A Comprehensive Analysis of 21 Actionable Pharmacogenes in the Spanish Population: From Genetic Characterisation to Clinical Impact
Abstract
1. Introduction
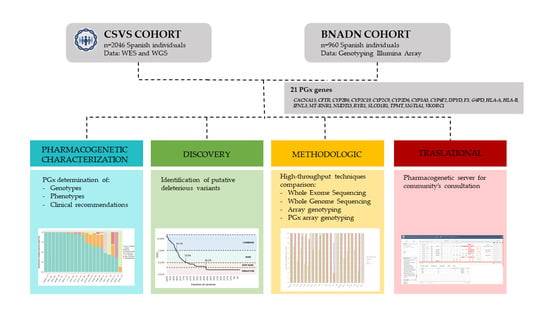
2. Materials and Methods
2.1. Subjects and Genotyping
2.2. Pharmacogenetic Gene Selection
2.3. Pharmacogenetic Data Interpretation
2.4. Allele Nomenclature Considerations
2.5. Allele Frequency Comparison
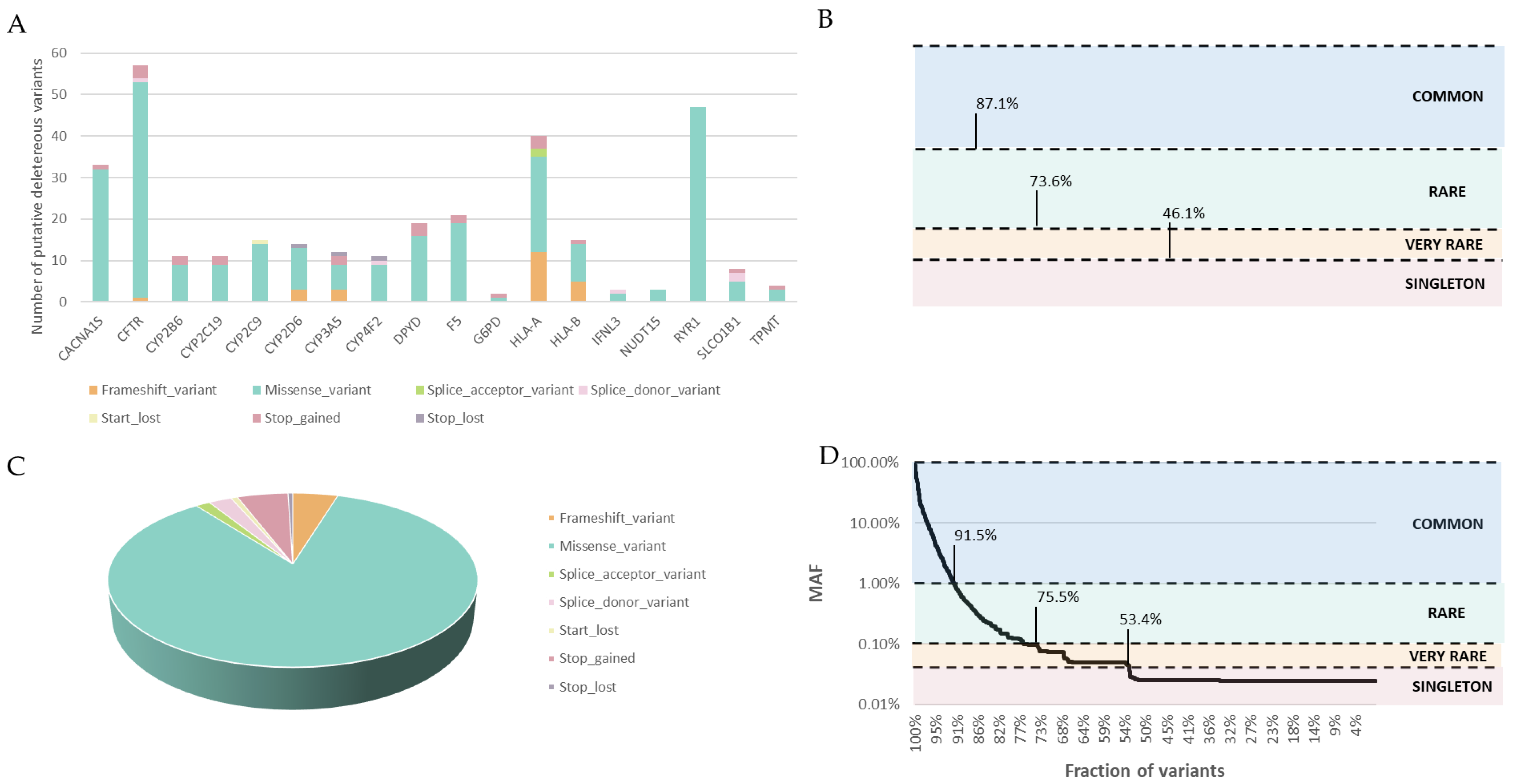
2.6. Putative Deleterious Variants
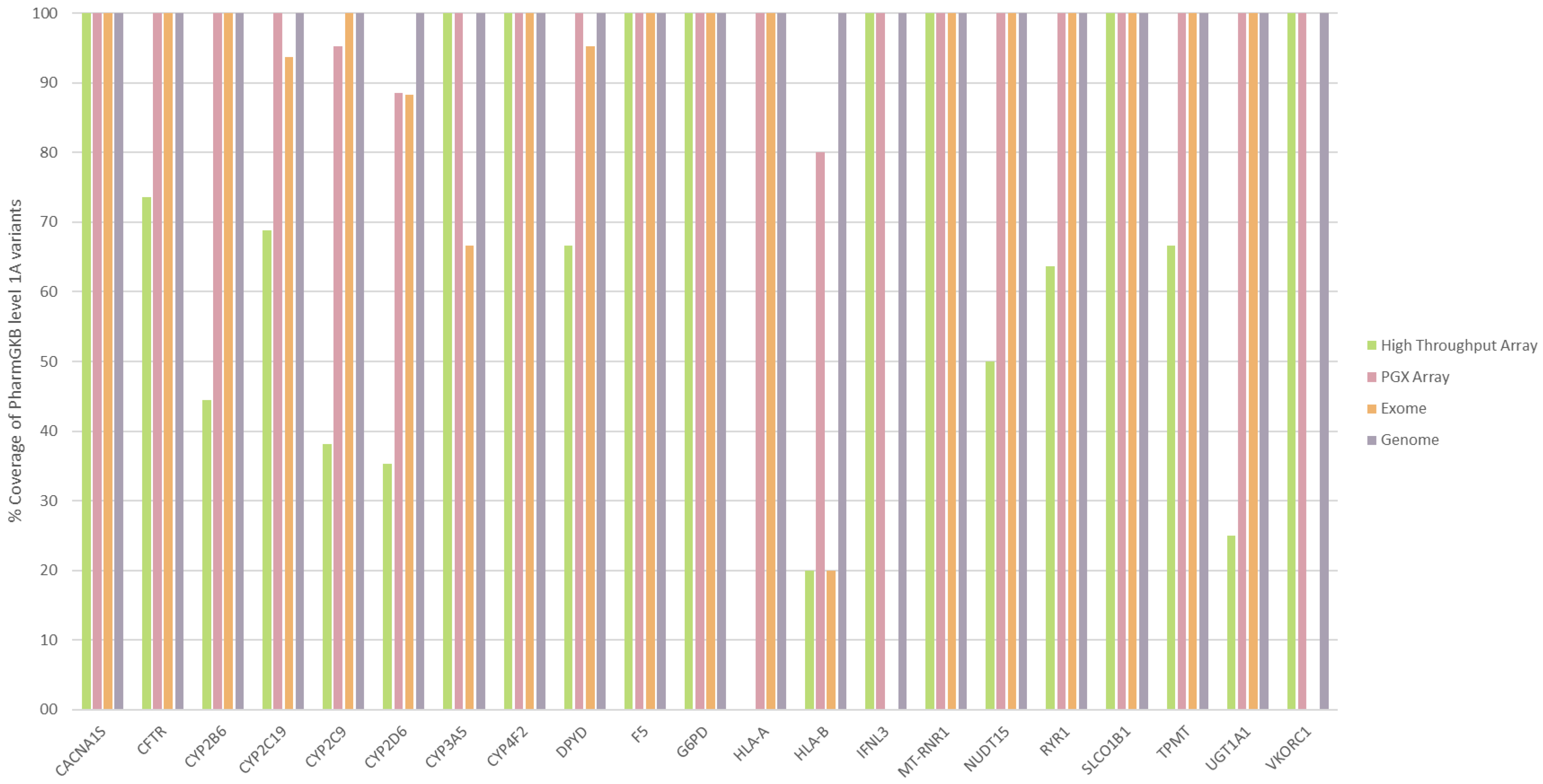
2.7. Comparison of Different Pharmacogenetic Diagnostic Techniques
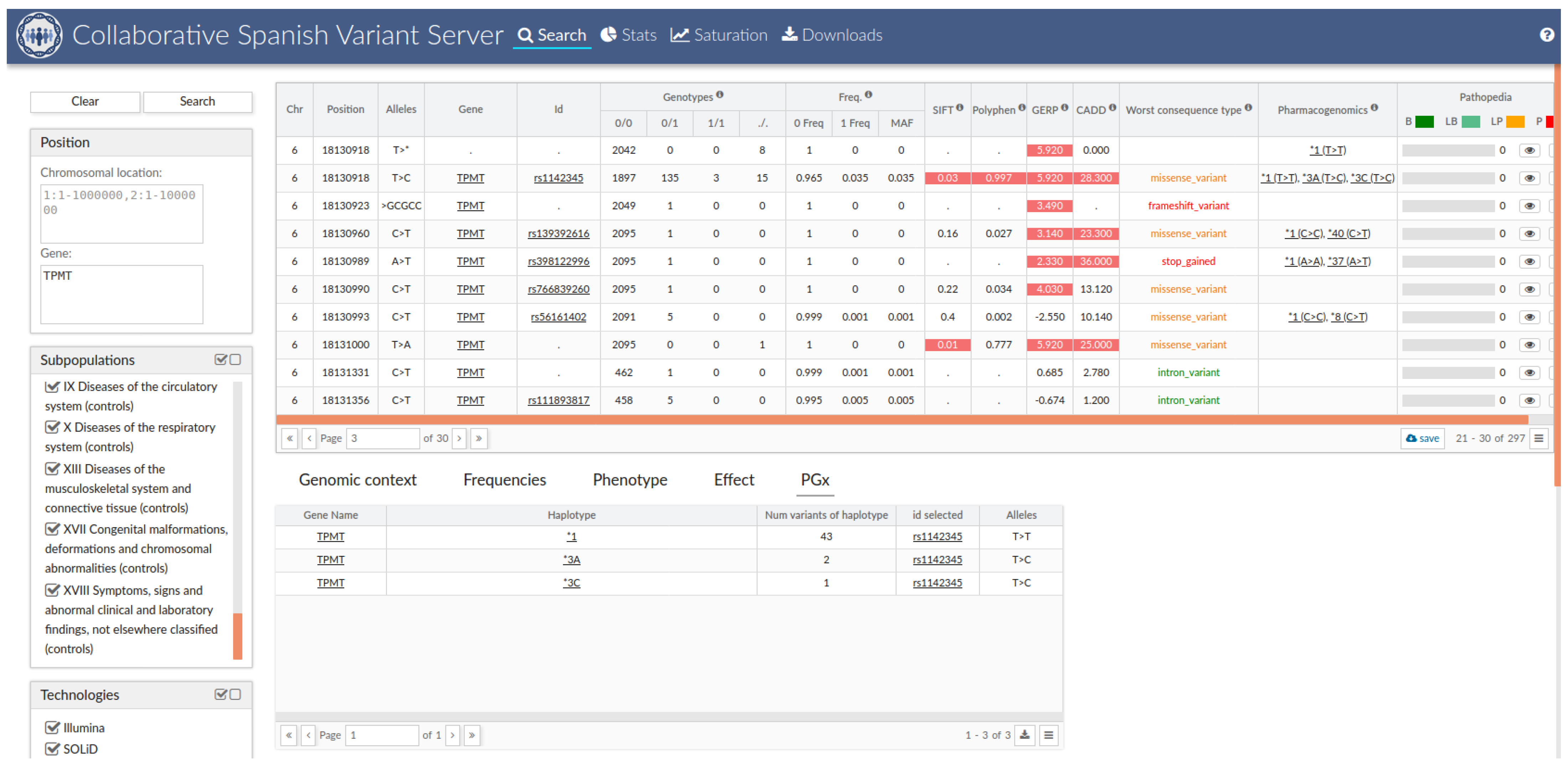
2.8. Collaborative Spanish Variant Server (CSVS) Integration
3. Results
3.1. Pharmacogenetic Genotypes and Phenotypes
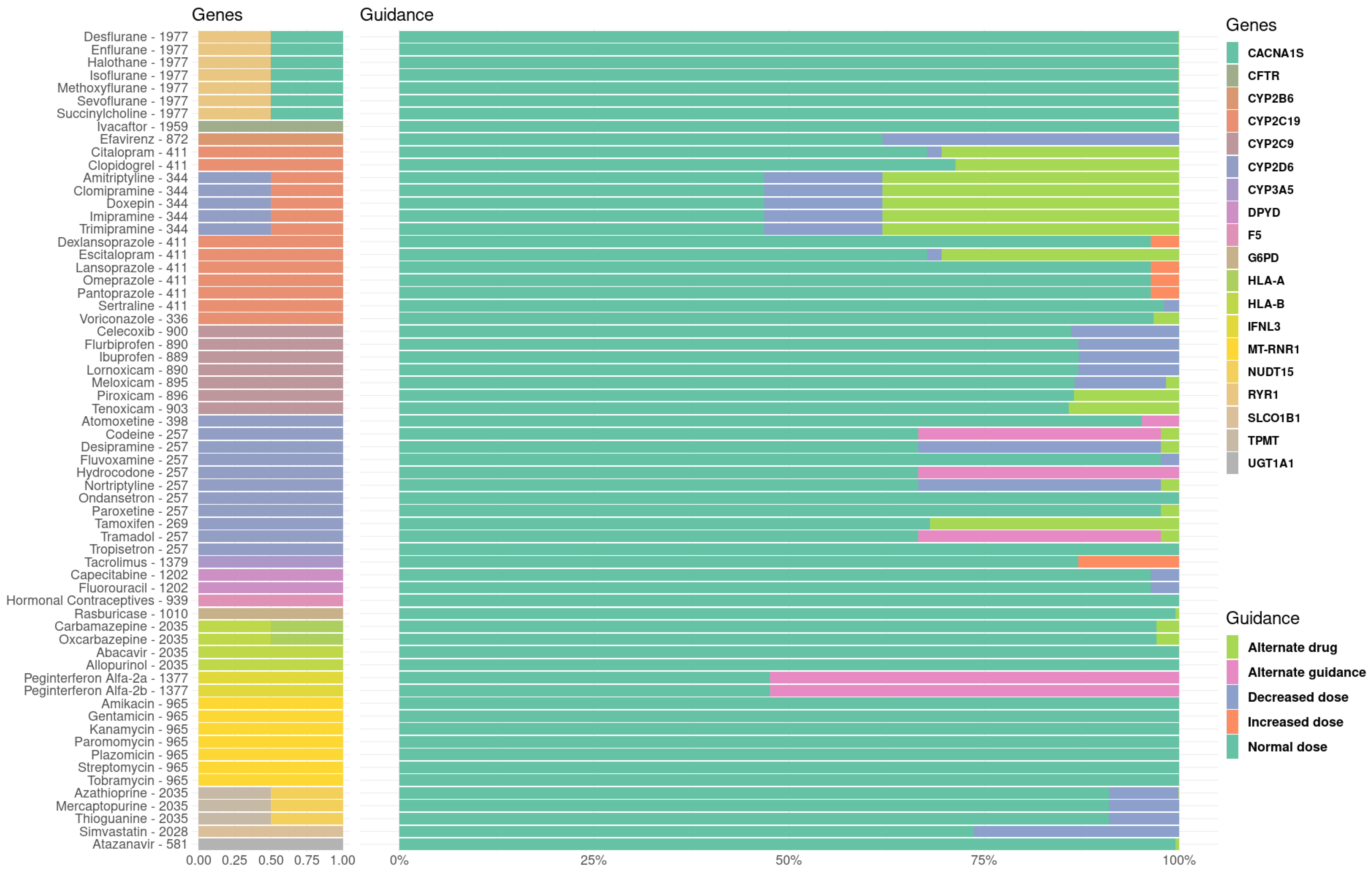
3.2. Therapeutical Impact of the Pharmacogenetic Landscape
3.3. Deleterious Variant Analysis
3.4. Comparison of Different PGx Diagnostic Techniques
3.5. The Spanish Pharmacogenetic Database
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Roden, D.M.; McLeod, H.L.; Relling, M.V.; Williams, M.S.; Mensah, G.A.; Peterson, J.F.; Van Driest, S.L. Pharmacogenomics. Lancet 2019, 394, 521–532. [Google Scholar] [CrossRef]
- Doble, B.; Schofield, D.J.; Roscioli, T.; Mattick, J.S. Prioritising the application of genomic medicine. Npj Genom. Med. 2017, 2, 35. [Google Scholar] [CrossRef] [PubMed]
- van der Wouden, C.; Cambon-Thomsen, A.; Cecchin, E.; Cheung, K.; Dávila-Fajardo, C.; Deneer, V.; Dolžan, V.; Ingelman-Sundberg, M.; Jönsson, S.; Karlsson, M.; et al. Implementing Pharmacogenomics in Europe: Design and Implementation Strategy of the Ubiquitous Pharmacogenomics Consortium. Clin. Pharmacol. Ther. 2017, 101, 341–358. [Google Scholar] [CrossRef]
