Abstract
A severe form of myopia defined as pathologic/high myopia is the main cause of visual impairment and one of the most frequent causes of blindness worldwide. It is characterized by at least 6 diopters or axial length (AL) of eyeball > 26 mm and choroidal neovascularization (CNV) in 5 to 10% of cases. Ranibizumab is a humanized recombinant monoclonal antibody fragment targeted against human vascular endothelial growth factor A (VEGF-A) used in the treatment of CNV. It acts by preventing VEGF-A from interacting with its receptors (VEGFR-1 and -2) encoded by the FLT1 and KDR genes. Several studies found that the KDR and FLT1 genotypes may represent predictive determinants of efficacy in ranibizumab-treated neovascular age-related macular degeneration (nAMD) patients. We performed a retrospective study to evaluate the association of single nucleotide polymorphisms (SNPs) in VEGFR coding genes with the response rate to ranibizumab in patients with high myopia and CNV. In the association study of genotypes in FLT1 with the response to ranibizumab, we found a significant association between two FLT1 variants (rs9582036, rs7993418) with ranibizumab efficacy at the 12-month follow-up. About the KDR gene, we found that two KDR variants (rs2305948, rs2071559) are associated with best-corrected visual acuity (BCVA) improvement and KDR (rs2239702) is associated with lower rates of BCVA worsening considering a 12-month follow-up period.
1. Introduction
Myopia is an eye disease with a varied geographic prevalence. In Asian countries, its prevalence is up to 70–90%, whereas in western Europe, it is 25–50% [1,2,3]. The most severe form of myopia, defined as pathological/high myopia, is the main cause of visual impairment and one of the most frequent causes of blindness worldwide [4]. Patients with pathological myopia are characterized by an axial length ≥26 mm or a refractive error of −6.0 diopters or more [1,5,6]. These patients have progressive and excessive globe elongation with retinal and choroidal thinning, peripheral retinal degeneration, and an increased risk of retinal detachment, cataracts, glaucoma, and myopic choroidal neovascularization (mCNV) [7,8].
mCNV develops in 5–10% of pathological myopia patients and it is one of the most vision-threatening complications of these patients [1,9,10]. Furthermore, pathologic/high myopia is the most common cause of CNV in individuals aged 50 years or younger and the second cause of CNV after nAMD [11].
mCNV is characterized clinically by retinal hemorrhage with or without exudation. This hemorrhage, after its resolution, may leave an area of chorioretinal atrophy or a grayish-white pigmented scar (Fuchs spot). mCNV is different in young or aged patients. Young patients usually show small classic lesions located close to the fovea. These lesions can cause rapid visual loss with or without metamorphopsia and/or central scotoma. Moreover, in aged individuals, mCNV is more extensive and exudative; it may lead to chorioretinal scar formation similar to nAMD. Some cases of mCNV may regress spontaneously with minimal impact on vision; however, without appropriate treatment, most eyes will have a poor visual outcome [11].
The most common treatment for mCNV includes thermal laser photocoagulation, verteporfin photodynamic therapy (vPDT), and surgery, specifically removing the mCNV [12,13,14]. Laser photocoagulation treatment can result in permanent retinal damage, causing scotoma increase, scarring, and visual loss over time. Moreover, recurrent CNV may arise at the margins of previously laser-treated areas. This may be due to rupture of the retinal pigment epithelium and Bruch’s membrane stimulating new CNV lesions in some patients [15,16].
In 2001, the results of the VIP study based on the use of vPDT in mCNV were published, which showed that it was capable of maintaining visual acuity (VA) at 12 months [17], although no significant difference was observed in the primary outcome at 24 months [18]. Furthermore, the development of long-term chorioretinal atrophy based on the use of vPDT in mCNV was observed in another study [19]. The results of the surgery were not good either [14].
The VEGF is a potent proangiogenic factor that stimulates CNV development. Following binding to VEGFR, endothelial cells are stimulated to proliferate, migrate, and express matrix-degrading proteases, causing vascular instability, leakage, and finally angiogenesis [11].
The introduction of monoclonal antibodies directed against VEGF in the treatment of mCNV was a real breakthrough. Ranibizumab (Lucentis®, Novartis Pharma AG; Stein, Switzerland) is a humanized recombinant monoclonal antibody fragment targeted against human VEGF-A. It binds with high affinity to VEGF-A, thereby preventing binding of VEGF-A to VEGFR-1 and -2 at the endothelial cell surface, thus inhibiting cell division, migration, and angiogenesis; increasing apoptosis; and reducing the permeability/leakage of the vasculature [20,21].
The RADIANCE, a phase III, 12-month study that evaluated Ranibizumab and PDT (verteporfin) in mCNV, proved that ranibizumab has good efficacy and safety, with a significant and maintained gain in VA. Furthermore, a relatively low number of injections are required to treat most patients with mCNV. Over 60% of patients did not require any injections from month 6 onwards, and more than 50% required only 1 or 2 injections over 12 months. Although, 34.5% required 3–5 injections, and 14.7% needed 6–12 injections during the 12-month study period. This suggests a need to understand factors such as genetic influence that may predict treatment response and the development of individualized treatment regimens [22].
Ranibizumab binds VEGF-A, impeding its action on VEGFR-1 and -2. VEGFR-1 and -2 are encoded by FLT1 and KDR, respectively (Figure 1).
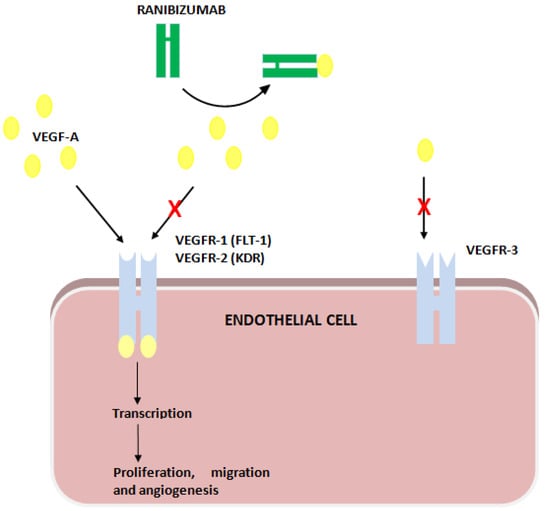
Figure 1.
