Metabolomics Profiling Reveals the Role of PEDF in Triple-Negative Breast Cancer Cell MDA-MB-231 under Glycaemic Loading
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Cell Line and Culture Conditions
2.3. Sample Extraction and Derivatisation
2.4. GC-Q MS Analysis
2.5. Data Processing and Statistical Analysis
3. Results
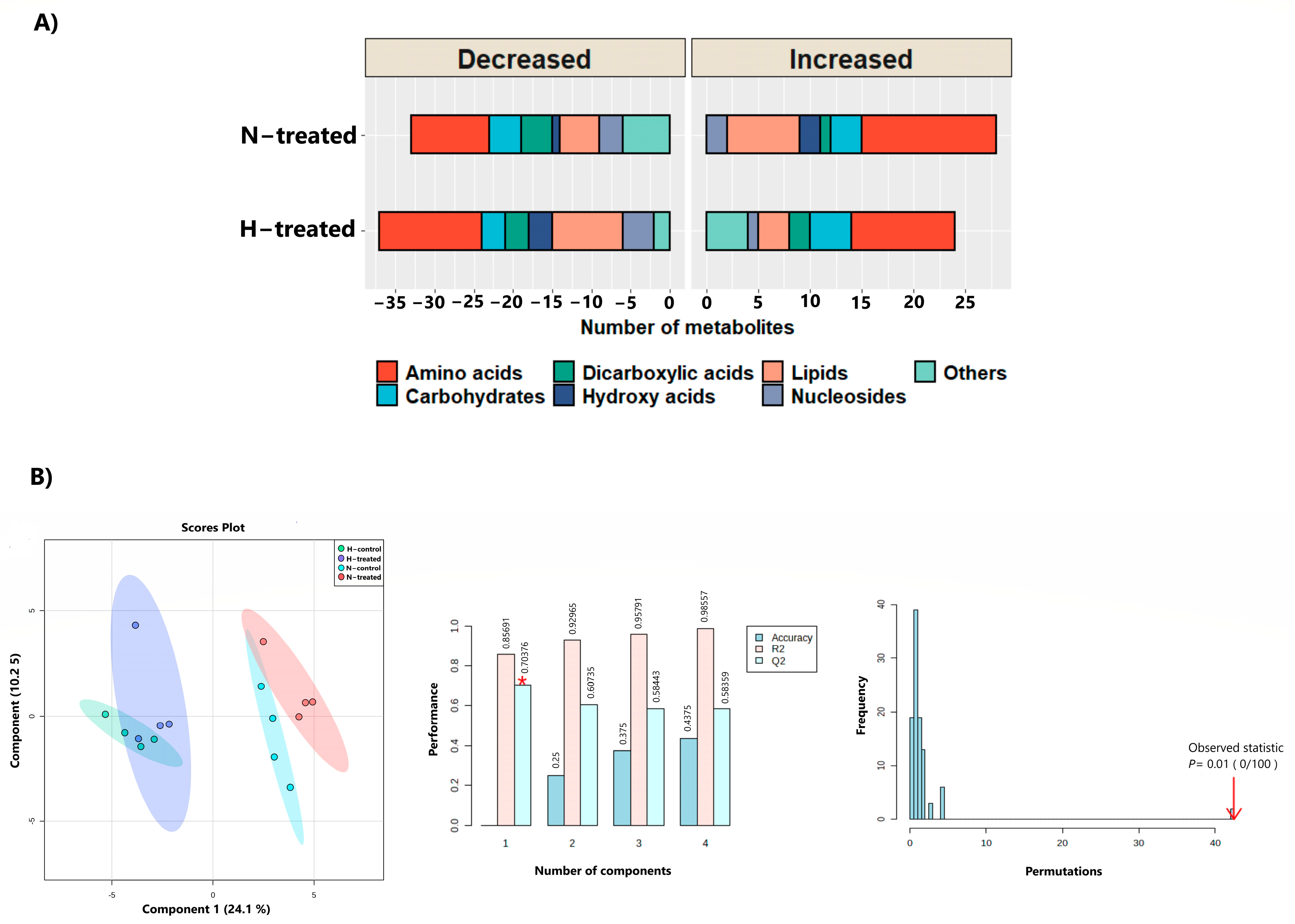
3.1. Untargeted Metabolomics Profiling
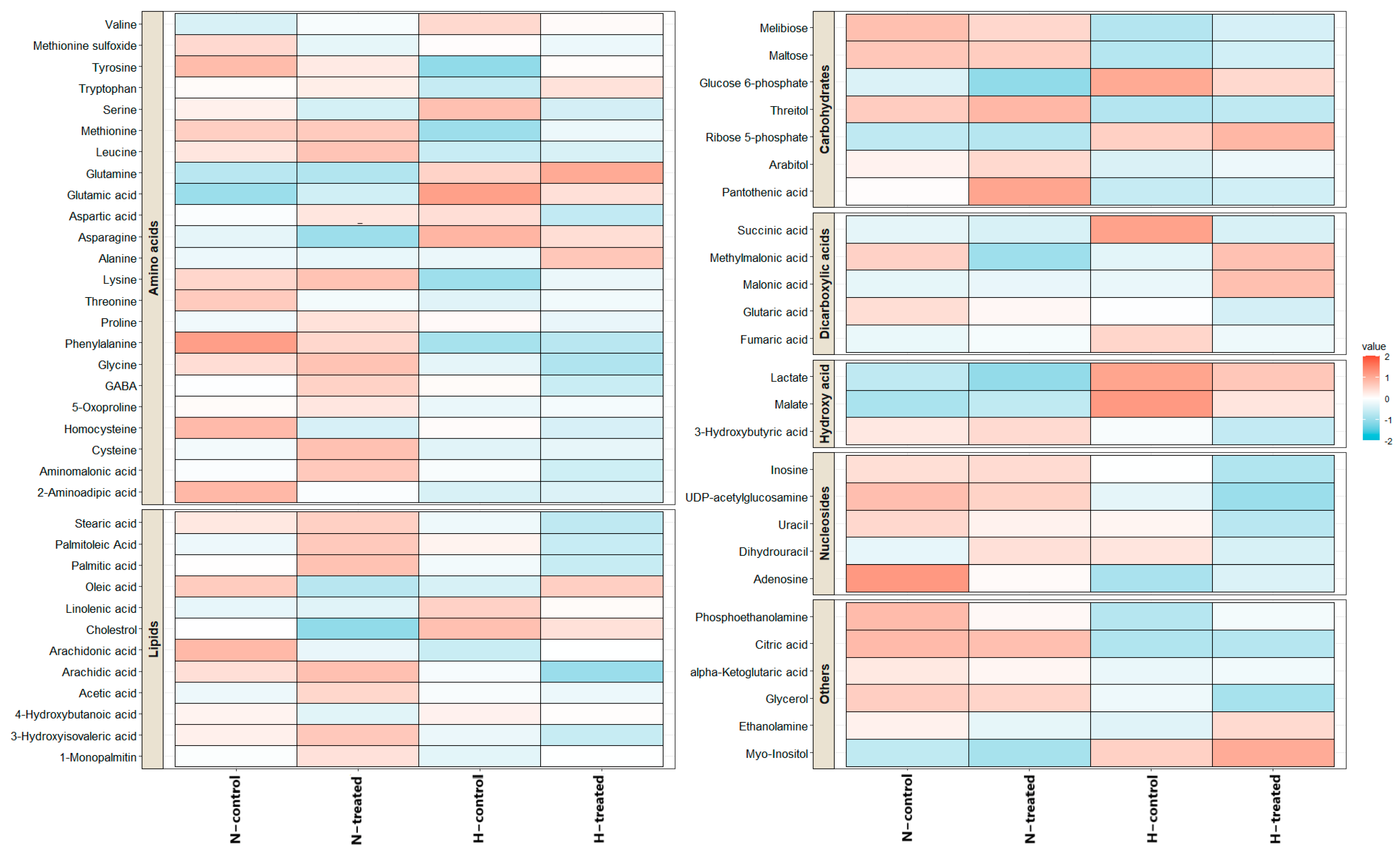
3.2. Effect of PEDF on MDA-MB-231 Metabolomes under Glycaemic Loading
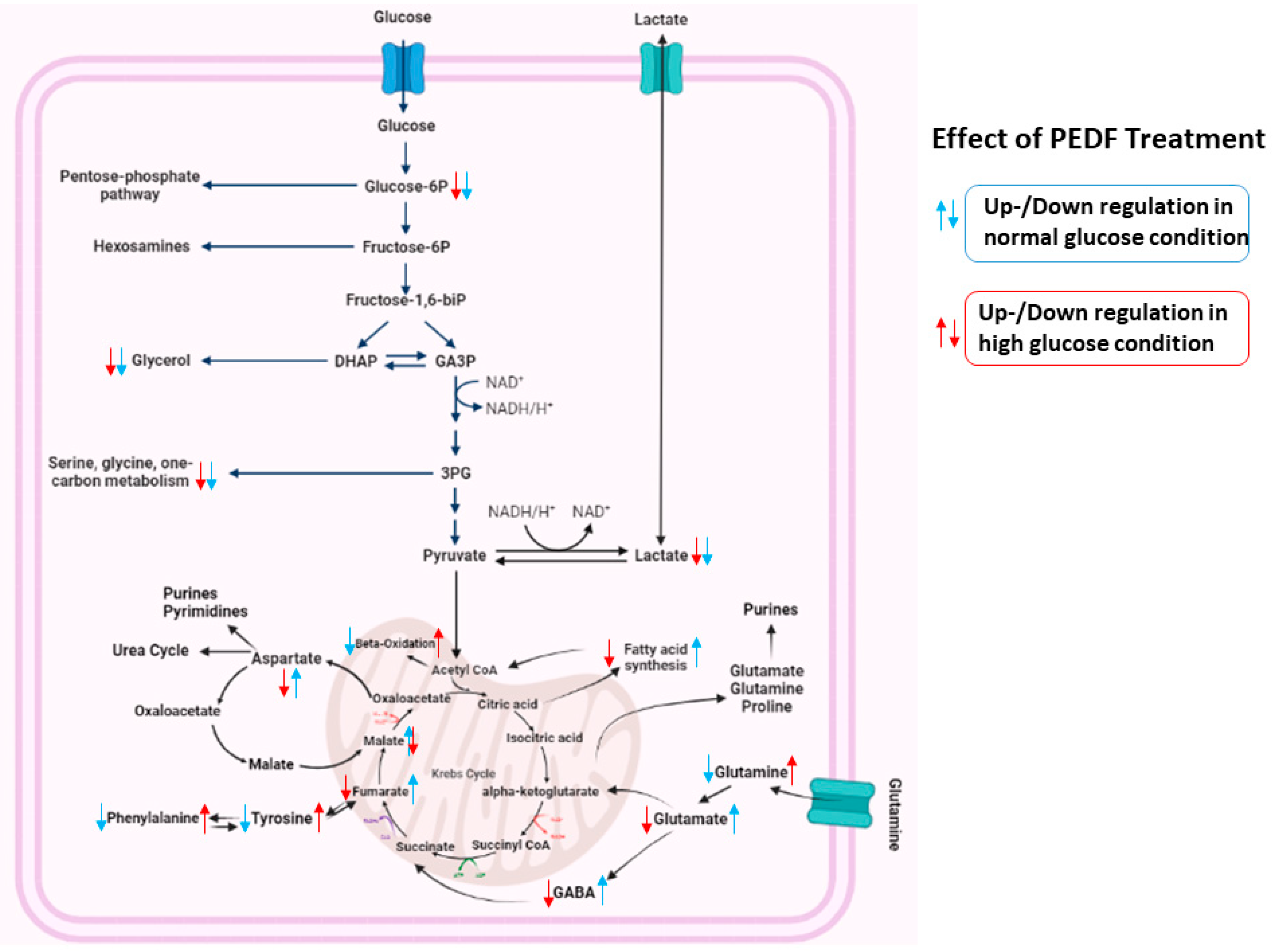
3.3. Pathway Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ferlay, J.; Ervik, M.; Lam, F.; Colombet, M.; Mery, L.; Piñeros, M.; Znaor, A.; Soerjomataram, I.; Bray, F. Global Cancer Observatory: Cancer Tomorrow; International Agency for Research on Cancer: Lyon, France, 2020. [Google Scholar]
- Turashvili, G.; Brogi, E. Tumor heterogeneity in breast cancer. Front. Med. 2017, 4, 227. [Google Scholar] [CrossRef] [PubMed]
- Eisen, M.B.; Spellman, P.T.; Brown, P.O.; Botstein, D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 1998, 95, 14863–14868. [Google Scholar] [CrossRef] [PubMed]
- Sørlie, T.; Perou, C.M.; Tibshirani, R.; Aas, T.; Geisler, S.; Johnsen, H.; Hastie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. USA 2001, 98, 10869–10874. [Google Scholar] [CrossRef] [PubMed]
- Pereira, A.; Siegrist, J.; Lizarraga, S.; Pérez-Medina, T. Clustering Molecular Subtypes in Breast Cancer, Immunohistochemical Parameters and Risk of Axillary Nodal Involvement. J. Pers. Med. 2022, 12, 1404. [Google Scholar] [CrossRef]
- Dent, R.; Trudeau, M.; Pritchard, K.I.; Hanna, W.M.; Kahn, H.K.; Sawka, C.A.; Lickley, L.A.; Rawlinson, E.; Sun, P.; Narod, S.A. Triple-negative breast cancer: Clinical features and patterns of recurrence. Clin. Cancer Res. 2007, 13, 4429–4434. [Google Scholar] [CrossRef]
