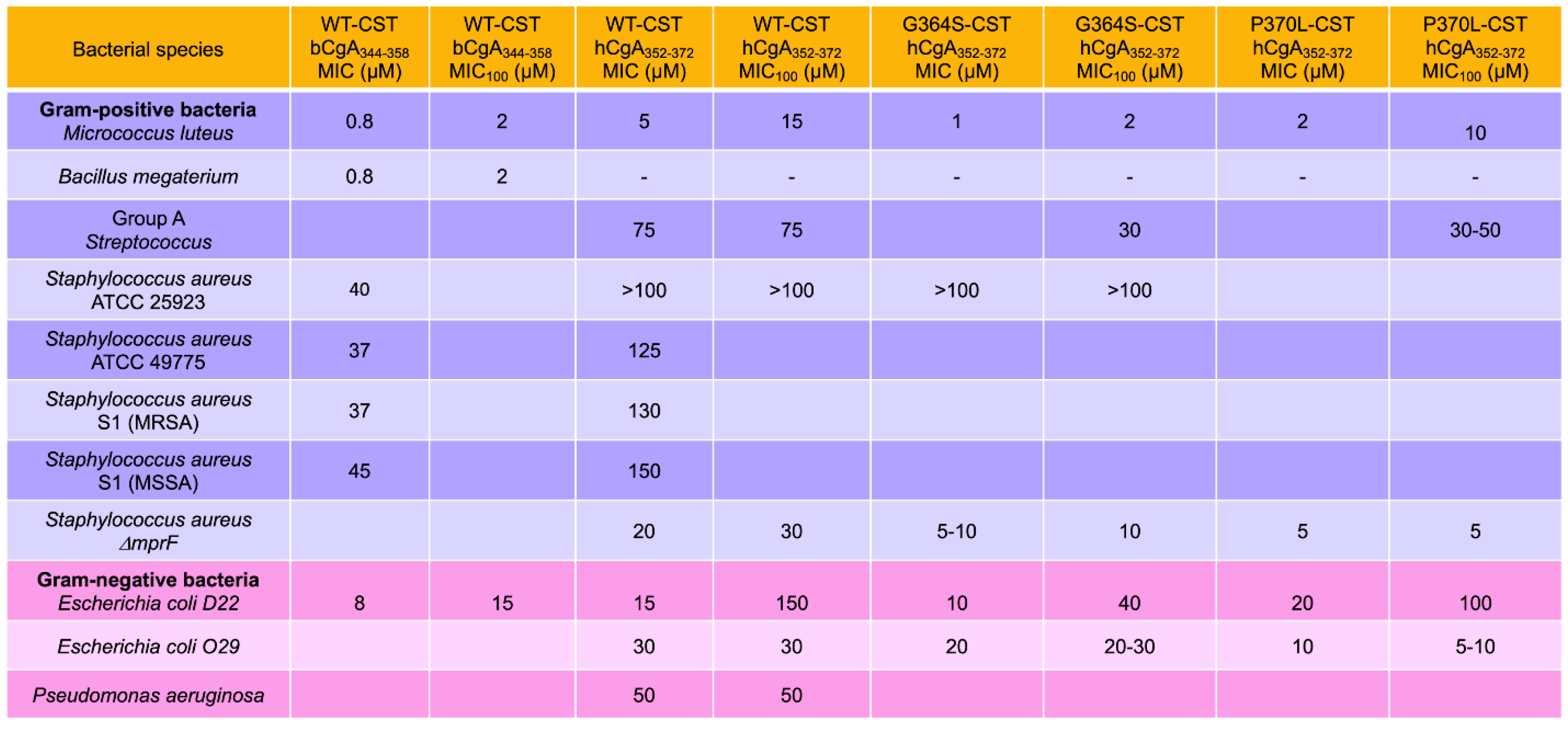
2. Antibacterial Effects of CST
2.1. Inhibition of Bacterial Growth by CST
2.2. Composition of Bacterial Membranes
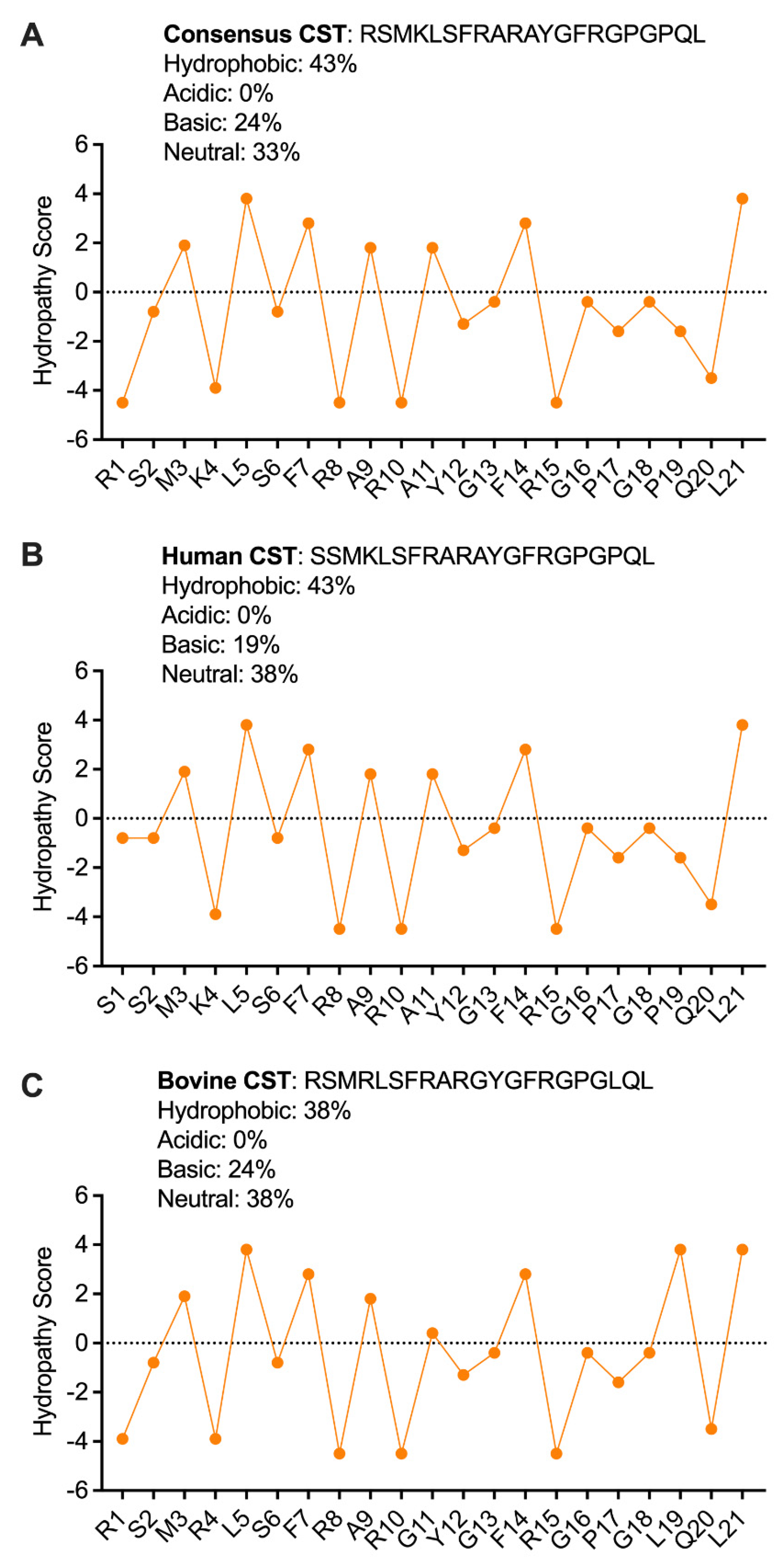
2.3. Secondary Structure of CST Explains the Antibacterial Effects of CST
2.4. CST as a Potential Therapy for Bacterial Diseases
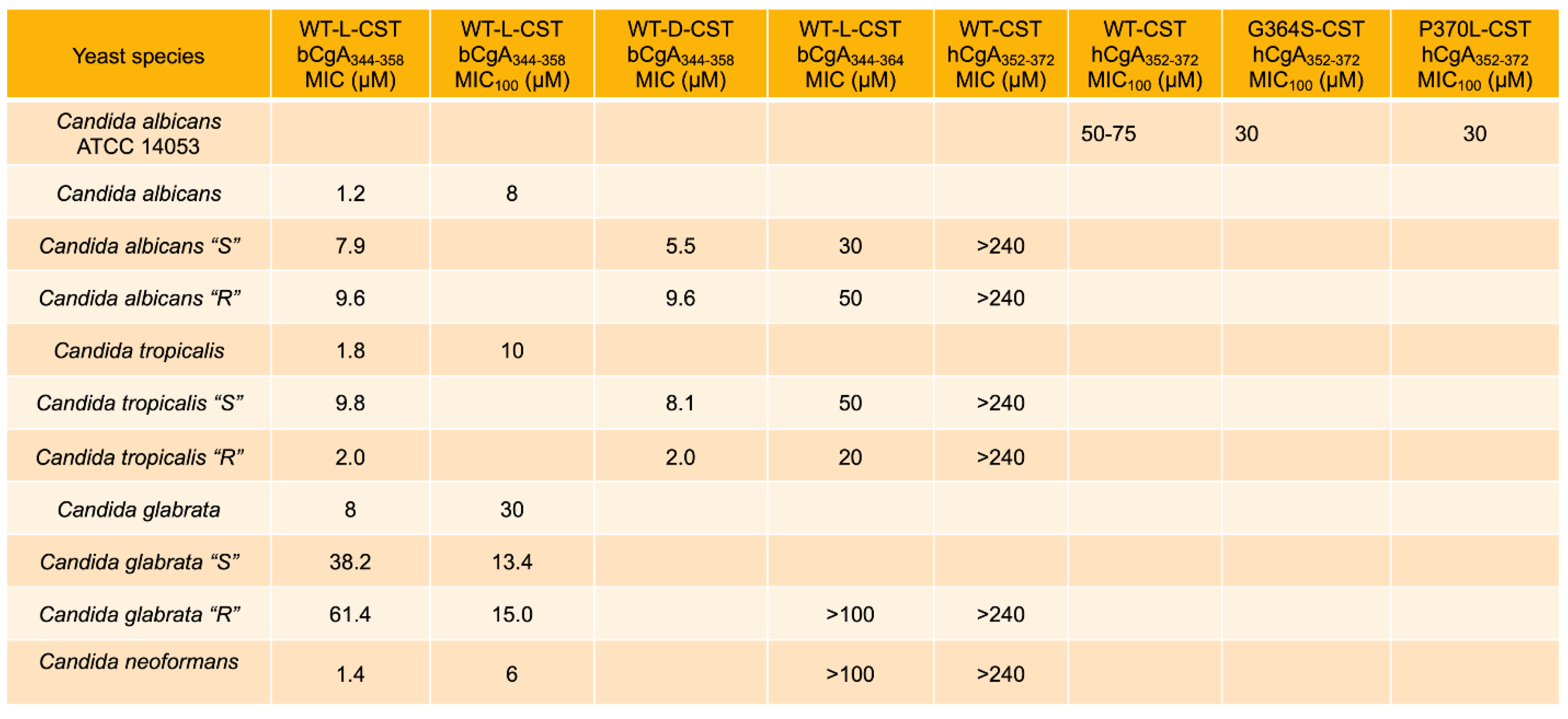
3. Antifungal and Anti-Yeast Effects of CST
3.1. Inhibition of Growth of Fungus and Yeast by CST
3.2. Mechanisms Underlying the Antifungal and Antiyeast Activities of CST
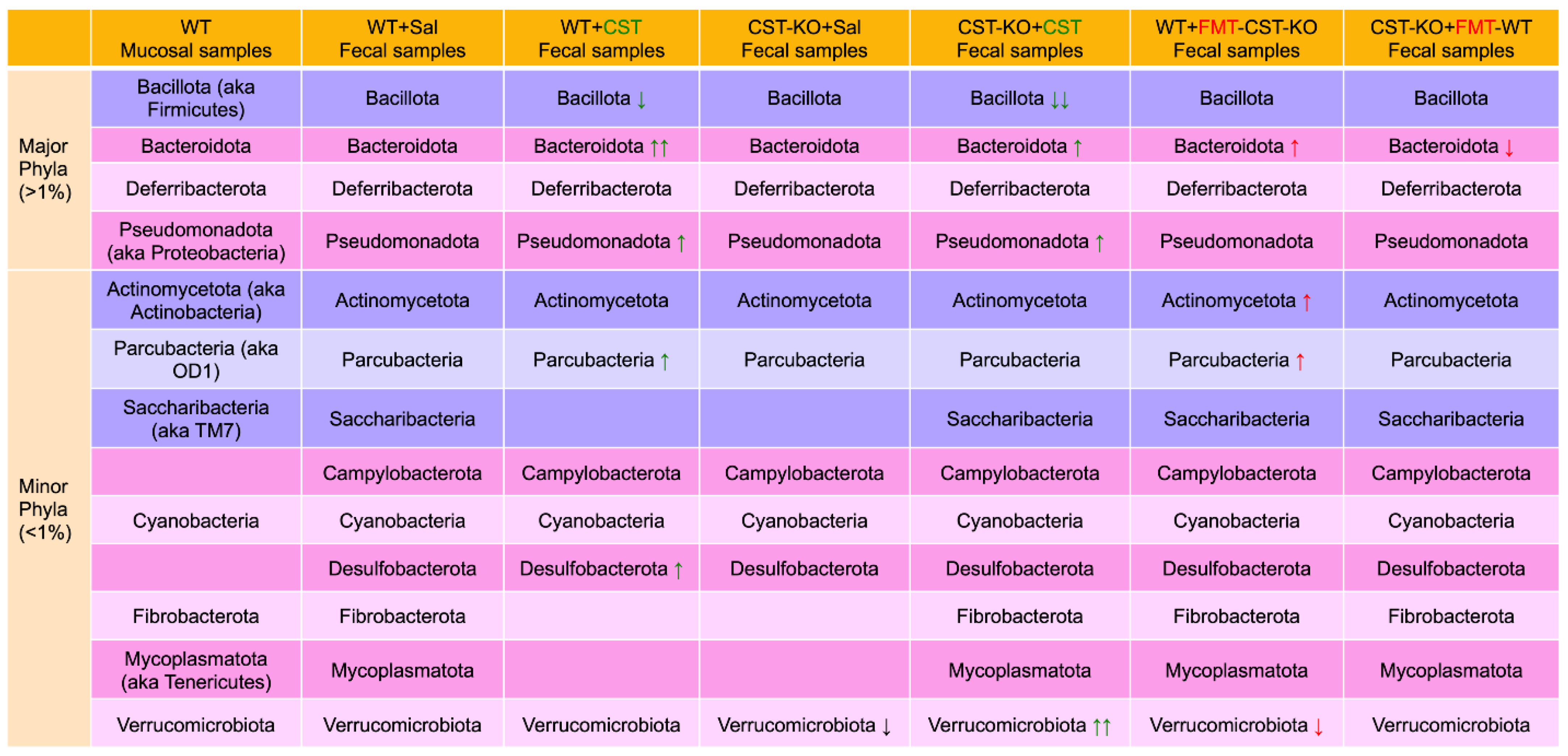
4. CST Regulation of Gut Microbiota
4.1. Microbiomes in Colonic Mucosa versus Feces
4.2. Microbiomes in CST Knockout (CST-KO) Mice and Inflammation
4.3. Alteration of Diversity and Composition of the Microbiota in the CST-KO after Supplementation with CST
4.4. Restoration of Microbial Dysbiosis in CST-KO Mice after Fecal Microbial Transplant (FMT) from WT Donor Mice
5. Catestatin and Innate Immunity
6. Evolutionary Conservation and Selection Pressure on CST in Mammals
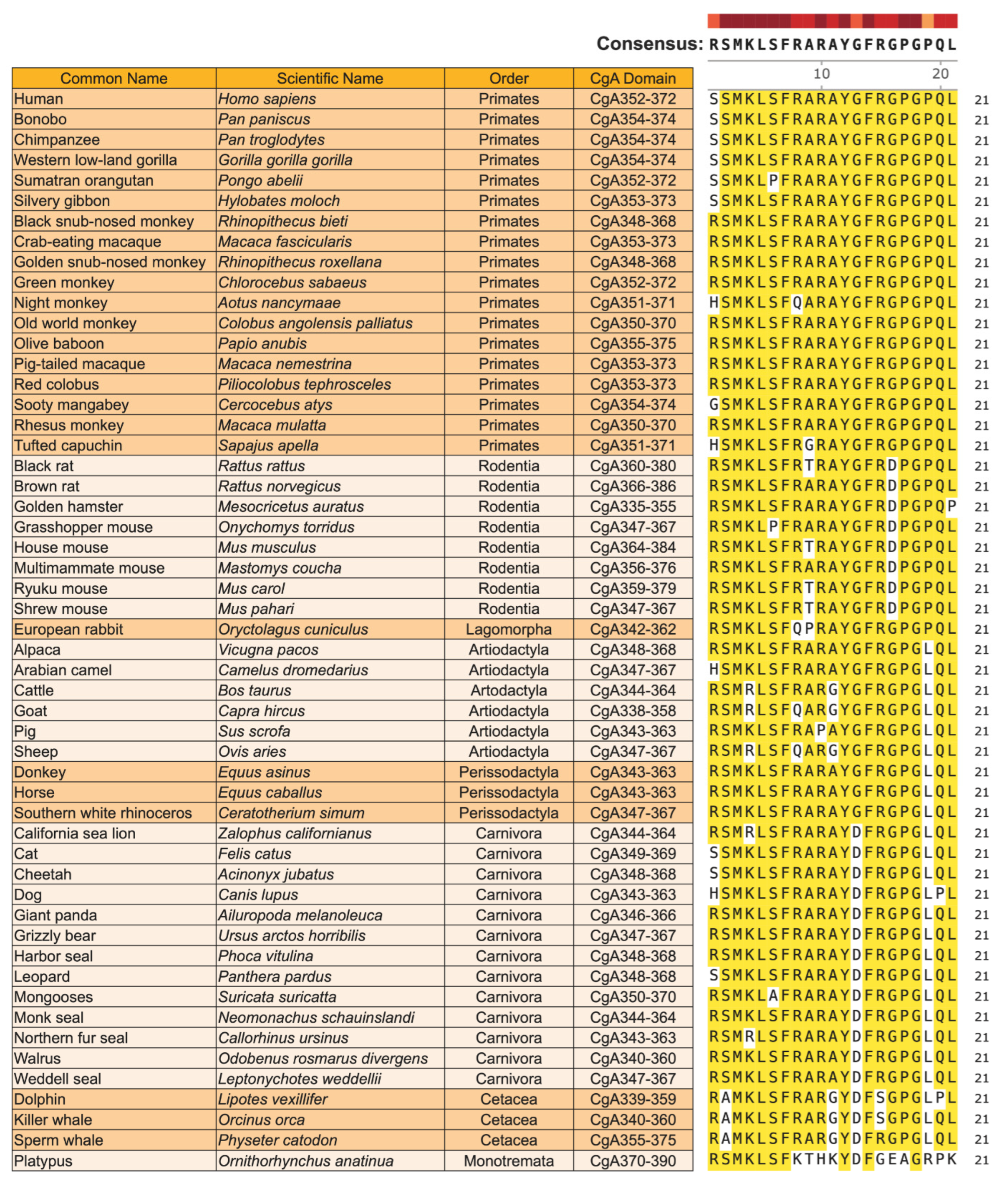
6.1. Homology of CST in Mammals
6.2. Single Nucleotide Polymorphisms (SNPs) in the CST Domain of Mammals
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zasloff, M. Antimicrobial peptides of multicellular organisms. Nature 2002, 415, 389–395. [Google Scholar] [CrossRef] [PubMed]
- Winkler, H.; Fischer-Colbrie, R. The Chromogranins A and B: The first 25 years and future perspectives. Neuroscience 1992, 49, 497–528. [Google Scholar] [CrossRef] [PubMed]
- Montero-Hadjadje, M.; Vaingankar, S.; Elias, S.; Tostivint, H.; Mahata, S.K.; Anouar, Y. Chromogranins A and B and secretogranin II: Evolutionary and functional aspects. Acta Physiol. (Oxf.) 2008, 192, 309–324. [Google Scholar] [CrossRef] [PubMed]
- Bartolomucci, A.; Possenti, R.; Mahata, S.K.; Fischer-Colbrie, R.; Loh, Y.P.; Salton, S.R. The extended granin family: Structure, function, and biomedical implications. Endocr. Rev. 2011, 32, 755–797. [Google Scholar] [CrossRef]
- Mahata, S.K.; O’Connor, D.T.; Mahata, M.; Yoo, S.H.; Taupenot, L.; Wu, H.; Gill, B.M.; Parmer, R.J. Novel autocrine feedback control of catecholamine release. A discrete Chromogranin A fragment is a noncompetitive nicotinic cholinergic antagonist. J. Clin. Investig. 1997, 100, 1623–1633. [Google Scholar] [CrossRef]
- Mahata, S.K.; Corti, A. Chromogranin A and its fragments in cardiovascular, immunometabolic, and cancer regulation. Ann. N. Y. Acad. Sci. 2019, 1455, 34–58. [Google Scholar] [CrossRef]
- Taylor, C.V.; Taupenot, L.; Mahata, S.K.; Mahata, M.; Wu, H.; Yasothornsrikul, S.; Toneff, T.; Caporale, C.; Jiang, Q.; Parmer, R.J.; et al. Formation of the catecholamine release-inhibitory peptide catestatin from Chromogranin A. Determination of proteolytic cleavage sites in hormone storage granules. J. Biol. Chem. 2000, 275, 22905–22915. [Google Scholar] [CrossRef]
- Parmer, R.J.; Mahata, M.; Gong, Y.; Mahata, S.K.; Jiang, Q.; O’Connor, D.T.; Xi, X.-P.; Miles, L.A. Processing of Chromogranin A by plasmin provides a novel mechanism for regulating catecholamine secretion. J. Clin. Investig. 2000, 106, 907–915. [Google Scholar] [CrossRef]
- Jiang, Q.; Taupenot, L.; Mahata, S.K.; Mahata, M.; O’Connor, D.T.; Miles, L.A.; Parmer, R.J. Proteolytic cleavage of Chromogranin A (CgA) by plasmin: Selective liberation of a specific bioactive CgA fragment that regulates catecholamine release. J. Biol. Chem. 2001, 276, 25022–25029. [Google Scholar] [CrossRef]
- Lee, J.C.; Taylor, C.V.; Gaucher, S.P.; Toneff, T.; Taupenot, L.; Yasothornsrikul, S.; Mahata, S.K.; Sei, C.; Parmer, R.J.; Neveu, J.M.; et al. Primary sequence characterization of catestatin intermediates and peptides defines proteolytic cleavage sites utilized for converting Chromogranin A into active catestatin secreted from neuroendocrine chromaffin cells. Biochemistry 2003, 42, 6938–6946. [Google Scholar] [CrossRef]
- Biswas, N.; Vaingankar, S.M.; Mahata, M.; Das, M.; Gayen, J.R.; Taupenot, L.; Torpey, J.W.; O’Connor, D.T.; Mahata, S.K. Proteolytic cleavage of human Chromogranin A containing naturally occurring catestatin variants: Differential processing at catestatin region by plasmin. Endocrinology 2008, 149, 749–757. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Biswas, N.; Rodriguez-Flores, J.L.; Courel, M.; Gayen, J.R.; Vaingankar, S.M.; Mahata, M.; Torpey, J.W.; Taupenot, L.; O’Connor, D.T.; Mahata, S.K. Cathepsin L Co-Localizes with Chromogranin A in Chromaffin Vesicles to Generate Active Peptides. Endocrinology 2009, 150, 3547–3557. [Google Scholar] [CrossRef] [PubMed]
- Mahata, S.K.; Mahata, M.; Parmer, R.J.; O’Connor, D.T. Desensitization of catecholamine release: The novel catecholamine release-inhibitory peptide catestatin (Chromogranin A344–364) acts at the receptor to prevent nicotinic cholinergic tolerance. J. Biol. Chem. 1999, 274, 2920–2928. [Google Scholar] [CrossRef] [PubMed]
