A Machine Learning Approach for PLGA Nanoparticles in Antiviral Drug Delivery
Abstract
1. Introduction
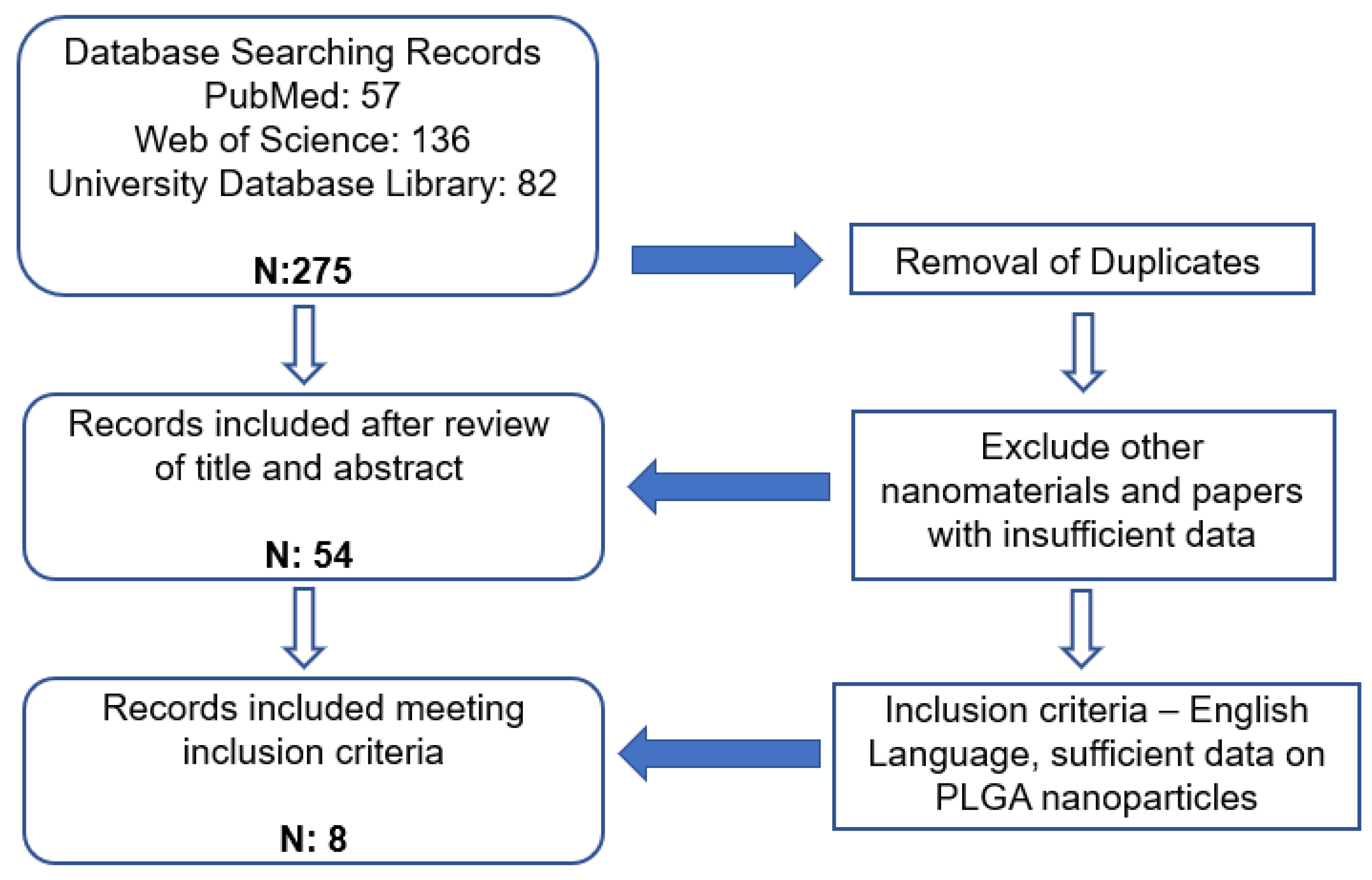
2. Materials and Methods
3. Results
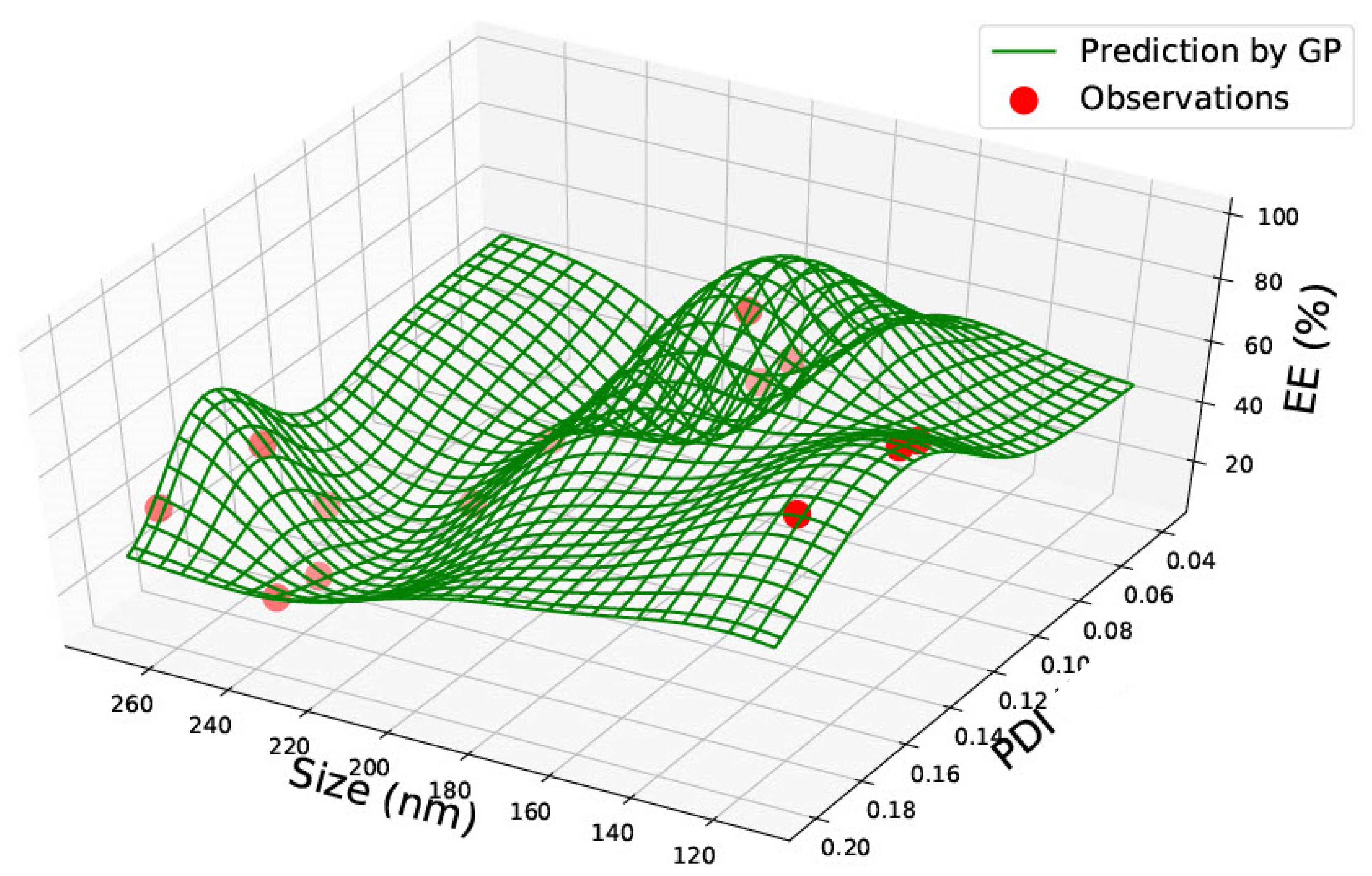
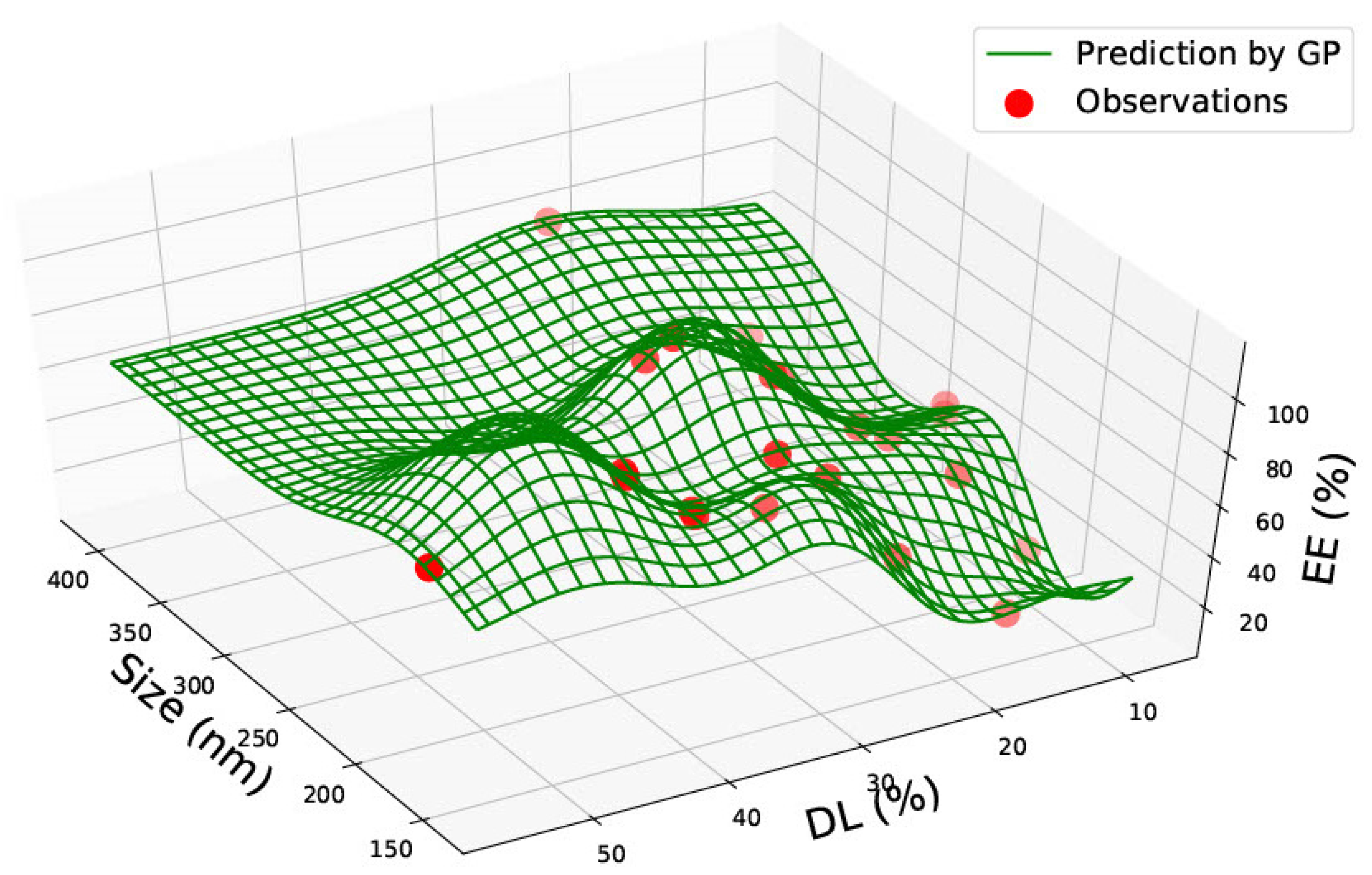
3.1. PLGA 50:50 Nanoparticles
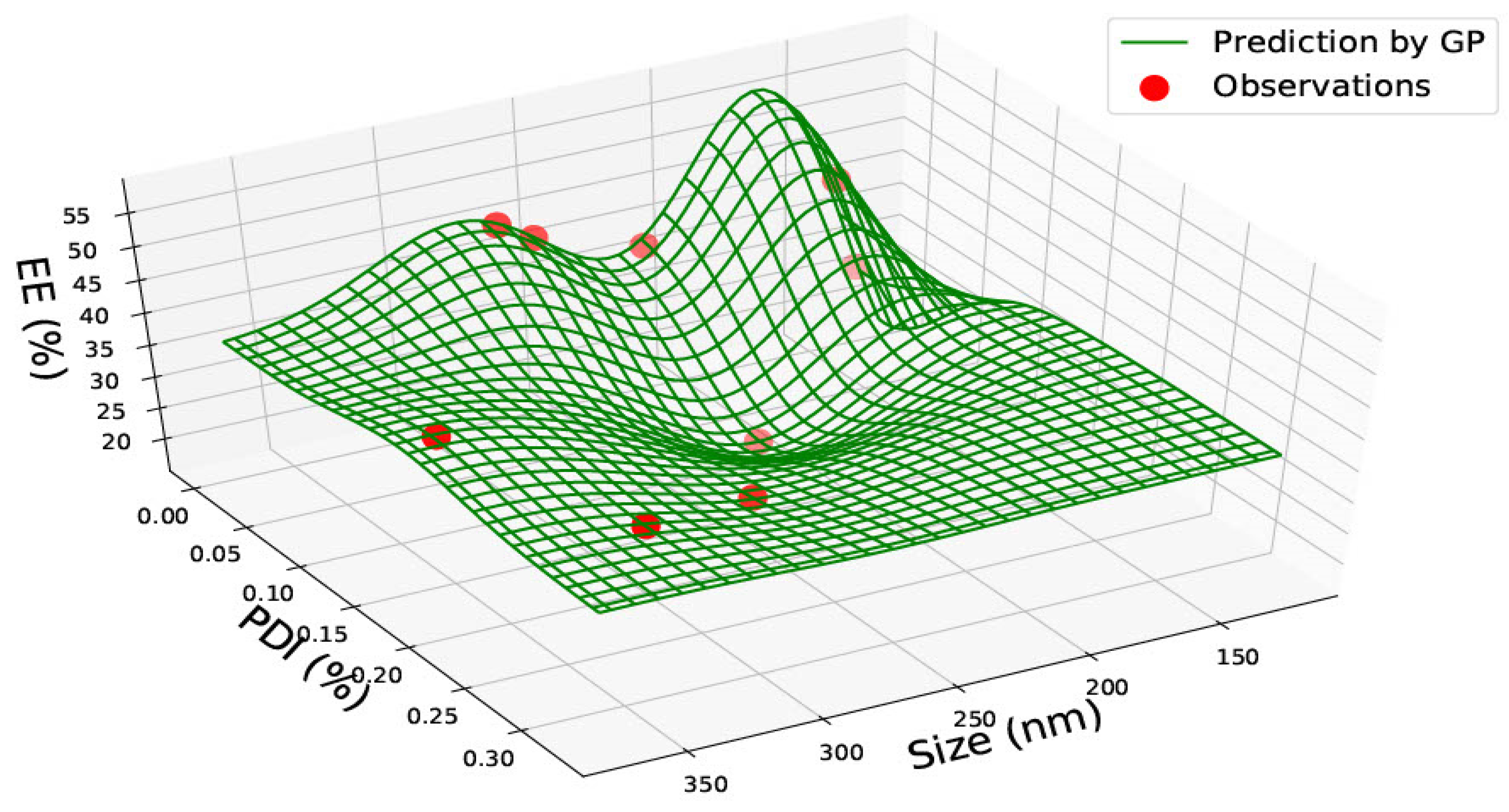
3.2. PLGA 65:15 Nanoparticles
3.3. PLGA 75:25 Nanoparticles
3.4. PLGA 85:15 Nanoparticles
4. Discussion
4.1. PLGA Nanoparticles
4.2. Current Research in this Field
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sun, D.; Gao, W.; Hu, H.; Zhou, S. Why 90% of clinical drug development fails and how to improve it? Acta Pharm. Sin. B 2022, 12, 3049–3062. [Google Scholar] [CrossRef]
- Taylor, D. The pharmaceutical industry and the future of drug development. In Pharmaceuticals in the Environment; Hester, R.E., Harrison, R.M., Eds.; The Royal Society of Chemistry: London, UK, 2015. [Google Scholar]
- Wong, H.H.; Jessup, A.; Sertkaya, A.; Birkenbach, A.; Berlind, A.; Eyraud, J. Examination of Clinical Trial Costs and Barriers for Drug Development Final; Office of the Assistant Secretary for Planning and Evaluation, U.S. Department of Health & Human Services: Washington, DC, USA, 2014.