Ranibizumab’s action on VEGF receptors (VEGFR).
Cobos et al. [23] found that FLT1 (VEGFR-1 coding gene) rs7993418 is associated with the ranibizumab treatment response in nAMD patients. Furthermore, Beuselinck B. et al. [24,25] and Dornbusch J. et al. [26] found that for sunitinib, used in the treatment of neoplastic pathologies such as metastatic kidney cancer by inhibiting neoangiogenic processes, FLT1 (rs9582036) is a predictive biomarker of treatment response.
The VEGFR-2 receptor, encoded by the KDR gene, is a high-affinity receptor tyrosine-kinase responsible for most of the angiogenic- and permeability-enhancing effects of VEGF-A. Thus, KDR (VEGFR-2 coding gene) variants are candidates for a possible genetic influence susceptibility to anti-VEGF therapy [27]. Even Lazzeri et al. [27] found that the KDR (VEGFR-2) rs2071559 genotype may be a predictive determinant of short- and long-term functional and anatomical outcomes in ranibizumab-treated nAMD patients. Hermann et al. [28] concluded that genetic polymorphisms in the KDR gene significantly influence the visual outcome in patients receiving ranibizumab treatment for nAMD.
Among our patients, when diagnosed with mCNV and treated with ranibizumab, ARMS2 (rs10490924) and CFH (rs1061170) SNPs were associated with response [29]. The ARMS2 (rs10490924) G allele and GG genotype led to a better response to ranibizumab at the 6-month follow-up. In contrast, the CFH (rs1061170) T allele and TT genotype were associated with higher rates of BCVA worsening at the 1-month follow-up.
In summary, ranibizumab is used in the treatment of high myopia and CNV patients. It binds VEGF-A, impeding its binding to VEGFR-1 and -2, encoded the by FLT1 and KDR genes. Thus, the genetic polymorphisms in both genes previously associated with interindividual differences in drug responses may also affect the ranibizumab response in mCNV.
This study aimed to evaluate the association of SNPs in the VEGFR (VEGFR-1 (FLT-1) and VEGFR-2 (KDR)) coding genes with the response rate to ranibizumab in patients with mCNV.
2. Materials and Methods
In this study, we assessed the same cohort of patients as the previous study by Blánquez-Martínez et al. [29] but with a 12-month follow-up period.
In summary, we carried out a retrospective study, recruiting patients between 2014 and 2019 with high myopia and CNV treated with ranibizumab at our hospital. Among these patients, we assessed the association of genetic polymorphisms in KDR and FLT1 with the response (BCVA improvement and worsening) to ranibizumab. We also recruited a control group with high myopia but no CNV to take care of the association of the included SNPs with the disease instead of the response to ranibizumab.
Specific characteristics about the inclusion/exclusion criteria, considered criteria for high myopia diagnosis, patient management, data management including definitions of BCVA “improvement” and “worsening”, and DNA extraction and genotyping have been previously published in the study by Blánquez-Martínez et al. [29].
This study was conducted in accordance with the Declaration of Helsinki and approved by the Ethics Committee of Granada (Spain) “CEIM/CEI Provincial de Granada” (approval code: 0085-N14; 26 May 2014).
2.1. Procedures for the Inclusion of Genetic Variants in the Study
We considered for inclusion those genetic variants in genes encoding the VEGFR-1 and -2 (KDR and FLT1) that had previously been proved to affect drug efficacy or toxicity. To do this, we searched PharmGKB for information about both genes. We considered for inclusion those genetic variants with a related annotation reporting the association with drug responses (efficacy or toxicity) in at least one publication and excluded those genetic variants with a minor allele frequency (MAF) lower than 10%. This means that we considered for inclusion the KDR rs2071559, rs2239702, rs1870377, rs34231037, rs2305948, and rs7667298 variants; and FLT1 rs664393, rs7993418, rs9554320, and rs9582036. Finally, we excluded KDR rs34231037 since it was the only one with a known MAF lower than 10% in the Iberian Peninsula population.
2.2. DNA Extraction and Genotyping
For genotyping, we took 4 saliva samples with sterile cotton swabs from each recruited patient. We isolated the DNA using standard procedures and DNA extraction was carried out following the method by Freeman et al. [30] with modifications by Gomez-Martín A. et al. [31]. We genotyped SNPs using KASP assay technology (LGC Genomics, Hoddesdon, Hertfordshire, UK) following the manufacturer’s instructions and these were analyzed with the KlusterCaller Software (LGC Genomics, Hoddesdon, Hertfordshire, UK). The call rate for all tested SNPs was >98%. Quality control for the genotyping results was achieved with negative controls and randomly selected samples included as duplicates.
2.3. Statistical Analysis
First, we carried out a descriptive analysis of the clinical parameters (Table 1) and calculated the distribution (number of patients and percentage) of genotypes and the minor allele frequency (MAF) of the included SNPs for both the control and study groups (Table 2). Then, we studied the Hardy–Weinberg (H-W) equilibrium of each SNP by group and for the total number of patients, and we performed an LD analysis of the genetic variants in each gene. Finally, the distribution of genotypes between the control and study groups was compared to assess the association of each SNP with CNV.

Table 1.
Baseline and follow-up descriptive analysis.

Table 2.
Genotype distribution, minor allele frequency, and Hardy–Weinberg equilibrium analysis for each studied SNP in our population.
The main aim was to study the association of included SNPs and BCVA improvement or worsening at the 12-month follow-up. In this regard, we carried out both allele and genotype comparison analyses. In the genotype analysis, we used each genetic model (recessive, dominant, co-dominant, and log-additive). We also performed a haplotype association study.