- Taupenot, L.; Mahata, S.K.; Mahata, M.; Parmer, R.J.; O’Connor, D.T. Interaction of the catecholamine release-inhibitory peptide catestatin (human Chromogranin A(352–372)) with the chromaffin cell surface and Torpedo electroplax: Implications for nicotinic cholinergic antagonism. Regul. Pept. 2000, 95, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Mahata, S.K.; Mahata, M.; Wakade, A.R.; O’Connor, D.T. Primary structure and function of the catecholamine release inhibitory peptide catestatin (Chromogranin A344–364): Identification of amino acid residues crucial for activity. Mol. Endocrinol. 2000, 14, 1525–1535. [Google Scholar] [PubMed]
- Preece, N.E.; Nguyen, M.; Mahata, M.; Mahata, S.K.; Mahapatra, N.R.; Tsigelny, I.; O’Connor, D.T. Conformational preferences and activities of peptides from the catecholamine release-inhibitory (catestatin) region of Chromogranin A. Regul. Pept. 2004, 118, 75–87. [Google Scholar] [CrossRef]
- Mahata, S.K. Catestatin—The catecholamine release inhibitory peptide: A structural and functional overview. Curr. Med. Chem. Immun. Endoc. Metab. Agents 2004, 4, 221–234. [Google Scholar] [CrossRef]
- Mahapatra, N.R.; Mahata, M.; Mahata, S.K.; O’Connor, D.T. The Chromogranin A fragment catestatin: Specificity, potency and mechanism to inhibit exocytotic secretion of multiple catecholamine storage vesicle co-transmitters. J. Hypertens. 2006, 24, 895–904. [Google Scholar] [CrossRef]
- Mahata, S.K.; Mahata, M.; Fung, M.M.; O’Connor, D.T. Catestatin: A multifunctional peptide from Chromogranin A. Regul. Pept. 2010, 162, 33–43. [Google Scholar] [CrossRef]
- Mahata, S.K.; Kiranmayi, M.; Mahapatra, N.R. Catestatin: A Master Regulator of Cardiovascular Functions. Curr. Med. Chem. 2018, 25, 1352–1374. [Google Scholar] [CrossRef]
- Briolat, J.; Wu, S.D.; Mahata, S.K.; Gonthier, B.; Bagnard, D.; Chasserot-Golaz, S.; Helle, K.B.; Aunis, D.; Metz-Boutigue, M.H. New antimicrobial activity for the catecholamine release-inhibitory peptide from Chromogranin A. Cell. Mol. Life Sci. 2005, 62, 377–385. [Google Scholar] [CrossRef] [PubMed]
- Wen, G.; Mahata, S.K.; Cadman, P.; Mahata, M.; Ghosh, S.; Mahapatra, N.R.; Rao, F.; Stridsberg, M.; Smith, D.W.; Mahboubi, P.; et al. Both rare and common polymorphisms contribute functional variation at CHGA, a regulator of catecholamine physiology. Am. J. Hum. Genet. 2004, 74, 197–207. [Google Scholar] [CrossRef] [PubMed]
- Zaet, A.; Dartevelle, P.; Daouad, F.; Ehlinger, C.; Quiles, F.; Francius, G.; Boehler, C.; Bergthold, C.; Frisch, B.; Prevost, G.; et al. D-Cateslytin, a new antimicrobial peptide with therapeutic potential. Sci. Rep. 2017, 7, 15199. [Google Scholar] [CrossRef]
- Radek, K.A.; Lopez-Garcia, B.; Hupe, M.; Niesman, I.R.; Elias, P.M.; Taupenot, L.; Mahata, S.K.; O’Connor, D.T.; Gallo, R.L. The neuroendocrine peptide catestatin is a cutaneous antimicrobial and induced in the skin after injury. J. Investig. Dermatol. 2008, 128, 1525–1534. [Google Scholar] [CrossRef] [PubMed]
- Scheffers, D.J.; Pinho, M.G. Bacterial cell wall synthesis: New insights from localization studies. Microbiol. Mol. Biol. Rev. 2005, 69, 585–607. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.R.; Gordon, R.A.; Hyland, S.N.; Siegrist, M.S.; Grimes, C.L. Chemical Biology Tools for Examining the Bacterial Cell Wall. Cell. Chem. Biol. 2020, 27, 1052–1062. [Google Scholar] [CrossRef]
- Lopez-Siles, M.; Corral-Lugo, A.; McConnell, M.J. Vaccines for multidrug resistant Gram negative bacteria: Lessons from the past for guiding future success. FEMS Microbiol. Rev. 2021, 45, fuaa054. [Google Scholar] [CrossRef]
- Welch, R.A. Pore-forming cytolysins of gram-negative bacteria. Mol. Microbiol. 1991, 5, 521–528. [Google Scholar] [CrossRef]
- Mahlapuu, M.; Hakansson, J.; Ringstad, L.; Bjorn, C. Antimicrobial Peptides: An Emerging Category of Therapeutic Agents. Front. Cell. Infect. Microbiol. 2016, 6, 194. [Google Scholar] [CrossRef]
- Lee, T.H.; Hirst, D.J.; Aguilar, M.I. New insights into the molecular mechanisms of biomembrane structural changes and interactions by optical biosensor technology. Biochim. Biophys. Acta 2015, 1848, 1868–1885. [Google Scholar] [CrossRef]
- Wadhwani, P.; Epand, R.F.; Heidenreich, N.; Burck, J.; Ulrich, A.S.; Epand, R.M. Membrane-active peptides and the clustering of anionic lipids. Biophys. J. 2012, 103, 265–274. [Google Scholar] [CrossRef] [PubMed]
- Bogdanov, M.; Pyrshev, K.; Yesylevskyy, S.; Ryabichko, S.; Boiko, V.; Ivanchenko, P.; Kiyamova, R.; Guan, Z.; Ramseyer, C.; Dowhan, W. Phospholipid distribution in the cytoplasmic membrane of Gram-negative bacteria is highly asymmetric, dynamic, and cell shape-dependent. Sci. Adv. 2020, 6, eaaz6333. [Google Scholar] [CrossRef] [PubMed]
