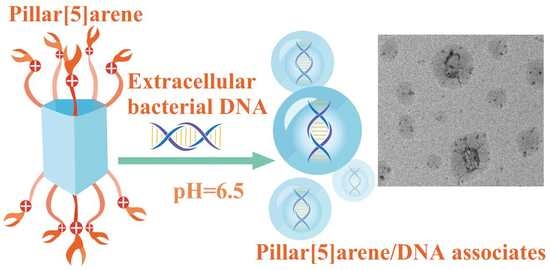
Toward Pathogenic Biofilm Suppressors: Synthesis of Amino Derivatives of Pillar[5]arene and Supramolecular Assembly with DNA
Abstract
1. Introduction
2. Materials and Methods
General Procedure for the Synthesis of Macrocycles 3–6
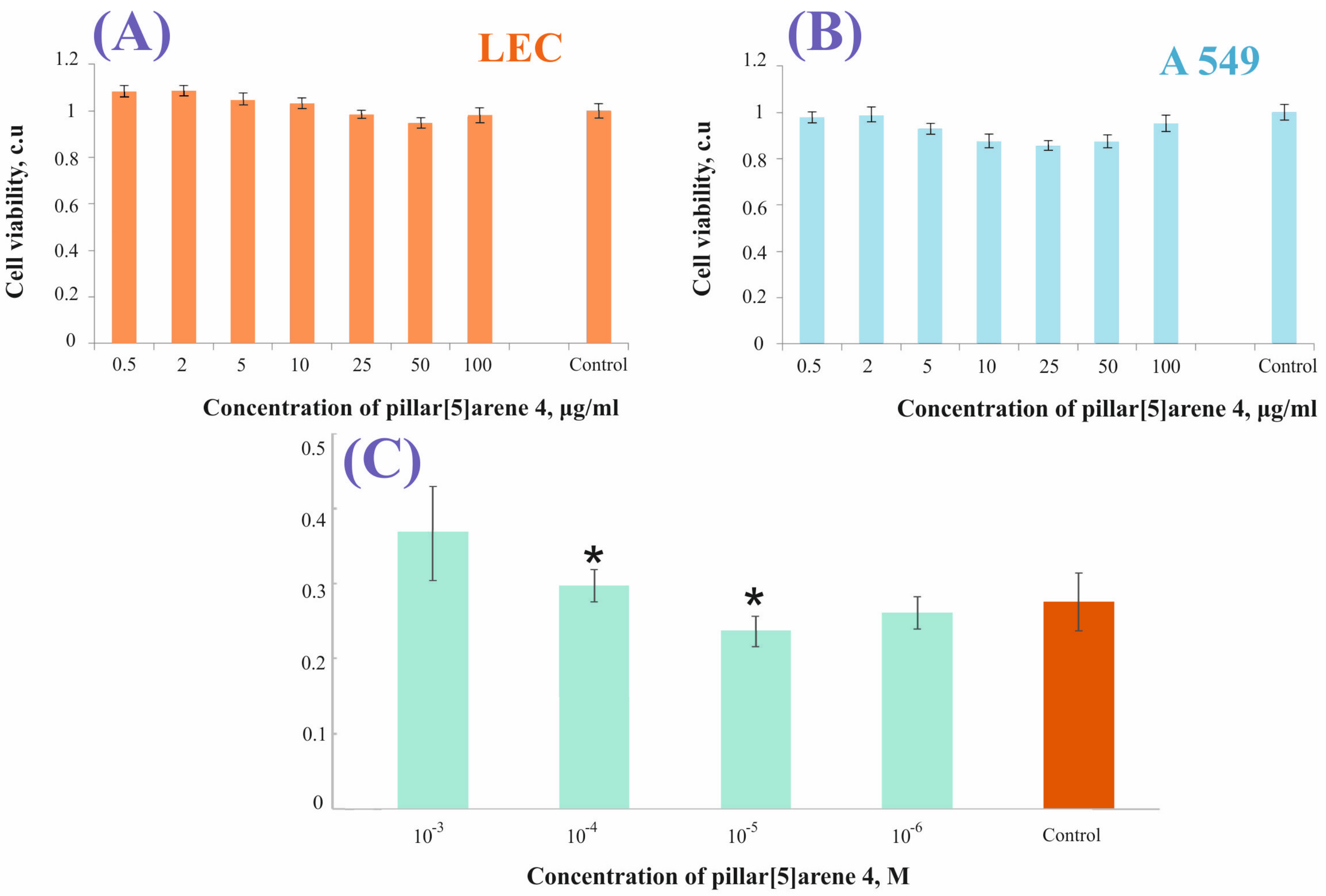
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Høiby, N.; Bjarnsholt, T.; Givskov, M.; Molin, S.; Ciofu, O. Antibiotic resistance of bacterial biofilms. Int. J. Antimicrob. Agents 2010, 35, 322–332. [Google Scholar] [CrossRef] [PubMed]
- Stewart, P.S. Mechanisms of antibiotic resistance in bacterial biofilms. Int. J. Med. Microbiol. 2002, 292, 107–113. [Google Scholar] [CrossRef] [PubMed]
- Kranjec, C.; Morales Angeles, D.; Torrissen Mårli, M.; Fernández, L.; García, P.; Kjos, M.; Diep, D.B. Staphylococcal biofilms: Challenges and novel therapeutic perspectives. Antibiotics 2021, 10, 131. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, D.A.; Shivapriya, P.M.; Gautam, P.K.; Misra, K.; Sahoo, A.K.; Samanta, S.K. Review on basic biology of bacterial biofilm infections and their Treatments by nanotechnology-based approaches. Proc. Natl. Acad. Sci. India B Biol. Sci. 2019, 90, 243–259. [Google Scholar] [CrossRef]
- Noor, Z.Z.; Rabiu, Z.; Sani, M.H.M.; Samad, A.F.A.; Kamaroddin, M.F.A.; Perez, M.F.; Dib, J.R.; Fatima, H.; Sinha, R.; Khare, S.K.; et al. A Review of bacterial antibiotic resistance genes and their removal strategies from wastewater. Curr. Pollut. Rep. 2021, 7, 494–509. [Google Scholar] [CrossRef]
- Sahli, C.; Moya, S.E.; Lomas, J.S.; Gravier-Pelletier, C.; Briandet, R.; Hémadi, M. Recent advances in nanotechnology for eradicating bacterial biofilm. Theranostics 2022, 12, 2383–2405. [Google Scholar] [CrossRef]