- Arruebo, M.; Vilaboa, N.; Sáez-Gutierrez, B.; Lambea, J.; Tres, A.; Valladares, M.; González-Fernández, Á. Assessment of the evolution of cancer treatment therapies. Cancers 2011, 3, 3279–3330. [Google Scholar] [CrossRef]
- Pavlova, N.N.; Thompson, C.B. The emerging hallmarks of cancer metabolism. Cell Metab. 2016, 23, 27–47. [Google Scholar] [CrossRef]
- Warburg, O. On the origin of cancer cells. Science 1956, 123, 309–314. [Google Scholar] [CrossRef]
- Michiels, C. Physiological and pathological responses to hypoxia. Am. J. Pathol. 2004, 164, 1875–1882. [Google Scholar] [CrossRef]
- Vigneri, P.; Frasca, F.; Sciacca, L.; Pandini, G.; Vigneri, R. Diabetes and cancer. Endocr. -Relat. Cancer 2009, 16, 1103–1123. [Google Scholar] [CrossRef]
- Qiu, J.; Zheng, Q.; Meng, X. Hyperglycemia and chemoresistance in breast cancer: From cellular mechanisms to treatment response. Front. Oncol. 2021, 11, 628359. [Google Scholar] [CrossRef] [PubMed]
- Becerra, S.P.; Sagasti, A.; Spinella, P.; Notario, V. Pigment Epithelium-derived Factor Behaves Like a Noninhibitory Serpin: NEUROTROPHIC ACTIVITY DOES NOT REQUIRE THE SERPIN REACTIVE LOOP (∗). J. Biol.Chem. 1995, 270, 25992–25999. [Google Scholar] [CrossRef] [PubMed]
- Abooshahab, R.; Al-Salami, H.; Dass, C.R. The increasing role of pigment epithelium-derived factor in metastasis: From biological importance to a promising target. Biochem. Pharmacol. 2021, 193, 114787. [Google Scholar] [CrossRef] [PubMed]
- Carnagarin, R.; Dharmarajan, A.M.; Dass, C.R. PEDF-induced alteration of metabolism leading to insulin resistance. Mol. Cell. Endocrinol. 2015, 401, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Abooshahab, R.; Dass, C.R. The biological relevance of pigment epithelium-derived factor on the path from aging to age-related disease. Mech. Ageing Dev. 2021, 196, 111478. [Google Scholar] [CrossRef]