We used the Chi-square test or Fischer exact test. We calculated the odds ratio (OR) and p-values. p-values < 0.05 were considered statistically significant. The Bayesian information criterion (BIC) and Akaike information criterion (AIC) were calculated for each genetic model and SNP–response association study.
The descriptive analysis (Table 1), MAFs, genotypic distribution, and its comparison among groups (Table 2) were performed using R commander. The association studies and H–W equilibrium analysis were conducted using the SNPstats online tool [32].
The sample size calculation was based on that shown in the previous research article evaluating the influence of VEGFR variants on ranibizumab response in non-high myopia patients [23,24,25,26,27,28]. Furthermore, we recruited the total number of patients with high myopia and treated with ranibizumab in our hospital for five years. Both the sample size and statistical influence of SNPs on genetic diseases were studied in several works that investigated the impact on pathological phenotypes [33,34,35,36].
3. Results
In this study, we assessed the same cohort of patients published by Blánquez-Martínez et al. [29], including new genetic variants and extending the follow-up period to 12 months.
As commented in this previous study [29], we recruited n = 100 patients and n = 113 eyes diagnosed with high myopia and CNV that were treated with ranibizumab. We did not find an adequate DNA concentration for genotyping in one patient (n = 1 eye). Finally, n = 99 patients and n = 112 eyes were included in the study. Among the patients in the study group, 75% were women, with a mean age of 57.5 ± 13.9 years. Most of the studied eyes showed a juxta foveal CNV location (66.1%) and were not previously treated with LP/PDT (92%). On the other hand, n = 30 (26.8%) and n = 8 (7.1%) eyes presented a sub-foveal and extra-foveal location, respectively, and n = 8 (7.1%) had been treated with LP and n = 1 (0.9%) with PDT. The mean BCVA (logMAR) at baseline was 0.62 ± 0.48 and 0.34 ± 0.38 at the 12-month follow-up.
Regarding the control group, we recruited n = 116 patients and n = 219 eyes. In this group, n = 7 eyes could not be assessed. Among them, in 4 patients (n = 4 eyes), we could not obtain an adequate DNA concentration to investigate the needed genotypes; in n = 2 patients (n = 2 eyes), saliva samples were lost; in and 1 patient (n = 1 eye), mCNV was diagnosed one month after being recruited. This patient was transferred from the control to the treatment group.
The baseline characteristics of the recruited patients (study and control group) and the follow-up characteristics of the study group are shown in Table 1.
3.1. Genotypic Distribution, H–W Equilibrium, Association of Genetic Variants with High Myopia and CNV, and Linkage Disequilibrium Analysis
Among the included genetic variants, FLT1 C > T (rs664393) was the only one that showed a minor allele frequency (MAF) lower than 0.1, with no patients carrying the recessive homozygous genotype in the study group. None of the studied SNPs showed significant differences in the H–W equilibrium analysis, and the FLT1 C > A (rs9554320) was the only one that showed differences in the comparison of the genotypic distribution between the control and study groups (Table 2).
In the linkage disequilibrium (LD) analysis, regarding the FLT1 variants, we found that rs664393 was not linked to any other variant; however, three other included variants were linked among them (D’ > 0.97; p < 0.001) (Figure S1). Among the KDR variants, rs2305948 was not linked to any other variant and the other four variants were linked among them (p < 0.01) (Figure S2).
3.2. Association of Genetic Polymorphisms with Response to Ranibizumab
3.2.1. Genotype Association Study with Response
In the association study of the genotypes in FLT1 with the response to ranibizumab (Table 3), we found that the FLT1 (rs9582036) CC genotype was associated with lower rates of BCVA improvement at the 12-month follow-up (recessive model: CC vs. CA or AA; OR = 0.2; 95%CI = 0.04–0.9; p = 0.032). On the other hand, we did not find a significant association between the FLT1 (rs9582036) CC genotype with higher rates of worsening. Additionally, in FLT1, we found that the FLT1 (rs7993418) AA genotype was associated with higher rates of worsening (dominant model: AA vs. AG or GG; p = 0.013). Finally, we found no significant association of FLT1 (rs9554320) with BCVA improvement or worsening at the 12-month follow-up. We do not provide results about FLT1 (rs664393) since we did not identify patients who were carrying the recessive homozygous genotype.

Table 3.
Association study of FLT1 genotypes with BCVA improvement/worsening at 12 months.
About the KDR gene (Table 4), KDR (rs2305948) showed a significant association with BCVA improvement in the log-additive model (p = 0.049). Based on the other genetic models, it seems that the CC genotype is related to higher rates of BCVA improvement (dominant model: CC vs. CT or TT; OR = 0.35; 95%CI = 0.11–1.11; p = 0.055). Additionally, KDR (rs2071559) was associated with BCVA improvement among ranibizumab-treated patients (over-dominant model: OR = 2.91; 95%CI= 1.19–7.08; p = 0.015). Regarding KDR, among our patients, the KDR (rs2239702) GG genotype was associated with lower rates of BCVA worsening (dominant model: GG vs. GA or AA; OR= 0.15; 95%CI= 0.02–1.31); p = 0.044).

Table 4.
Association study of KDR genotypes with BCVA improvement/worsening at 12 months.
3.2.2. Alleles Association Study with Response
In the allele association study with the response, we found that the KDR (rs2239702) A allele was associated with higher rates of BCVA worsening (OR = 3.33; 95%CI = 1.03–10.78; p = 0.035) (Table 5). In contrast, it was the only SNP found to be related to ranibizumab response (BCVA improvement or worsening) among our patients. On the other hand, the KDR (rs2305948) T allele and (rs1870377) A allele showed close-to-significant results (p = 0.073 and p = 0.097, respectively) for their association with BCVA improvement and ranibizumab treatment.

Table 5.
Alleles association study with BCVA improvement/worsening at 12 months.
