Repurposing Drugs via Network Analysis: Opportunities for Psychiatric Disorders
Abstract
1. Challenges of Drug Research for Psychiatric Disorders
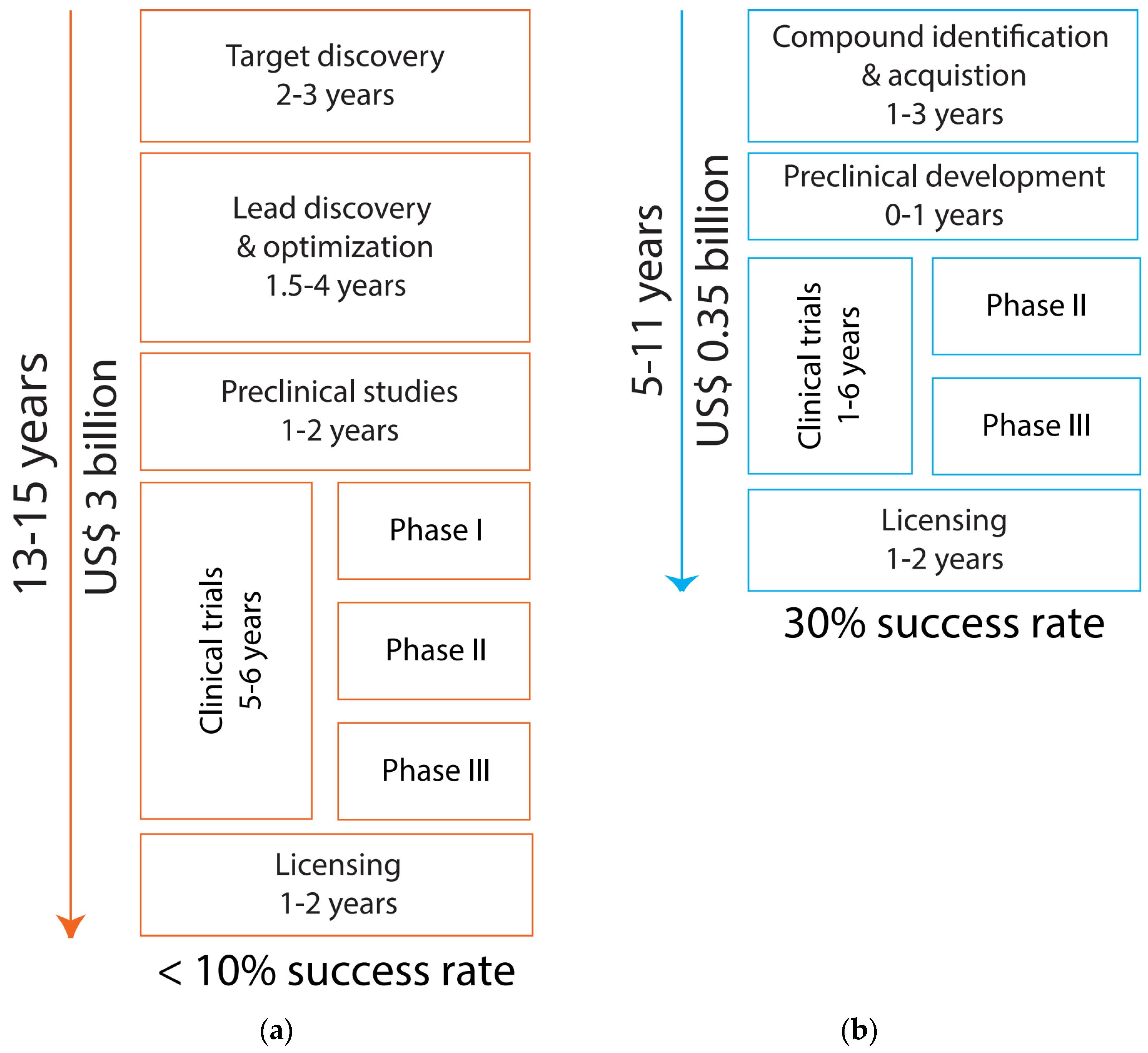
2. Drug Repurposing—An Accelerated Framework for Psychiatric Drug Development
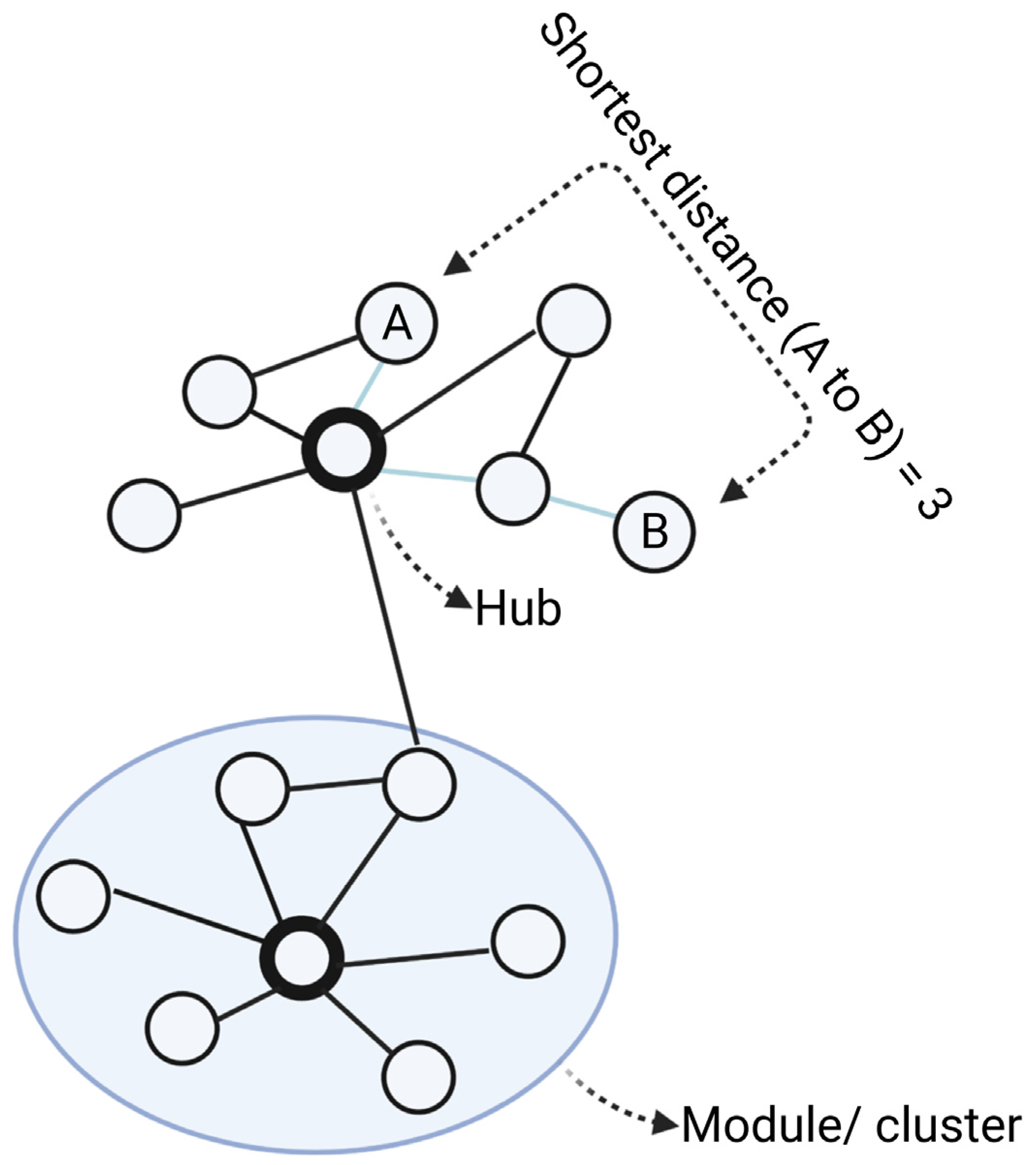
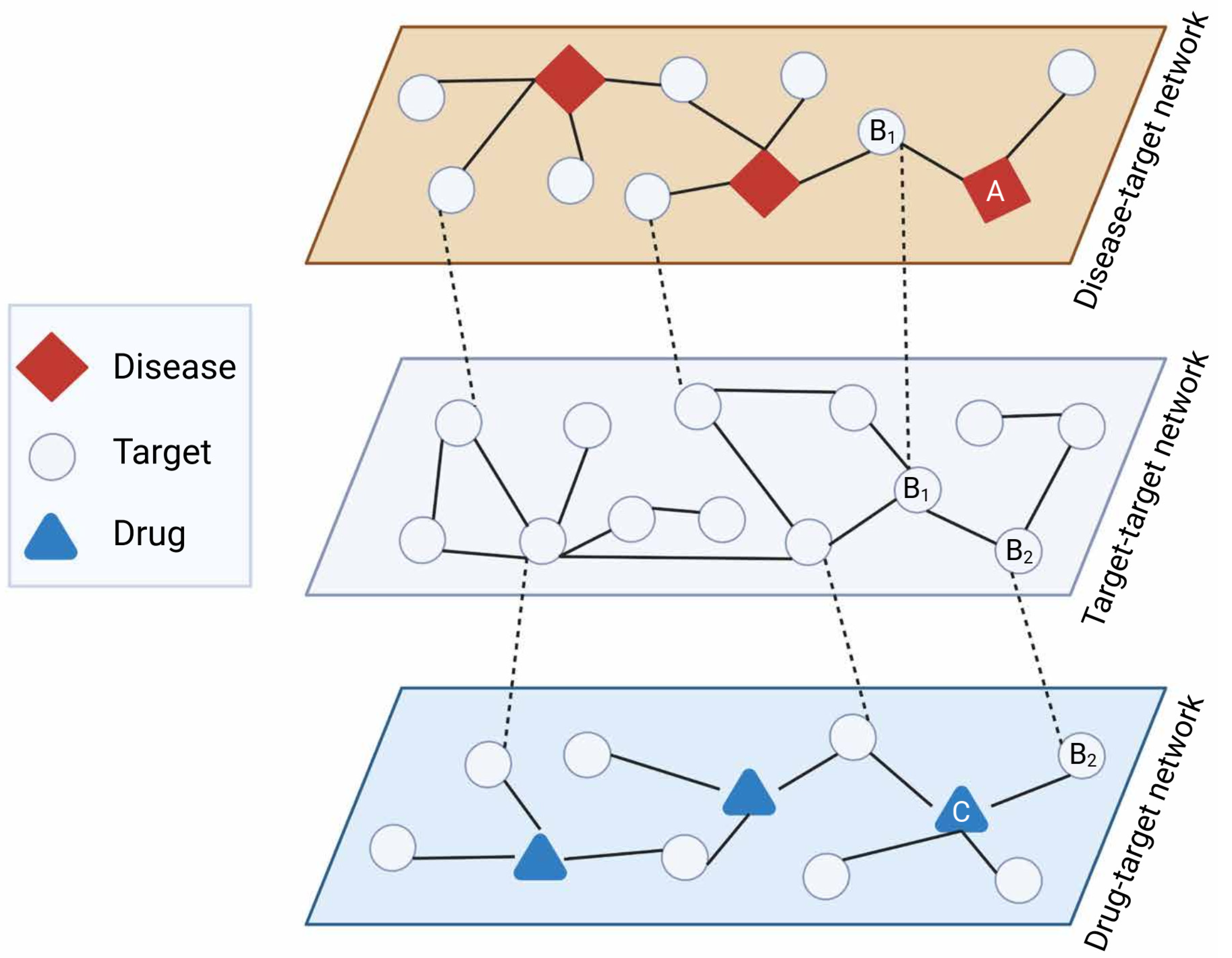
3. Why Networks Matter for Psychiatric Drug Research
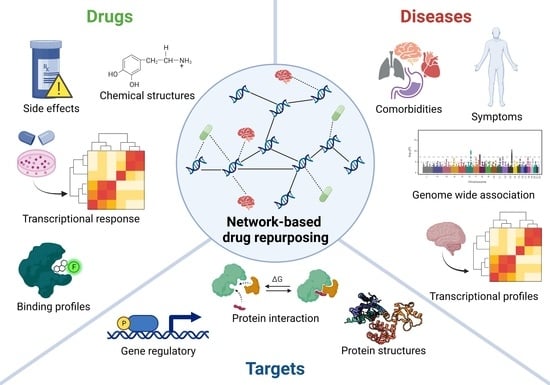
4. Network-Based Drug Repurposing in Psychiatry
4.1. Structural Data (Structome)
4.2. Genome
4.3. Transcriptome
4.4. Interactome
4.5. Phenome
4.6. Network-Based Drug Repurposing Platforms
5. Challenges of Network-Based Drug Repurposing in Psychiatry
6. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Vos, T.; Abajobir, A.A.; Abate, K.H.; Abbafati, C.; Abbas, K.M.; Abd-Allah, F.; Abdulkader, R.S.; Abdulle, A.M.; Abebo, T.A.; Abera, S.F. Global, regional, and national incidence, prevalence, and years lived with disability for 328 diseases and injuries for 195 countries, 1990–2016: A systematic analysis for the Global Burden of Disease Study 2016. Lancet 2017, 390, 1211–1259. [Google Scholar] [CrossRef]
- Scott, K.M.; de Jonge, P.; Stein, D.J.; Kessler, R.C. Mental Disorders around the World: Facts and Figures from the WHO World Mental Health Surveys; Cambridge University Press: Cambridge, UK, 2018. [Google Scholar]
- U.S. Food and Drug Administration. New Molecular Entity (NME) Drug and New Biologic Approvals; U.S. Food and Drug Administration: Silver Spring, MD, USA, 2019.
- U.S. Food and Drug Administration. New Molecular Entity (NME) Drug and New Biologic Approvals; U.S. Food and Drug Administration: Silver Spring, MD, USA, 2020.