- Manson, L.E.; van der Wouden, C.H.; Swen, J.J.; Guchelaar, H.-J. The Ubiquitous Pharmacogenomics consortium: Making effective treatment optimization accessible to every European citizen. Pharmacogenomics 2017, 18, 1041–1045. [Google Scholar] [CrossRef]
- Dunnenberger, H.M.; Crews, K.R.; Hoffman, J.M.; Caudle, K.E.; Broeckel, U.; Howard, S.C.; Hunkler, R.J.; Klein, T.E.; Evans, W.E.; Relling, M.V. Preemptive clinical pharmacogenetics implementation: Current programs in five us medical centers. Annu. Rev. Pharmacol. Toxicol. 2015, 55, 89–106. [Google Scholar] [CrossRef] [PubMed]
- Giacomini, K.M.; Karnes, J.H.; Crews, K.R.; Monte, A.A.; Murphy, W.A.; Oni-Orisan, A.; Ramsey, L.B.; Yang, J.J.; Whirl-Carrillo, M. Advancing Precision Medicine through the New Pharmacogenomics Global Research Network. Clin. Pharmacol. Ther. 2021, 110, 559–562. [Google Scholar] [CrossRef] [PubMed]
- Verbelen, M.; Weale, M.E.; Lewis, C.M. Cost-effectiveness of pharmacogenetic-guided treatment: Are we there yet? Pharm. J. 2017, 17, 395–402. [Google Scholar] [CrossRef] [PubMed]
- Relling, M.V.; Klein, T.E. CPIC: Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network. Clin. Pharmacol. Ther. 2011, 89, 464–467. [Google Scholar] [CrossRef]
- Swen, J.J.; Nijenhuis, M.; de Boer, A.; Grandia, L.; Maitland-van der Zee, A.H.; Mulder, H.; Rongen, G.A.P.J.M.; Van Schaik, R.H.N.; Schalekamp, T.; Touw, D.J.; et al. Pharmacogenetics: From bench to byte—An update of guidelines. Clin. Pharmacol. Ther. 2011, 89, 662–673. [Google Scholar] [CrossRef]