3.3. Haplotype Association Study with Response
In the haplotype association analysis with response to ranibizumab, we found that no haplotypes for FLT1 were associated with BCVA improvement nor with worsening (Table S1). For KDR, and according to the allele association study with the response, we found the most common haplotype (rs2239702 G; rs2305948: C; rs7667298: C; rs1870377: T and rs2071559: T) was associated with lower rates of worsening (OR= 0.09; 95% CI= 0.01–0.86; p = 0.039) compared to the ACTTC haplotype, carried by 21.9% of the patients (Table S2).
4. Discussion
Ranibizumab is an anti-VEGF drug used in the treatment of mCNV among other pathologies. Many genetic polymorphisms have been associated with the response to this drug. Among AMD patients, CFH (rs1061170) and ARMS2 rs10490924 were related to interindividual differences in the response to anti-VEGF drugs [37,38]. Additionally, among AMD patients, genetic polymorphisms in VEGFA (rs3025000, 833069, and rs699947) [37,39,40,41], NRP1 (rs2070296) [42], and CXCL8 (rs4073) [27] are associated with interindividual differences in the response to ranibizumab. Polypoidal choroidal neovascularization is usually considered a sub-type of CNV. In patients with CNV and/or PCV, VEGFA (rs2010963) [43], HTRA1 (rs11200638) [44], and especially CFH (rs1061170) and ARMS2 (rs10490924) [45] were also found to be associated with ranibizumab efficacy.
The influence of genetic polymorphisms on the ranibizumab response among mCNV patients has not been widely studied. Among our patients, we found in a previous study [29] that CFH (rs1061170) and ARMS2 (rs10490924) are related to ranibizumab efficacy at the 1- and 6-month follow-up. The CFH (rs1061170) C allele was associated with lower BCVA worsening and BCVA improvement, the TT genotype was associated with BCVA worsening, and the CC genotype was associated with BCVA improvement. About ARMS2 (rs10490924), the G allele and GG genotype showed an association with BCVA improvement.
On the other hand, in this previous study, we still found patients that did not meet the expected progress of the illness based on our results. Thus, we considered a study of the genetic polymorphism in KDR and FLT1 considering a larger follow-up time (12 months). SNPs in these genes have not been previously studied in mCNV patients treated with ranibizumab.
This study had some limitations. It is a retrospective observational study; this means that we did not assess the clinical impact of the studied genetic polymorphisms on the ranibizumab response in daily clinical conditions. However, this would not be ethical since these genetic polymorphisms have not been associated with differences in the ranibizumab response. We recruited a control group without collecting data about BCVA improvement/worsening because this was only to control the possible association of genetic polymorphisms with mCNV, not with ranibizumab efficacy. We recruited n= 100 patients; the study cohort is low, and it should be increased in further studies. On the other hand, this was the total number of patients with mCNV treated with ranibizumab in our hospital in a 5-year period. In the same regard, we did not perform a multivariant analysis considering the combined genetic and clinical parameters. We did not perform a multiple test analysis since it would increase the risk of type I errors and the low number of recruited patients did not allow a Bonferroni correction to be performed. Regarding the assessment of eyes/patients, we included both eyes of patients with bilateral treatment considering that they might progress in a different way depending on the expression/silencing of PGx variants.
Our results still support the need for further studies, considering a clinical trial including combined PGx, clinical parameters (e.g., previous treatments such as LP and PDT), in a multivariant analysis, and in a larger cohort that includes patients from different populations.
4.1. FLT1 Genetic Polymorphisms and Ranibizumab
FLT1 (fms-related receptor tyrosine kinase 1) is a protein-coding gene encoding a member of the VEGFR family that binds to VEGFR-A and VEGFR-B with an important role in angiogenesis and vasculogenesis. This receptor is expressed in vascular endothelial and placental trophoblast cells and peripheral blood monocytes. Ranibizumab binds VEGFA; thus, genetic variants in FLT1 may result in conformational changes in the receptor or differences in VEGFR1 administration and expression. FLT1 (rs9582036) is an intron variant and FLT1 (rs7993418) is a synonymous variant. Thus, in both cases, they may affect the administration or expression of VEGFR1, affecting how VEGFA binds this receptor and, in some way, how ranibizumab impedes the effect of VEGFA on VEGFR1.
Many genetic variants in FLT1 are associated with interindividual differences in the response to different treatments, especially in oncological patients [24,25]; however, there is only one study that has reported results about FLT1 SNPs’ influence on ranibizumab efficacy [46]. In this study, Lotery et al. assessed the association of FLT1 (rs12877323), among more than 450 SNPs in different genes with changes in the total retinal thickness (TRT) at the 3-, 6-, 9-, and 12-month follow-up in AMD patients without finding significant results. Among our patients, we did not study this SNP since this had not been previously associated with any drug response.
As commented above in the results section, among the included SNPs, we found a significant association between two FLT1 variants (rs9582036, rs7993418) with ranibizumab efficacy in mCNV at the 12-month follow-up in the genotype association study. Even FLT1 (rs7993418) showed a significant association with ranibizumab response in the allele association study (p = 0). In this regard, we did not find patients carrying the FLT1 (rs7993418) G allele and BCVA worsening.
As we can see (Table 2), none of these two SNPs showed differences between the study and control group and there is a theoretical background supporting the correlation between this gene and ranibizumab efficacy, thus FLT1 (rs9582036, rs7993418) may be considered a genetic marker of ranibizumab efficacy in mCNV patients.
4.2. KDR Genetic Polymorphisms and Ranibizumab
KDR (kinase insert domain receptor) is another protein-coding gene encoding a member of the VEGFR family. This receptor works as the main mediator of VEGF-induced endothelial proliferation, survival, migration, tubular morphogenesis, and sprouting. The KDR (rs2305948) is a missense variant and KDR (rs2239702) is an upstream gene variant. As ranibizumab binds VEGFA, these variants may result in conformational changes or differences in VEGFR1 administration and expression, respectively, leading to interindividual differences in the response to ranibizumab.
