Computation-Aided Design of Albumin Affibody-Inserted Antibody Fragment for the Prolonged Serum Half-Life
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Design and Plasmid Construction
2.3. Structural Computation
2.4. Expression and Purification
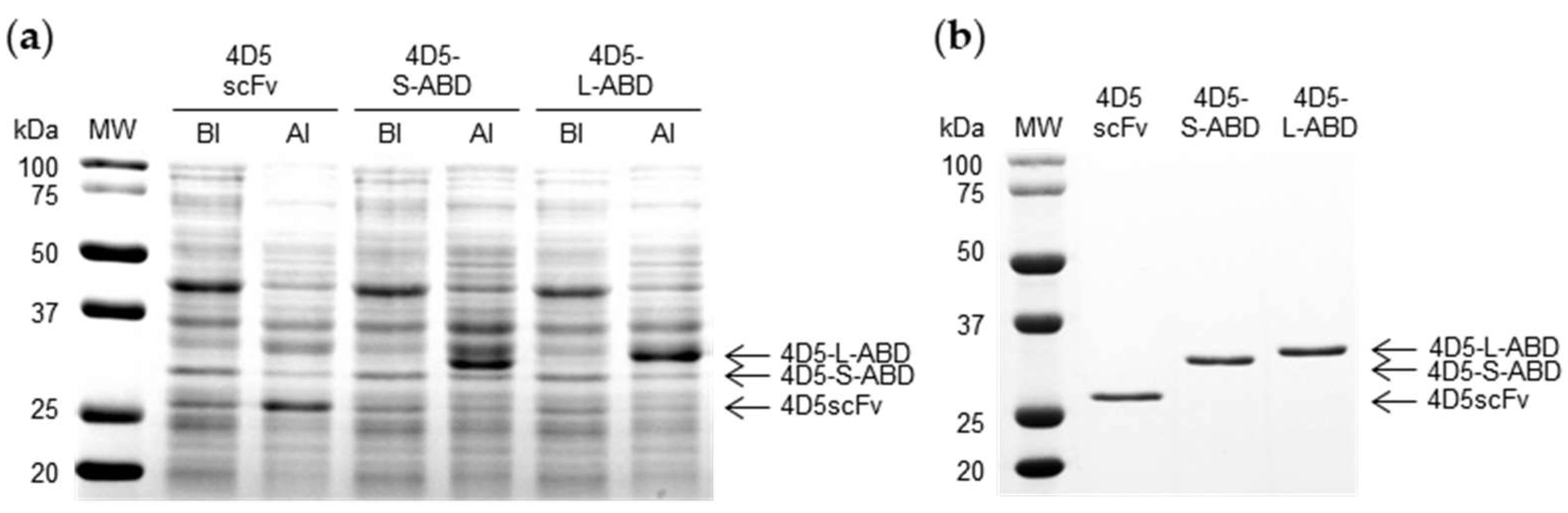
2.5. Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis (SDS-PAGE) Analysis
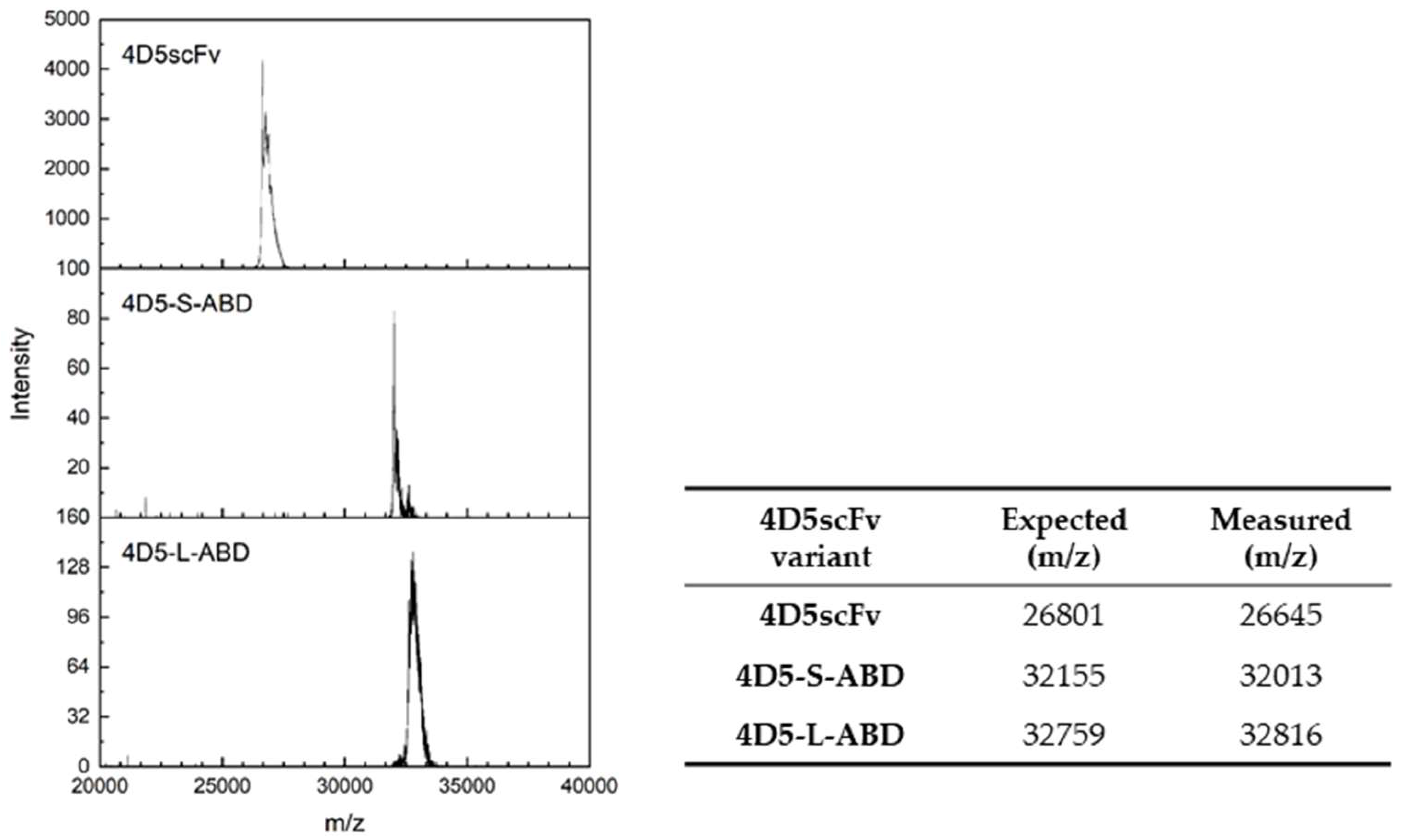
2.6. Matrix-Assisted Laser Desorption/Ionization Time-of-Flight (MALDI-TOF) Analysis
2.7. Size Exclusion Chromatography (SEC)
2.8. Enzyme-Linked Immunosorbent Assay (ELISA)
2.9. Pharmacokinetic Study
3. Results and Discussion
3.1. Protein Design and Computation
3.2. Expression and Purification
3.3. Binding Affinity Assays
3.4. Pharmacokinetic Study
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lu, R.M.; Hwang, Y.C.; Liu, I.J.; Lee, C.C.; Tsai, H.Z.; Li, H.J.; Wu, H.C. Development of Therapeutic Antibodies for the Treatment of Diseases. J. Biomed. Sci. 2020, 27, 1. [Google Scholar] [CrossRef] [PubMed]
- Bates, A.; Power, C.A. David vs. Goliath: The Structure, Function, and Clinical Prospects of Antibody Fragments. Antibodies 2019, 8, 28. [Google Scholar] [CrossRef] [PubMed]
- Bird, R.E.; Hardman, K.D.; Jacobson, J.W.; Johnson, S.Y.D.; Kaufman, B.M.; Lee, S.; Lee, T.; Pope, S.H.; Riordan, G.S.; Whitlow, M. Single-Chain Antigen-Binding Proteins. Science 1988, 242, 423–426. [Google Scholar] [CrossRef] [PubMed]
- The Antibody Society Therapeutic Monoclonal Antibodies Approved or in Review in the EU or US. Available online: www.antibodysociety.org/resources/approved-antibodies. (accessed on 5 July 2022).