- Malaekeh-Nikouei, B.; Fazly Bazzaz, B.S.; Mirhadi, E.; Tajani, A.S.; Khameneh, B. The role of nanotechnology in combating biofilm-based antibiotic resistance. J. Drug Deliv. Sci. Technol. 2020, 60, 101880. [Google Scholar] [CrossRef]
- Talapko, J.; Škrlec, I. The principles, mechanisms, and benefits of unconventional agents in the treatment of biofilm infection. Pharmaceuticals 2020, 13, 299. [Google Scholar] [CrossRef]
- Okshevsky, M.; Regina, V.R.; Meyer, R.L. Extracellular DNA as a target for biofilm control. Curr. Opin. Biotechnol. 2015, 33, 73–80. [Google Scholar] [CrossRef]
- Tetz, G.V.; Artemenko, N.K.; Tetz, V.V. Effect of DNase and antibiotics on biofilm characteristics. Antimicrob. Agents Chemother. 2009, 53, 1204–1209. [Google Scholar] [CrossRef]
- Simpson, G.; Roomes, D.; Heron, M. Effects of streptokinase and deoxyribonuclease on viscosity of human surgical and empyema pus. Chest 2000, 117, 1728–1733. [Google Scholar] [CrossRef]
- Gao, L.; Wang, H.; Zheng, B.; Huang, F. Combating antibiotic resistance: Current strategies for the discovery of novel antibacterial materials based on macrocycle supramolecular chemistry. Giant 2021, 7, 100066. [Google Scholar] [CrossRef]
- Li, X.; Bai, H.; Yang, Y.; Yoon, J.; Wang, S.; Zhang, X. Supramolecular antibacterial materials for combatting antibiotic resistance. Adv. Mater. 2018, 31, 1805092. [Google Scholar] [CrossRef] [PubMed]
- Consoli, G.M.L.; Granata, G.; Ginestra, G.; Marino, A.; Toscano, G.; Nostro, A. Antibacterial nanoassembled calix[4]arene exposing choline units inhibits biofilm and motility of gram negative bacteria. ACS Med. Chem. Lett. 2022, 13, 916–922. [Google Scholar] [CrossRef]
- Shurpik, D.N.; Padnya, P.L.; Stoikov, I.I.; Cragg, P.J. Antimicrobial activity of calixarenes and related macrocycles. Molecules 2020, 25, 5145. [Google Scholar] [CrossRef]