KDR variants, similar to FLT1 variants, were also associated with interindividual differences in the response to oncological treatments [47,48,49,50,51] and also to clopidogrel response [52]. About ranibizumab, Lazzeri et al. [27] concluded that the KDR (rs2071559) CC genotype revealed a better functional response as measured by the mean retinal sensitivity (p = 0.034) in AMD patients. On the other hand, Smailhodzic et al. [37] did not find significant results about the association of KDR (rs2071559, rs7671745) SNPs with ranibizumab response at a 3-month follow-up, but the discrepancies with our results may be explained by the follow-up period (3 vs. 12 months).
Among our patients, considering the 12-month follow-up period, we found that the KDR (rs2305948) CC genotype was associated with BCVA improvement (log-additive model: p-value= 0.049; dominant model: CC vs. CT or TT, OR= 0.35; 95%CI= 0.11–1.11; p = 0.055). Additionally, the KDR (rs2239702) GG genotype was associated with lower rates of BCVA worsening (dominant model: GG vs. GA or AA; OR= 0.15; 95%CI= 0.02–1.31; p = 0.044).
Furthermore, in the allele association study with the response, we found some results supporting this. The rs2239702 A allele was associated with higher rates of BCVA worsening (p = 0.035), the rs2305948 T allele was almost associated with BCVA improvement (p = 0.073), and the haplotype combining the rs2239702 A allele and rs2305948 C allele was related to higher rates of worsening (p = 0.039).
The rs2305948 was the only variant not linked to any of the others included and it was related to ranibizumab efficacy. Among the other SNPs in KDR included in the analysis, they were linked among them and we observed different associations with ranibizumab efficacy. Additionally, in the haplotypes study, we found that the most common haplotype (rs2239702 G; rs2305948: C; rs7667298: C; rs1870377: T and rs2071559: T) was associated with lower rates of BCVA worsening (OR = 0.09; 95% CI = 0.01–0.86; p = 0.039) compared to the ACTTC haplotype, carried by 21.9% of patients. Because of this, we might consider the ACTTC haplotype as a predictor of a worse response to ranibizumab, not a single analysis of SNPs.
5. Conclusions
Based on our results, FLT1 (rs9582036, rs7993418) variants may be genetic markers of ranibizumab efficacy in mCNV patients, and KDR (rs2305948, rs2239702) SNPs may be single predictors of ranibizumab efficacy in mCNV patients; however, the analysis of the ACTTC haplotype should be considered.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/pharmaceutics14081555/s1, Figure S1: FLT1 linkage disequilibrium analysis; Figure S2: KDR linkage disequilibrium analysis; Table S1: Haplotype frequencies of FLT1 variants and association with response; Table S2: Haplotype frequencies of KDR variants and association with response.
Author Contributions
Conceptualization, D.B.-M., X.D.-V. and C.L.D.-F.; methodology, D.B.-M., X.D.-V. and J.I.M.-Á.; validation, C.L.D.-F. and L.J.M.-G.; formal analysis, D.B.-M., S.G.-R. and X.D.-V.; investigation, resources and data curation, S.G.-R., A.A.-R. and A.P.-A.; writing—original draft preparation, D.B.-M. and X.D.-V.; writing—review and editing, J.I.M.-Á., L.J.M.-G. and C.L.D.-F.; visualization, D.B.-M. and X.D.-V.; supervision, C.L.D.-F. and L.J.M.-G.; project administration, C.L.D.-F. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and approved by the Ethics Committee of Granada (Spain) “CEIM/CEI Provincial de Granada” (0085-N14; 26 May 2014).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available due to containing clinical and personal information.
Acknowledgments
This study includes results from a doctoral thesis being developed and not yet published of the doctoral program in Pharmacy at the University of Granada (Spain), to whom we thank for their collaboration. We also would like to thank recruited patients for their consent and collaboration; nurses and personnel of the ophthalmology, and pharmacy units in the “Hospital San Cecilio” (Granada) for their kindly collaboration; and personnel of the Genomics Unit in the Pfizer—University of Granada—Junta de Andalucía Centre for Genomics and Oncological Research (GENYO) for their technical assistance.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Fredrick, D.R. Myopia. BMJ 2002, 324, 1195–1199. [Google Scholar] [CrossRef] [PubMed]
- Kempen, J.H.; Mitchell, P.; Lee, K.E.; Tielsch, J.M.; Broman, A.T.; Taylor, H.R.; Ikram, M.K.; Congdon, N.G.; O’Colmain, B.J.; Eye Diseases Prevalence Research Group. The prevalence of refractive errors among adults in the United States, Western Europe, and Australia. Arch. Ophthalmol. 2004, 122, 495–505. [Google Scholar] [PubMed]
- Wong, T.Y.; Foster, P.J.; Hee, J.; Ng, T.P.; Tielsch, J.M.; Chew, S.J.; Johnson, G.J.; Seah, S.K. Prevalence and risk factors for refractive errors in adult Chinese in Singapore. Investig. Ophthalmol. Vis. Sci. 2000, 41, 2486–2494. [Google Scholar]
