Curcumin Disrupts a Positive Feedback Loop between ADMSCs and Cancer Cells in the Breast Tumor Microenvironment via the CXCL12/CXCR4 Axis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture and Collection of Conditioned Medium (CM)
2.2. Cell Viability Assay
2.3. mRNA Extraction and Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
2.4. Western Blotting Analysis
2.5. Immunocytochemistry Staining
2.6. Enzyme-Linked Immunosorbent Assay (ELISA)
2.7. Cell Migration and Invasion Assays
2.8. Statistical Analysis
3. Results
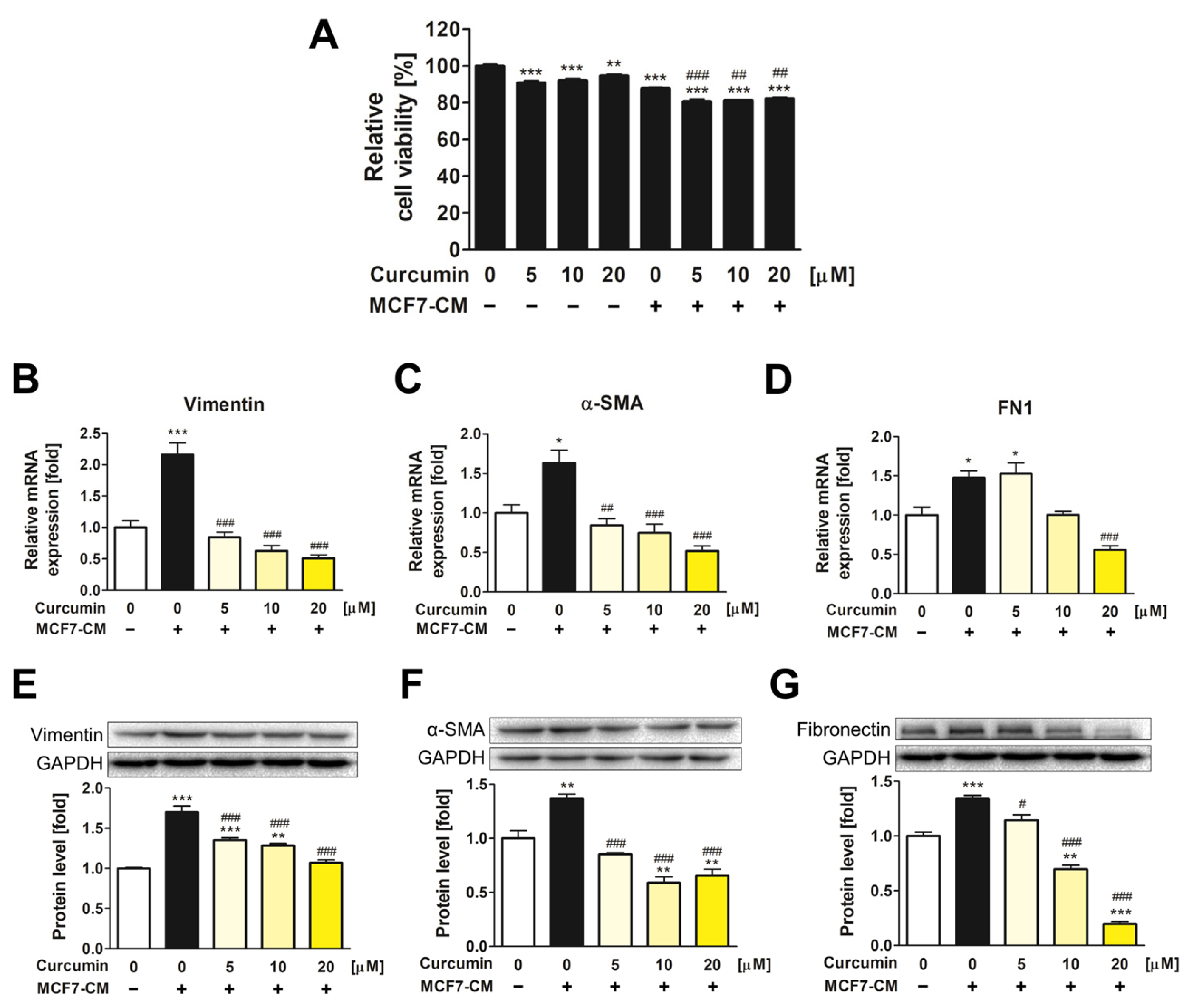
3.1. The Inhibitory Effect of Curcumin on the Transformation of ADMSCs into CAFs
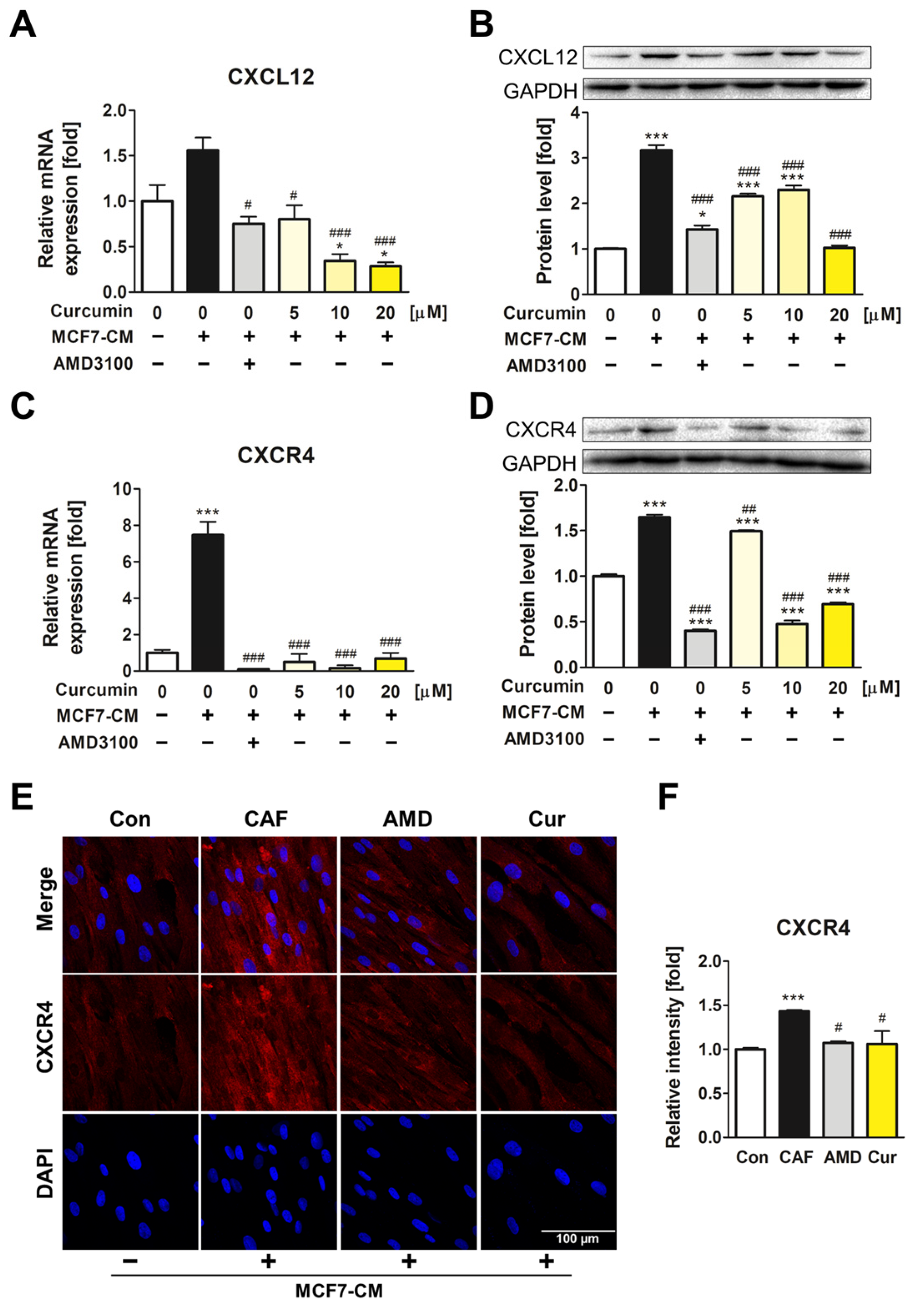
3.2. The Downregulatory Impact of Curcumin on CXCL12 and CXCR4 Expression of ADMSCs during CAF Transformation
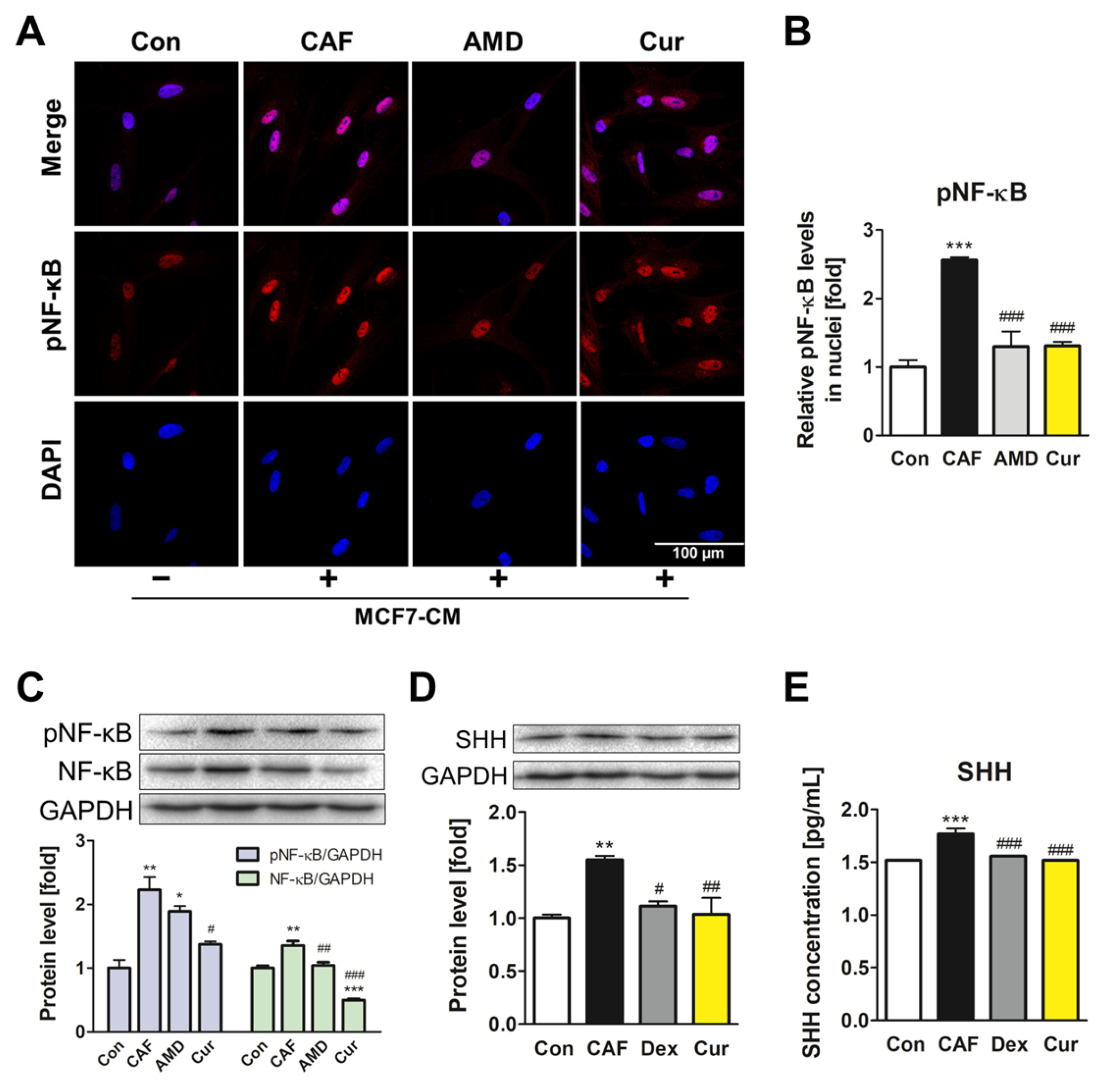
3.3. Repression of the NF-κB Signaling Pathway and SHH Expression by Curcumin during the ADMSC-CAF Transformation
3.4. Suppression of CXCL12/CXCR4 Axis and Its Downstream Effects in MCF7 by Curcumin
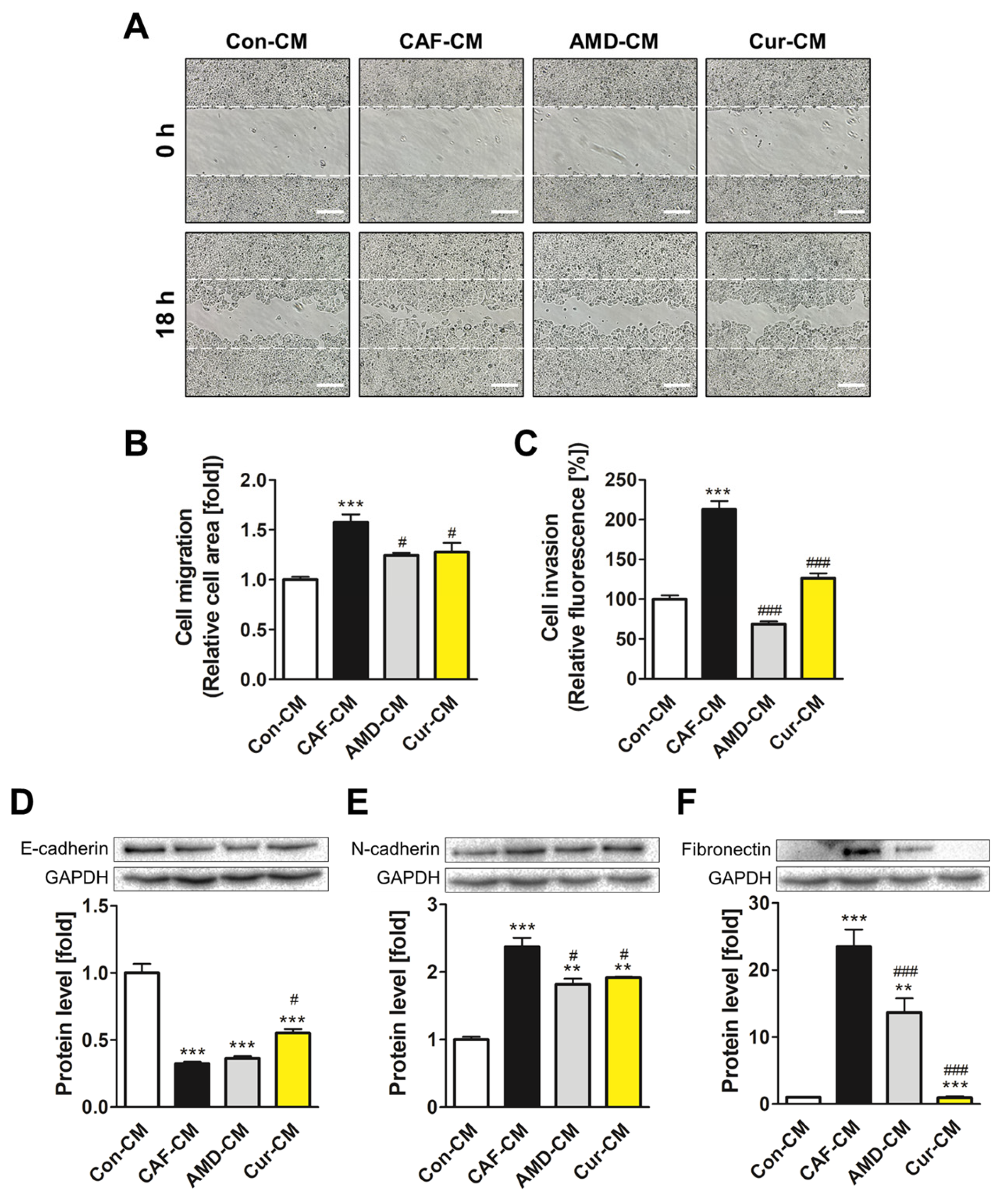
3.5. Inhibition of CAF Transformation by Curcumin Suppresses the CXCL12/CXCR4 Axis-Mediated Positive Interaction between CAFs and MCF7 Cells
3.6. Inhibiting CXCL12/CXCR4 Axis-Mediated Transformation of ADMSCs by Curcumin Reduced the Metastatic Potential of MCF7 Cells
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fletcher, S.J.; Sacca, P.A.; Pistone-Creydt, M.; Coló, F.A.; Serra, M.F.; Santino, F.E.; Sasso, C.V.; Lopez-Fontana, C.M.; Carón, R.W.; Calvo, J.C.; et al. Human breast adipose tissue: Characterization of factors that change during tumor progression in human breast cancer. J. Exp. Clin. Cancer Res. 2017, 36, 26. [Google Scholar] [CrossRef] [PubMed]
- Kothari, C.; Diorio, C.; Durocher, F. The Importance of Breast Adipose Tissue in Breast Cancer. Int. J. Mol. Sci. 2020, 21, 5760. [Google Scholar] [CrossRef] [PubMed]