- Whirl-Carrillo, M.; Huddart, R.; Gong, L.; Sangkuhl, K.; Thorn, C.F.; Whaley, R.; Klein, T.E. An Evidence-Based Framework for Evaluating Pharmacogenomics Knowledge for Personalized Medicine. Clin. Pharmacol. Ther. 2021, 110, 563–572. [Google Scholar] [CrossRef]
- McInnes, G.; Lavertu, A.; Sangkuhl, K.; Klein, T.E.; Whirl-Carrillo, M.; Altman, R.B. Pharmacogenetics at Scale: An Analysis of the UK Biobank. Clin. Pharmacol. Ther. 2021, 109, 1528–1537. [Google Scholar] [CrossRef] [PubMed]
- Yasuda, S.; Zhang, L.; Huang, S.-M. The Role of Ethnicity in Variability in Response to Drugs: Focus on Clinical Pharmacology Studies. Clin. Pharmacol. Ther. 2008, 84, 417–423. [Google Scholar] [CrossRef]
- Huang, S.-M.; Temple, R. Is This the Drug or Dose for You?: Impact and Consideration of Ethnic Factors in Global Drug Development, Regulatory Review, and Clinical Practice. Clin. Pharmacol. Ther. 2008, 84, 287–294. [Google Scholar] [CrossRef]
- Ramamoorthy, A.; Pacanowski, M.; Bull, J.; Zhang, L. Racial/ethnic differences in drug disposition and response: Review of recently approved drugs. Clin. Pharmacol. Ther. 2015, 97, 263–273. [Google Scholar] [CrossRef] [PubMed]
- Goljan, E.; Abouelhoda, M.; ElKalioby, M.M.; Jabaan, A.; Alghithi, N.; Meyer, B.F.; Monies, D. Identification of pharmacogenetic variants from large scale next generation sequencing data in the Saudi population. PLoS ONE 2022, 17, e0263137. [Google Scholar] [CrossRef]
- Jithesh, P.V.; Abuhaliqa, M.; Syed, N.; Ahmed, I.; El Anbari, M.; Bastaki, K.; Sherif, S.; Umlai, U.-K.; Jan, Z.; Gandhi, G.; et al. A population study of clinically actionable genetic variation affecting drug response from the Middle East. Npj Genom. Med. 2022, 7, 10. [Google Scholar] [CrossRef] [PubMed]