- Li, Q.; Zhu, H.; Huang, F. Pillararene-based supramolecular functional materials. Trends Chem. 2020, 2, 850–864. [Google Scholar] [CrossRef]
- Guo, S.; Huang, Q.; Chen, Y.; Wei, J.; Zheng, J.; Wang, L.; Wang, Y.; Wang, R. Synthesis and bioactivity of guanidinium-functionalized pillar[5]arene as a biofilm disruptor. Angew. Chem. Int. Ed. 2020, 60, 618–623. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Xie, B.; Yang, X.; Dai, J.; Wei, G.; He, Y. Pillar[5]arene-based, dual pH and enzyme responsive supramolecular vesicles for targeted antibiotic delivery against intracellular MRSA. Chem. Commun. 2020, 56, 8115–8118. [Google Scholar] [CrossRef]
- Yan, S.; Chen, S.; Gou, X.; Yang, J.; An, J.; Jin, X.; Yang, Y.W.; Chen, L.; Gao, H. Biodegradable supramolecular materials based on cationic polyaspartamides and pillar[5]arene for targeting gram-positive bacteria and mitigating antimicrobial resistance. Adv. Funct. Mater. 2019, 29, 1904683. [Google Scholar] [CrossRef]
- Joseph, R.; Naugolny, A.; Feldman, M.; Herzog, I.M.; Fridman, M.; Cohen, Y. Cationic pillararenes potently inhibit biofilm formation without affecting bacterial growth and viability. J. Am. Chem. Soc. 2016, 138, 754–757. [Google Scholar] [CrossRef]
- Nierengarten, I.; Nothisen, M.; Sigwalt, D.; Biellmann, T.; Holler, M.; Remy, J.-S.; Nierengarten, J.-F. Polycationic pillar[5]arene derivatives: Interaction with DNA and Biological Applications. Chem. Eur. J. 2013, 19, 17552–17558. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Zhang, F.; Quan, J.; Li, J.; Li, H. Construction of pillar[5]arene-based nanochannels with chiral-enhanced DNA translocation ability. ChemNanoMat 2022, 8, 202200342. [Google Scholar] [CrossRef]