- Huston, J.S.; Levinson, D.; Mudgett-Hunter, M.; Tai, M.S.; Novotny, J.; Margolies, M.N.; Ridge, R.J.; Bruccoleri, R.E.; Haber, E.; Crea, R.; et al. Protein Engineering of Antibody Binding Sites: Recovery of Specific Activity in an Anti-Digoxin Single-Chain Fv Analogue Produced in Escherichia Coli. Proc. Natl. Acad. Sci. USA 1988, 85, 5879–5883. [Google Scholar] [CrossRef]
- Chen, X.; Zaro, J.L.; Shen, W.C. Fusion Protein Linkers: Property, Design and Functionality. Adv. Drug Deliv. Rev. 2013, 65, 1357–1369. [Google Scholar] [CrossRef]
- Gupta, S.K.; Shukla, P. Microbial Platform Technology for Recombinant Antibody Fragment Production: A Review. Crit. Rev. Microbiol. 2017, 43, 31–42. [Google Scholar] [CrossRef]
- Wang, X.; Mathieu, M.; Brezski, R.J. IgG Fc Engineering to Modulate Antibody Effector Functions. Protein Cell 2018, 9, 63–73. [Google Scholar] [CrossRef]
- Yokota, T.; Milenic, D.E.; Whitlow, M.; Schlom, J. Rapid Tumor Penetration of a Single-Chain Fv and Comparison with Other Immunoglobulin Forms. Cancer Res. 1992, 52, 3402–3408. [Google Scholar]
- Graff, C.P.; Wittrup, K.D. Theoretical Analysis of Antibody Targeting of Tumor Spheroids: Importance of Dosage for Penetration, and Affinity for Retention. Cancer Res. 2003, 63, 1288–1296. [Google Scholar]
- Batra, S.K.; Jain, M.; Wittel, U.A.; Chauhan, S.C.; Colcher, D. Pharmacokinetics and Biodistribution of Genetically Engineered Antibodies. Curr. Opin. Biotechnol. 2002, 13, 603–608. [Google Scholar] [CrossRef]
- Kleinová, V.; Švecová, H.; Chaloupková, H.; Kranda, K.; Fišer, M. Biodistribution of the Radiolabeled Anti III Β-Tubulin ScFv Fragment in Mice. AIP Conf. Proc. 2007, 958, 288. [Google Scholar] [CrossRef]
- Kang, T.H.; Seong, B.L. Solubility, Stability, and Avidity of Recombinant Antibody Fragments Expressed in Microorganisms. Front. Microbiol. 2020, 11, 1927. [Google Scholar] [CrossRef] [PubMed]
- Eigenbrot, C.; Randal, M.; Presta, L.; Carter, P.; Kossiakoff, A. X-Ray Structures of the Antigen-Binding Domains from Three Variants of Humanized Anti-P185HER2 Antibody 4D5 and Comparison with Molecular Modeling. J. Mol. Biol. 1993, 229, 969–995. [Google Scholar] [CrossRef]
- Rahimizadeh, P.; Yang, S.; Lim, S.I. Albumin: An Emerging Opportunity in Drug Delivery. Biotechnol. Bioprocess Eng. 2020, 25, 985–995. [Google Scholar] [CrossRef]
- Chaudhury, C.; Mehnaz, S.; Robinson, J.M.; Hayton, W.L.; Pearl, D.K.; Roopenian, D.C.; Anderson, C.L. The Major Histocompatibility Complex–Related Fc Receptor for IgG (FcRn) Binds Albumin and Prolongs Its Lifespan. J. Exp. Med. 2003, 197, 315–322. [Google Scholar] [CrossRef]
- Zaman, R.; Islam, R.A.; Ibnat, N.; Othman, I.; Zaini, A. Current Strategies in Extending Half-Lives of Therapeutic Proteins. J. Control. Release 2019, 301, 176–189. [Google Scholar] [CrossRef]
- Tao, H.; Wang, R.; Sheng, W.; Zhen, Y. The Development of Human Serum Albumin-Based Drugs and Relevant Fusion Proteins for Cancer Therapy. Int. J. Biol. Macromol. 2021, 187, 24–34. [Google Scholar] [CrossRef]