- Chen, C.; Yaari, Z.; Apfelbaum, E.; Grodzinski, P.; Shamay, Y.; Heller, D.A. Merging Data Curation and Machine Learning to Improve Nanomedicines. Adv. Drug Deliv. Rev. 2022, 183, 114172. [Google Scholar] [CrossRef]
- Dara, S.; Dhamercherla, S.; Jadav, S.S.; Babu, C.M.; Ahsan, M.J. Machine learning in drug discovery: A review. Artif. Intell. Rev. 2022, 55, 1947–1999. [Google Scholar] [CrossRef]
- Wang, M.; Wang, T.; Cai, P.; Chen, X. Nanomaterials Discovery and Design through Machine Learning. Small Methods 2019, 3, 1900025. [Google Scholar] [CrossRef]
- Champion, J.A.; Katare, Y.K.; Mitragotri, S. Particle shape: A new design parameter for micro- and nanoscale drug delivery carriers. J. Control. Release 2007, 121, 3–9. [Google Scholar] [CrossRef]
- Ren, J.; Hong, H.; Song, J.; Ren, T. Particle size and distribution of biodegradable poly-D,L-lactide-co-poly(ethylene glycol) block polymer nanoparticles prepared by nanoprecipitation. J. Appl. Polym. Sci. 2005, 98, 1884–1890. [Google Scholar] [CrossRef]