- Miyazaki, Y.; Oda, T.; Inagaki, Y.; Kushige, H.; Saito, Y.; Mori, N.; Takayama, Y.; Kumagai, Y.; Mitsuyama, T.; Kida, Y.S. Adipose-derived mesenchymal stem cells differentiate into heterogeneous cancer-associated fibroblasts in a stroma-rich xenograft model. Sci. Rep. 2021, 11, 4690. [Google Scholar] [CrossRef] [PubMed]
- Miyazaki, Y.; Oda, T.; Mori, N.; Kida, Y.S. Adipose-derived mesenchymal stem cells differentiate into pancreatic cancer-associated fibroblasts in vitro. FEBS Open Bio 2020, 10, 2268–2281. [Google Scholar] [CrossRef] [PubMed]
- Strong, A.L.; Pei, D.T.; Hurst, C.G.; Gimble, J.M.; Burow, M.E.; Bunnell, B.A. Obesity Enhances the Conversion of Adipose-Derived Stromal/Stem Cells into Carcinoma-Associated Fibroblast Leading to Cancer Cell Proliferation and Progression to an Invasive Phenotype. Stem Cells Int. 2017, 2017, 9216502. [Google Scholar] [CrossRef] [PubMed]
- Xue, C.; Gao, Y.; Li, X.; Zhang, M.; Yang, Y.; Han, Q.; Sun, Z.; Bai, C.; Zhao, R.C. Mesenchymal stem cells derived from adipose accelerate the progression of colon cancer by inducing a MT-CAFs phenotype via TRPC3/NF-KB axis. Stem Cell Res. Ther. 2022, 13, 335. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.T.; Tan, Y.J.; Falasca, M.; Oon, C.E. Cancer-Associated Fibroblasts: Epigenetic Regulation and Therapeutic Intervention in Breast Cancer. Cancers 2020, 12, 2949. [Google Scholar] [CrossRef]
- Ping, Q.; Yan, R.; Cheng, X.; Wang, W.; Zhong, Y.; Hou, Z.; Shi, Y.; Wang, C.; Li, R. Cancer-associated fibroblasts: Overview, progress, challenges, and directions. Cancer Gene Ther. 2021, 28, 984–999. [Google Scholar] [CrossRef]
- Louault, K.; Li, R.-R.; Declerck, Y.A. Cancer-Associated Fibroblasts: Understanding Their Heterogeneity. Cancers 2020, 12, 3108. [Google Scholar] [CrossRef]