- Lanillos, J.; Carcajona, M.; Maietta, P.; Alvarez, S.; Rodriguez-Antona, C. Clinical pharmacogenetic analysis in 5001 individuals with diagnostic Exome Sequencing data. Npj Genom. Med. 2022, 7, 12. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.H.C.; Chan, M.C.Y.; Chung, C.C.Y.; Li, A.W.T.; Yip, C.Y.W.; Mak, C.C.Y.; Chau, J.F.T.; Lee, M.; Fung, J.L.F.; Tsang, M.H.Y.; et al. Actionable pharmacogenetic variants in Hong Kong Chinese exome sequencing data and projected prescription impact in the Hong Kong population. PLoS Genet. 2021, 17, e1009323. [Google Scholar] [CrossRef]
- Lunenburg, C.A.T.C.; Thirstrup, J.P.; Bybjerg-Grauholm, J.; Bækvad-Hansen, M.; Hougaard, D.M.; Nordentoft, M.; Werge, T.; Børglum, A.D.; Mors, O.; Mortensen, P.B.; et al. Pharmacogenetic genotype and phenotype frequencies in a large Danish population-based case-cohort sample. Transl. Psychiatry 2021, 11, 294. [Google Scholar] [CrossRef] [PubMed]
- Gayán, J.; Galan, J.J.; González-Pérez, A.; Sáez, M.E.; Martínez-Larrad, M.T.; Zabena, C.; Rivero, M.C.; Salinas, A.; Ramírez-Lorca, R.; Morón, F.J.; et al. Genetic Structure of the Spanish Population. BMC Genom. 2010, 11, 326. [Google Scholar] [CrossRef]
- Nagar, S.D.; Conley, A.B.; Jordan, I.K. Population structure and pharmacogenomic risk stratification in the United States. BMC Biol. 2020, 18, 140. [Google Scholar] [CrossRef] [PubMed]
- Peña-Chilet, M.; Roldán, G.; Perez-Florido, J.; Ortuño, F.M.; Carmona, R.; Aquino, V.; Lopez-Lopez, D.; Loucera, C.; Fernandez-Rueda, J.L.; Gallego, A.; et al. CSVS, a crowdsourcing database of the Spanish population genetic variability. Nucleic Acids Res. 2021, 49, D1130–D1137. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Fairley, S.; Lowy-Gallego, E.; Perry, E.; Flicek, P. The International Genome Sample Resource (IGSR) collection of open human genomic variation resources. Nucleic Acids Res. 2020, 48, D941–D947. [Google Scholar] [CrossRef]