- Vollmer, W.; Seligman, S.J. Architecture of peptidoglycan: More data and more models. Trends Microbiol. 2010, 18, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Rojas, E.R.; Billings, G.; Odermatt, P.D.; Auer, G.K.; Zhu, L.; Miguel, A.; Chang, F.; Weibel, D.B.; Theriot, J.A.; Huang, K.C. The outer membrane is an essential load-bearing element in Gram-negative bacteria. Nature 2018, 559, 617–621. [Google Scholar] [CrossRef] [PubMed]
- Percy, M.G.; Grundling, A. Lipoteichoic acid synthesis and function in gram-positive bacteria. Annu. Rev. Microbiol. 2014, 68, 81–100. [Google Scholar] [CrossRef]
- Bowdish, D.M.; Davidson, D.J.; Hancock, R.E. A re-evaluation of the role of host defence peptides in mammalian immunity. Curr. Protein. Pept. Sci. 2005, 6, 35–51. [Google Scholar] [CrossRef]
- Sahu, B.S.; Mohan, J.; Sahu, G.; Singh, P.K.; Sonawane, P.J.; Sasi, B.K.; Allu, P.K.; Maji, S.K.; Bera, A.K.; Senapati, S.; et al. Molecular interactions of the physiological anti-hypertensive peptide catestatin with the neuronal nicotinic acetylcholine receptor. J. Cell Sci. 2012, 125, 2323–2337. [Google Scholar] [CrossRef]
- Tsigelny, I.; Mahata, S.K.; Taupenot, L.; Preece, N.E.; Mahata, M.; Khan, I.; Parmer, R.J.; O’Connor, D.T. Mechanism of action of Chromogranin A on catecholamine release: Molecular modeling of the catestatin region reveals a b-strand/loop/b-strand structure secured by hydrophobic interactions and predictive of activity. Regul. Pept. 1998, 77, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Dathe, M.; Nikolenko, H.; Klose, J.; Bienert, M. Cyclization increases the antimicrobial activity and selectivity of arginine- and tryptophan-containing hexapeptides. Biochemistry 2004, 43, 9140–9150. [Google Scholar] [CrossRef]
- Lazzaro, B.P.; Zasloff, M.; Rolff, J. Antimicrobial peptides: Application informed by evolution. Science 2020, 368, eaau5480. [Google Scholar] [CrossRef]
- Wessolowski, A.; Bienert, M.; Dathe, M. Antimicrobial activity of arginine- and tryptophan-rich hexapeptides: The effects of aromatic clusters, D-amino acid substitution and cyclization. J. Pept. Res. 2004, 64, 159–169. [Google Scholar] [CrossRef] [PubMed]
- Jean-Francois, F.; Castano, S.; Desbat, B.; Odaert, B.; Roux, M.; Metz-Boutigue, M.H.; Dufourc, E.J. Aggregation of cateslytin beta-sheets on negatively charged lipids promotes rigid membrane domains. A new mode of action for antimicrobial peptides? Biochemistry 2008, 47, 6394–6402. [Google Scholar] [CrossRef] [PubMed]
- Epand, R.M.; Epand, R.F. Lipid domains in bacterial membranes and the action of antimicrobial agents. Biochim. Biophys. Acta 2009, 1788, 289–294. [Google Scholar] [CrossRef]
- Scavello, F.; Mutschler, A.; Helle, S.; Schneider, F.; Chasserot-Golaz, S.; Strub, J.M.; Cianferani, S.; Haikel, Y.; Metz-Boutigue, M.H. Catestatin in innate immunity and Cateslytin-derived peptides against superbugs. Sci. Rep. 2021, 11, 15615. [Google Scholar] [CrossRef] [PubMed]
- Costa, T.R.; Felisberto-Rodrigues, C.; Meir, A.; Prevost, M.S.; Redzej, A.; Trokter, M.; Waksman, G. Secretion systems in Gram-negative bacteria: Structural and mechanistic insights. Nat. Rev. Microbiol. 2015, 13, 343–359. [Google Scholar] [CrossRef] [PubMed]
- Bonten, M.; Johnson, J.R.; van den Biggelaar, A.H.J.; Georgalis, L.; Geurtsen, J.; de Palacios, P.I.; Gravenstein, S.; Verstraeten, T.; Hermans, P.; Poolman, J.T. Epidemiology of Escherichia coli Bacteremia: A Systematic Literature Review. Clin. Infect. Dis. 2021, 72, 1211–1219. [Google Scholar] [CrossRef] [PubMed]
- Pechous, R.D.; Sivaraman, V.; Stasulli, N.M.; Goldman, W.E. Pneumonic Plague: The Darker Side of Yersinia pestis. Trends Microbiol. 2016, 24, 190–197. [Google Scholar] [CrossRef]
- Kim, S.H.; Chelliah, R.; Ramakrishnan, S.R.; Perumal, A.S.; Bang, W.S.; Rubab, M.; Daliri, E.B.; Barathikannan, K.; Elahi, F.; Park, E.; et al. Review on Stress Tolerance in Campylobacter jejuni. Front. Cell. Infect. Microbiol. 2020, 10, 596570. [Google Scholar] [CrossRef]
- Cunha, B.A.; Burillo, A.; Bouza, E. Legionnaires’ disease. Lancet 2016, 387, 376–385. [Google Scholar] [CrossRef]
- Coburn, B.; Grassl, G.A.; Finlay, B.B. Salmonella, the host and disease: A brief review. Immunol. Cell Biol. 2007, 85, 112–118. [Google Scholar] [CrossRef]
- Lee, G.C.; Burgess, D.S. Treatment of Klebsiella pneumoniae carbapenemase (KPC) infections: A review of published case series and case reports. Ann. Clin. Microbiol. Antimicrob. 2012, 11, 32. [Google Scholar] [CrossRef] [PubMed]
- Capatina, D.; Feier, B.; Hosu, O.; Tertis, M.; Cristea, C. Analytical methods for the characterization and diagnosis of infection with Pseudomonas aeruginosa: A critical review. Anal. Chim. Acta 2022, 1204, 339696. [Google Scholar] [CrossRef] [PubMed]