- Wani, N.A.; Nasser, M.W.; Ahirwar, D.K.; Zhao, H.; Miao, Z.; Shilo, K.; Ganju, R.K. C-X-C motif chemokine 12/C-X-C chemokine receptor type 7 signaling regulates breast cancer growth and metastasis by modulating the tumor microenvironment. Breast Cancer Res. 2014, 16, R54. [Google Scholar] [CrossRef]
- Zhou, W.; Guo, S.; Liu, M.; Burow, M.E.; Wang, G. Targeting CXCL12/CXCR4 Axis in Tumor Immunotherapy. Curr. Med. Chem. 2019, 26, 3026–3041. [Google Scholar] [CrossRef] [PubMed]
- Guo, F.; Wang, Y.; Liu, J.; Mok, S.C.; Xue, F.; Zhang, W. CXCL12/CXCR4: A symbiotic bridge linking cancer cells and their stromal neighbors in oncogenic communication networks. Oncogene 2016, 35, 816–826. [Google Scholar] [CrossRef] [PubMed]
- Boimel, P.J.; Smirnova, T.; Zhou, Z.N.; Wyckoff, J.; Park, H.; Coniglio, S.J.; Qian, B.-Z.; Stanley, E.R.; Cox, D.; Pollard, J.W.; et al. Contribution of CXCL12 secretion to invasion of breast cancer cells. Breast Cancer Res. 2012, 14, R23. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Cheng, G.; Hao, M.; Zheng, J.; Zhou, X.; Zhang, J.; Taichman, R.S.; Pienta, K.J.; Wang, J. CXCL12/CXCR4/CXCR7 chemokine axis and cancer progression. Cancer Metastasis Rev. 2010, 29, 709–722. [Google Scholar] [CrossRef] [PubMed]
- Santagata, S.; Ieranò, C.; Trotta, A.M.; Capiluongo, A.; Auletta, F.; Guardascione, G.; Scala, S. CXCR4 and CXCR7 Signaling Pathways: A Focus on the Cross-Talk Between Cancer Cells and Tumor Microenvironment. Front. Oncol. 2021, 11, 591386. [Google Scholar] [CrossRef] [PubMed]
- Khare, T.; Bissonnette, M.; Khare, S. CXCL12-CXCR4/CXCR7 Axis in Colorectal Cancer: Therapeutic Target in Preclinical and Clinical Studies. Int. J. Mol. Sci. 2021, 22, 7371. [Google Scholar] [CrossRef]