- Huang, K.-T.; Lin, C.-C.; Tsai, M.-C.; Chen, K.-D.; Chiu, K.-W. Pigment epithelium-derived factor in lipid metabolic disorders. Biomed. J. 2018, 41, 102–108. [Google Scholar] [CrossRef]
- Palmieri, D.; Fitzgerald, D.; Shreeve, S.M.; Hua, E.; Bronder, J.L.; Weil, R.J.; Davis, S.; Stark, A.M.; Merino, M.J.; Kurek, R.; et al. Analyses of Resected Human Brain Metastases of Breast Cancer Reveal the Association between Up-Regulation of Hexokinase 2 and Poor PrognosisUp-Regulation of HK2 Is Associated with Brain Metastasis. Mol. Cancer Res. 2009, 7, 1438–1445. [Google Scholar] [CrossRef]
- Zhou, D.; Cheng, S.-Q.; Ji, H.-F.; Wang, J.-S.; Xu, H.-T.; Zhang, G.-Q.; Pang, D. Evaluation of protein pigment epithelium-derived factor (PEDF) and microvessel density (MVD) as prognostic indicators in breast cancer. J. Cancer Res. Clin. Oncol. 2010, 136, 1719–1727. [Google Scholar] [CrossRef]
- Fitzgerald, D.P.; Subramanian, P.; Deshpande, M.; Graves, C.; Gordon, I.; Qian, Y.; Snitkovsky, Y.; Liewehr, D.J.; Steinberg, S.M.; Paltán-Ortiz, J.D.; et al. Opposing Effects of Pigment Epithelium–Derived Factor on Breast Cancer Cell versus Neuronal Survival: Implication for Brain Metastasis and Metastasis-Induced Brain Damage. Cancer Res. 2012, 72, 144–153. [Google Scholar] [CrossRef]
- Hong, H.; Zhou, T.; Fang, S.; Jia, M.; Xu, Z.; Dai, Z.; Li, C.; Li, S.; Li, L.; Zhang, T.; et al. Pigment epithelium-derived factor (PEDF) inhibits breast cancer metastasis by down-regulating fibronectin. Breast Cancer Res. Treat. 2014, 148, 61–72. [Google Scholar] [CrossRef]
- Schmidt, D.R.; Patel, R.; Kirsch, D.G.; Lewis, C.A.; Vander Heiden, M.G.; Locasale, J.W. Metabolomics in cancer research and emerging applications in clinical oncology. CA A Cancer J. Clin. 2021, 71, 333–358. [Google Scholar] [CrossRef]