- Hennebique, A.; Boisset, S.; Maurin, M. Tularemia as a waterborne disease: A review. Emerg. Microbes Infect. 2019, 8, 1027–1042. [Google Scholar] [CrossRef] [PubMed]
- Butler, T. Treatment of typhoid fever in the 21st century: Promises and shortcomings. Clin. Microbiol. Infect. 2011, 17, 959–963. [Google Scholar] [CrossRef]
- Mazzariol, A.; Bazaj, A.; Cornaglia, G. Multi-drug-resistant Gram-negative bacteria causing urinary tract infections: A review. J. Chemother. 2017, 29, 2–9. [Google Scholar] [CrossRef]
- Blair, J.M.; Richmond, G.E.; Piddock, L.J. Multidrug efflux pumps in Gram-negative bacteria and their role in antibiotic resistance. Future Microbiol. 2014, 9, 1165–1177. [Google Scholar] [CrossRef]
- Vasoo, S.; Barreto, J.N.; Tosh, P.K. Emerging issues in gram-negative bacterial resistance: An update for the practicing clinician. Mayo Clin. Proc. 2015, 90, 395–403. [Google Scholar] [CrossRef]
- Sizemore, T.C. Rheumatologic manifestations of histoplasmosis: A review. Rheumatol. Int. 2013, 33, 2963–2965. [Google Scholar] [CrossRef]
- Jude, C.M.; Nayak, N.B.; Patel, M.K.; Deshmukh, M.; Batra, P. Pulmonary coccidioidomycosis: Pictorial review of chest radiographic and CT findings. Radiographics 2014, 34, 912–925. [Google Scholar] [CrossRef]
- Linder, K.A.; Kauffman, C.A.; Miceli, M.H. Blastomycosis: A Review of Mycological and Clinical Aspects. J. Fungi 2023, 9, 117. [Google Scholar] [CrossRef]
- Mahdavinia, M.; Grammer, L.C. Management of allergic bronchopulmonary aspergillosis: A review and update. Ther. Adv. Respir. Dis. 2012, 6, 173–187. [Google Scholar] [CrossRef]
- Griffith, N.; Danziger, L. Candida auris Urinary Tract Infections and Possible Treatment. Antibiotics 2020, 9, 898. [Google Scholar] [CrossRef]
- Ben-Ami, R. Treatment of Invasive Candidiasis: A Narrative Review. J. Fungi 2018, 4, 97. [Google Scholar] [CrossRef]
- McCarty, T.P.; Pappas, P.G. Invasive Candidiasis. Infect. Dis. Clin. N. Am. 2016, 30, 103–124. [Google Scholar] [CrossRef]
- de Boer, M.G.; de Fijter, J.W.; Kroon, F.P. Outbreaks and clustering of Pneumocystis pneumonia in kidney transplant recipients: A systematic review. Med. Mycol. 2011, 49, 673–680. [Google Scholar]
- Jeong, W.; Keighley, C.; Wolfe, R.; Lee, W.L.; Slavin, M.A.; Kong, D.C.M.; Chen, S.C. The epidemiology and clinical manifestations of mucormycosis: A systematic review and meta-analysis of case reports. Clin. Microbiol. Infect. 2019, 25, 26–34. [Google Scholar] [CrossRef]
- Nair, A.G.; Dave, T.V. Transcutaneous retrobulbar injection of amphotericin B in rhino-orbital-cerebral mucormycosis: A review. Orbit 2022, 41, 275–286. [Google Scholar] [CrossRef]
- Setianingrum, F.; Rautemaa-Richardson, R.; Denning, D.W. Pulmonary cryptococcosis: A review of pathobiology and clinical aspects. Med. Mycol. 2019, 57, 133–150. [Google Scholar] [CrossRef]
- Montoya, M.C.; Magwene, P.M.; Perfect, J.R. Associations between Cryptococcus Genotypes, Phenotypes, and Clinical Parameters of Human Disease: A Review. J. Fungi 2021, 7, 260. [Google Scholar] [CrossRef]
- Bermas, A.; Geddes-McAlister, J. Combatting the evolution of antifungal resistance in Cryptococcus neoformans. Mol. Microbiol. 2020, 114, 721–734. [Google Scholar] [CrossRef]
- Wall, G.; Lopez-Ribot, J.L. Current Antimycotics, New Prospects, and Future Approaches to Antifungal Therapy. Antibiotics 2020, 9, 445. [Google Scholar] [CrossRef] [PubMed]
- Buda De Cesare, G.; Cristy, S.A.; Garsin, D.A.; Lorenz, M.C. Antimicrobial Peptides: A New Frontier in Antifungal Therapy. mBio 2020, 11, e02123-20. [Google Scholar] [CrossRef] [PubMed]
- Biswas, N.; Gayen, J.; Mahata, M.; Su, Y.; Mahata, S.K.; O’Connor, D.T. Novel peptide isomer strategy for stable inhibition of catecholamine release: Application to hypertension. Hypertension 2012, 60, 1552–1559. [Google Scholar] [CrossRef] [PubMed]
- Aslam, R.; Marban, C.; Corazzol, C.; Jehl, F.; Delalande, F.; Van Dorsselaer, A.; Prevost, G.; Haikel, Y.; Taddei, C.; Schneider, F.; et al. Cateslytin, a Chromogranin A derived peptide is active against Staphylococcus aureus and resistant to degradation by its proteases. PLoS ONE 2013, 8, e68993. [Google Scholar] [CrossRef] [PubMed]
- Mancino, D.; Kharouf, N.; Scavello, F.; Helle, S.; Salloum-Yared, F.; Mutschler, A.; Mathieu, E.; Lavalle, P.; Metz-Boutigue, M.H.; Haikel, Y. The Catestatin-Derived Peptides Are New Actors to Fight the Development of Oral Candidosis. Int. J. Mol. Sci. 2022, 23, 2066. [Google Scholar] [CrossRef]
- Rios-Covian, D.; Salazar, N.; Gueimonde, M.; de Los Reyes-Gavilan, C.G. Shaping the Metabolism of Intestinal Bacteroides Population through Diet to Improve Human Health. Front. Microbiol. 2017, 8, 376. [Google Scholar] [CrossRef]
- Rabbi, M.F.; Eissa, N.; Munyaka, P.M.; Kermarrec, L.; Elgazzar, O.; Khafipour, E.; Bernstein, C.N.; Ghia, J.E. Reactivation of Intestinal Inflammation Is Suppressed by Catestatin in a Murine Model of Colitis via M1 Macrophages and Not the Gut Microbiota. Front. Immunol. 2017, 8, 985. [Google Scholar] [CrossRef]