- Lu, D.-Y.; Tang, C.-H.; Yeh, W.-L.; Wong, K.-L.; Lin, C.-P.; Chen, Y.-H.; Lai, C.-H.; Chen, Y.-F.; Leung, Y.-M.; Fu, W.-M. SDF-1alpha up-regulates interleukin-6 through CXCR4, PI3K/Akt, ERK, and NF-kappaB-dependent pathway in microglia. Eur. J. Pharmacol. 2009, 613, 146–154. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.-Y.; Lee, C.-Y.; Chen, M.-Y.; Yang, W.-H.; Chen, Y.-H.; Chang, C.-H.; Hsu, H.-C.; Fong, Y.-C.; Tang, C.-H. Stromal cell-derived factor-1/CXCR4 enhanced motility of human osteosarcoma cells involves MEK1/2, ERK and NF-κB-dependent pathways. J. Cell. Physiol. 2009, 221, 204–212. [Google Scholar] [CrossRef]
- Xie, Z.; Zhang, M.; Zhou, G.; Lin, L.; Han, J.; Wang, Y.; Li, L.; He, Y.; Zeng, Z.; Chen, M.; et al. Emerging roles of the Hedgehog signaling pathway in inflammatory bowel disease. Cell Death Discov. 2021, 7, 314. [Google Scholar] [CrossRef]
- Douceau, S.; Deutsch Guerrero, T.; Ferent, J. Establishing Hedgehog Gradients during Neural Development. Cells 2023, 12, 225. [Google Scholar] [CrossRef]
- Patel, T.N.; Dhanyamraju, P.K. Role of aberrant Sonic hedgehog signaling pathway in cancers and developmental anomalies. J. Biomed. Res. 2021, 36, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Sleightholm, R.L.; Neilsen, B.K.; Li, J.; Steele, M.M.; Singh, R.K.; Hollingsworth, M.A.; Oupicky, D. Emerging roles of the CXCL12/CXCR4 axis in pancreatic cancer progression and therapy. Pharmacol. Ther. 2017, 179, 158–170. [Google Scholar] [CrossRef] [PubMed]
- Kotha, R.R.; Luthria, D.L. Curcumin: Biological, Pharmaceutical, Nutraceutical, and Analytical Aspects. Molecules 2019, 24, 2930. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Hu, Y.; Zhao, Y.; Cheng, M.; Qin, H.; Cheng, T.; Wang, Q.; Peng, X.; Zhang, X. Curcumin Attenuates Titanium Particle-Induced Inflammation by Regulating Macrophage Polarization In Vitro and In Vivo. Front. Immunol. 2017, 8, 55. [Google Scholar] [CrossRef] [PubMed]