- Barbarino, J.M.; Whirl-Carrillo, M.; Altman, R.B.; Klein, T.E. PharmGKB: A worldwide resource for pharmacogenomic information. Wiley Interdiscip. Rev. Syst. Biol. Med. 2018, 10, e1417. [Google Scholar] [CrossRef]
- Sangkuhl, K.; Whirl-Carrillo, M.; Whaley, R.M.; Woon, M.; Lavertu, A.; Altman, R.; Carter, L.; Verma, A.; Ritchie, M.D.; Klein, T.E. Pharmacogenomics Clinical Annotation Tool (PharmCAT). Clin. Pharmacol. Ther. 2020, 107, 203–210. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.B.; Wheeler, M.M.; Patterson, K.; McGee, S.; Dalton, R.; Woodahl, E.L.; Gaedigk, A.; Thummel, K.E.; Nickerson, D.A. Stargazer: A software tool for calling star alleles from next-generation sequencing data using CYP2D6 as a model. Genet Med. 2019, 21, 361–372. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Shen, F.; Gonzaludo, N.; Malhotra, A.; Rogert, C.; Taft, R.J.; Bentley, D.R.; Eberle, M.A. Cyrius: Accurate CYP2D6 genotyping using whole-genome sequencing data. Pharm. J. 2021, 21, 251–261. [Google Scholar] [CrossRef]
- Luo, Y.; Kanai, M.; Choi, W.; Li, X.; Sakaue, S.; Yamamoto, K.; Ogawa, K.; Gutierrez-Arcelus, M.; Gregersen, P.K.; Stuart, P.E.; et al. A high-resolution HLA reference panel capturing global population diversity enables multi-ancestry fine-mapping in HIV host response. Nat. Genet. 2021, 53, 1504–1516. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; Available online: https://ggplot2.tidyverse.org (accessed on 3 November 2021).