- Rabbi, M.F.; Munyaka, P.M.; Eissa, N.; Metz-Boutigue, M.H.; Khafipour, E.; Ghia, J.E. Human Catestatin Alters Gut Microbiota Composition in Mice. Front. Microbiol. 2016, 7, 2151. [Google Scholar] [CrossRef]
- Horvath, T.D.; Ihekweazu, F.D.; Haidacher, S.J.; Ruan, W.; Engevik, K.A.; Fultz, R.; Hoch, K.M.; Luna, R.A.; Oezguen, N.; Spinler, J.K.; et al. Bacteroides ovatus colonization influences the abundance of intestinal short chain fatty acids and neurotransmitters. iScience 2022, 25, 104158. [Google Scholar] [CrossRef]
- Fernandez-Julia, P.J.; Munoz-Munoz, J.; van Sinderen, D. A comprehensive review on the impact of beta-glucan metabolism by Bacteroides and Bifidobacterium species as members of the gut microbiota. Int. J. Biol. Macromol. 2021, 181, 877–889. [Google Scholar] [CrossRef]
- Bornet, E.; Westermann, A.J. The ambivalent role of Bacteroides in enteric infections. Trends Microbiol. 2022, 30, 104–108. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.M.; de Souza, R.; Kendall, C.W.; Emam, A.; Jenkins, D.J. Colonic health: Fermentation and short chain fatty acids. J. Clin. Gastroenterol. 2006, 40, 235–243. [Google Scholar] [CrossRef]
- Cummings, J.H.; Macfarlane, G.T. Role of intestinal bacteria in nutrient metabolism. JPEN J. Parenter. Enteral Nutr. 1997, 21, 357–365. [Google Scholar] [CrossRef]
- Reigstad, C.S.; Salmonson, C.E.; Rainey, J.F., 3rd; Szurszewski, J.H.; Linden, D.R.; Sonnenburg, J.L.; Farrugia, G.; Kashyap, P.C. Gut microbes promote colonic serotonin production through an effect of short-chain fatty acids on enterochromaffin cells. FASEB J. 2015, 29, 1395–1403. [Google Scholar] [CrossRef] [PubMed]
- Ge, X.; Pan, J.; Liu, Y.; Wang, H.; Zhou, W.; Wang, X. Intestinal Crosstalk between Microbiota and Serotonin and its Impact on Gut Motility. Curr. Pharm. Biotechnol. 2018, 19, 190–195. [Google Scholar] [CrossRef]
- Christiansen, C.B.; Gabe, M.B.N.; Svendsen, B.; Dragsted, L.O.; Rosenkilde, M.M.; Holst, J.J. The impact of short-chain fatty acids on GLP-1 and PYY secretion from the isolated perfused rat colon. Am. J. Physiol. Gastrointest. Liver Physiol. 2018, 315, G53–G65. [Google Scholar] [CrossRef]
- Psichas, A.; Sleeth, M.L.; Murphy, K.G.; Brooks, L.; Bewick, G.A.; Hanyaloglu, A.C.; Ghatei, M.A.; Bloom, S.R.; Frost, G. The short chain fatty acid propionate stimulates GLP-1 and PYY secretion via free fatty acid receptor 2 in rodents. Int. J. Obes. (Lond.) 2015, 39, 424–429. [Google Scholar] [CrossRef]
- Ma, Y.; Lee, E.; Yoshikawa, H.; Noda, T.; Miyamoto, J.; Kimura, I.; Hatano, R.; Miki, T. Phloretin suppresses carbohydrate-induced GLP-1 secretion via inhibiting short chain fatty acid release from gut microbiome. Biochem. Biophys. Res. Commun. 2022, 621, 176–182. [Google Scholar] [CrossRef]
- Canfora, E.E.; Jocken, J.W.; Blaak, E.E. Short-chain fatty acids in control of body weight and insulin sensitivity. Nat. Rev. Endocrinol. 2015, 11, 577–591. [Google Scholar] [CrossRef]
- Muller, M.; Hernandez, M.A.G.; Goossens, G.H.; Reijnders, D.; Holst, J.J.; Jocken, J.W.E.; van Eijk, H.; Canfora, E.E.; Blaak, E.E. Circulating but not faecal short-chain fatty acids are related to insulin sensitivity, lipolysis and GLP-1 concentrations in humans. Sci. Rep. 2019, 9, 12515. [Google Scholar] [CrossRef]
- Hernandez, M.A.G.; Canfora, E.E.; Jocken, J.W.E.; Blaak, E.E. The Short-Chain Fatty Acid Acetate in Body Weight Control and Insulin Sensitivity. Nutrients 2019, 11, 1943. [Google Scholar] [CrossRef] [PubMed]
- Wexler, H.M. Bacteroides: The good, the bad, and the nitty-gritty. Clin. Microbiol. Rev. 2007, 20, 593–621. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.Y.; Lee, Y.S.; Kim, Y.; Lee, S.H.; Ryu, S.; Fukuda, S.; Hase, K.; Yang, C.S.; Lim, H.S.; Kim, M.S.; et al. Gut commensal Bacteroides acidifaciens prevents obesity and improves insulin sensitivity in mice. Mucosal Immunol. 2017, 10, 104–116. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, N.; Yamashita, T.; Osone, T.; Hosooka, T.; Shinohara, M.; Kitahama, S.; Sasaki, K.; Sasaki, D.; Yoneshiro, T.; Suzuki, T.; et al. Bacteroides spp. promotes branched-chain amino acid catabolism in brown fat and inhibits obesity. iScience 2021, 24, 103342. [Google Scholar] [CrossRef] [PubMed]
- Mazmanian, S.K.; Kasper, D.L. The love-hate relationship between bacterial polysaccharides and the host immune system. Nat. Rev. Immunol. 2006, 6, 849–858. [Google Scholar] [CrossRef] [PubMed]
- Round, J.L.; Mazmanian, S.K. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc. Natl. Acad. Sci. USA 2010, 107, 12204–12209. [Google Scholar] [CrossRef] [PubMed]