- Ojo, O.A.; Adeyemo, T.R.; Rotimi, D.; Batiha, G.E.-S.; Mostafa-Hedeab, G.; Iyobhebhe, M.E.; Elebiyo, T.C.; Atunwa, B.; Ojo, A.B.; Lima, C.M.G.; et al. Anticancer Properties of Curcumin Against Colorectal Cancer: A Review. Front. Oncol. 2022, 12, 881641. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Zhang, Q.; Zhang, W.; Zhu, L. Curcumin inhibits liver metastasis of gastric cancer through reducing circulating tumor cells. Aging 2019, 11, 1501–1509. [Google Scholar] [CrossRef] [PubMed]
- Zhao, R.; Liu, J.; Li, Z.; Zhang, W.; Wang, F.; Zhang, B. Recent Advances in CXCL12/CXCR4 Antagonists and Nano-Based Drug Delivery Systems for Cancer Therapy. Pharmaceutics 2022, 14, 1541. [Google Scholar] [CrossRef] [PubMed]
- Riggio, A.I.; Varley, K.E.; Welm, A.L. The lingering mysteries of metastatic recurrence in breast cancer. Br. J. Cancer 2021, 124, 13–26. [Google Scholar] [CrossRef]
- Czerwiec, K.; Zawrzykraj, M.; Deptuła, M.; Skoniecka, A.; Tymińska, A.; Zieliński, J.; Kosiński, A.; Pikuła, M. Adipose-Derived Mesenchymal Stromal Cells in Basic Research and Clinical Applications. Int. J. Mol. Sci. 2023, 24, 3888. [Google Scholar] [CrossRef]
- Frisbie, L.; Buckanovich, R.J.; Coffman, L. Carcinoma-Associated Mesenchymal Stem/Stromal Cells: Architects of the Pro-tumorigenic Tumor Microenvironment. Stem Cells 2022, 40, 705–715. [Google Scholar] [CrossRef]
- Zhang, C.; Fei, Y.; Wang, H.; Hu, S.; Liu, C.; Hu, R.; Du, Q. CAFs orchestrates tumor immune microenvironment—A new target in cancer therapy? Front. Pharmacol. 2023, 14, 1113378. [Google Scholar] [CrossRef] [PubMed]
- Geng, X.; Chen, H.; Zhao, L.; Hu, J.; Yang, W.; Li, G.; Cheng, C.; Zhao, Z.; Zhang, T.; Li, L.; et al. Cancer-Associated Fibroblast (CAF) Heterogeneity and Targeting Therapy of CAFs in Pancreatic Cancer. Front. Cell Dev. Biol. 2021, 9, 655152. [Google Scholar] [CrossRef] [PubMed]