- Rizvi, S.A.; Saleh, A.M. Applications of nanoparticle systems in drug delivery technology. Saudi Pharm. J. 2018, 26, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Patra, J.K.; Das, G.; Fraceto, L.F.; Campos, E.V.R.; Rodriguez-Torres, M.D.P.; Acosta-Torres, L.S.; Diaz-Torres, L.A.; Grillo, R.; Swamy, M.K.; Sharma, S.; et al. Nano based drug delivery systems: Recent developments and future prospects. J. Nanobiotechnol. 2018, 16, 71. [Google Scholar] [CrossRef] [PubMed]
- Danaei, M.; Dehghankhold, M.; Ataei, S.; Hasanzadeh Davarani, F.; Javanmard, R.; Dokhani, A.; Khorasani, S.; Mozafari, M.R. Impact of particle size and polydispersity index on the clinical applications of lipidic nanocarrier systems. Pharmaceutics 2018, 10, 57. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, B.; Santra, K.; Pattnaik, G.; Ghosh, S. Preparation, characterization and in-vitro evaluation of sustained release protein-loaded nanoparticles based on biodegradable polymers. Int. J. Nanomed. 2008, 3, 487–496. [Google Scholar] [CrossRef] [PubMed]
- Williams, C.K.; Rasmussen, C.E. Gaussian Processes for Machine Learning; MIT Press: Cambridge, MA, USA, 2006; 4p. [Google Scholar]
- Nguyen, V.; Deisenroth, M.P.; Osborne, M. Gaussian Process Sampling and Optimization with Approximate Upper and Lower Bounds. Available online: https://arxiv.org/abs/2110.12087 (accessed on 8 November 2022).