- Ying, W.; Mahata, S.; Bandyopadhyay, G.K.; Zhou, Z.; Wollam, J.; Vu, J.; Mayoral, R.; Chi, N.W.; Webster, N.J.G.; Corti, A.; et al. Catestatin Inhibits Obesity-Induced Macrophage Infiltration and Inflammation in the Liver and Suppresses Hepatic Glucose Production, Leading to Improved Insulin Sensitivity. Diabetes 2018, 67, 841–848. [Google Scholar] [CrossRef] [PubMed]
- Ying, W.; Tang, K.; Avolio, E.; Schilling, J.M.; Pasqua, T.; Liu, M.A.; Cheng, H.; Gao, H.; Zhang, J.; Mahata, S.; et al. Immunosuppression of Macrophages Underlies the Cardioprotective Effects of CST (Catestatin). Hypertension 2021, 77, 1670–1682. [Google Scholar] [CrossRef]
- Muntjewerff, E.M.; Tang, K.; Lutter, L.; Christoffersson, G.; Nicolasen, M.J.T.; Gao, H.; Katkar, G.D.; Das, S.; Ter Beest, M.; Ying, W.; et al. Chromogranin A regulates gut permeability via the antagonistic actions of its proteolytic peptides. Acta Physiol. (Oxf.) 2021, 232, e13655. [Google Scholar] [CrossRef]
- Gonzalez-Davila, P.; Schwalbe, M.; Danewalia, A.; Dalile, B.; Verbeke, K.; Mahata, S.K.; El Aidy, S. Catestatin selects for colonization of antimicrobial-resistant gut bacterial communities. ISME J. 2022, 16, 1873–1882. [Google Scholar] [CrossRef]
- Corb Aron, R.A.; Abid, A.; Vesa, C.M.; Nechifor, A.C.; Behl, T.; Ghitea, T.C.; Munteanu, M.A.; Fratila, O.; Andronie-Cioara, F.L.; Toma, M.M.; et al. Recognizing the Benefits of Pre-/Probiotics in Metabolic Syndrome and Type 2 Diabetes Mellitus Considering the Influence of Akkermansia muciniphila as a Key Gut Bacterium. Microorganisms 2021, 9, 618. [Google Scholar] [CrossRef] [PubMed]
- Bhutiani, N.; Schucht, J.E.; Miller, K.R.; McClave, S.A. Technical Aspects of Fecal Microbial Transplantation (FMT). Curr. Gastroenterol. Rep. 2018, 20, 30. [Google Scholar] [CrossRef] [PubMed]
- Mattila, E.; Uusitalo-Seppala, R.; Wuorela, M.; Lehtola, L.; Nurmi, H.; Ristikankare, M.; Moilanen, V.; Salminen, K.; Seppala, M.; Mattila, P.S.; et al. Fecal transplantation, through colonoscopy, is effective therapy for recurrent Clostridium difficile infection. Gastroenterology 2012, 142, 490–496. [Google Scholar] [CrossRef] [PubMed]
- Friedman-Korn, T.; Livovsky, D.M.; Maharshak, N.; Aviv Cohen, N.; Paz, K.; Bar-Gil Shitrit, A.; Goldin, E.; Koslowsky, B. Fecal Transplantation for Treatment of Clostridium Difficile Infection in Elderly and Debilitated Patients. Dig. Dis. Sci. 2018, 63, 198–203. [Google Scholar] [CrossRef]
- Cohen, N.A.; Maharshak, N. Novel Indications for Fecal Microbial Transplantation: Update and Review of the Literature. Dig. Dis. Sci. 2017, 62, 1131–1145. [Google Scholar] [CrossRef]
- Winslet, M.C.; Andrews, H.; Allan, R.N.; Keighley, M.R. Fecal diversion in the management of Crohn’s disease of the colon. Dis. Colon Rectum 1993, 36, 757–762. [Google Scholar] [CrossRef]
- Grehan, M.J.; Borody, T.J.; Leis, S.M.; Campbell, J.; Mitchell, H.; Wettstein, A. Durable alteration of the colonic microbiota by the administration of donor fecal flora. J. Clin. Gastroenterol. 2010, 44, 551–561. [Google Scholar] [CrossRef]
- Bakken, J.S.; Borody, T.; Brandt, L.J.; Brill, J.V.; Demarco, D.C.; Franzos, M.A.; Kelly, C.; Khoruts, A.; Louie, T.; Martinelli, L.P.; et al. Treating Clostridium difficile infection with fecal microbiota transplantation. Clin. Gastroenterol. Hepatol. 2011, 9, 1044–1049. [Google Scholar] [CrossRef]
- Gonzalez-Davila, P.; Schwalbe, M.; Danewalia, A.; Wardenaar, R.; Dalile, B.; Verbeke, K.; Mahata, S.K.; El Aidy, S. Gut microbiota transplantation drives the adoptive transfer of colonic genotype-phenotype characteristics between mice lacking catestatin and their wild type counterparts. Gut Microbes 2022, 14, 2081476. [Google Scholar] [CrossRef]
- Schneeberger, M.; Everard, A.; Gomez-Valades, A.G.; Matamoros, S.; Ramirez, S.; Delzenne, N.M.; Gomis, R.; Claret, M.; Cani, P.D. Akkermansia muciniphila inversely correlates with the onset of inflammation, altered adipose tissue metabolism and metabolic disorders during obesity in mice. Sci. Rep. 2015, 5, 16643. [Google Scholar] [CrossRef]
- Zhou, Q.; Pang, G.; Zhang, Z.; Yuan, H.; Chen, C.; Zhang, N.; Yang, Z.; Sun, L. Association Between Gut Akkermansia and Metabolic Syndrome is Dose-Dependent and Affected by Microbial Interactions: A Cross-Sectional Study. Diabetes Metab. Syndr. Obes. 2021, 14, 2177–2188. [Google Scholar] [CrossRef] [PubMed]
- Earley, H.; Lennon, G.; Balfe, A.; Coffey, J.C.; Winter, D.C.; O’Connell, P.R. The abundance of Akkermansia muciniphila and its relationship with sulphated colonic mucins in health and ulcerative colitis. Sci. Rep. 2019, 9, 15683. [Google Scholar] [CrossRef]
- Glassner, K.L.; Abraham, B.P.; Quigley, E.M.M. The microbiome and inflammatory bowel disease. J. Allergy Clin. Immunol. 2020, 145, 16–27. [Google Scholar] [CrossRef] [PubMed]
- den Besten, G.; van Eunen, K.; Groen, A.K.; Venema, K.; Reijngoud, D.J.; Bakker, B.M. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J. Lipid Res. 2013, 54, 2325–2340. [Google Scholar] [CrossRef]