- Tan, H.-X.; Xiao, Z.-G.; Huang, T.; Fang, Z.-X.; Liu, Y.; Huang, Z.-C. CXCR4/TGF-β1 mediated self-differentiation of human mesenchymal stem cells to carcinoma-associated fibroblasts and promoted colorectal carcinoma development. Cancer Biol. Ther. 2020, 21, 248–257. [Google Scholar] [CrossRef] [PubMed]
- Hussain, S.; Peng, B.; Cherian, M.; Song, J.W.; Ahirwar, D.K.; Ganju, R.K. The Roles of Stroma-Derived Chemokine in Different Stages of Cancer Metastases. Front. Immunol. 2020, 11, 598532. [Google Scholar] [CrossRef] [PubMed]
- Otsuka, S.; Klimowicz, A.C.; Kopciuk, K.; Petrillo, S.K.; Konno, M.; Hao, D.; Muzik, H.; Stolte, E.; Boland, W.; Morris, D.; et al. CXCR4 Overexpression Is Associated with Poor Outcome in Females Diagnosed with Stage IV Non-small Cell Lung Cancer. J. Thorac. Oncol. 2011, 6, 1169–1178. [Google Scholar] [CrossRef] [PubMed]
- Batra, P.; Sharma, A.K. Anti-cancer potential of flavonoids: Recent trends and future perspectives. 3 Biotech 2013, 3, 439–459. [Google Scholar] [CrossRef]
- Sharma, T.; Singh, D.; Mahapatra, A.; Mohapatra, P.; Sahoo, S.; Sahoo, S.K. Advancements in clinical translation of flavonoid nanoparticles for cancer treatment. OpenNano 2022, 8, 100074. [Google Scholar] [CrossRef]
- Zhang, Z.; Chen, H.; Xu, C.; Song, L.; Huang, L.; Lai, Y.; Wang, Y.; Chen, H.; Gu, D.; Ren, L.; et al. Curcumin inhibits tumor epithelial-mesenchymal transition by downregulating the Wnt signaling pathway and upregulating NKD2 expression in colon cancer cells. Oncol. Rep. 2016, 35, 2615–2623. [Google Scholar] [CrossRef]
- Domingo-Domenech, J.; Mellado, B.; Ferrer, B.; Truan, D.; Codony-Servat, J.; Sauleda, S.; Alcover, J.; Campo, E.; Gascon, P.; Rovira, A.; et al. Activation of nuclear factor-κB in human prostate carcinogenesis and association to biochemical relapse. Br. J. Cancer 2005, 93, 1285–1294. [Google Scholar] [CrossRef]
- Li, Q.; Yang, G.; Feng, M.; Zheng, S.; Cao, Z.; Qiu, J.; You, L.; Zheng, L.; Hu, Y.; Zhang, T.; et al. NF-κB in pancreatic cancer: Its key role in chemoresistance. Cancer Lett. 2018, 421, 127–134. [Google Scholar] [CrossRef]