- R Core Team. R: A Language and Environment for Statistical Computing. 2021. Available online: https://www.r-project.org/ (accessed on 3 November 2021).
- Gaedigk, A.; Ingelman-Sundberg, M.; Miller, N.A.; Leeder, J.S.; Whirl-Carrillo, M.; Klein, T.E. The Pharmacogene Variation (PharmVar) Consortium: Incorporation of the Human Cytochrome P450 (CYP) Allele Nomenclature Database. Clin. Pharmacol. Ther. 2018, 103, 399–401. [Google Scholar] [CrossRef]
- McLaren, W.; Gil, L.; Hunt, S.E.; Riat, H.S.; Ritchie, G.R.S.; Thormann, A.; Flicek, P.; Cunningham, F. The Ensembl Variant Effect Predictor. Genome Biol. 2016, 17, 122. [Google Scholar] [CrossRef] [PubMed]
- Ng, P.C. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003, 31, 3812–3814. [Google Scholar] [CrossRef]
- Adzhubei, I.A.; Schmidt, S.; Peshkin, L.; Ramensky, V.E.; Gerasimova, A.; Bork, P.; Kondrashov, A.S.; Sunyaev, S.R. A method and server for predicting damaging missense mutations. Nat. Methods 2010, 7, 248–249. [Google Scholar] [CrossRef] [PubMed]
- Kircher, M.; Witten, D.M.; Jain, P.; O’Roak, B.J.; Cooper, G.M.; Shendure, J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014, 46, 310–315. [Google Scholar] [CrossRef] [PubMed]
- Cooper, G.M.; Stone, E.A.; Asimenos, G.; Green, E.D.; Batzoglou, S.; Sidow, A. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 2005, 15, 901–913. [Google Scholar] [CrossRef]
- Karczewski, K.J.; Francioli, L.C.; Tiao, G.; Cummings, B.B.; Alföldi, J.; Wang, Q.; Collins, R.L.; Laricchia, K.M.; Ganna, A.; Birnbaum, D.P.; et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2020, 581, 434–443. [Google Scholar] [CrossRef] [PubMed]
- Higgins, J.; Dalgleish, R.; Dunnen, J.T.D.; Barsh, G.; Freeman, P.J.; Cooper, D.N.; Cullinan, S.; Davies, K.E.; Dorkins, H.; Gong, L.; et al. Verifying nomenclature of DNA variants in submitted manuscripts: Guidance for journals. Hum. Mutat. 2021, 42, 3–7. [Google Scholar] [CrossRef]
- Gong, L.; Whirl-Carrillo, M.; Klein, T.E. PharmGKB, an Integrated Resource of Pharmacogenomic Knowledge. Curr. Protoc. 2021, 1, e226. [Google Scholar] [CrossRef]
- Birdwell, K.; Decker, B.; Barbarino, J.; Peterson, J.; Stein, C.; Sadee, W.; Wang, D.; Vinks, A.; He, Y.; Swen, J.; et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guidelines for CYP3A5 Genotype and Tacrolimus Dosing. Clin. Pharmacol. Ther. 2015, 98, 19–24. [Google Scholar] [CrossRef]
- Yu, M.; Liu, M.; Zhang, W.; Ming, Y. Pharmacokinetics, Pharmacodynamics and Pharmacogenetics of Tacrolimus in Kidney Transplantation. Curr. Drug Metab. 2018, 19, 513–522. [Google Scholar] [CrossRef] [PubMed]
- Dennis, B.B.; Naji, L.; Jajarmi, Y.; Ahmed, A.; Kim, D. New hope for hepatitis C virus: Summary of global epidemiologic changes and novel innovations over 20 years. World J. Gastroenterol. 2021, 27, 4818–4830. [Google Scholar] [CrossRef]
- Barrios, V.; Escobar, C.; Prieto, L.; Lobos, J.M.; Polo, J.; Vargas, D. Control de la anticoagulación con warfarina o acenocumarol en España. ¿Hay diferencias? Rev. Española Cardiol. 2015, 68, 1181–1182. [Google Scholar] [CrossRef]
- Mega, J.L.; Simon, T.; Collet, J.-P.; Anderson, J.L.; Antman, E.M.; Bliden, K.; Cannon, C.P.; Danchin, N.; Giusti, B.; Gurbel, P.; et al. Reduced-Function CYP2C19 Genotype and Risk of Adverse Clinical Outcomes among Patients Treated with Clopidogrel Predominantly for PCI. JAMA 2010, 304, 1821. [Google Scholar] [CrossRef] [PubMed]
- Hicks, J.K.; Bishop, J.R.; Gammal, R.; Sangkuhl, K.; Bousman, C.A.; Leeder, J.S.; Llerena, A.; Mueller, D.J.; Ramsey, L.B.; Scott, S.A.; et al. A Call for Clear and Consistent Communications Regarding the Role of Pharmacogenetics in Antidepressant Pharmacotherapy. Clin. Pharmacol. Ther. 2020, 107, 50–52. [Google Scholar] [CrossRef] [PubMed]
- Bousman, C.A.; Bengesser, S.A.; Aitchison, K.J.; Amare, A.T.; Aschauer, H.; Baune, B.T.; Asl, B.B.; Bishop, J.R.; Burmeister, M.; Chaumette, B.; et al. Review and Consensus on Pharmacogenomic Testing in Psychiatry. Pharmacopsychiatry 2021, 54, 5–17. [Google Scholar] [CrossRef]