- Bolognini, D.; Tobin, A.B.; Milligan, G.; Moss, C.E. The Pharmacology and Function of Receptors for Short-Chain Fatty Acids. Mol. Pharmacol. 2016, 89, 388–398. [Google Scholar] [CrossRef] [PubMed]
- Morrison, D.J.; Preston, T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes 2016, 7, 189–200. [Google Scholar] [CrossRef]
- De Preter, V.; Geboes, K.P.; Bulteel, V.; Vandermeulen, G.; Suenaert, P.; Rutgeerts, P.; Verbeke, K. Kinetics of butyrate metabolism in the normal colon and in ulcerative colitis: The effects of substrate concentration and carnitine on the beta-oxidation pathway. Aliment. Pharmacol. Ther. 2011, 34, 526–532. [Google Scholar] [CrossRef]
- Huda-Faujan, N.; Abdulamir, A.S.; Fatimah, A.B.; Anas, O.M.; Shuhaimi, M.; Yazid, A.M.; Loong, Y.Y. The impact of the level of the intestinal short chain Fatty acids in inflammatory bowel disease patients versus healthy subjects. Open Biochem. J. 2010, 4, 53–58. [Google Scholar] [CrossRef]
- Wang, H.B.; Wang, P.Y.; Wang, X.; Wan, Y.L.; Liu, Y.C. Butyrate enhances intestinal epithelial barrier function via up-regulation of tight junction protein Claudin-1 transcription. Dig. Dis. Sci. 2012, 57, 3126–3135. [Google Scholar] [CrossRef]
- Arpaia, N.; Campbell, C.; Fan, X.; Dikiy, S.; van der Veeken, J.; deRoos, P.; Liu, H.; Cross, J.R.; Pfeffer, K.; Coffer, P.J.; et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 2013, 504, 451–455. [Google Scholar] [CrossRef]
- Furusawa, Y.; Obata, Y.; Fukuda, S.; Endo, T.A.; Nakato, G.; Takahashi, D.; Nakanishi, Y.; Uetake, C.; Kato, K.; Kato, T.; et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 2013, 504, 446–450. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Sun, M.; Wu, W.; Yang, W.; Huang, X.; Xiao, Y.; Ma, C.; Xu, L.; Yao, S.; Liu, Z.; et al. Microbiota Metabolite Butyrate Differentially Regulates Th1 and Th17 Cells’ Differentiation and Function in Induction of Colitis. Inflamm. Bowel. Dis. 2019, 25, 1450–1461. [Google Scholar] [CrossRef] [PubMed]
- Fregeau, C.J.; Helgason, C.D.; Bleackley, R.C. Two cytotoxic cell proteinase genes are differentially sensitive to sodium butyrate. Nucleic Acids Res. 1992, 20, 3113–3119. [Google Scholar] [CrossRef]
- Tsuda, H.; Ochiai, K.; Suzuki, N.; Otsuka, K. Butyrate, a bacterial metabolite, induces apoptosis and autophagic cell death in gingival epithelial cells. J. Periodontal Res. 2010, 45, 626–634. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, B.P.; Mahata, S.K.; O’Connor, D.T.; Ziegler, M.G. Mechanism of cardiovascular actions of the Chromogranin A fragment catestatin in vivo. Peptides 1998, 19, 1241–1248. [Google Scholar] [CrossRef]
- Kruger, P.G.; Mahata, S.K.; Helle, K.B. Catestatin (CgA344–364) stimulates rat mast cell release of histamine in a manner comparable to mastoparan and other cationic charged neuropeptides. Regul. Pept. 2003, 114, 29–35. [Google Scholar] [CrossRef]
- Kojima, M.; Ozawa, N.; Mori, Y.; Takahashi, Y.; Watanabe-Kominato, K.; Shirai, R.; Watanabe, R.; Sato, K.; Matsuyama, T.A.; Ishibashi-Ueda, H.; et al. Catestatin Prevents Macrophage-Driven Atherosclerosis but Not Arterial Injury-Induced Neointimal Hyperplasia. Thromb. Haemost. 2018, 118, 182–194. [Google Scholar] [CrossRef] [PubMed]
- Aung, G.; Niyonsaba, F.; Ushio, H.; Kajiwara, N.; Saito, H.; Ikeda, S.; Ogawa, H.; Okumura, K. Catestatin, a neuroendocrine antimicrobial peptide, induces human mast cell migration, degranulation and production of cytokines and chemokines. Immunology 2011, 132, 527–539. [Google Scholar] [CrossRef]
- Kljakovic-Gaspic, T.; Tokic, D.; Martinovic, D.; Kumric, M.; Supe-Domic, D.; Stojanovic Stipic, S.; Delic, N.; Vrdoljak, J.; Vilovic, M.; Ticinovic Kurir, T.; et al. Prognostic Value of Catestatin in Severe COVID-19: An ICU-Based Study. J. Clin. Med. 2022, 11, 4496. [Google Scholar] [CrossRef]
- Jati, S.; Kundu, S.; Chakraborty, A.; Mahata, S.K.; Nizet, V.; Sen, M. Wnt5A Signaling Promotes Defense Against Bacterial Pathogens by Activating a Host Autophagy Circuit. Front. Immunol. 2018, 9, 679. [Google Scholar] [CrossRef]
- Zhang, W.; Carravetta, V.; Plekan, O.; Feyer, V.; Richter, R.; Coreno, M.; Prince, K.C. Electronic structure of aromatic amino acids studied by soft x-ray spectroscopy. J. Chem. Phys. 2009, 131, 035103. [Google Scholar] [CrossRef] [PubMed]
- Matta, C.F.; Hernandez-Trujillo, J.; Tang, T.H.; Bader, R.F. Hydrogen-hydrogen bonding: A stabilizing interaction in molecules and crystals. Chemistry (Easton) 2003, 9, 1940–1951. [Google Scholar] [CrossRef] [PubMed]
- Scheiner, S.; Kar, T.; Pattanayak, J. Comparison of various types of hydrogen bonds involving aromatic amino acids. J. Am. Chem. Soc. 2002, 124, 13257–13264. [Google Scholar] [CrossRef] [PubMed]
- Dougherty, D.A. Cation-pi interactions involving aromatic amino acids. J. Nutr. 2007, 137, 1504S–1508S. [Google Scholar] [CrossRef] [PubMed]