- Cai, Z.; Tchou-Wong, K.-M.; Rom, W.N. NF-kappaB in Lung Tumorigenesis. Cancers 2011, 3, 4258–4268. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.M.; Lyu, Y.L.; Cai, L. NF-κB Affects Proliferation and Invasiveness of Breast Cancer Cells by Regulating CD44 Expression. PLoS ONE 2014, 9, e106966. [Google Scholar] [CrossRef] [PubMed]
- Olivera, A.; Moore, T.W.; Hu, F.; Brown, A.P.; Sun, A.; Liotta, D.C.; Snyder, J.P.; Yoon, Y.; Shim, H.; Marcus, A.I.; et al. Inhibition of the NF-κB signaling pathway by the curcumin analog, 3,5-bis(2-pyridinylmethylidene)-4-piperidone (EF31): Anti-inflammatory and anti-cancer properties. Int. Immunopharmacol. 2012, 12, 368–377. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Riese, D.J.; Shen, J. The Role of the CXCL12/CXCR4/CXCR7 Chemokine Axis in Cancer. Front. Pharmacol. 2020, 11, 574667. [Google Scholar] [CrossRef] [PubMed]
- Zielińska, K.A.; Katanaev, V.L. The Signaling Duo CXCL12 and CXCR4: Chemokine Fuel for Breast Cancer Tumorigenesis. Cancers 2020, 12, 3071. [Google Scholar] [CrossRef] [PubMed]
- Huber, M.A.; Azoitei, N.; Baumann, B.; Grünert, S.; Sommer, A.; Pehamberger, H.; Kraut, N.; Beug, H.; Wirth, T. NF-κB is essential for epithelial-mesenchymal transition and metastasis in a model of breast cancer progression. J. Clin. Investig. 2004, 114, 569–581. [Google Scholar] [CrossRef] [PubMed]
- Georgakopoulos-Soares, I.; Chartoumpekis, D.V.; Kyriazopoulou, V.; Zaravinos, A. EMT Factors and Metabolic Pathways in Cancer. Front. Oncol. 2020, 10, 499. [Google Scholar] [CrossRef]
- Tan, B.L.; Norhaizan, M.E. Curcumin Combination Chemotherapy: The Implication and Efficacy in Cancer. Molecules 2019, 24, 2527. [Google Scholar] [CrossRef]