- Kuzin, Y.; Kappo, D.; Porfireva, A.; Shurpik, D.; Stoikov, I.; Evtugyn, G.; Hianik, T. Electrochemical DNA sensor based on carbon black—Poly(Neutral Red) composite for detection of oxidative DNA damage. Sensors 2018, 18, 3489. [Google Scholar] [CrossRef] [PubMed]
- Qian, X.; Yang, H.; Liu, S.; Yang, L.; Li, J.; Gao, W.; Du, G.; Qu, Q.; Ran, X. Supramolecular DNA sensor based on the integration of host-guest immobilization strategy and WP5-Ag/PEHA supramolecular aggregates. Anal. Chim. Acta 2022, 1220, 340077. [Google Scholar] [CrossRef]
- Cao, S.; Zhou, L.; Liu, C.; Zhang, H.; Zhao, Y. Pillararene-based self-assemblies for electrochemical biosensors. Biosens. Bioelectron. 2021, 181, 113164. [Google Scholar] [CrossRef]
- Yakimova, L.S.; Shurpik, D.N.; Guralnik, E.G.; Evtugyn, V.G.; Osin, Y.N.; Stoikov, I.I. Fluorescein-loaded solid lipid nanoparticles based on monoamine pillar[5]arene: Synthesis and interaction with DNA. ChemNanoMat 2018, 4, 919–923. [Google Scholar] [CrossRef]
- Yang, K.; Chang, Y.; Wen, J.; Lu, Y.; Pei, Y.; Cao, S.; Wang, F.; Pei, Z. Supramolecular vesicles based on complex of trp-modified pillar[5]arene and galactose derivative for synergistic and targeted drug delivery. Chem. Mater. 2016, 28, 1990–1993. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, Z.; Xin, F.; Zhao, Y. Metal-ligated pillararene materials: From chemosensors to multidimensional self-assembled architectures. Coord. Chem. Rev. 2020, 420, 213425. [Google Scholar] [CrossRef]
- Zhang, S.; Xu, Y.; Wang, B.; Qiao, W.; Liu, D.; Li, Z. Cationic compounds used in lipoplexes and polyplexes for gene delivery. J. Control. Release 2004, 100, 165–180. [Google Scholar] [CrossRef]
- Shurpik, D.N.; Mostovaya, O.A.; Sevastyanov, D.A.; Lenina, O.A.; Sapunova, A.S.; Voloshina, A.D.; Petrov, K.A.; Kovyazina, I.V.; Cragg, P.J.; Stoikov, I.I. Supramolecular neuromuscular blocker inhibition by a pillar[5]arene through aqueous inclusion of rocuronium bromide. Org. Biomol. Chem. 2019, 17, 9951–9959. [Google Scholar] [CrossRef]