- Petersen, S.V.; Valnickova, Z.; Enghild, J.J. Pigment-epithelium-derived factor (PEDF) occurs at a physiologically relevant concentration in human blood: Purification and characterization. Biochem. J. 2003, 374, 199–206. [Google Scholar] [CrossRef]
- Gatenby, R.A.; Gillies, R.J. Why do cancers have high aerobic glycolysis? Nat. Rev. Cancer 2004, 4, 891–899. [Google Scholar] [CrossRef]
- Cho, E.S.; Cha, Y.H.; Kim, H.S.; Kim, N.H.; Yook, J.I. The pentose phosphate pathway as a potential target for cancer therapy. Biomol. Ther. 2018, 26, 29. [Google Scholar] [CrossRef] [PubMed]
- Porporato, P.E.; Filigheddu, N.; Pedro, J.M.B.-S.; Kroemer, G.; Galluzzi, L. Mitochondrial metabolism and cancer. Cell Res. 2018, 28, 265–280. [Google Scholar] [CrossRef]
- Wei, Z.; Liu, X.; Cheng, C.; Yu, W.; Yi, P. Metabolism of amino acids in cancer. Front. Cell Dev. Biol. 2021, 8, 603837. [Google Scholar] [CrossRef]
- DeBerardinis, R.J.; Cheng, T. Q’s next: The diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene 2010, 29, 313–324. [Google Scholar] [CrossRef]
- Cha, Y.J.; Kim, E.-S.; Koo, J.S. Amino acid transporters and glutamine metabolism in breast cancer. Int. J. Mol. Sci. 2018, 19, 907. [Google Scholar] [CrossRef]
- Altman, B.J.; Stine, Z.E.; Dang, C.V. From Krebs to clinic: Glutamine metabolism to cancer therapy. Nat. Rev. Cancer 2016, 16, 619–634. [Google Scholar] [CrossRef]
- Felig, P.; Wahren, J.; Karl, I.; Cerasi, E.; Luft, R.; Kipnis, D.M. Glutamine and glutamate metabolism in normal and diabetic subjects. Diabetes 1973, 22, 573–576. [Google Scholar] [CrossRef]
- Li, P.-A.; Shuaib, A.; Miyashita, H.; He, Q.-P.; Siesjö, B.K. Hyperglycemia enhances extracellular glutamate accumulation in rats subjected to forebrain ischemia. Stroke 2000, 31, 183–192. [Google Scholar] [CrossRef] [PubMed]