- Nguyen, V.; Osborne, M.A. Knowing the What but Not the Where in Bayesian Optimization. In Proceedings of the 37th International Conference on Machine Learning, Virtual Event, 13–18 July 2020; pp. 7317–7326. [Google Scholar]
- Venkatesh, D.N.; Baskaran, M.; Karri, V.V.S.R.; Mannemala, S.S.; Radhakrishna, K.; Goti, S. Fabrication and in vivo evaluation of Nelfinavir loaded PLGA nanoparticles for enhancing oral bioavailability and therapeutic effect. Saudi Pharm. J. 2015, 23, 667–674. [Google Scholar] [CrossRef] [PubMed]
- Tyo, K.M.; Duan, J.; Kollipara, P.; dela Cerna, M.V.C.; Lee, D.; Palmer, K.E.; Steinbach-Rankins, J.M. pH-responsive delivery of Griffithsin from electrospun fibers. Eur. J. Pharm. Biopharm. 2019, 138, 64–74. [Google Scholar] [CrossRef] [PubMed]
- Dhoke, D.M.; Basaiyye, S.S.; Khedekar, P.B. Development and characterization of L-HSA conjugated PLGA nanoparticle for hepatocyte targeted delivery of antiviral drug. J. Drug Deliv. Sci. Technol. 2018, 47, 77–94. [Google Scholar] [CrossRef]
- Bhosale, U.V.; Kusum Devi, V.; Jain, N. Formulation and optimization of mucoadhesive nanodrug delivery system of acyclovir. J. Young Pharm. 2011, 3, 275–283. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Li, J.; Patel, S.K.; Palmer, K.E.; Devlin, B.; Rohan, L.C. Design of poly(Lactic-co-glycolic acid) (plga) nanoparticles for vaginal co-delivery of griffthsin and dapivirine and their synergistic effect for hiv prophylaxis. Pharmaceutics 2019, 11, 184. [Google Scholar] [CrossRef] [PubMed]
- Öztürk, A.A.; Yurtdaş Kirimlioğlu, G. Preparation and in vitro characterization of lamivudine loaded nanoparticles prepared by acid and/or ester terminated PLGA for effective oral anti-retroviral therapy. J. Res. Pharm. 2019, 23, 897–913. [Google Scholar] [CrossRef]