- Shurpik, D.N.; Aleksandrova, Y.I.; Zelenikhin, P.V.; Subakaeva, E.V.; Cragg, P.J.; Stoikov, I.I. Towards new nanoporous biomaterials: Self-assembly of sulfopillar[5]arenes with vitamin D3 into supramolecular polymers. Org. Biomol. Chem. 2020, 18, 4210–4216. [Google Scholar] [CrossRef] [PubMed]
- Shurpik, D.N.; Aleksandrova, Y.I.; Mostovaya, O.A.; Nazmutdinova, V.A.; Tazieva, R.E.; Murzakhanov, F.F.; Gafurov, M.R.; Zelenikhin, P.V.; Subakaeva, E.V.; Sokolova, E.A.; et al. Self-healing thiolated pillar[5]arene films containing moxifloxacin suppress the development of bacterial biofilms. Nanomaterials 2022, 12, 1604. [Google Scholar] [CrossRef] [PubMed]
- Shurpik, D.N.; Aleksandrova, Y.I.; Mostovaya, O.A.; Nazmutdinova, V.A.; Zelenikhin, P.V.; Subakaeva, E.V.; Mukhametzyanov, T.A.; Cragg, P.J.; Stoikov, I.I. Water-soluble pillar[5]arene sulfo-derivatives self-assemble into biocompatible nanosystems to stabilize therapeutic proteins. Bioorg. Chem. 2021, 117, 105415. [Google Scholar] [CrossRef] [PubMed]
- Maurizi, L.; Forte, J.; Ammendolia, M.G.; Hanieh, P.N.; Conte, A.L.; Relucenti, M.; Donfrancesco, O.; Ricci, C.; Rinaldi, F.; Marianecci, C.; et al. Effect of Ciprofloxacin-loaded niosomes on escherichia coli and staphylococcus aureus biofilm formation. Pharmaceutics 2022, 14, 2662. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Gias, E.L.M.; Jones, M.N. The adsorption of cationic liposomes to Staphylococcus aureus biofilms. Colloids Surf. A Physicochem. Eng. Asp. 1999, 149, 561–570. [Google Scholar] [CrossRef]
- Yao, Y.; Xue, M.; Chi, X.; Ma, Y.; He, J.; Abliz, Z.; Huang, F. A new water-soluble pillar[5]arene: Synthesis and application in the preparation of gold nanoparticles. Chem. Commun. 2012, 48, 6505. [Google Scholar] [CrossRef]