- Stumvoll, M.; Perriello, G.; Meyer, C.; Gerich, J. Role of glutamine in human carbohydrate metabolism in kidney and other tissues. Kidney Int. 1999, 55, 778–792. [Google Scholar] [CrossRef] [PubMed]
- Adams, S.H. Emerging perspectives on essential amino acid metabolism in obesity and the insulin-resistant state. Adv. Nutr. 2011, 2, 445–456. [Google Scholar] [CrossRef]
- Miyagi, Y.; Higashiyama, M.; Gochi, A.; Akaike, M.; Ishikawa, T.; Miura, T.; Saruki, N.; Bando, E.; Kimura, H.; Imamura, F.; et al. Plasma free amino acid profiling of five types of cancer patients and its application for early detection. PLoS ONE 2011, 6, e24143. [Google Scholar] [CrossRef]
- Yuan, B.; Schafferer, S.; Tang, Q.; Scheffler, M.; Nees, J.; Heil, J.; Schott, S.; Golatta, M.; Wallwiener, M.; Sohn, C.; et al. A plasma metabolite panel as biomarkers for early primary breast cancer detection. Int. J. Cancer 2019, 144, 2833–2842. [Google Scholar] [CrossRef]
- Zhou, Q.; Sun, W.-W.; Chen, J.-C.; Zhang, H.-L.; Liu, J.; Lin, Y.; Lin, P.-C.; Wu, B.-X.; An, Y.-P.; Huang, L.; et al. Phenylalanine impairs insulin signaling and inhibits glucose uptake through modification of IRβ. Nat. Commun. 2022, 13, 1–19. [Google Scholar] [CrossRef]
- Haskó, G.; Antonioli, L.; Cronstein, B.N. Adenosine metabolism, immunity and joint health. Biochem. Pharmacol. 2018, 151, 307–313. [Google Scholar] [CrossRef]
- Fredholm, B.B.; Ijzerman, A.P.; Jacobson, K.A.; Klotz, K.N.; Linden, J. International Union of Pharmacology. XXV. Nomenclature and classification of adenosine receptors. Pharmacol. Rev. 2001, 53, 527–552. [Google Scholar]
- Leone, R.D.; Emens, L.A. Targeting adenosine for cancer immunotherapy. J. Immunother. Cancer 2018, 6, 1–9. [Google Scholar] [CrossRef]
- Fernandez-Gallardo, M.; González-Ramírez, R.; Sandoval, A.; Felix, R.; Monjaraz, E. Adenosine stimulate proliferation and migration in triple negative breast cancer cells. PLoS ONE 2016, 11, e0167445. [Google Scholar] [CrossRef]