- Tang, X.; Liang, Y.; Liu, X.; Zhou, S.; Liu, L.; Zhang, F.; Xie, C.; Cai, S.; Wei, J.; Zhu, Y.; et al. PLGA-PEG Nanoparticles Coated with Anti-CD45RO and Loaded with HDAC Plus Protease Inhibitors Activate Latent HIV and Inhibit Viral Spread. Nanoscale Res. Lett. 2015, 10, 413. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Shah, S.J.; Wang, Z.; Agrahari, V.; Pal, D.; Mitra, A.K. Nanoparticle-based topical ophthalmic formulation for sustained release of stereoisomeric dipeptide prodrugs of ganciclovir. Drug Deliv. 2016, 23, 2399–2409. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Kosorok, M.R.; Zeng, D. Reinforcement learning design for cancer clinical trials. Stat. Med. 2009, 28, 3294–3315. [Google Scholar] [CrossRef] [PubMed]
- Findlay, M.R.; Freitas, D.N.; Mobed-Miremadi, M.; Wheeler, K.E. Machine learning provides predictive analysis into silver nanoparticle protein corona formation from physicochemical properties. Environ. Sci. Nano 2018, 5, 64–71. [Google Scholar] [CrossRef] [PubMed]
- Pfeiffer, C.; Rehbock, C.; Hühn, D.; Carrillo-Carrion, C.; De Aberasturi, D.J.; Merk, V.; Barcikowski, S.; Parak, W.J. Interaction of colloidal nanoparticles with their local environment: The (ionic) nanoenvironment around nanoparticles is different from bulk and determines the physico-chemical properties of the nanoparticles. J. R. Soc. Interface 2014, 11, 20130931. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.L.; Lee, S.B.; Jeong, H.J.; Kim, D.W. Enhanced tumor targetability of PEGylated mesoporous silica nanoparticles on in vivo optical imaging according to their size. RSC Adv. 2014, 4, 31318–31322. [Google Scholar] [CrossRef]