- Stahl, S.M. Stahl’s Essential Psychopharmacology: Neuroscientific Basis and Practical Applications; Cambridge University Press: Cambridge, UK, 2013. [Google Scholar]
- Lee, H.-M.; Kim, Y. Drug Repurposing Is a New Opportunity for Developing Drugs against Neuropsychiatric Disorders. Schizophr. Res. Treat. 2016, 2016, 6378137. [Google Scholar] [CrossRef] [PubMed]
- Scannell, J.W.; Blanckley, A.; Boldon, H.; Warrington, B. Diagnosing the decline in pharmaceutical R&D efficiency. Nat. Rev. Drug Discov. 2012, 11, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Akhondzadeh, S. The Importance of Clinical Trials in Drug Development. Avicenna J. Med. Biotechnol. 2016, 8, 151. [Google Scholar] [PubMed]
- Hughes, J.P.; Rees, S.; Kalindjian, S.B.; Philpott, K.L. Principles of early drug discovery. Br. J. Pharmacol. 2011, 162, 1239–1249. [Google Scholar] [CrossRef]
- Blokhin, I.O.; Khorkova, O.; Saveanu, R.V.; Wahlestedt, C. Molecular mechanisms of psychiatric diseases. Neurobiol. Dis. 2020, 146, 105136. [Google Scholar] [CrossRef]
- Gribkoff, V.K.; Kaczmarek, L.K. The need for new approaches in CNS drug discovery: Why drugs have failed, and what can be done to improve outcomes. Neuropharmacology 2017, 120, 11–19. [Google Scholar] [CrossRef]
- Pushpakom, S.; Iorio, F.; Eyers, P.A.; Escott, K.J.; Hopper, S.; Wells, A.; Doig, A.; Guilliams, T.; Latimer, J.; McNamee, C.; et al. Drug repurposing: Progress, challenges and recommendations. Nat. Rev. Drug Discov. 2019, 18, 41–58. [Google Scholar] [CrossRef]
- Ashburn, T.T.; Thor, K.B. Drug repositioning: Identifying and developing new uses for existing drugs. Nat. Rev. Drug Discov. 2004, 3, 673–683. [Google Scholar] [CrossRef]