- Wolf, S.; Balciuniene, V.J.; Laganovska, G.; Menchini, U.; Ohno-Matsui, K.; Sharma, T.; Wong, T.Y.; Silva, R.; Pilz, S.; Gekkieva, M.; et al. RADIANCE: A randomized controlled study of ranibizumab in patients with choroidal neovascularization secondary to pathologic myopia. Ophthalmology 2014, 121, 682–692.e2. [Google Scholar] [CrossRef]
- Miller, D.G.; Singerman, L.J. Natural history of choroidal neovascularization in high myopia. Curr. Opin. Ophthalmol. 2001, 12, 222–224. [Google Scholar] [CrossRef] [PubMed]
- Chung, Y.W.; Choi, M.Y.; Kim, J.S.; Kwon, J.W. The Association between Macular Thickness and Axial Length in Myopic Eyes. Biomed. Res. Int. 2019, 2019, 8913582. [Google Scholar] [CrossRef] [PubMed]
- Saw, S.M.; Gazzard, G.; Shih-Yen, E.C.; Chua, W.H. Myopia and associated pathological complications. Ophthalmic Physiol. Opt. 2005, 25, 381–391. [Google Scholar] [CrossRef]
- Chuck, R.S.; Jacobs, D.S.; Lee, J.K.; Afshari, N.A.; Vitale, S.; Shen, T.T.; Keenan, J.D.; American Academy of Ophthalmology Preferred Practice Pattern Refractive Management/Intervention Panel. Refractive Errors & Refractive Surgery Preferred Practice Pattern®. Ophthalmology 2018, 125, P1–P104. [Google Scholar] [PubMed]
- Wong, T.Y.; Ferreira, A.; Hughes, R.; Carter, G.; Mitchell, P. Epidemiology and disease burden of pathologic myopia and myopic choroidal neovascularization: An evidence-based systematic review. Am. J. Ophthalmol. 2014, 157, 9–25.e12. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, K.; Ohno-Matsui, K.; Shimada, N.; Moriyama, M.; Kojima, A.; Hayashi, W.; Yasuzumi, K.; Nagaoka, N.; Saka, N.; Yoshida, T.; et al. Long-term pattern of progression of myopic maculopathy: A natural history study. Ophthalmology 2010, 117, 1595–1611, 1611.e1–4. [Google Scholar] [CrossRef]
- Ng, D.; Fung, N.; Yip, F.; Lai, T. Ranibizumab for myopic choroidal neovascularization. Expert Opin. Biol. Ther. 2020, 20, 1385–1393. [Google Scholar] [CrossRef]
- Chan, W.M.; Ohji, M.; Lai, T.Y.; Liu, D.T.; Tano, Y.; Lam, D.S. Choroidal neovascularisation in pathological myopia: An update in management. Br. J. Ophthalmol. 2005, 89, 1522–1528. [Google Scholar] [CrossRef]
- Soubrane, G. Choroidal neovascularization in pathologic myopia: Recent developments in diagnosis and treatment. Surv. Ophthalmol. 2008, 53, 121–138. [Google Scholar] [CrossRef] [PubMed]
- Uemura, A.; Thomas, M.A. Subretinal surgery for choroidal neovascularization in patients with high myopia. Arch. Ophthalmol. 2000, 118, 344–350. [Google Scholar] [CrossRef]
- Ruiz-Moreno, J.M.; Montero, J.A. Long-term visual acuity after argon green laser photocoagulation of juxtafoveal choroidal neovascularization in highly myopic eyes. Eur. J. Ophthalmol. 2002, 12, 117–122. [Google Scholar] [CrossRef]
- Parodi, M.B.; Iacono, P.; Papayannis, A.; Sheth, S.; Bandello, F. Laser photocoagulation, photodynamic therapy, and intravitreal bevacizumab for the treatment of juxtafoveal choroidal neovascularization secondary to pathologic myopia. Arch. Ophthalmol. 2010, 128, 437–442. [Google Scholar] [CrossRef] [PubMed]
- Verteporfin in Photodynamic Therapy Study Group. Photodynamic therapy of subfoveal choroidal neovascularization in pathologic myopia with verteporfin. 1-year results of a randomized clinical trial—VIP report no. 1. Ophthalmology 2001, 108, 841–852. [Google Scholar] [CrossRef]
- Blinder, K.J.; Blumenkranz, M.S.; Bressler, N.M.; Bressler, S.B.; Donato, G.; Lewis, H.; Lim, J.I.; Menchini, U.; Miller, J.W.; Mones, J.M.; et al. Verteporfin therapy of subfoveal choroidal neovascularization in pathologic myopia: 2-year results of a randomized clinical trial—VIP report no. 3. Ophthalmology 2003, 110, 667–673. [Google Scholar]
- Giansanti, F.; Virgili, G.; Donati, M.C.; Giuntoli, M.; Pieretti, G.; Abbruzzese, G.; Menchini, U. Long-term results of photodynamic therapy for subfoveal choroidal neovascularization with pathologic myopia. Retina 2012, 32, 1547–1552. [Google Scholar] [CrossRef] [PubMed]
- Claxton, L.; Malcolm, B.; Taylor, M.; Haig, J.; Leteneux, C. Ranibizumab, verteporfin photodynamic therapy or observation for the treatment of myopic choroidal neovascularization: Cost effectiveness in the UK. Drugs Aging 2014, 31, 837–848. [Google Scholar] [CrossRef][Green Version]
- Deeks, E.D. Ranibizumab: A review of its use in myopic choroidal neovascularization. BioDrugs 2014, 28, 403–410. [Google Scholar] [CrossRef]
- Holz, F.G.; Tufail, A.; Leveziel, N.; Lai, T.Y.; Lanzetta, P.; Wong, T.Y.; Yu, H.G.; Chen, Y.X.; Heinrichs, N.; Pilz, S.; et al. Ranibizumab in Myopic Choroidal Neovascularization: A Subgroup Analysis by Ethnicity, Age, and Ocular Characteristics in RADIANCE. Ophthalmologica 2016, 236, 19–28. [Google Scholar] [CrossRef]