- Asadi, H.; Rostamizadeh, K.; Salari, D.; Hamidi, M. Preparation of biodegradable nanoparticles of tri-block PLA-PEG-PLA copolymer and determination of factors controlling the particle size using artificial neural network. J. Microencapsul. 2011, 28, 406–416. [Google Scholar] [CrossRef] [PubMed]
- Shalaby, K.S.; Soliman, M.E.; Casettari, L.; Bonacucina, G.; Cespi, M.; Palmieri, G.F.; Sammour, O.A.; El Shamy, A.A. Determination of factors controlling the particle size and entrapment efficiency of noscapine in PEG/PLA nanoparticles using artificial neural networks. Int. J. Nanomed. 2014, 9, 4953–4964. [Google Scholar]
- Yamankurt, G.; Berns, E.J.; Xue, A.; Lee, A.; Bagheri, N.; Mrksich, M.; Mirkin, C.A. Exploration of the nanomedicine-design space with high-throughput screening and machine learning. Nat. Biomed. Eng. 2019, 3, 318–327. [Google Scholar] [CrossRef] [PubMed]
- Winkler, D.A.; Burden, F.R.; Yan, B.; Weissleder, R.; Tassa, C.; Shaw, S.; Epa, V. Modelling and predicting the biological effects of nanomaterials. SAR QSAR Environ. Res. 2014, 25, 161–172. [Google Scholar] [CrossRef] [PubMed]

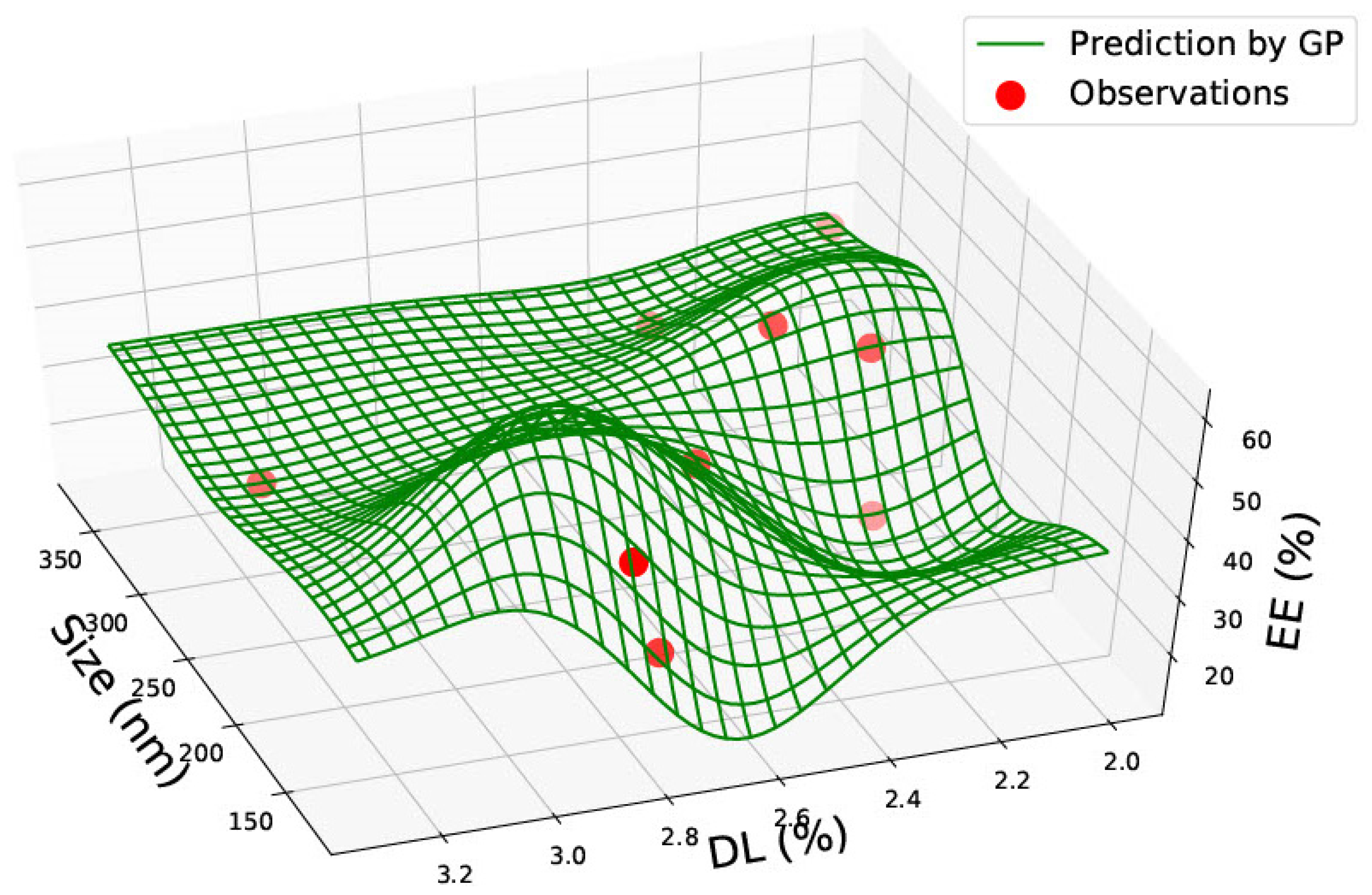

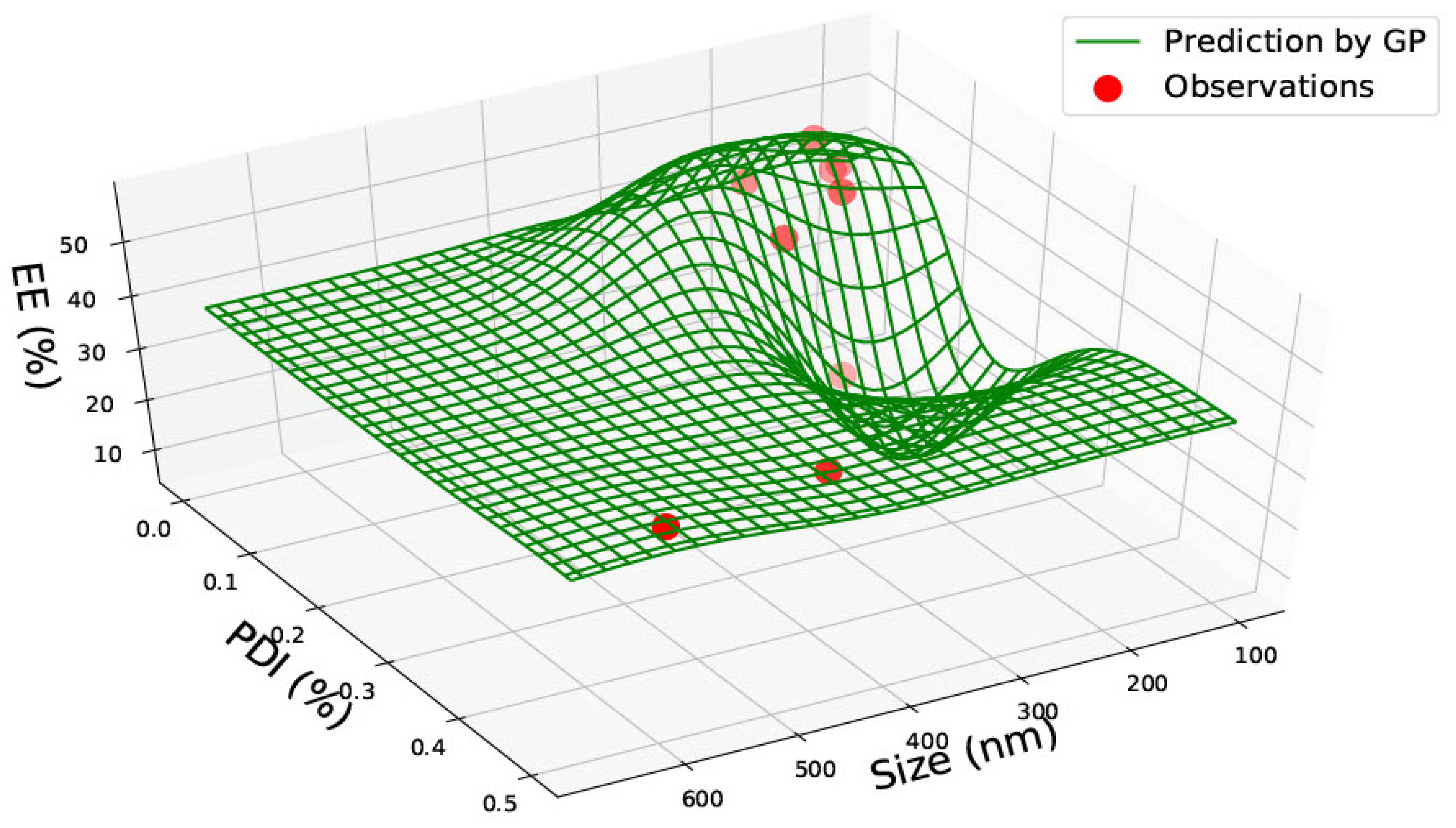
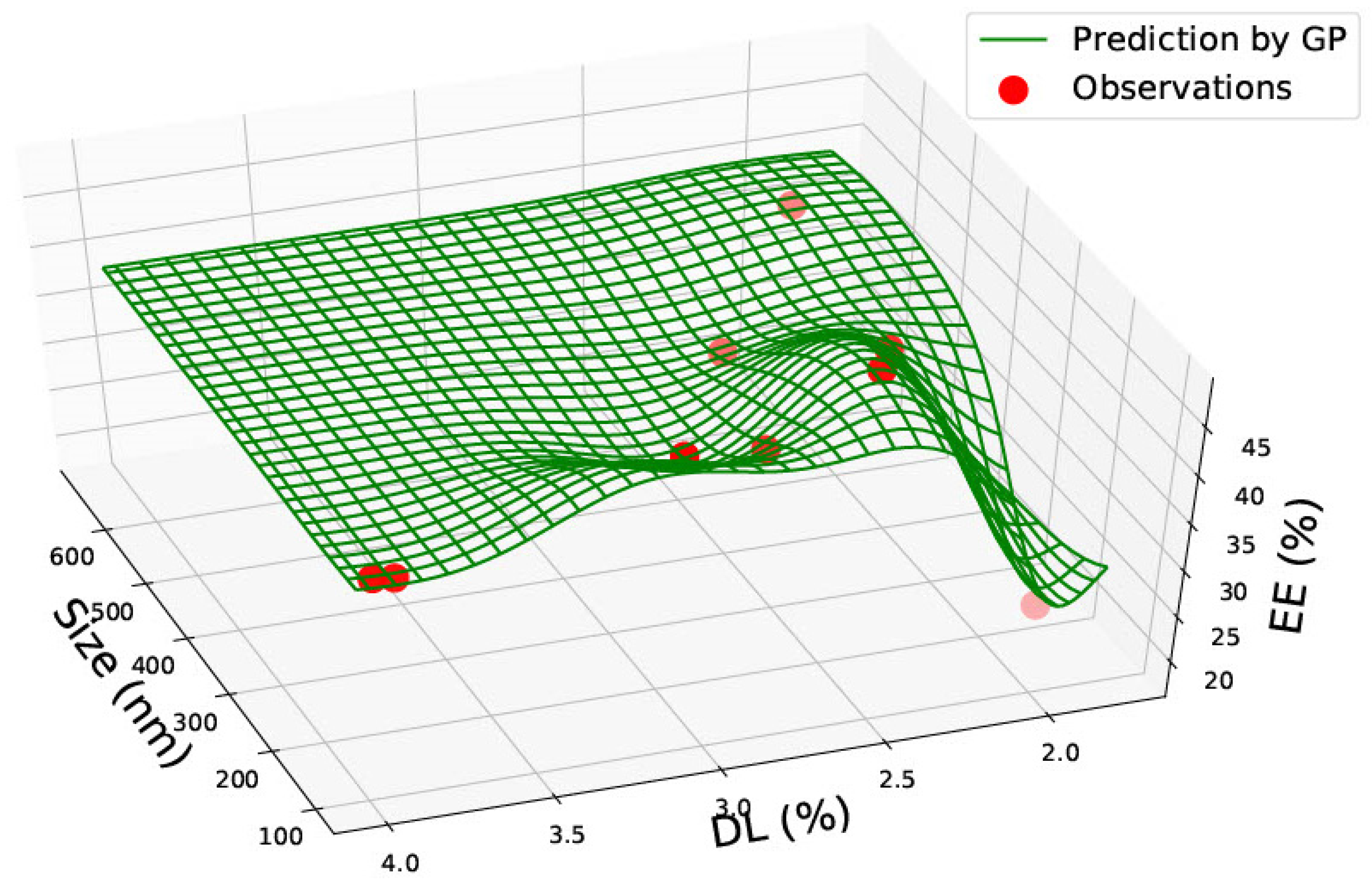
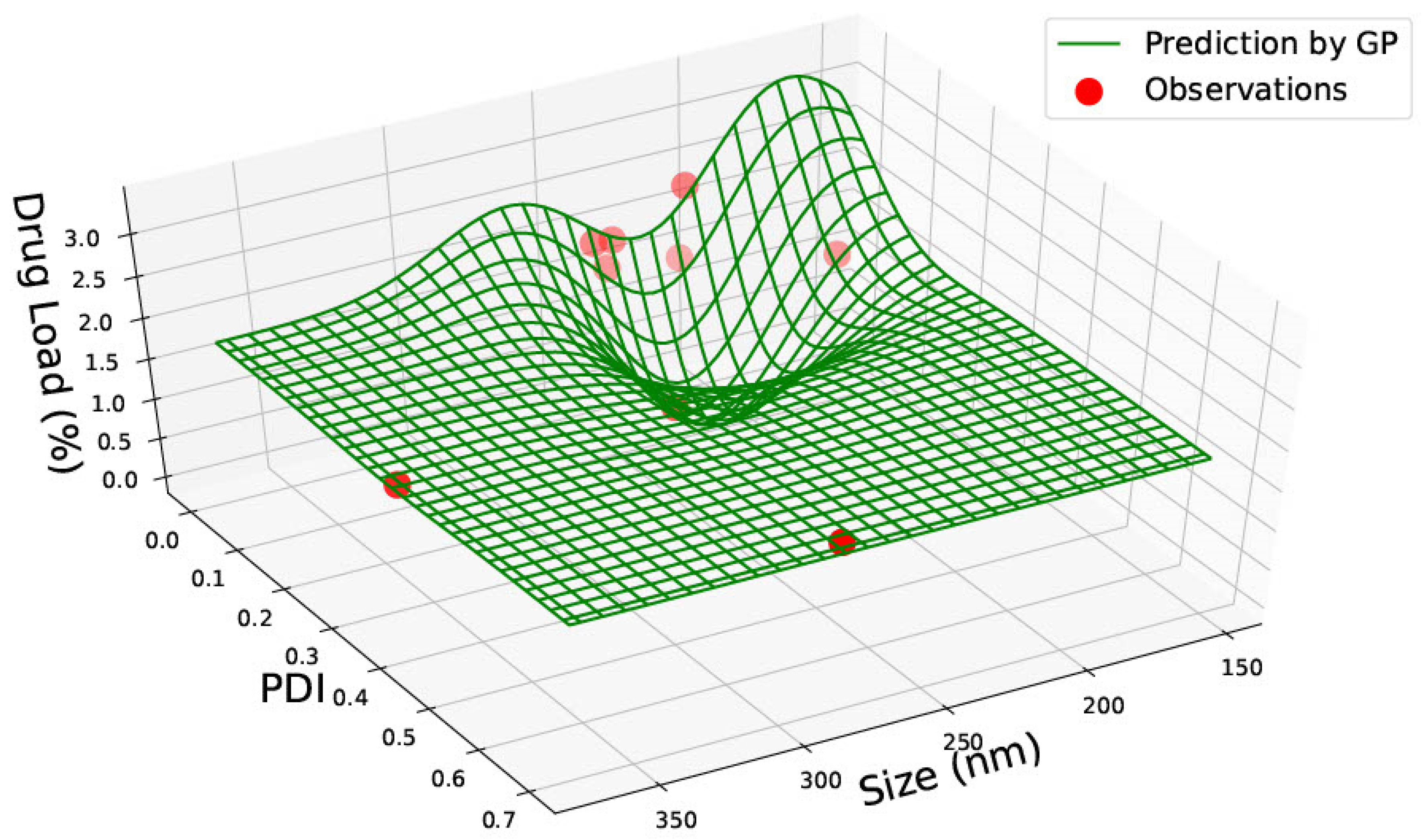
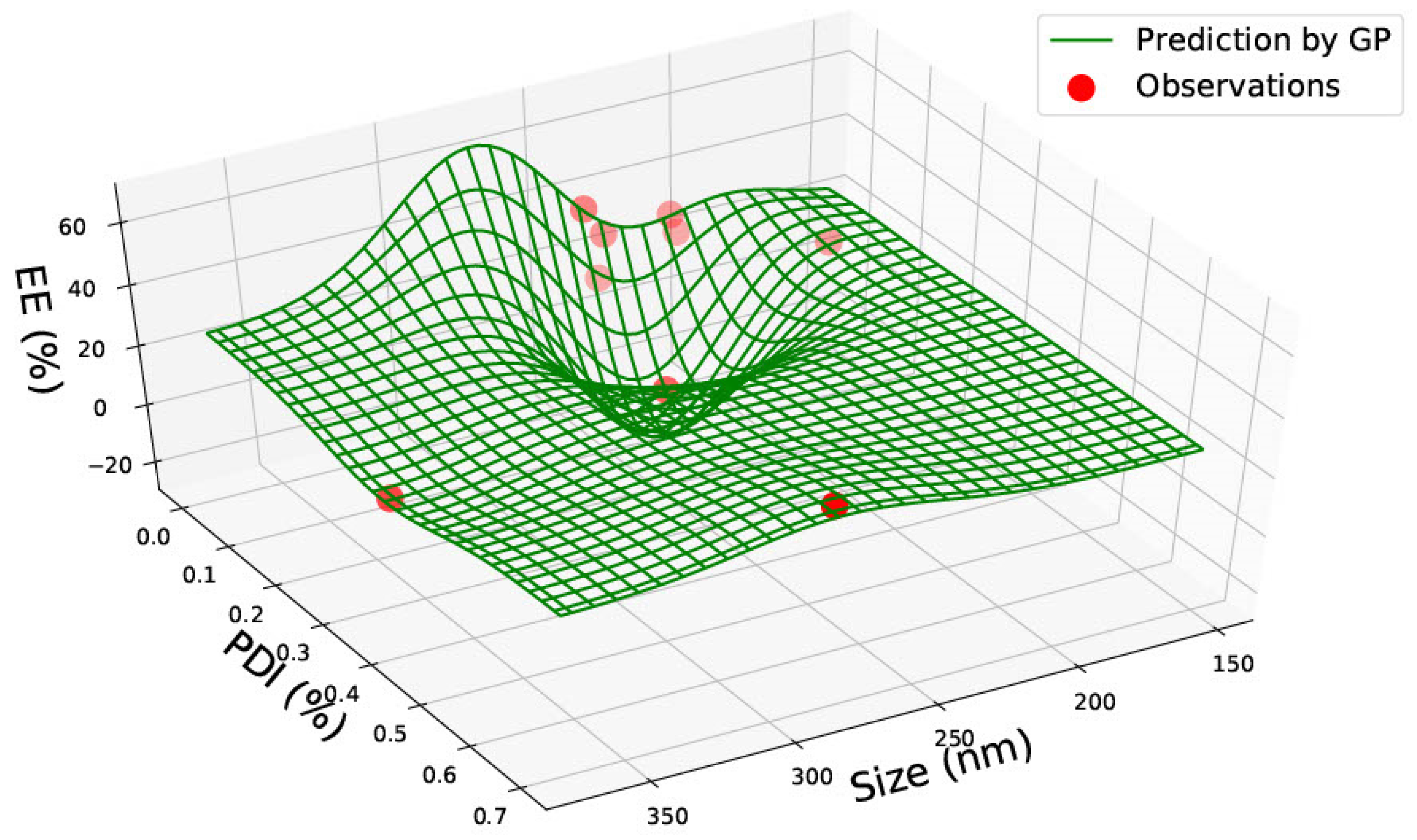
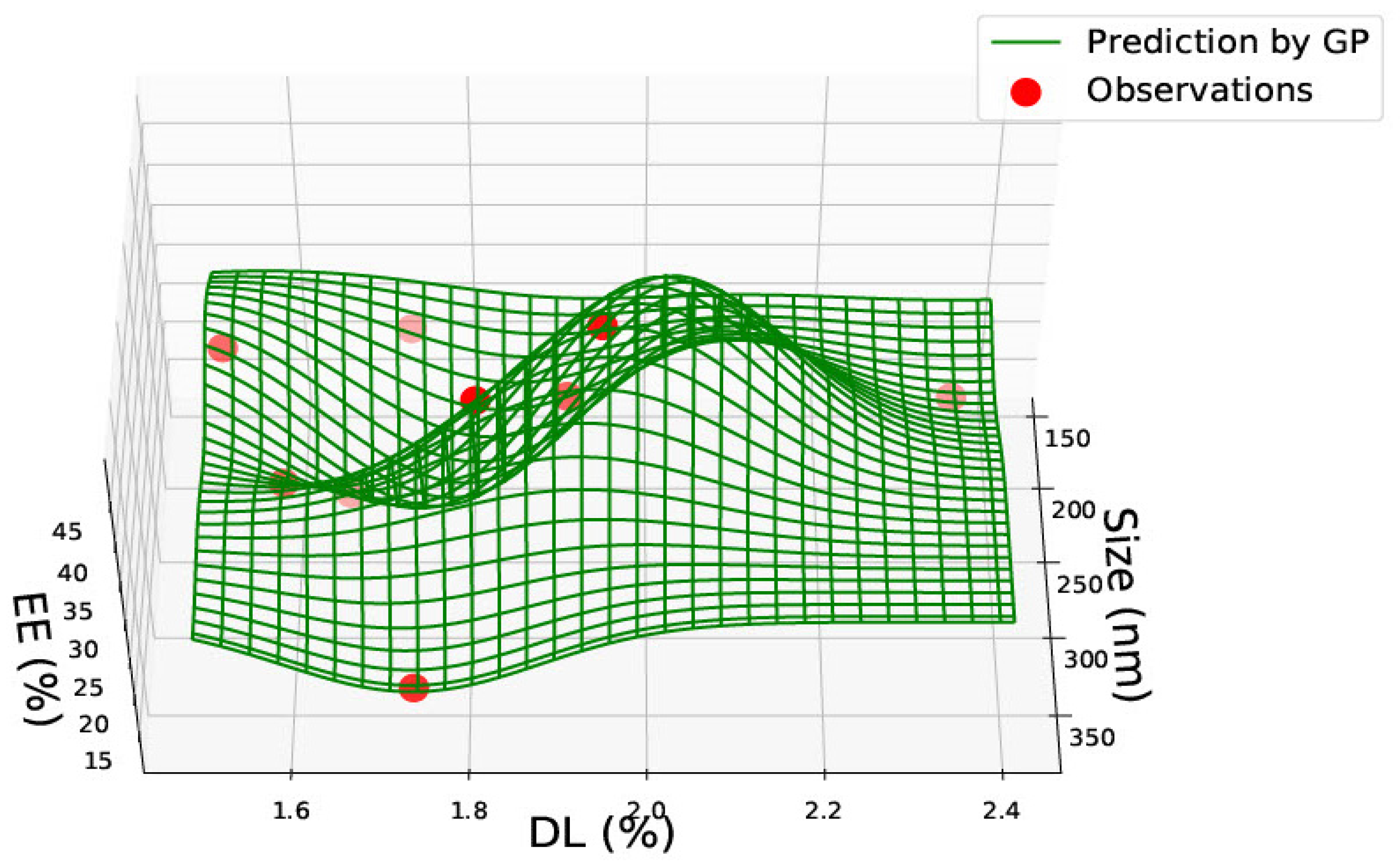
| PLGA 50:50 | ||||
|---|---|---|---|---|
| Size (nm) | PDI | %DL | EE% | Reference |
| 118 | 0.14 | 84 | [16,17,18,19,20,21,22] | |
| 119 | 0.13 | 80 | ||
| 125 | 0.17 | 78 | ||
| 140.3 | 17.50 | 27.50 | ||
| 167 | 23 | 46 | ||
| 171.8 | 30.72 | |||
| 174.2 | 55.84 | 84.30 | ||
| 180.3 | 36.75 | 77.50 | ||