- Ko, Y. Computational Drug Repositioning: Current Progress and Challenges. Appl. Sci. 2020, 10, 5076. [Google Scholar] [CrossRef]
- Graul, A.I.; Pina, P.; Cruces, E.; Stringer, M. The year’s new drugs and biologics 2018: Part I. Drugs Today 2019, 55, 35–87. [Google Scholar] [CrossRef] [PubMed]
- Sardana, D.; Zhu, C.; Zhang, M.; Gudivada, R.C.; Yang, L.; Jegga, A.G. Drug repositioning for orphan diseases. Brief. Bioinform. 2011, 12, 346–356. [Google Scholar] [CrossRef] [PubMed]
- Power, A.; Berger, A.C.; Ginsburg, G.S. Genomics-enabled drug repositioning and repurposing: Insights from an IOM Roundtable activity. JAMA 2014, 311, 2063–2064. [Google Scholar] [CrossRef] [PubMed]
- Caban, A.; Pisarczyk, K.; Kopacz, K.; Kapuśniak, A.; Toumi, M.; Rémuzat, C.; Kornfeld, A. Filling the gap in CNS drug development: Evaluation of the role of drug repurposing. J. Mark Access Health Policy 2017, 5, 1299833. [Google Scholar] [CrossRef] [PubMed]
- Yildiz, A.; Aydin, B.; Gökmen, N.; Yurt, A.; Cohen, B.; Keskinoglu, P.; Öngür, D.; Renshaw, P. Antimanic Treatment With Tamoxifen Affects Brain Chemistry: A Double-Blind, Placebo-Controlled Proton Magnetic Resonance Spectroscopy Study. Biol. Psychiatry Cogn. Neurosci. Neuroimaging 2016, 1, 125–131. [Google Scholar] [CrossRef][Green Version]
- Pharmaceuticals, A. A Multicenter, Randomized, Double-blind, Placebo-Controlled, Parallel-Arm Study to Assess the Efficacy, Safety, and Tolerability of AVP-786 (Deudextromethorphan Hydrobromide [d6-DM]/Quinidine Sulfate [Q]) for the Treatment of Negative Symptoms of Schizophrenia. Available online: https://www.clinicaltrials.gov/ct2/show/study/NCT03896945 (accessed on 16 May 2022).