- Andersen, J.T.; Cameron, J.; Plumridge, A.; Evans, L.; Sleep, D.; Sandlie, I. Single-Chain Variable Fragment Albumin Fusions Bind the Neonatal Fc Receptor (FcRn) in a Species-Dependent Manner: Implications for in Vivo Half-Life Evaluation of Albumin Fusion Therapeutics. J. Biol. Chem. 2013, 288, 24277–24285. [Google Scholar] [CrossRef]
- Müller, D.; Karle, A.; Meißburger, B.; Höfig, I.; Stork, R.; Kontermann, R.E. Improved Pharmacokinetics of Recombinant Bispecific Antibody Molecules by Fusion to Human Serum Albumin. J. Biol. Chem. 2007, 282, 12650–12660. [Google Scholar] [CrossRef]
- Liu, Z.; Chen, X. Simple Bioconjugate Chemistry Serves Great Clinical Advances: Albumin as a Versatile Platform for Diagnosis and Precision Therapy. Chem. Soc. Rev. 2016, 45, 1432–1456. [Google Scholar] [CrossRef]
- Hoogenboezem, E.N.; Duvall, C.L. Harnessing Albumin as a Carrier for Cancer Therapies. Adv. Drug Deliv. Rev. 2018, 130, 73–89. [Google Scholar] [CrossRef] [PubMed]
- Forte, N.; Livanos, M.; Miranda, E.; Morais, M.; Yang, X.; Rajkumar, V.S.; Chester, K.A.; Chudasama, V.; Baker, J.R. Tuning the Hydrolytic Stability of Next Generation Maleimide Cross-Linkers Enables Access to Albumin-Antibody Fragment Conjugates and Tri-ScFvs. Bioconjug. Chem. 2018, 29, 486–492. [Google Scholar] [CrossRef] [Green Version]
- Cho, J.; Kim, S.H.; Yang, B.; Jung, J.M.; Kwon, I.; Lee, D.S. Albumin Affibody-Outfitted Injectable Gel Enabling Extended Release of Urate Oxidase-Albumin Conjugates for Hyperuricemia Treatment. J. Control. Release 2020, 324, 532–544. [Google Scholar] [CrossRef]
- Andersen, J.T.; Pehrson, R.; Tolmachev, V.; Daba, M.B.; Abrahmsén, L.; Ekblad, C. Extending Half-Life by Indirect Targeting of the Neonatal Fc Receptor (FcRn) Using a Minimal Albumin Binding Domain. J. Biol. Chem. 2011, 286, 5234–5241. [Google Scholar] [CrossRef]
- Johansson, M.U.; Frick, I.M.; Nilsson, H.; Kraulis, P.J.; Hober, S.; Jonasson, P.; Linhult, M.; Nygren, P.Å.; Uhlén, M.; Björck, L.; et al. Structure, Specificity, and Mode of Interaction for Bacterial Albumin-Binding Modules. J. Biol. Chem. 2002, 277, 8114–8120. [Google Scholar] [CrossRef] [PubMed]
- Jonsson, A.; Dogan, J.; Herne, N.; Abrahmsén, L.; Nygren, P.Å. Engineering of a Femtomolar Affinity Binding Protein to Human Serum Albumin. Protein Eng. Des. Sel. 2008, 21, 515–527. [Google Scholar] [CrossRef] [PubMed]
- Stork, R.; Müller, D.; Kontermann, R.E. A Novel Tri-Functional Antibody Fusion Protein with Improved Pharmacokinetic Properties Generated by Fusing a Bispecific Single-Chain Diabody with an Albumin-Binding Domain from Streptococcal Protein G. Protein Eng. Des. Sel. 2007, 20, 569–576. [Google Scholar] [CrossRef]
- Hopp, J.; Hornig, N.; Zettlitz, K.A.; Schwarz, A.; Fuß, N.; Müller, D.; Kontermann, R.E. The Effects of Affinity and Valency of an Albumin-Binding Domain (ABD) on the Half-Life of a Single-Chain Diabody-ABD Fusion Protein. Protein Eng. Des. Sel. 2010, 23, 827–834. [Google Scholar] [CrossRef]
- Kenanova, V.E.; Olafsen, T.; Salazar, F.B.; Williams, L.E.; Knowles, S.; Wu, A.M. Tuning the Serum Persistence of Human Serum Albumin Domain III: Diabody Fusion Proteins. Protein Eng. Des. Sel. 2010, 23, 789–798. [Google Scholar] [CrossRef]