- Cobos, E.; Recalde, S.; Anter, J.; Hernandez-Sanchez, M.; Barreales, C.; Olavarrieta, L.; Valverde, A.; Suarez-Figueroa, M.; Cruz, F.; Abraldes, M.; et al. Association between CFH, CFB, ARMS2, SERPINF1, VEGFR1 and VEGF polymorphisms and anatomical and functional response to ranibizumab treatment in neovascular age-related macular degeneration. Acta Ophthalmol. 2018, 96, e201–e212. [Google Scholar] [CrossRef] [PubMed]
- Beuselinck, B.; Jean-Baptiste, J.; Schöffski, P.; Couchy, G.; Meiller, C.; Rolland, F.; Allory, Y.; Joniau, S.; Verkarre, V.; Elaidi, R.; et al. Validation of VEGFR1 rs9582036 as predictive biomarker in metastatic clear-cell renal cell carcinoma patients treated with sunitinib. BJU Int. 2016, 118, 890–901. [Google Scholar] [CrossRef]
- Beuselinck, B.; Karadimou, A.; Lambrechts, D.; Claes, B.; Wolter, P.; Couchy, G.; Berkers, J.; van Poppel, H.; Paridaens, R.; Schöffski, P.; et al. VEGFR1 single nucleotide polymorphisms associated with outcome in patients with metastatic renal cell carcinoma treated with sunitinib—A multicentric retrospective analysis. Acta Oncol. 2014, 53, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Dornbusch, J.; Walter, M.; Gottschalk, A.; Obaje, A.; Junker, K.; Ohlmann, C.H.; Meinhardt, M.; Zacharis, A.; Zastrow, S.; Schoffer, O.; et al. Evaluation of polymorphisms in angiogenesis-related genes as predictive and prognostic markers for sunitinib-treated metastatic renal cell carcinoma patients. J. Cancer Res. Clin. Oncol. 2016, 142, 1171–1182. [Google Scholar] [CrossRef] [PubMed]
- Lazzeri, S.; Orlandi, P.; Piaggi, P.; Sartini, M.S.; Casini, G.; Guidi, G.; Figus, M.; Fioravanti, A.; Di Desidero, T.; Ripandelli, G.; et al. IL-8 and VEGFR-2 polymorphisms modulate long-term functional response to intravitreal ranibizumab in exudative age-related macular degeneration. Pharmacogenomics 2016, 17, 35–39. [Google Scholar] [CrossRef] [PubMed]
- Hermann, M.M.; van Asten, F.; Muether, P.S.; Smailhodzic, D.; Lichtner, P.; Hoyng, C.B.; Kirchhof, B.; Grefkes, C.; den Hollander, A.I.; Fauser, S. Polymorphisms in vascular endothelial growth factor receptor 2 are associated with better response rates to ranibizumab treatment in age-related macular degeneration. Ophthalmology 2014, 121, 905–910. [Google Scholar] [CrossRef]
- Blánquez-Martínez, D.; Díaz-Villamarín, X.; Antúnez-Rodríguez, A.; Pozo-Agundo, A.; Muñoz-Ávila, J.I.; Martínez-González, L.J.; Dávila-Fajardo, C.L. Genetic Polymorphisms Affecting Ranibizumab Response in High Myopia Patients. Pharmaceutics 2021, 13, 1973. [Google Scholar] [CrossRef]
- Freeman, B.; Smith, N.; Curtis, C.; Huckett, L.; Mill, J.; Craig, I. DNA from buccal swabs recruited by mail: Evaluation of storage effects on long-term stability and suitability for multiplex polymerase chain reaction genotyping. Behav. Genet. 2003, 33, 67–72. [Google Scholar] [CrossRef]
- Gómez-Martín, A.; Hernández, A.F.; Martínez-González, L.J.; González-Alzaga, B.; Rodríguez-Barranco, M.; López-Flores, I.; Aguilar-Garduno, C.; Lacasana, M. Polymorphisms of pesticide-metabolizing genes in children living in intensive farming communities. Chemosphere 2015, 139, 534–540. [Google Scholar] [CrossRef]
- Solé, X.; Guinó, E.; Valls, J.; Iniesta, R.; Moreno, V. SNPStats: A web tool for the analysis of association studies. Bioinformatics 2006, 22, 1928–19289. [Google Scholar] [CrossRef] [PubMed]
- Rinaldi, C.; Bramanti, P.; Scimone, C.; Donato, L.; Alafaci, C.; D’Angelo, R.; Sidoti, A. Relevance of CCM gene polymorphisms for clinical management of sporadic cerebral cavernous malformations. J. Neurol. Sci. 2017, 380, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Scimone, C.; Donato, L.; Katsarou, Z.; Bostantjopoulou, S.; D’Angelo, R.; Sidoti, A. Two Novel KRIT1 and CCM2 Mutations in Patients Affected by Cerebral Cavernous Malformations: New Information on CCM2 Penetrance. Front. Neurol. 2018, 9, 953. [Google Scholar] [CrossRef]
- Scimone, C.; Bramanti, P.; Ruggeri, A.; Donato, L.; Alafaci, C.; Crisafulli, C.; Mucciardi, M.; Rinaldi, C.; Sidoti, A.; D’Angelo, R. CCM3/SERPINI1 bidirectional promoter variants in patients with cerebral cavernous malformations: A molecular and functional study. BMC Med. Genet. 2016, 17, 74. [Google Scholar] [CrossRef] [PubMed]
- Scimone, C.; Bramanti, P.; Ruggeri, A.; Katsarou, Z.; Donato, L.; Sidoti, A.; D’Angelo, R. Detection of Novel Mutation in Ccm3 Causes Familial Cerebral Cavernous Malformations. J. Mol. Neurosci. 2015, 57, 400–403. [Google Scholar] [CrossRef] [PubMed]
- Smailhodzic, D.; Muether, P.S.; Chen, J.; Kwestro, A.; Zhang, A.Y.; Omar, A.; Van de Ven, J.P.; Keunen, J.E.; Kirchhof, B.; Hoyng, C.B.; et al. Cumulative effect of risk alleles in CFH, ARMS2, and VEGFA on the response to ranibizumab treatment in age-related macular degeneration. Ophthalmology 2012, 119, 2304–2311. [Google Scholar] [CrossRef]