- Bowden, C. The effectiveness of divalproate in all forms of mania and the broader bipolar spectrum: Many questions, few answers. J. Affect. Disord. 2004, 79, 9–14. [Google Scholar] [CrossRef]
- Schwartz, J.; Murrough, J.W.; Iosifescu, D.V. Ketamine for treatment-resistant depression: Recent developments and clinical applications. Evid. Based Ment. Health 2016, 19, 35. [Google Scholar] [CrossRef]
- Maron, B.A.; Altucci, L.; Balligand, J.-L.; Baumbach, J.; Ferdinandy, P.; Filetti, S.; Parini, P.; Petrillo, E.; Silverman, E.K.; Barabási, A.-L.; et al. A global network for network medicine. NPJ Syst. Biol. Appl. 2020, 6, 29. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, A.L. Network pharmacology: The next paradigm in drug discovery. Nat. Chem. Biol. 2008, 4, 682–690. [Google Scholar] [CrossRef]
- Bianchi, M.T.; Botzolakis, E.J. Targeting ligand-gated ion channels in neurology and psychiatry: Is pharmacological promiscuity an obstacle or an opportunity? BMC Pharmacol. 2010, 10, 3. [Google Scholar] [CrossRef]
- Cross-Disorder Group of the Psychiatric Genomics Consortium. Identification of risk loci with shared effects on five major psychiatric disorders: A genome-wide analysis. Lancet 2013, 381, 1371–1379. [Google Scholar] [CrossRef]
- Anttila, V.; Bulik-Sullivan, B.; Finucane, H.K.; Walters, R.K.; Bras, J.; Duncan, L.; Escott-Price, V.; Falcone, G.J.; Gormley, P.; Malik, R.; et al. Analysis of shared heritability in common disorders of the brain. Science 2018, 360, eaap875. [Google Scholar] [CrossRef]
- Gandal Michael, J.; Haney Jillian, R.; Parikshak Neelroop, N.; Leppa, V.; Ramaswami, G.; Hartl, C.; Schork Andrew, J.; Appadurai, V.; Buil, A.; Werge Thomas, M.; et al. Shared molecular neuropathology across major psychiatric disorders parallels polygenic overlap. Science 2018, 359, 693–697. [Google Scholar] [CrossRef]
- Gandal, M.J.; Zhang, P.; Hadjimichael, E.; Walker, R.L.; Chen, C.; Liu, S.; Won, H.; van Bakel, H.; Varghese, M.; Wang, Y.; et al. Transcriptome-wide isoform-level dysregulation in ASD, schizophrenia, and bipolar disorder. Science 2018, 362, eaat8127. [Google Scholar] [CrossRef] [PubMed]
- Jacobi, F.; Wittchen, H.U.; HÖLting, C.; Höfler, M.; Pfister, H.; Müller, N.; Lieb, R. Prevalence, co-morbidity and correlates of mental disorders in the general population: Results from the German Health Interview and Examination Survey (GHS). Psychol. Med. 2004, 34, 597–611. [Google Scholar] [CrossRef]
- Andrews, G.; Henderson, S.; Hall, W. Prevalence, comorbidity, disability and service utilisation: Overview of the Australian National Mental Health Survey. Br. J. Psychiatry 2001, 178, 145–153. [Google Scholar] [CrossRef]
- Kessler, R.C.; McGonagle, K.A.; Zhao, S.; Nelson, C.B.; Hughes, M.; Eshleman, S.; Wittchen, H.-U.; Kendler, K.S. Lifetime and 12-Month Prevalence of DSM-III-R Psychiatric Disorders in the United States: Results From the National Comorbidity Survey. Arch. Gen. Psychiatry 1994, 51, 8–19. [Google Scholar] [CrossRef]
- Merikangas, K.R.; Angst, J.; Eaton, W.; Canino, G.; Rubio-Stipec, M.; Wacker, H.; Wittchen, H.U.; Andrade, L.; Essau, C.; Whitaker, A.; et al. Comorbidity and boundaries of affective disorders with anxiety disorders and substance misuse: Results of an international task force. Br. J. Psychiatry Suppl. 1996, 168, 58–67. [Google Scholar] [CrossRef]
- Qu, X.A.; Gudivada, R.C.; Jegga, A.G.; Neumann, E.K.; Aronow, B.J. Inferring novel disease indications for known drugs by semantically linking drug action and disease mechanism relationships. BMC Bioinform. 2009, 10 (Suppl. S5), S4. [Google Scholar] [CrossRef]
- Barabási, A.-L. Network Science; Cambridge University Press: Cambridge, UK, 2016. [Google Scholar]
- Recanatini, M.; Cabrelle, C. Drug Research Meets Network Science: Where Are We? J. Med. Chem. 2020, 63, 8653–8666. [Google Scholar] [CrossRef]
- Csermely, P.; Korcsmáros, T.; Kiss, H.J.M.; London, G.; Nussinov, R. Structure and dynamics of molecular networks: A novel paradigm of drug discovery: A comprehensive review. Pharmacol. Ther. 2013, 138, 333–408. [Google Scholar] [CrossRef] [PubMed]
- Albert, R.K. Network Inference, Analysis, and Modeling in Systems Biology. Plant Cell 2007, 19, 3327–3338. [Google Scholar] [CrossRef]
- Kitano, H. Systems Biology: A Brief Overview. Science 2002, 295, 1662–1664. [Google Scholar] [CrossRef] [PubMed]
- Vidal, M.; Cusick, M.E.; Barabási, A.-L. Interactome Networks and Human Disease. Cell 2011, 144, 986–998. [Google Scholar] [CrossRef] [PubMed]
- Network Medicine: Complex Systems in Human Disease and Therapeutics; Harvard University Press: Cambridge, MA, USA, 2017.