| 182.7 | 34.49 | |||
| 184.3 | 0.06 | 45.90 | ||
| 185.3 | 36 | 72 | ||
| 186.6 | 0.08 | 70.10 | ||
| 188.8 | 0.07 | 40.70 | ||
| 188.9 | 40.61 | 91.24 | ||
| 192.5 | 29.43 | 83.41 | ||
| 192.8 | 10 | 20 | ||
| 195.9 | 25.30 | 67.75 | ||
| 198.1 | 39.81 | 88.19 | ||
| 203.2 | 29 | 58 | ||
| 204 | 14.50 | 51.30 | ||
| 220.6 | 33.25 | |||
| 221 | 0.10 | 30.28 | ||
| 223.4 | 0.13 | 25.37 | ||
| 230.7 | 42.18 | |||
| 232.7 | 40.19 | |||
| 234.2 | 0.20 | 28.25 | ||
| 236 | 18.70 | 62.30 | ||
| 236.7 | 0.18 | 21.88 | ||
| 239 | 16 | 53.40 | ||
| 244.3 | 46.64 | |||
| 248.5 | 40.18 | |||
| 250.1 | 0.15 | 25.71 | ||
| 251.2 | 23.55 | 81.95 | ||
| 256.3 | 39.27 | |||
| 256.9 | 51.64 | |||
| 258 | 0.16 | 9.62 | 49.90 | |
| 258.6 | 41.57 | |||
| 259.4 | 37.76 | |||
| 259.4 | 41.85 | |||
| 260.7 | 49.46 | |||
| 263 | 29.60 | 98.70 | ||
| 268.7 | 0.19 | 8.34 | 40.27 | |
| 272.4 | 30.64 | 88.34 | ||
| 274.3 | 47.25 | |||
| 275.6 | 20.31 | 67.95 | ||
| 276.9 | 46.81 | |||
| 281.4 | 53.44 | |||
| 284.2 | 51.64 | |||
| 292.4 | 58.54 | |||
| 299.8 | 60.91 | |||
| 311.1 | 57.57 | |||
| 336 | 16.20 | 54 | ||
| 407 | 24 | 80 | ||
| 740 | 80.16 | |||
| 810 | 80.09 | |||
| 870 | 84.12 | |||
| 914 | 86.26 | |||
| 1107 | 80.59 | |||
| 1210 | 89.90 | |||
| 1420 | 88.12 | |||
| 1580 | 93.70 | |||
| 1630 | 94.10 | |||
| PLGA 65:35 | ||||
|---|---|---|---|---|
| Size (nm) | PDI | %DL | EE% | Reference |
| 134.7 | 0.005 | 2.76 | 27.6 | [23] |
| 143 | 0.005 | 2.79 | 41.8 | |
| 215.7 | 0.005 | 2.54 | 38.1 | |
| 223.6 | 0.123 | 2.2 | 22 | |
| 256.4 | 0.005 | 2.15 | 42.9 | |
| 269.5 | 0.005 | 2.3 | 46 | |
| 273.5 | 0.233 | 3.2 | 32 | |
| 323.8 | 0.260 | 2.41 | 36.1 | |
| 356.3 | 0.16 | 2.02 | 40.4 | |
| PLGA 75:25 | ||||