- Mironova, K.E.; Proshkina, G.M.; Ryabova, A.V.; Stremovskiy, O.A.; Lukyanov, S.A.; Petrov, R.V.; Deyev, S.M. Genetically Encoded Immunophotosensitizer 4D5scFv-MiniSOG Is a Highly Selective Agent for Targeted Photokilling of Tumor Cells In Vitro. Theranostics 2013, 3, 831. [Google Scholar] [CrossRef]
- Agha Amiri, S.; Shahhosseini, S.; Zarei, N.; Khorasanizadeh, D.; Aminollahi, E.; Rezaie, F.; Zargari, M.; Azizi, M.; Khalaj, V. A Novel Anti-CD22 ScFv–Apoptin Fusion Protein Induces Apoptosis in Malignant B-Cells. AMB Express 2017, 7, 112. [Google Scholar] [CrossRef] [PubMed]
- Silver, A.B.; Leonard, E.K.; Gould, J.R.; Spangler, J.B. Engineered Antibody Fusion Proteins for Targeted Disease Therapy. Trends Pharmacol. Sci. 2021, 42, 1064–1081. [Google Scholar] [CrossRef]
- Ahamadi-Fesharaki, R.; Fateh, A.; Vaziri, F.; Solgi, G.; Siadat, S.D.; Mahboudi, F.; Rahimi-Jamnani, F. Single-Chain Variable Fragment-Based Bispecific Antibodies: Hitting Two Targets with One Sophisticated Arrow. Mol. Ther. Oncolytics 2019, 14, 38–56. [Google Scholar] [CrossRef] [PubMed]
- Malm, M.; Bass, T.; Gudmundsdotter, L.; Lord, M.; Frejd, F.Y.; Ståhl, S.; Löfblom, J. Engineering of a Bispecific Affibody Molecule towards HER2 and HER3 by Addition of an Albumin-Binding Domain Allows for Affinity Purification and in Vivo Half-Life Extension. Biotechnol. J. 2014, 9, 1215–1222. [Google Scholar] [CrossRef] [PubMed]
- AlQuraishi, M. Machine Learning in Protein Structure Prediction. Curr. Opin. Chem. Biol. 2021, 65, 1–8. [Google Scholar] [CrossRef]
- Pakhrin, S.C.; Shrestha, B.; Adhikari, B.; KC, D.B. Deep Learning-Based Advances in Protein Structure Prediction. Int. J. Mol. Sci. 2021, 22, 5553. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Pearce, R.; Zhang, Y. Deep Learning Techniques Have Significantly Impacted Protein Structure Prediction and Protein Design. Curr. Opin. Struct. Biol. 2021, 68, 194–207. [Google Scholar] [CrossRef]
- Cho, H.S.; Mason, K.; Ramyar, K.X.; Stanley, A.M.; Gabelli, S.B.; Denney, D.W.; Leahy, D.J. Structure of the Extracellular Region of HER2 Alone and in Complex with the Herceptin Fab. Nature 2003, 421, 756–760. [Google Scholar] [CrossRef]
- Mirdita, M.; Schütze, K.; Moriwaki, Y.; Heo, L. ColabFold: Making Protein Folding Accessible to All. Nat. Methods 2022, 19, 679–682. [Google Scholar] [CrossRef]
- Burley, S.K.; Bhikadiya, C.; Bi, C.; Bittrich, S.; Chen, L.; Crichlow, G.V.; Christie, C.H.; Dalenberg, K.; Di Costanzo, L.; Duarte, J.M.; et al. RCSB Protein Data Bank: Powerful New Tools for Exploring 3D Structures of Biological Macromolecules for Basic and Applied Rsearch and Education in Fundamental Biology, Biomedicine, Biotechnology, Bioengineering and Energy Sciences. Nucleic Acids Res. 2021, 49, D437–D451. [Google Scholar] [CrossRef] [PubMed]
- Koçer, İ.; Cox, E.C.; DeLisa, M.P.; Çelik, E. Effects of Variable Domain Orientation on Anti-HER2 Single-Chain Variable Fragment Antibody Expressed in the Escherichia Coli Cytoplasm. Biotechnol. Prog. 2021, 37, e3102. [Google Scholar] [CrossRef] [PubMed]