- Barabási, A.-L.; Albert, R. Emergence of Scaling in Random Networks. Science 1999, 286, 509–512. [Google Scholar] [CrossRef]
- Seebacher, J.; Gavin, A.C. SnapShot: Protein-protein interaction networks. Cell 2011, 144, 1000. [Google Scholar] [CrossRef]
- Barabási, A.-L.; Oltvai, Z.N. Network biology: Understanding the cell’s functional organization. Nat. Rev. Genet. 2004, 5, 101–113. [Google Scholar] [CrossRef]
- Penrod, N.M.; Cowper-Sal-lari, R.; Moore, J.H. Systems genetics for drug target discovery. Trends Pharmacol. Sci. 2011, 32, 623–630. [Google Scholar] [CrossRef]
- Swanson, D.R. Fish oil, Raynaud’s syndrome, and undiscovered public knowledge. Perspect. Biol. Med. 1986, 30, 7–18. [Google Scholar] [CrossRef]
- Baek, S.H.; Lee, D.; Kim, M.; Lee, J.H.; Song, M. Enriching plausible new hypothesis generation in PubMed. PLoS ONE 2017, 12, e0180539. [Google Scholar] [CrossRef]
- Weeber, M.; Klein, H.; de Jong-van den Berg, L.T.W.; Vos, R. Using concepts in literature-based discovery: Simulating Swanson’s Raynaud–fish oil and migraine–magnesium discoveries. J. Am. Soc. Inf. Sci. Technol. 2001, 52, 548–557. [Google Scholar] [CrossRef]
- Chiang, A.P.; Butte, A.J. Systematic evaluation of drug-disease relationships to identify leads for novel drug uses. Clin. Pharm. 2009, 86, 507–510. [Google Scholar] [CrossRef] [PubMed]
- Andronis, C.; Sharma, A.; Virvilis, V.; Deftereos, S.; Persidis, A. Literature mining, ontologies and information visualization for drug repurposing. Brief. Bioinform. 2011, 12, 357–368. [Google Scholar] [CrossRef] [PubMed]
- Lekka, E.; Deftereos, S.N.; Persidis, A.; Persidis, A.; Andronis, C. Literature analysis for systematic drug repurposing: A case study from Biovista. Drug Discov. Today Ther. Strateg. 2011, 8, 103–108. [Google Scholar] [CrossRef]
- Krassowski, M.; Das, V.; Sahu, S.K.; Misra, B.B. State of the Field in Multi-Omics Research: From Computational Needs to Data Mining and Sharing. Front. Genet. 2020, 11, 610798. [Google Scholar] [CrossRef]
- Mendez, D.; Gaulton, A.; Bento, A.P.; Chambers, J.; De Veij, M.; Félix, E.; Magariños, M.P.; Mosquera, J.F.; Mutowo, P.; Nowotka, M.; et al. ChEMBL: Towards direct deposition of bioassay data. Nucleic Acids Res. 2019, 47, D930–D940. [Google Scholar] [CrossRef]
- Pence, H.E.; Williams, A. ChemSpider: An Online Chemical Information Resource. J. Chem. Educ. 2010, 87, 1123–1124. [Google Scholar] [CrossRef]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z.; et al. DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Res. 2018, 46, D1074–D1082. [Google Scholar] [CrossRef]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem 2019 update: Improved access to chemical data. Nucleic Acids Res. 2019, 47, D1102–D1109. [Google Scholar] [CrossRef]
- Burley, S.K.; Bhikadiya, C.; Bi, C.; Bittrich, S.; Chen, L.; Crichlow, G.V.; Christie, C.H.; Dalenberg, K.; Di Costanzo, L.; Duarte, J.M.; et al. RCSB Protein Data Bank: Powerful new tools for exploring 3D structures of biological macromolecules for basic and applied research and education in fundamental biology, biomedicine, biotechnology, bioengineering and energy sciences. Nucleic Acids Res. 2021, 49, D437–D451. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Tan, F.; Yang, R.; Xu, X.; Chen, X.; Wang, Y.; Ma, H.; Liu, X.; Wu, X.; Chen, Y.; Liu, L.; et al. Drug repositioning by applying ‘expression profiles’ generated by integrating chemical structure similarity and gene semantic similarity. Mol. Biosyst. 2014, 10, 1126–1138. [Google Scholar] [CrossRef]
- Watanabe, K.; Stringer, S.; Frei, O.; Umićević Mirkov, M.; de Leeuw, C.; Polderman, T.J.C.; van der Sluis, S.; Andreassen, O.A.; Neale, B.M.; Posthuma, D. A global overview of pleiotropy and genetic architecture in complex traits. Nat. Genet. 2019, 51, 1339–1348. [Google Scholar] [CrossRef] [PubMed]
- Tryka, K.A.; Hao, L.; Sturcke, A.; Jin, Y.; Wang, Z.Y.; Ziyabari, L.; Lee, M.; Popova, N.; Sharopova, N.; Kimura, M.; et al. NCBI’s Database of Genotypes and Phenotypes: dbGaP. Nucleic Acids Res. 2014, 42, D975–D979. [Google Scholar] [CrossRef] [PubMed]
- Home|NRGR. NIMH Repository and Genomics Resource. Available online: https://www.nimhgenetics.org/ (accessed on 1 June 2022).