- Chen, L.; Cai, Y.; Feng, W.; Yuan, L. Pillararenes as macrocyclic hosts: A rising star in metal ion separation. Chem. Commun. 2019, 55, 7883–7898. [Google Scholar] [CrossRef]
- Fang, Y.; Deng, Y.; Dehaen, W. Tailoring pillararene-based receptors for specific metal ion binding: From recognition to supramolecular assembly. Coord. Chem. Rev. 2020, 415, 213313. [Google Scholar] [CrossRef]
- Gao, Y.; Wang, J.; Chai, M.; Li, X.; Deng, Y.; Jin, Q.; Ji, J. Size and charge adaptive clustered nanoparticles targeting the biofilm microenvironment for chronic lung infection management. ACS Nano 2020, 14, 5686–5699. [Google Scholar] [CrossRef]
- Liu, Y.; Shi, L.; Su, L.; Van der Mei, H.C.; Jutte, P.C.; Ren, Y.; Busscher, H.J. Nanotechnology-based antimicrobials and delivery systems for biofilm-infection control. Chem. Soc. Rev. 2019, 48, 428–446. [Google Scholar] [CrossRef]
- Chen, M.; Wei, J.; Xie, S.; Tao, X.; Zhang, Z.; Ran, P.; Li, X. Bacterial biofilm destruction by size/surface charge-adaptive micelles. Nanoscale 2019, 11, 1410–1422. [Google Scholar] [CrossRef] [PubMed]
- Brynn Hibbert, D.; Thordarson, P. The death of the Job plot, transparency, open science and online tools, uncertainty estimation methods and other developments in supramolecular chemistry data analysis. Chem. Commun. 2016, 52, 12792–12805. [Google Scholar] [CrossRef] [PubMed]
- Bindfit. Available online: http://supramolecular.org (accessed on 1 September 2022).
- Skvortsova, P.; Shurpik, D.; Stoikov, I.; Khairutdinov, B. Pillar[5]arene-induced DNA condensation: Liquid–liquid phase separation in pillar[5]arene-oligonucleotide system. J. Mol. Liquids 2022, 368, 120683. [Google Scholar] [CrossRef]