- Sakowicz, M.; Szutowicz, A.; Pawelczyk, T. Differential effect of insulin and elevated glucose level on adenosine transport in rat B lymphocytes. Int. Immunol. 2005, 17, 145–154. [Google Scholar] [CrossRef] [PubMed]
- Phillis, J.W.; Simpson, R.; Walter, G. The effect of hyperglycemia on extracellular levels of adenosine in the hypoxic rat cerebral cortex. Brain Res. 1990, 524, 336–338. [Google Scholar] [CrossRef]
- Dong, Q.; Ginsberg, H.; Erlanger, B. Overexpression of the A1 adenosine receptor in adipose tissue protects mice from obesity-related insulin resistance. Diabetes Obes. Metab. 2001, 3, 360–366. [Google Scholar] [CrossRef]
- Vannucci, S.J.; Nishimura, H.; Satoh, S.; Cushman, S.W.; Holman, G.; A Simpson, I. Cell surface accessibility of GLUT4 glucose transporters in insulin-stimulated rat adipose cells. Modulation by isoprenaline and adenosine. Biochem. J. 1992, 288, 325–330. [Google Scholar]
- Mayengbam, S.S.; Singh, A.; Pillai, A.D.; Bhat, M.K. Influence of cholesterol on cancer progression and therapy. Transl. Oncol. 2021, 14, 101043. [Google Scholar] [CrossRef]
- Ding, X.; Zhang, W.; Li, S.; Yang, H. The role of cholesterol metabolism in cancer. Am. J. Cancer Res. 2019, 9, 219. [Google Scholar]


| Metabolites | RT | VIP | One-Way ANOVA | Multiple Comparisons Tukey HSD (a p-Value) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ap-Value | FDR | HT vs. HC | NC vs. HC | NT vs. HC | NC vs. HT | NT vs. HT | NC vs. NT | |||
| Lactic acid | 5.58 | 1.9908 | 3.63 × 10−5 | 0.002216 | 0.488 | 0.0002 ↓↑ | 4.20 × 10−5 ↓↑ | 0.002 ↓↑ | 0.0003 ↓↑ | 0.56 |
| Phenylalanine | 9.56 | 1.532 | 0.00052 | 0.015849 | 0.983 | 0.001 ↑↓ | 0.025 ↑↓ | 0.001 ↑↓ | 0.047 ↑↓ | 0.304 |
| Malate | 8.65 | 1.7187 | 0.001395 | 0.023962 | 0.213 | 0.001 ↓↑ | 0.003 ↓↑ | 0.055 | 0.121 | 0.966 |