- The Psychiatric, G.C.S.C. A framework for interpreting genome-wide association studies of psychiatric disorders. Mol. Psychiatry 2009, 14, 10–17. [Google Scholar] [CrossRef]
- Buxbaum, J.D.; Daly, M.J.; Devlin, B.; Lehner, T.; Roeder, K.; State, M.W. The autism sequencing consortium: Large-scale, high-throughput sequencing in autism spectrum disorders. Neuron 2012, 76, 1052–1056. [Google Scholar] [CrossRef]
- Sanders, S.J.; Neale, B.M.; Huang, H.; Werling, D.M.; An, J.-Y.; Dong, S.; Abecasis, G.; Arguello, P.A.; Blangero, J.; Boehnke, M.; et al. Whole genome sequencing in psychiatric disorders: The WGSPD consortium. Nat. Neurosci. 2017, 20, 1661–1668. [Google Scholar] [CrossRef]
- Akbarian, S.; Liu, C.; Knowles, J.A.; Vaccarino, F.M.; Farnham, P.J.; Crawford, G.E.; Jaffe, A.E.; Pinto, D.; Dracheva, S.; Geschwind, D.H.; et al. The PsychENCODE project. Nat. Neurosci. 2015, 18, 1707–1712. [Google Scholar] [CrossRef]
- McConnell, M.J.; Moran, J.V.; Abyzov, A.; Akbarian, S.; Bae, T.; Cortes-Ciriano, I.; Erwin, J.A.; Fasching, L.; Flasch, D.A.; Freed, D.; et al. Intersection of diverse neuronal genomes and neuropsychiatric disease: The Brain Somatic Mosaicism Network. Science 2017, 356, eaal1641. [Google Scholar] [CrossRef]
- Hoffman, G.E.; Bendl, J.; Voloudakis, G.; Montgomery, K.S.; Sloofman, L.; Wang, Y.-C.; Shah, H.R.; Hauberg, M.E.; Johnson, J.S.; Girdhar, K.; et al. CommonMind Consortium provides transcriptomic and epigenomic data for Schizophrenia and Bipolar Disorder. Sci. Data 2019, 6, 180. [Google Scholar] [CrossRef]
- Sunkin, S.M.; Ng, L.; Lau, C.; Dolbeare, T.; Gilbert, T.L.; Thompson, C.L.; Hawrylycz, M.; Dang, C. Allen Brain Atlas: An integrated spatio-temporal portal for exploring the central nervous system. Nucleic Acids Res. 2013, 41, D996–D1008. [Google Scholar] [CrossRef] [PubMed]
- Lamb, J. The Connectivity Map: A new tool for biomedical research. Nat. Rev. Cancer 2007, 7, 54–60. [Google Scholar] [CrossRef]
- Subramanian, A.; Narayan, R.; Corsello, S.M.; Peck, D.D.; Natoli, T.E.; Lu, X.; Gould, J.; Davis, J.F.; Tubelli, A.A.; Asiedu, J.K.; et al. A Next Generation Connectivity Map: L1000 Platform and the First 1,000,000 Profiles. Cell 2017, 171, 1437–1452.e1417. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; He, E.; Sani, K.; Jagodnik, K.M.; Silverstein, M.C.; Ma’ayan, A. Drug Gene Budger (DGB): An application for ranking drugs to modulate a specific gene based on transcriptomic signatures. Bioinformatics 2019, 35, 1247–1248. [Google Scholar] [CrossRef] [PubMed]
- Gaspar, H.A.; Gerring, Z.; Hübel, C.; Middeldorp, C.M.; Derks, E.M.; Breen, G.; Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium. Using genetic drug-target networks to develop new drug hypotheses for major depressive disorder. Transl. Psychiatry 2019, 9, 117. [Google Scholar] [CrossRef]
- Rodriguez-López, J.; Arrojo, M.; Paz, E.; Páramo, M.; Costas, J. Identification of relevant hub genes for early intervention at gene coexpression modules with altered predicted expression in schizophrenia. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2020, 98, 109815. [Google Scholar] [CrossRef]
- Cabrera-Mendoza, B.; Martínez-Magaña, J.J.; Monroy-Jaramillo, N.; Genis-Mendoza, A.D.; Fresno, C.; Fries, G.R.; Walss-Bass, C.; López Armenta, M.; García-Dolores, F.; Díaz-Otañez, C.E.; et al. Candidate pharmacological treatments for substance use disorder and suicide identified by gene co-expression network-based drug repositioning. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2021, 186, 193–206. [Google Scholar] [CrossRef]
- Gao, H.; Ni, Y.; Mo, X.; Li, D.; Teng, S.; Huang, Q.; Huang, S.; Liu, G.; Zhang, S.; Tang, Y.; et al. Drug repositioning based on network-specific core genes identifies potential drugs for the treatment of autism spectrum disorder in children. Comput. Struct. Biotechnol. J. 2021, 19, 3908–3921. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
- Peri, S.; Navarro, J.D.; Kristiansen, T.Z.; Amanchy, R.; Surendranath, V.; Muthusamy, B.; Gandhi, T.K.B.; Chandrika, K.N.; Deshpande, N.; Suresh, S.; et al. Human protein reference database as a discovery resource for proteomics. Nucleic Acids Res. 2004, 32, D497–D501. [Google Scholar] [CrossRef]
- Croft, D.; O’Kelly, G.; Wu, G.; Haw, R.; Gillespie, M.; Matthews, L.; Caudy, M.; Garapati, P.; Gopinath, G.; Jassal, B.; et al. Reactome: A database of reactions, pathways and biological processes. Nucleic Acids Res. 2011, 39, D691–D697. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Lambert, S.A.; Jolma, A.; Campitelli, L.F.; Das, P.K.; Yin, Y.; Albu, M.; Chen, X.; Taipale, J.; Hughes, T.R.; Weirauch, M.T. The Human Transcription Factors. Cell 2018, 172, 650–665. [Google Scholar] [CrossRef]