- Tian, J.; Qin, X.; Fang, K.; Chen, Q.; Hou, J.; Li, J.; Yu, W.; Chen, D.; Hu, Y.; Li, X. Association of genetic polymorphisms with response to bevacizumab for neovascular age-related macular degeneration in the Chinese population. Pharmacogenomics 2012, 13, 779–787. [Google Scholar] [CrossRef]
- Abedi, F.; Wickremasinghe, S.; Richardson, A.J.; Makalic, E.; Schmidt, D.F.; Sandhu, S.S.; Baird, P.N.; Guymer, R.H. Variants in the VEGFA gene and treatment outcome after anti-VEGF treatment for neovascular age-related macular degeneration. Ophthalmology 2013, 120, 115–121. [Google Scholar] [CrossRef]
- Chang, W.; Noh, D.H.; Sagong, M.; Kim, I.T. Pharmacogenetic association with early response to intravitreal ranibizumab for age-related macular degeneration in a Korean population. Mol. Vis. 2013, 19, 702–709. [Google Scholar]
- Lazzeri, S.; Figus, M.; Orlandi, P.; Fioravanti, A.; Di Desidero, T.; Agosta, E.; Sartini, M.S.; Posarelli, C.; Nardi, M.; Danesi, R.; et al. VEGF-A polymorphisms predict short-term functional response to intravitreal ranibizumab in exudative age-related macular degeneration. Pharmacogenomics 2013, 14, 623–630. [Google Scholar] [CrossRef] [PubMed]
- Lorés-Motta, L.; van Asten, F.; Muether, P.S.; Smailhodzic, D.; Groenewoud, J.M.; Omar, A.; Chen, J.; Koenekoop, R.K.; Fauser, S.; Hoyng, C.B.; et al. A genetic variant in NRP1 is associated with worse response to ranibizumab treatment in neovascular age-related macular degeneration. Pharmacogenet. Genom. 2016, 26, 20–27. [Google Scholar] [CrossRef]
- Miyake, M.; Yamashiro, K.; Akagi-Kurashige, Y.; Kumagai, K.; Nakata, I.; Nakanishi, H.; Oishi, A.; Tsujikawa, A.; Yamada, R.; Matsuda, F.; et al. Vascular endothelial growth factor gene and the response to anti-vascular endothelial growth factor treatment for choroidal neovascularization in high myopia. Ophthalmology 2014, 121, 225–233. [Google Scholar] [CrossRef] [PubMed]
- Park, U.C.; Shin, J.Y.; Chung, H.; Yu, H.G. Association of ARMS2 genotype with response to anti-vascular endothelial growth factor treatment in polypoidal choroidal vasculopathy. BMC Ophthalmol. 2017, 17, 241. [Google Scholar] [CrossRef]
- Díaz-Villamarín, X.; Blánquez-Martínez, D.; Pozo-Agundo, A.; Pérez-Gutiérrez, A.M.; Muñoz-Ávila, J.I.; Antúnez-Rodríguez, A.; Fernández-Gómez, A.E.; García-Navas, P.; Martínez-González, L.J.; Dávila-Fajardo, C.L. Genetic Variants Affecting Anti-VEGF Drug Response in Polypoidal Choroidal Vasculopathy Patients: A Systematic Review and Meta-Analysis. Genes 2020, 11, 1335. [Google Scholar] [CrossRef]
- Lotery, A.J.; Gibson, J.; Cree, A.J.; Downes, S.M.; Harding, S.P.; Rogers, C.A.; Reeves, B.C.; Ennis, S.; Chakravarthy, U.; Alternative Treatments to Inhibit VEGF in Patients with Age-Related Choroidal Neovascularisation (IVAN) Study Group. Pharmacogenetic associations with vascular endothelial growth factor inhibition in participants with neovascular age-related macular degeneration in the IVAN Study. Ophthalmology 2013, 120, 2637–2643. [Google Scholar] [CrossRef]
- Apellániz-Ruiz, M.; Diekstra, M.H.; Roldán, J.M.; Boven, E.; Castellano, D.; Gelderblom, H.; Mathijssen, R.; Swen, J.J.; Böhringer, S.; García-Donás, J.; et al. Evaluation of KDR rs34231037 as a predictor of sunitinib efficacy in patients with metastatic renal cell carcinoma. Pharmacogenet. Genom. 2017, 27, 227–231. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.B.; Zhan, M.X.; Zhao, W.; Liu, B.; Huang, J.W.; He, X.; Fu, S.R.; Zhao, Y.; Li, Y.; Hu, B.S.; et al. The relationship of kinase insert domain receptor gene polymorphisms and clinical outcome in advanced hepatocellular carcinoma patients treated with sorafenib. Med. Oncol. 2014, 31, 209. [Google Scholar] [CrossRef]
- Scartozzi, M.; Faloppi, L.; Svegliati Baroni, G.; Loretelli, C.; Piscaglia, F.; Iavarone, M.; Toniutto, P.; Fava, G.; De Minicis, S.; Mandolesi, A.; et al. VEGF and VEGFR genotyping in the prediction of clinical outcome for HCC patients receiving sorafenib: The ALICE-1 study. Int. J. Cancer 2014, 135, 1247–1256. [Google Scholar] [CrossRef]
- Escudier, B.; Rini, B.I.; Motzer, R.J.; Tarazi, J.; Kim, S.; Huang, X.; Rosbrook, B.; English, P.A.; Loomis, A.K.; Williams, J.A. Genotype Correlations With Blood Pressure and Efficacy From a Randomized Phase III Trial of Second-Line Axitinib Versus Sorafenib in Metastatic Renal Cell Carcinoma. Clin. Genitourin. Cancer 2015, 13, 328–337.e3. [Google Scholar] [CrossRef]
- Jain, L.; Sissung, T.M.; Danesi, R.; Kohn, E.C.; Dahut, W.L.; Kummar, S.; Venzon, D.; Liewehr, D.; English, B.C.; Baum, C.E.; et al. Hypertension and hand-foot skin reactions related to VEGFR2 genotype and improved clinical outcome following bevacizumab and sorafenib. J. Exp. Clin. Cancer Res. 2010, 29, 95. [Google Scholar] [CrossRef]
- Zhang, L.J.; Zhang, Y.Q.; Han, X.; Zhang, Z.T.; Zhang, Z.Q. Association of VEGFR-2 Gene Polymorphisms with Clopidogrel Resistance in Patients with Coronary Heart Disease. Am. J. Ther. 2016, 23, e1663–e1670. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).