- Chen, J.F.; Lin, Q.; Zhang, Y.M.; Yao, H.; Wei, T.B. Pillararene-based fluorescent chemosensors: Recent advances and perspectives. Chem. Commun. 2017, 53, 13296–13311. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Hu, X.Y.; Chen, D.; Shi, J.; Dong, Y.; Lin, C.; Pan, Y.; Wang, L. Pillar[5]arene-based side-chain polypseudorotaxanes as an anion-responsive fluorescent sensor. Polym. Chem. 2013, 4, 2224. [Google Scholar] [CrossRef]
- Cao, D.; Meier, H. Pillararene-based fluorescent sensors for the tracking of organic compounds. Chin. Chem. Lett. 2019, 30, 1758–1766. [Google Scholar] [CrossRef]
- Li, X.S.; Li, Y.F.; Wu, J.R.; Lou, X.Y.; Han, J.; Qin, J.; Yang, Y.W. A color-tunable fluorescent pillararene coordination polymer for efficient pollutant detection. J. Mater. Chem. A 2020, 8, 3651–3657. [Google Scholar] [CrossRef]
- Yang, H.L.; Li, Z.H.; Liu, P.P.; Sun, X.W.; Wang, Z.H.; Yao, H.; Zhang, Y.M.; Wei, T.B.; Lin, Q. Metal-free white light-emitting fluorescent material based on simple pillar[5]arene-tripodal amide system and theoretical insights on its assembly and fluorescent properties. Langmuir 2020, 36, 13469–13476. [Google Scholar] [CrossRef]
- Zhu, H.; Li, Q.; Zhu, W.; Huang, F. Pillararenes as versatile building blocks for fluorescent materials. Acc. Mater. Res. 2022, 3, 658–668. [Google Scholar] [CrossRef]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Huttner, A.; Bielicki, J.; Clements, M.N.; Frimodt-Møller, N.; Muller, A.E.; Paccaud, J.P.; Mouton, J.W. Oral amoxicillin and amoxicillin–clavulanic acid: Properties, indications and usage. Clin. Microbiol. Infect. 2020, 26, 871–879. [Google Scholar] [CrossRef] [PubMed]


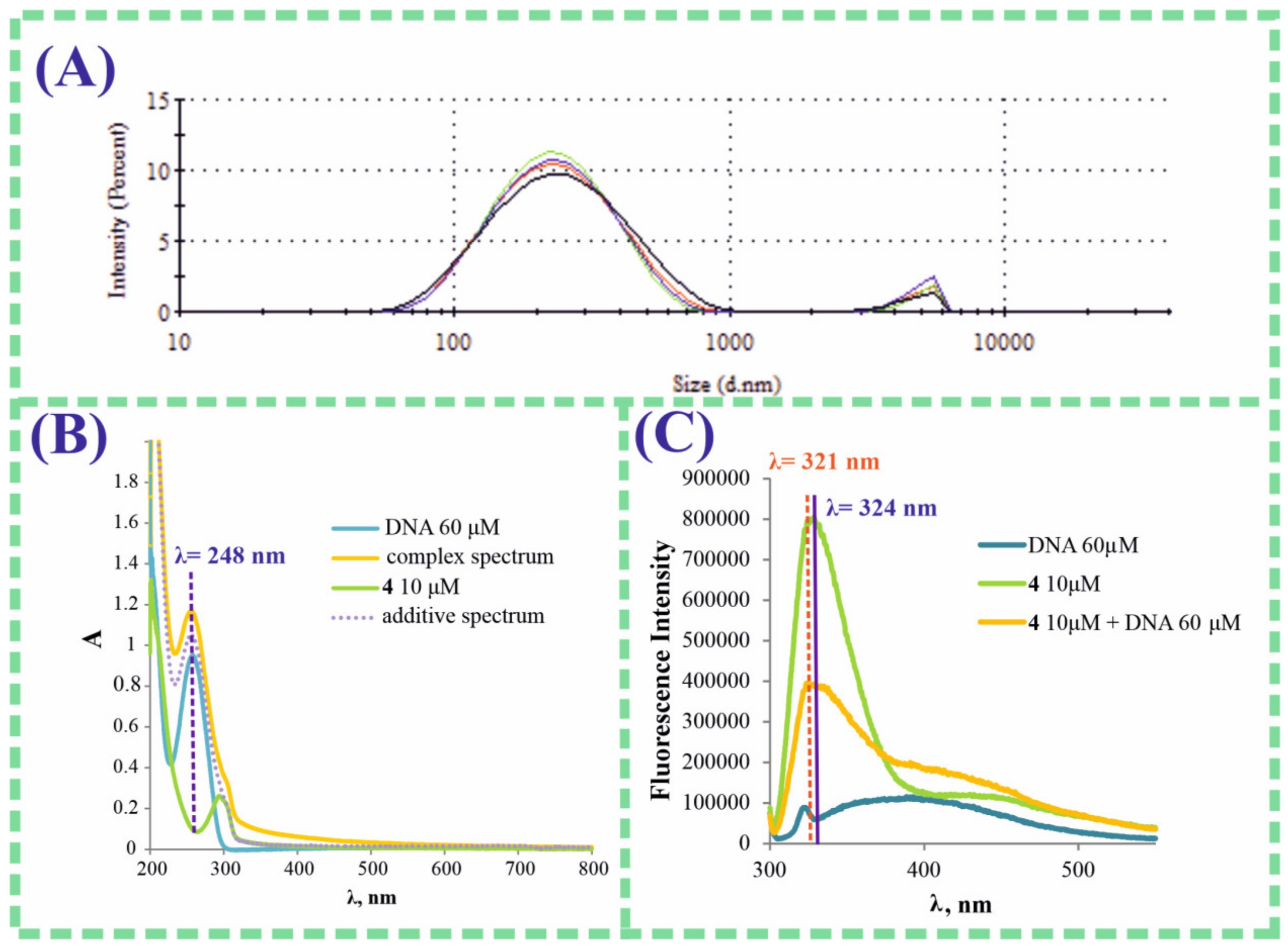

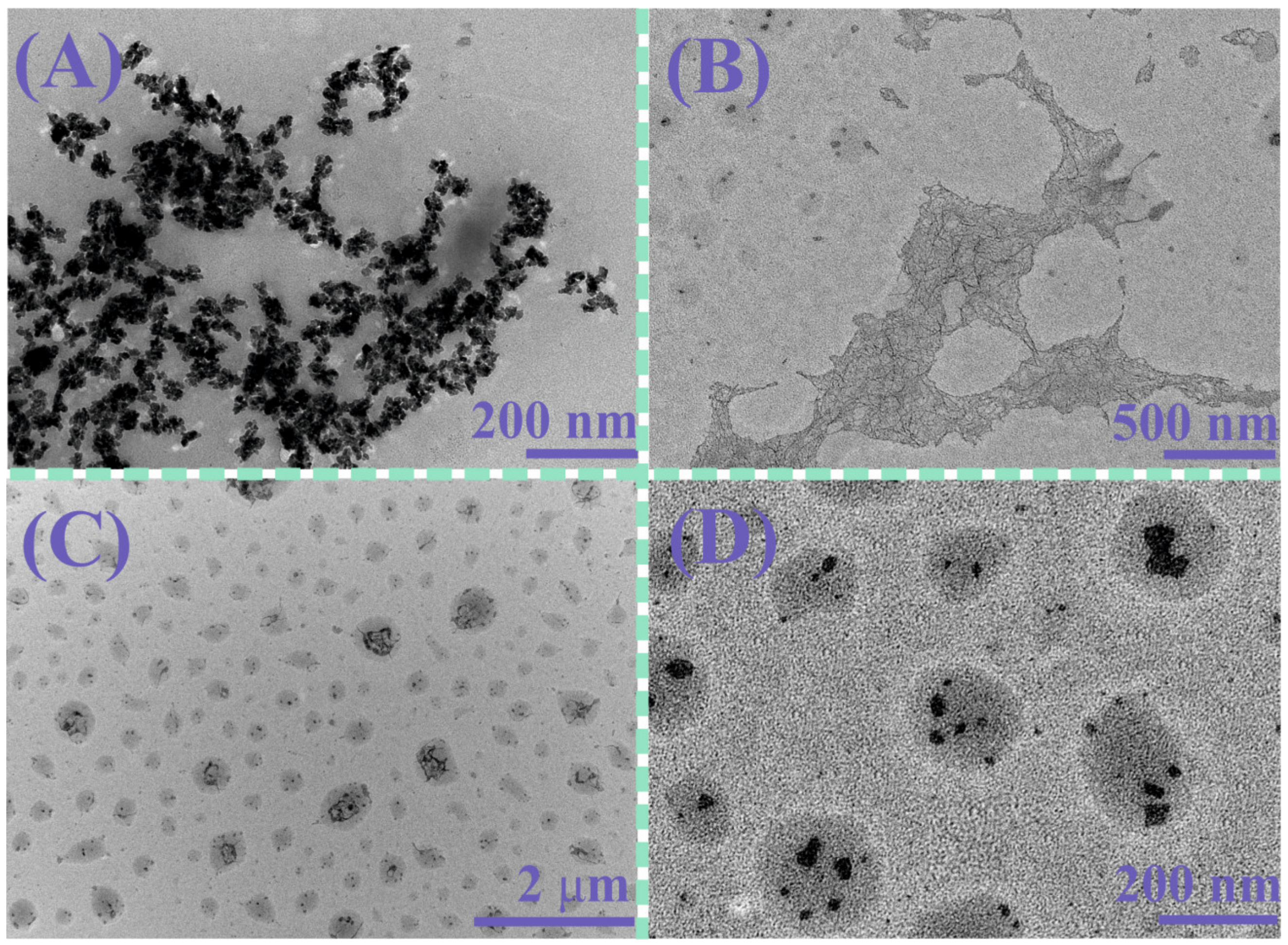
| Sample | CH, M | CDNA salmon sperm, M Base Pairs | daverage, nm | PDI | ζ-Potential, mV |
|---|---|---|---|---|---|
| 4 | 10−5 | 0 | 348 ± 2 | 0.09 ± 0.03 | |