- Crews, K.R.; Monte, A.A.; Huddart, R.; Caudle, K.E.; Kharasch, E.D.; Gaedigk, A.; Dunnenberger, H.M.; Leeder, J.S.; Callaghan, J.T.; Samer, C.F.; et al. Clinical Pharmacogenetics Implementation Consortium Guideline for CYP2D6, OPRM1, and COMT Genotypes and Select Opioid Therapy. Clin. Pharmacol. Ther. 2021, 110, 888–896. [Google Scholar] [CrossRef] [PubMed]
- Herrera-Gómez, F.; Gutierrez-Abejón, E.; Ayestarán, I.; Criado-Espegel, P.; Álvarez, F.J. The Trends in Opioid Use in Castile and Leon, Spain: A Population-Based Registry Analysis of Dispensations in 2015 to 2018. J. Clin. Med. 2019, 8, 2148. [Google Scholar] [CrossRef]
- Province, M.A.; Altman, R.B.; Klein, T.E. Interpreting the CYP2D6 Results from the International Tamoxifen Pharmacogenetics Consortium. Clin. Pharmacol. Ther. 2014, 96, 144–146. [Google Scholar] [CrossRef]
- Goetz, M.P.; Sangkuhl, K.; Guchelaar, H.-J.; Schwab, M.; Province, M.; Whirl-Carrillo, M.; Symmans, W.F.; McLeod, H.L.; Ratain, M.J.; Zembutsu, H.; et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2D6 and Tamoxifen Therapy. Clin. Pharmacol. Ther. 2018, 103, 770–777. [Google Scholar] [CrossRef]
- van Rensburg, R.; Nightingale, S.; Brey, N.; Albertyn, C.H.; Kellermann, T.A.; Taljaard, J.J.; Esterhuizen, T.M.; Sinxadi, P.Z.; Decloedt, E.H. Pharmacogenetics of the Late-Onset Efavirenz Neurotoxicity Syndrome (LENS). Clin. Infect. Dis. 2021, 75, 399–405. [Google Scholar] [CrossRef]
- Ramsey, L.B.; Johnson, S.G.; Caudle, K.E.; Haidar, C.E.; Voora, D.; Wilke, R.A.; Maxwell, W.D.; McLeod, H.L.; Krauss, R.M.; Roden, D.M.; et al. The Clinical Pharmacogenetics Implementation Consortium Guideline for SLCO1B1 and Simvastatin-Induced Myopathy: 2014 Update. Clin. Pharmacol. Ther. 2014, 96, 423–428. [Google Scholar] [CrossRef] [PubMed]
- Cooper-DeHoff, R.M.; Niemi, M.; Ramsey, L.B.; Luzum, J.A.; Tarkiainen, E.K.; Straka, R.J.; Gong, L.; Tuteja, S.; Wilke, R.A.; Wadelius, M.; et al. The Clinical Pharmacogenetics Implementation Consortium Guideline for SLCO1B1, ABCG2, and CYP2C9 genotypes and Statin-Associated Musculoskeletal Symptoms. Clin. Pharmacol. Ther. 2022, 111, 1007–1021. [Google Scholar] [CrossRef]
- Relling, M.V.; Schwab, M.; Whirl-Carrillo, M.; Suarez-Kurtz, G.; Pui, C.-H.; Stein, C.M.; Moyer, A.M.; Evans, W.E.; Klein, T.E.; Antillon-Klussmann, F.G.; et al. Clinical Pharmacogenetics Implementation Consortium Guideline for Thiopurine Dosing Based on TPMT and NUDT 15 Genotypes: 2018 Update. Clin. Pharmacol. Ther. 2019, 105, 1095–1105. [Google Scholar] [CrossRef]
- Meulendijks, D.; Henricks, L.; Sonke, G.; Deenen, M.J.; Froehlich, T.K.; Amstutz, U.; Largiader, C.; Jennings, B.; Marinaki, A.M.; Sanderson, J.D.; et al. Clinical relevance of DPYD variants c.1679T > G, c.1236G > A/HapB3, and c.1601G > A as predictors of severe fluoropyrimidine-associated toxicity: A systematic review and meta-analysis of individual patient data. Lancet Oncol. 2015, 16, 1639–1650. [Google Scholar] [CrossRef] [PubMed]
- Amstutz, U.; Henricks, L.; Offer, S.M.; Barbarino, J.; Schellens, J.H.; Swen, J.; Klein, T.E.; McLeod, H.L.; Caudle, K.E.; Diasio, R.B.; et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for Dihydropyrimidine Dehydrogenase Genotype and Fluoropyrimidine Dosing: 2017 Update. Clin. Pharmacol. Ther. 2018, 103, 210–216. [Google Scholar] [CrossRef]
- Lunenburg, C.A.T.C.; van der Wouden, C.H.; Nijenhuis, M.; Rhenen, M.H.C.-V.; de Boer-Veger, N.J.; Buunk, A.M.; Houwink, E.J.F.; Mulder, H.; Rongen, G.A.; van Schaik, R.H.N.; et al. Dutch Pharmacogenetics Working Group (DPWG) guideline for the gene-drug interaction of DPYD and fluoropyrimidines. Eur. J. Hum. Genet. 2019, 28, 508–517. [Google Scholar] [CrossRef]
- EMA. EMA Recommendations on DPD Testing Prior to Treatment with Fluorouracil, Capecitabine, Tegafur and Flucytosine. Available online: https://www.ema.europa.eu/en/news/ema-recommendations-dpd-testing-prior-treatment-fluorouracil-capecitabine-tegafur-flucytosine (accessed on 5 May 2022).