- Dougherty, D.A. The cation-pi interaction. Acc. Chem. Res. 2013, 46, 885–893. [Google Scholar] [CrossRef] [PubMed]
- Chelli, R.; Gervasio, F.L.; Procacci, P.; Schettino, V. Stacking and T-shape competition in aromatic-aromatic amino acid interactions. J. Am. Chem. Soc. 2002, 124, 6133–6143. [Google Scholar] [CrossRef]
- Yajima, T.; Takamido, R.; Shimazaki, Y.; Odani, A.; Nakabayashi, Y.; Yamauchi, O. π-π stacking assisted binding of aromatic amino acids by copper(II)-aromatic diimine complexes. Effects of ring substituents on ternary complex stability. Dalton Trans. 2007, 3, 299–307. [Google Scholar] [CrossRef]
- Kelkar, D.A.; Chattopadhyay, A. Membrane interfacial localization of aromatic amino acids and membrane protein function. J. Biosci. 2006, 31, 297–302. [Google Scholar] [CrossRef]
- Brocchieri, L.; Karlin, S. Geometry of interplanar residue contacts in protein structures. Proc. Natl. Acad. Sci. USA 1994, 91, 9297–9301. [Google Scholar] [CrossRef]
- Meyer, E.A.; Castellano, R.K.; Diederich, F. Interactions with aromatic rings in chemical and biological recognition. Angew. Chem. Int. Ed. Engl. 2003, 42, 1210–1250. [Google Scholar] [CrossRef]
- Espinoza-Fonseca, L.M. Aromatic residues link binding and function of intrinsically disordered proteins. Mol. Biosyst. 2012, 8, 237–246. [Google Scholar] [CrossRef] [PubMed]
- Loladze, V.V.; Ermolenko, D.N.; Makhatadze, G.I. Thermodynamic consequences of burial of polar and non-polar amino acid residues in the protein interior. J. Mol. Biol. 2002, 320, 343–357. [Google Scholar] [CrossRef] [PubMed]
- Kellis, J.T., Jr.; Nyberg, K.; Sali, D.; Fersht, A.R. Contribution of hydrophobic interactions to protein stability. Nature 1988, 333, 784–786. [Google Scholar] [CrossRef] [PubMed]
- Zhao, N.; Pang, B.; Shyu, C.R.; Korkin, D. Charged residues at protein interaction interfaces: Unexpected conservation and orchestrated divergence. Protein Sci. 2011, 20, 1275–1284. [Google Scholar] [CrossRef]
- Sahu, B.S.; Obbineni, J.M.; Sahu, G.; Allu, P.K.; Subramanian, L.; Sonawane, P.J.; Singh, P.K.; Sasi, B.K.; Senapati, S.; Maji, S.K.; et al. Functional genetic variants of the catecholamine-release-inhibitory peptide catestatin in an Indian population: Allele-specific effects on metabolic traits. J. Biol. Chem. 2012, 287, 43840–43852. [Google Scholar] [CrossRef]
- Choi, Y.; Miura, M.; Nakata, Y.; Sugasawa, T.; Nissato, S.; Otsuki, T.; Sugawara, J.; Iemitsu, M.; Kawakami, Y.; Shimano, H.; et al. A common genetic variant of the Chromogranin A-derived peptide catestatin is associated with atherogenesis and hypertension in a Japanese population. Endocr. J. 2015, 62, 797–804. [Google Scholar] [CrossRef]
- Dhindsa, R.S.; Copeland, B.R.; Mustoe, A.M.; Goldstein, D.B. Natural Selection Shapes Codon Usage in the Human Genome. Am. J. Hum. Genet. 2020, 107, 83–95. [Google Scholar] [CrossRef]
- McGarrah, R.W.; White, P.J. Branched-chain amino acids in cardiovascular disease. Nat. Rev. Cardiol. 2023, 20, 77–89. [Google Scholar] [CrossRef]
- Rees, J.S.; Castellano, S.; Andres, A.M. The Genomics of Human Local Adaptation. Trends Genet. 2020, 36, 415–428. [Google Scholar] [CrossRef]
- Brunner, J.S.; Finley, L.W.S. Metabolic determinants of tumour initiation. Nat. Rev. Endocrinol. 2023, 19, 134–150. [Google Scholar] [CrossRef]
- Muntjewerff, E.M.; Christoffersson, G.; Mahata, S.K.; van den Bogaart, G. Putative regulation of macrophage-mediated inflammation by catestatin. Trends Immunol. 2022, 43, 41–50. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Shooshtarizadeh, P.; Laventie, B.J.; Colin, D.A.; Chich, J.F.; Vidic, J.; de Barry, J.; Chasserot-Golaz, S.; Delalande, F.; Van Dorsselaer, A.; et al. Two Chromogranin A-derived peptides induce calcium entry in human neutrophils by calmodulin-regulated calcium independent phospholipase A2. PLoS ONE 2009, 4, e4501. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jati, S.; Mahata, S.; Das, S.; Chatterjee, S.; Mahata, S.K. Catestatin: Antimicrobial Functions and Potential Therapeutics. Pharmaceutics 2023, 15, 1550. https://doi.org/10.3390/pharmaceutics15051550
Jati S, Mahata S, Das S, Chatterjee S, Mahata SK. Catestatin: Antimicrobial Functions and Potential Therapeutics. Pharmaceutics. 2023; 15(5):1550. https://doi.org/10.3390/pharmaceutics15051550
Chicago/Turabian StyleJati, Suborno, Sumana Mahata, Soumita Das, Saurabh Chatterjee, and Sushil K. Mahata. 2023. "Catestatin: Antimicrobial Functions and Potential Therapeutics" Pharmaceutics 15, no. 5: 1550. https://doi.org/10.3390/pharmaceutics15051550
APA StyleJati, S., Mahata, S., Das, S., Chatterjee, S., & Mahata, S. K. (2023). Catestatin: Antimicrobial Functions and Potential Therapeutics. Pharmaceutics, 15(5), 1550. https://doi.org/10.3390/pharmaceutics15051550