- Xiao, B.; Si, X.; Han, M.K.; Viennois, E.; Zhang, M.; Merlin, D. Co-delivery of camptothecin and curcumin by cationic polymeric nanoparticles for synergistic colon cancer combination chemotherapy. J. Mater. Chem. B 2015, 3, 7724–7733. [Google Scholar] [CrossRef]
- Banerjee, S.; Singh, S.K.; Chowdhury, I.; Lillard, J.W., Jr.; Singh, R. Combinatorial effect of curcumin with docetaxel modulates apoptotic and cell survival molecules in prostate cancer. FBE 2017, 9, 235–245. [Google Scholar] [CrossRef]
- Zhou, X.; Wang, W.; Li, P.; Zheng, Z.; Tu, Y.; Zhang, Y.; You, T. Curcumin Enhances the Effects of 5-Fluorouracil and Oxaliplatin in Inducing Gastric Cancer Cell Apoptosis Both In Vitro and In Vivo. Oncol. Res. Featur. Preclin. Clin. Cancer Ther. 2016, 23, 29–34. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jang, B.-Y.; Shin, M.K.; Han, D.-H.; Sung, J.-S. Curcumin Disrupts a Positive Feedback Loop between ADMSCs and Cancer Cells in the Breast Tumor Microenvironment via the CXCL12/CXCR4 Axis. Pharmaceutics 2023, 15, 2627. https://doi.org/10.3390/pharmaceutics15112627
Jang B-Y, Shin MK, Han D-H, Sung J-S. Curcumin Disrupts a Positive Feedback Loop between ADMSCs and Cancer Cells in the Breast Tumor Microenvironment via the CXCL12/CXCR4 Axis. Pharmaceutics. 2023; 15(11):2627. https://doi.org/10.3390/pharmaceutics15112627
Chicago/Turabian StyleJang, Bo-Young, Min Kyoung Shin, Dong-Hee Han, and Jung-Suk Sung. 2023. "Curcumin Disrupts a Positive Feedback Loop between ADMSCs and Cancer Cells in the Breast Tumor Microenvironment via the CXCL12/CXCR4 Axis" Pharmaceutics 15, no. 11: 2627. https://doi.org/10.3390/pharmaceutics15112627
APA StyleJang, B.-Y., Shin, M. K., Han, D.-H., & Sung, J.-S. (2023). Curcumin Disrupts a Positive Feedback Loop between ADMSCs and Cancer Cells in the Breast Tumor Microenvironment via the CXCL12/CXCR4 Axis. Pharmaceutics, 15(11), 2627. https://doi.org/10.3390/pharmaceutics15112627