- Reizine, N.M.; Danahey, K.; Truong, T.M.; George, D.; House, L.K.; Karrison, T.G.; Wijk, X.M.R.; Yeo, K.J.; Ratain, M.J.; O’donnell, P.H. Clinically actionable genotypes for anticancer prescribing among >1500 patients with pharmacogenomic testing. Cancer 2022, 128, 1649–1657. [Google Scholar] [CrossRef]
- Ingelman-Sundberg, M.; Mkrtchian, S.; Zhou, Y.; Lauschke, V.M. Integrating rare genetic variants into pharmacogenetic drug response predictions. Hum. Genom. 2018, 12, 26. [Google Scholar] [CrossRef]
- Ahn, E.; Park, T. Analysis of population-specific pharmacogenomic variants using next-generation sequencing data. Sci. Rep. 2017, 7, 8416. [Google Scholar] [CrossRef] [PubMed]
- Morris, S.A.; Alsaidi, A.T.; Verbyla, A.; Cruz, A.; Macfarlane, C.; Bauer, J.; Patel, J.N. Cost Effectiveness of Pharmacogenetic Testing for Drugs with Clinical Pharmacogenetics Implementation Consortium (CPIC) Guidelines: A Systematic Review. Clin. Pharmacol. Ther. 2022, 112, 1318–1328. [Google Scholar] [CrossRef]
- Schwarze, K.; Buchanan, J.; Taylor, J.C.; Wordsworth, S. Are whole-exome and whole-genome sequencing approaches cost-effective? A systematic review of the literature. Genet. Med. 2018, 20, 1122–1130. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, R.A.; Belmont, J.W.; Hardenbol, P.; Willis, T.D.; Yu, F.L.; Yang, H.M.; Ch’ang, L.Y.; Huang, W.; Liu, B.; Shen, Y.; et al. The International HapMap Project. Nature 2003, 426, 789–796. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nunez-Torres, R.; Pita, G.; Peña-Chilet, M.; López-López, D.; Zamora, J.; Roldán, G.; Herráez, B.; Álvarez, N.; Alonso, M.R.; Dopazo, J.; et al. A Comprehensive Analysis of 21 Actionable Pharmacogenes in the Spanish Population: From Genetic Characterisation to Clinical Impact. Pharmaceutics 2023, 15, 1286. https://doi.org/10.3390/pharmaceutics15041286
Nunez-Torres R, Pita G, Peña-Chilet M, López-López D, Zamora J, Roldán G, Herráez B, Álvarez N, Alonso MR, Dopazo J, et al. A Comprehensive Analysis of 21 Actionable Pharmacogenes in the Spanish Population: From Genetic Characterisation to Clinical Impact. Pharmaceutics. 2023; 15(4):1286. https://doi.org/10.3390/pharmaceutics15041286
Chicago/Turabian StyleNunez-Torres, Rocio, Guillermo Pita, María Peña-Chilet, Daniel López-López, Jorge Zamora, Gema Roldán, Belén Herráez, Nuria Álvarez, María Rosario Alonso, Joaquín Dopazo, and et al. 2023. "A Comprehensive Analysis of 21 Actionable Pharmacogenes in the Spanish Population: From Genetic Characterisation to Clinical Impact" Pharmaceutics 15, no. 4: 1286. https://doi.org/10.3390/pharmaceutics15041286
APA StyleNunez-Torres, R., Pita, G., Peña-Chilet, M., López-López, D., Zamora, J., Roldán, G., Herráez, B., Álvarez, N., Alonso, M. R., Dopazo, J., & Gonzalez-Neira, A. (2023). A Comprehensive Analysis of 21 Actionable Pharmacogenes in the Spanish Population: From Genetic Characterisation to Clinical Impact. Pharmaceutics, 15(4), 1286. https://doi.org/10.3390/pharmaceutics15041286