- Boyle, A.P.; Hong, E.L.; Hariharan, M.; Cheng, Y.; Schaub, M.A.; Kasowski, M.; Karczewski, K.J.; Park, J.; Hitz, B.C.; Weng, S.; et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012, 22, 1790–1797. [Google Scholar] [CrossRef] [PubMed]
- Weirauch, M.T.; Yang, A.; Albu, M.; Cote, A.G.; Montenegro-Montero, A.; Drewe, P.; Najafabadi, H.S.; Lambert, S.A.; Mann, I.; Cook, K.; et al. Determination and inference of eukaryotic transcription factor sequence specificity. Cell 2014, 158, 1431–1443. [Google Scholar] [CrossRef] [PubMed]
- Castro-Mondragon, J.A.; Riudavets-Puig, R.; Rauluseviciute, I.; Berhanu Lemma, R.; Turchi, L.; Blanc-Mathieu, R.; Lucas, J.; Boddie, P.; Khan, A.; Manosalva Pérez, N.; et al. JASPAR 2022: The 9th release of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2022, 50, D165–D173. [Google Scholar] [CrossRef] [PubMed]
- Hume, M.A.; Barrera, L.A.; Gisselbrecht, S.S.; Bulyk, M.L. UniPROBE, update 2015: New tools and content for the online database of protein-binding microarray data on protein-DNA interactions. Nucleic Acids Res. 2015, 43, D117–D122. [Google Scholar] [CrossRef] [PubMed]
- Matys, V.; Kel-Margoulis, O.V.; Fricke, E.; Liebich, I.; Land, S.; Barre-Dirrie, A.; Reuter, I.; Chekmenev, D.; Krull, M.; Hornischer, K.; et al. TRANSFAC and its module TRANSCompel: Transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006, 34, D108–D110. [Google Scholar] [CrossRef]
- Türei, D.; Korcsmáros, T.; Saez-Rodriguez, J. OmniPath: Guidelines and gateway for literature-curated signaling pathway resources. Nat. Methods 2016, 13, 966–967. [Google Scholar] [CrossRef]
- Ganapathiraju, M.K.; Thahir, M.; Handen, A.; Sarkar, S.N.; Sweet, R.A.; Nimgaonkar, V.L.; Loscher, C.E.; Bauer, E.M.; Chaparala, S. Schizophrenia interactome with 504 novel protein–protein interactions. NPJ Schizophr. 2016, 2, 16012. [Google Scholar] [CrossRef]
- Kauppi, K.; Rosenthal, S.B.; Lo, M.-T.; Sanyal, N.; Jiang, M.; Abagyan, R.; McEvoy, L.K.; Andreassen, O.A.; Chen, C.-H. Revisiting Antipsychotic Drug Actions Through Gene Networks Associated With Schizophrenia. Am. J. Psychiatry 2018, 175, 674–682. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhou, D.-S.; Chang, H.; Wang, L.; Liu, W.; Dai, S.-X.; Zhang, C.; Cai, J.; Liu, W.; Li, X.; et al. Interactome Analyses implicated CAMK2A in the genetic predisposition and pharmacological mechanism of Bipolar Disorder. J. Psychiatr. Res. 2019, 115, 165–175. [Google Scholar] [CrossRef] [PubMed]
- De Bastiani, M.A.; Pfaffenseller, B.; Klamt, F. Master Regulators Connectivity Map: A Transcription Factors-Centered Approach to Drug Repositioning. Front. Pharmacol. 2018, 9, 697. [Google Scholar] [CrossRef]
- Kuhn, M.; Campillos, M.; Letunic, I.; Jensen, L.J.; Bork, P. A side effect resource to capture phenotypic effects of drugs. Mol. Syst. Biol. 2010, 6, 343. [Google Scholar] [CrossRef] [PubMed]
- Whirl-Carrillo, M.; Huddart, R.; Gong, L.; Sangkuhl, K.; Thorn, C.F.; Whaley, R.; Klein, T.E. An Evidence-Based Framework for Evaluating Pharmacogenomics Knowledge for Personalized Medicine. Clin Pharm. 2021, 110, 563–572. [Google Scholar] [CrossRef]
- Freshour, S.L.; Kiwala, S.; Cotto, K.C.; Coffman, A.C.; McMichael, J.F.; Song, J.J.; Griffith, M.; Griffith, O.L.; Wagner, A.H. Integration of the Drug–Gene Interaction Database (DGIdb 4.0) with open crowdsource efforts. Nucleic Acids Res. 2021, 49, D1144–D1151. [Google Scholar] [CrossRef]
- Avram, S.; Bologa, C.G.; Holmes, J.; Bocci, G.; Wilson, T.B.; Nguyen, D.-T.; Curpan, R.; Halip, L.; Bora, A.; Yang, J.J.; et al. DrugCentral 2021 supports drug discovery and repositioning. Nucleic Acids Res. 2021, 49, D1160–D1169. [Google Scholar] [CrossRef]
- Mitsopoulos, C.; Di Micco, P.; Fernandez, E.V.; Dolciami, D.; Holt, E.; Mica, I.L.; Coker, E.A.; Tym, J.E.; Campbell, J.; Che, K.H.; et al. canSAR: Update to the cancer translational research and drug discovery knowledgebase. Nucleic Acids Res. 2021, 49, D1074–D1082. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef]
- Harding, S.D.; Armstrong, J.F.; Faccenda, E.; Southan, C.; Alexander, S.P.H.; Davenport, A.P.; Pawson, A.J.; Spedding, M.; Davies, J.A.; Nc, I. The IUPHAR/BPS guide to pharmacology in 2022: Curating pharmacology for COVID-19, malaria and antibacterials. Nucleic Acids Res. 2022, 50, D1282–D1294. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Santos, A.; von Mering, C.; Jensen, L.J.; Bork, P.; Kuhn, M. STITCH 5: Augmenting protein–chemical interaction networks with tissue and affinity data. Nucleic Acids Res. 2016, 44, D380–D384. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-W.; Diamante, G.; Ding, J.; Nghiem, T.X.; Yang, J.; Ha, S.-M.; Cohn, P.; Arneson, D.; Blencowe, M.; Garcia, J.; et al. PharmOmics: A species- and tissue-specific drug signature database and gene-network-based drug repositioning tool. iScience 2022, 25, 104052. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhang, Y.; Lian, X.; Li, F.; Wang, C.; Zhu, F.; Qiu, Y.; Chen, Y. Therapeutic target database update 2022: Facilitating drug discovery with enriched comparative data of targeted agents. Nucleic Acids Res. 2022, 50, D1398–D1407. [Google Scholar] [CrossRef] [PubMed]