| 5 | 10−5 | 0 | 449 ± 21 | 0.45 ± 0.06 | |
| 6 | 10−5 | 0 | 909 ± 45 | 0.38 ± 0.05 | |
| DNA | 0 | 6 × 10−5 | 1311 ± 113 | 0.59 ± 0.04 | |
| 4:DNA | 10−5 | 6 × 10−5 | 220 ± 10 | 0.24 ± 0.02 | 19.3 ± 0.5 |
| 5:DNA | 10−5 | 6 × 10−5 | 334 ± 4 | 0.30 ± 0.07 | 20.0 ± 0.8 |
| 6:DNA | 10−5 | 6 × 10−5 | 345 ± 25 | 0.42 ± 0.04 | 19.8 ± 0.5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aleksandrova, Y.I.; Shurpik, D.N.; Nazmutdinova, V.A.; Mostovaya, O.A.; Subakaeva, E.V.; Sokolova, E.A.; Zelenikhin, P.V.; Stoikov, I.I. Toward Pathogenic Biofilm Suppressors: Synthesis of Amino Derivatives of Pillar[5]arene and Supramolecular Assembly with DNA. Pharmaceutics 2023, 15, 476. https://doi.org/10.3390/pharmaceutics15020476
Aleksandrova YI, Shurpik DN, Nazmutdinova VA, Mostovaya OA, Subakaeva EV, Sokolova EA, Zelenikhin PV, Stoikov II. Toward Pathogenic Biofilm Suppressors: Synthesis of Amino Derivatives of Pillar[5]arene and Supramolecular Assembly with DNA. Pharmaceutics. 2023; 15(2):476. https://doi.org/10.3390/pharmaceutics15020476
Chicago/Turabian StyleAleksandrova, Yulia I., Dmitriy N. Shurpik, Viktoriya A. Nazmutdinova, Olga A. Mostovaya, Evgenia V. Subakaeva, Evgenia A. Sokolova, Pavel V. Zelenikhin, and Ivan I. Stoikov. 2023. "Toward Pathogenic Biofilm Suppressors: Synthesis of Amino Derivatives of Pillar[5]arene and Supramolecular Assembly with DNA" Pharmaceutics 15, no. 2: 476. https://doi.org/10.3390/pharmaceutics15020476
APA StyleAleksandrova, Y. I., Shurpik, D. N., Nazmutdinova, V. A., Mostovaya, O. A., Subakaeva, E. V., Sokolova, E. A., Zelenikhin, P. V., & Stoikov, I. I. (2023). Toward Pathogenic Biofilm Suppressors: Synthesis of Amino Derivatives of Pillar[5]arene and Supramolecular Assembly with DNA. Pharmaceutics, 15(2), 476. https://doi.org/10.3390/pharmaceutics15020476