- Goulet, A.; Cambillau, C. Present Impact of AlphaFold2 Revolution on Structural Biology, and an Illustration with the Structure Prediction of the Bacteriophage J-1 Host Adhesion Device. Front. Mol. Biosci. 2022, 9, 907452. [Google Scholar] [CrossRef] [PubMed]
- Bouatta, N.; Sorger, P.; AlQuraishi, M. Protein Structure Prediction by AlphaFold2: Are Attention and Symmetries All You Need? Acta Crystallogr. Sect. D Struct. Biol. 2021, 77, 982–991. [Google Scholar] [CrossRef]
- Giavazzi, R.; Yokota, T.; Filpula, D.R.; Finkelman, M.A.J.; Dodd, S.W.; Wood, J.F.; Whitlow, M.; Snoy, P.; Schlom, J. Construction, Binding Properties, Metabolism, and Tumor Targeting of a Single-Chain Fv Derived from the Pancarcinoma Monoclonal Antibody CC49. Cancer Res. 1991, 51, 6363–6371. [Google Scholar]
- Baek, M.; DiMaio, F.; Anishchenko, I.; Dauparas, J.; Ovchinnikov, S.; Lee, G.R.; Wang, J.; Cong, Q.; Kinch, L.N.; Dustin Schaeffer, R.; et al. Accurate Prediction of Protein Structures and Interactions Using a Three-Track Neural Network. Science 2021, 373, 871–876. [Google Scholar] [CrossRef]
- Leman, J.K.; Weitzner, B.D.; Lewis, S.M.; Adolf-Bryfogle, J.; Alam, N.; Alford, R.F.; Aprahamian, M.; Baker, D.; Barlow, K.A.; Barth, P.; et al. Macromolecular Modeling and Design in Rosetta: Recent Methods and Frameworks. Nat. Methods 2020, 17, 665–680. [Google Scholar] [CrossRef]
- Kratz, F. Albumin as a Drug Carrier: Design of Prodrugs, Drug Conjugates and Nanoparticles. J. Control. Release 2008, 132, 171–183. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, Y.; Wu, Y.; Jiang, X.; Tao, Y.; Yao, Y.; Peng, Y.; Chen, X.; Fu, Y.; Yu, L.; et al. Therapeutic Potential of an Anti-HER2 Single Chain Antibody–DM1 Conjugates for the Treatment of HER2-Positive Cancer. Signal Transduct. Target. Ther. 2017, 2, e17015. [Google Scholar] [CrossRef] [Green Version]



| 4D5scFv Variant | EC50 (nM) | ||
|---|---|---|---|
| Anti-HER2 HSA (−) | Anti-HER2 HSA (+) | Anti-HSA | |
| 4D5scFv | 1.20 ± 0.05 | 1.06 ± 0.21 | - |
| 4D5-S-ABD | 1.95 ± 0.05 | 4.07 ± 0.54 | 1.34 ± 0.05 |
| 4D5-L-ABD | 1.50 ± 0.06 | 3.96 ± 0.98 | 1.08 ± 0.22 |
| 4D5scFv Variant | t1/2 (h) | AUC (%∙h) |
|---|---|---|
| 4D5scFv | 0.30 ± 0.25 | 42 1 |
| 4D5-S-ABD | 34.19 ± 0.05 | 2887 2 |
| 4D5-L-ABD | 34.29 ± 0.05 | 3008 2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kwon, N.H.; Lee, J.H.; Kwon, I. Computation-Aided Design of Albumin Affibody-Inserted Antibody Fragment for the Prolonged Serum Half-Life. Pharmaceutics 2022, 14, 1769. https://doi.org/10.3390/pharmaceutics14091769
Kwon NH, Lee JH, Kwon I. Computation-Aided Design of Albumin Affibody-Inserted Antibody Fragment for the Prolonged Serum Half-Life. Pharmaceutics. 2022; 14(9):1769. https://doi.org/10.3390/pharmaceutics14091769
Chicago/Turabian StyleKwon, Na Hyun, Jae Hun Lee, and Inchan Kwon. 2022. "Computation-Aided Design of Albumin Affibody-Inserted Antibody Fragment for the Prolonged Serum Half-Life" Pharmaceutics 14, no. 9: 1769. https://doi.org/10.3390/pharmaceutics14091769
APA StyleKwon, N. H., Lee, J. H., & Kwon, I. (2022). Computation-Aided Design of Albumin Affibody-Inserted Antibody Fragment for the Prolonged Serum Half-Life. Pharmaceutics, 14(9), 1769. https://doi.org/10.3390/pharmaceutics14091769