|---|---|---|---|---|
| Size (nm) | PDI | %DL | EE% | Reference |
| 110.6 | 0.005 | 3.94 | 39.4 | [23] |
| 116.6 | 0.005 | 3.87 | 38.7 | |
| 134.1 | 0.005 | 3.02 | 45.3 | |
| 181.8 | 0.112 | 2.39 | 47.7 | |
| 198 | 0.007 | 2.69 | 40.4 | |
| 205.6 | 0.155 | 1.82 | 18.2 | |
| 251.4 | 0.14 | 2.26 | 45.2 | |
| 401 | 0.422 | 2.52 | 37.8 | |
| 569.4 | 0.467 | 2.07 | 41.3 | |
| PLGA 85:15 | ||||
|---|---|---|---|---|
| Size (nm) | PDI | %DL | EE% | Reference |
| 173.6 | 0.115 | 1.73 | 25.9 | [23] |
| 207.8 | 0.005 | 2.35 | 23.5 | |
| 210.6 | 0.009 | 1.52 | 30.4 | |
| 233.5 | 0.005 | 1.91 | 28.6 | |
| 237.8 | 0.018 | 1.66 | 16.6 | |
| 240.8 | 0.009 | 1.95 | 38.9 | |
| 266.6 | 0.315 | 1.59 | 23.8 | |
| 275.5 | 0.679 | 1.81 | 36.2 | |
| 367.5 | 0.353 | 1.74 | 17.4 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Noorain, L.; Nguyen, V.; Kim, H.-W.; Nguyen, L.T.B. A Machine Learning Approach for PLGA Nanoparticles in Antiviral Drug Delivery. Pharmaceutics 2023, 15, 495. https://doi.org/10.3390/pharmaceutics15020495
Noorain L, Nguyen V, Kim H-W, Nguyen LTB. A Machine Learning Approach for PLGA Nanoparticles in Antiviral Drug Delivery. Pharmaceutics. 2023; 15(2):495. https://doi.org/10.3390/pharmaceutics15020495
Chicago/Turabian StyleNoorain, Labiba, Vu Nguyen, Hae-Won Kim, and Linh T. B. Nguyen. 2023. "A Machine Learning Approach for PLGA Nanoparticles in Antiviral Drug Delivery" Pharmaceutics 15, no. 2: 495. https://doi.org/10.3390/pharmaceutics15020495
APA StyleNoorain, L., Nguyen, V., Kim, H.-W., & Nguyen, L. T. B. (2023). A Machine Learning Approach for PLGA Nanoparticles in Antiviral Drug Delivery. Pharmaceutics, 15(2), 495. https://doi.org/10.3390/pharmaceutics15020495