| Glutamic acid | 9.46 | 1.6052 | 0.001571 | 0.023962 | 0.285 | 0.001 ↓↑ | 0.006 ↓↑ | 0.026 ↓↑ | 0.171 | 0.68 |
| Glucose-6-phosphate | 13.01 | 1.834 | 0.00251 | 0.030615 | 0.601 | 0.031 ↓↑ | 0.0024 ↓↑ | 0.248 | 0.021 | 0.481 |
| Myo-inositol | 13.5 | 1.5331 | 0.003189 | 0.03242 | 0.794 | 0.087 | 0.037 ↓↑ | 0.017 ↓↑ | 0.007 ↓↑ | 0.957 |
| Glutamine | 10.33 | 1.4553 | 0.00443 | 0.035565 | 0.735 | 0.067 | 0.054 | 0.011 ↓↑ | 0.009 ↓↑ | 0.999 |
| Citric acid | 10.62 | 1.5986 | 0.004664 | 0.035565 | 0.999 | 0.030 ↑↓ | 0.035 ↑↓ | 0.032 ↑↓ | 0.038 ↑↓ | 0.999 |
| Adenosine | 14.29 | 1.1194 | 0.006105 | 0.041381 | 0.785 | 0.009 ↑↓ | 0.399 | 0.046 ↑↓ | 0.898 | 0.148 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abooshahab, R.; Hooshmand, K.; Luna, G.; Al-Salami, H.; Dass, C.R. Metabolomics Profiling Reveals the Role of PEDF in Triple-Negative Breast Cancer Cell MDA-MB-231 under Glycaemic Loading. Pharmaceutics 2023, 15, 543. https://doi.org/10.3390/pharmaceutics15020543
Abooshahab R, Hooshmand K, Luna G, Al-Salami H, Dass CR. Metabolomics Profiling Reveals the Role of PEDF in Triple-Negative Breast Cancer Cell MDA-MB-231 under Glycaemic Loading. Pharmaceutics. 2023; 15(2):543. https://doi.org/10.3390/pharmaceutics15020543
Chicago/Turabian StyleAbooshahab, Raziyeh, Kourosh Hooshmand, Giuseppe Luna, Hani Al-Salami, and Crispin R. Dass. 2023. "Metabolomics Profiling Reveals the Role of PEDF in Triple-Negative Breast Cancer Cell MDA-MB-231 under Glycaemic Loading" Pharmaceutics 15, no. 2: 543. https://doi.org/10.3390/pharmaceutics15020543
APA StyleAbooshahab, R., Hooshmand, K., Luna, G., Al-Salami, H., & Dass, C. R. (2023). Metabolomics Profiling Reveals the Role of PEDF in Triple-Negative Breast Cancer Cell MDA-MB-231 under Glycaemic Loading. Pharmaceutics, 15(2), 543. https://doi.org/10.3390/pharmaceutics15020543