- Yoo, M.; Shin, J.; Kim, J.; Ryall, K.A.; Lee, K.; Lee, S.; Jeon, M.; Kang, J.; Tan, A.C. DSigDB: Drug signatures database for gene set analysis. Bioinformatics 2015, 31, 3069–3071. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.-T.; Mathias, S.; Bologa, C.; Brunak, S.; Fernandez, N.; Gaulton, A.; Hersey, A.; Holmes, J.; Jensen, L.J.; Karlsson, A.; et al. Pharos: Collating protein information to shed light on the druggable genome. Nucleic Acids Res. 2017, 45, D995–D1002. [Google Scholar] [CrossRef] [PubMed]
- Jensen, H.N.; Roth, L.B. Massively Parallel Screening of the Receptorome. Comb. Chem. High Throughput Screen. 2008, 11, 420–426. [Google Scholar] [CrossRef] [PubMed]
- Gilson, M.K.; Liu, T.; Baitaluk, M.; Nicola, G.; Hwang, L.; Chong, J. BindingDB in 2015: A public database for medicinal chemistry, computational chemistry and systems pharmacology. Nucleic Acids Res. 2016, 44, D1045–D1053. [Google Scholar] [CrossRef]
- Amberger, J.S.; Bocchini, C.A.; Scott, A.F.; Hamosh, A. OMIM.org: Leveraging knowledge across phenotype–gene relationships. Nucleic Acids Res. 2019, 47, D1038–D1043. [Google Scholar] [CrossRef]
- Landrum, M.J.; Chitipiralla, S.; Brown, G.R.; Chen, C.; Gu, B.; Hart, J.; Hoffman, D.; Jang, W.; Kaur, K.; Liu, C.; et al. ClinVar: Improvements to accessing data. Nucleic Acids Res. 2020, 48, D835–D844. [Google Scholar] [CrossRef]
- Rappaport, N.; Twik, M.; Plaschkes, I.; Nudel, R.; Iny Stein, T.; Levitt, J.; Gershoni, M.; Morrey, C.P.; Safran, M.; Lancet, D. MalaCards: An amalgamated human disease compendium with diverse clinical and genetic annotation and structured search. Nucleic Acids Res. 2017, 45, D877–D887. [Google Scholar] [CrossRef]
- Piñero, J.; Ramírez-Anguita, J.M.; Saüch-Pitarch, J.; Ronzano, F.; Centeno, E.; Sanz, F.; Furlong, L.I. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res. 2020, 48, D845–D855. [Google Scholar] [CrossRef] [PubMed]
- Köhler, S.; Carmody, L.; Vasilevsky, N.; Jacobsen, J.O.B.; Danis, D.; Gourdine, J.-P.; Gargano, M.; Harris, N.L.; Matentzoglu, N.; McMurry, J.A.; et al. Expansion of the Human Phenotype Ontology (HPO) knowledge base and resources. Nucleic Acids Res. 2019, 47, D1018–D1027. [Google Scholar] [CrossRef] [PubMed]
- Shefchek, K.A.; Harris, N.L.; Gargano, M.; Matentzoglu, N.; Unni, D.; Brush, M.; Keith, D.; Conlin, T.; Vasilevsky, N.; Zhang, X.A.; et al. The Monarch Initiative in 2019: An integrative data and analytic platform connecting phenotypes to genotypes across species. Nucleic Acids Res. 2020, 48, D704–D715. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Wang, Z.; Chen, Q.; Li, K.; Wang, X.; Wang, Y.; Zeng, Q.; Han, Y.; Lu, B.; Zhao, Y.; et al. GPCards: An integrated database of genotype–phenotype correlations in human genetic diseases. Comput. Struct. Biotechnol. J. 2021, 19, 1603–1611. [Google Scholar] [CrossRef]
- Zhou, X.; Menche, J.; Barabási, A.-L.; Sharma, A. Human symptoms–disease network. Nat. Commun. 2014, 5, 4212. [Google Scholar] [CrossRef]
- Xu, R.; Li, L.; Wang, Q. Towards building a disease-phenotype knowledge base: Extracting disease-manifestation relationship from literature. Bioinformatics 2013, 29, 2186–2194. [Google Scholar] [CrossRef]
- Gillen, J.E.; Tse, T.; Ide, N.C.; McCray, A.T. Design, implementation and management of a web-based data entry system for ClinicalTrials.gov. Stud. Health Technol. Inform. 2004, 107, 1466–1470. [Google Scholar]
- Zhou, M.; Wang, Q.; Zheng, C.; John Rush, A.; Volkow, N.D.; Xu, R. Drug repurposing for opioid use disorders: Integration of computational prediction, clinical corroboration, and mechanism of action analyses. Mol. Psychiatry 2021, 26, 5286–5296. [Google Scholar] [CrossRef]
- Huang, L.-C.; Soysal, E.; Zheng, W.J.; Zhao, Z.; Xu, H.; Sun, J. A weighted and integrated drug-target interactome: Drug repurposing for schizophrenia as a use case. BMC Syst. Biol. 2015, 9, S2. [Google Scholar] [CrossRef]
- Lüscher Dias, T.; Schuch, V.; Beltrão-Braga, P.C.B.; Martins-de-Souza, D.; Brentani, H.P.; Franco, G.R.; Nakaya, H.I. Drug repositioning for psychiatric and neurological disorders through a network medicine approach. Transl. Psychiatry 2020, 10, 141. [Google Scholar] [CrossRef]
- Ben Guebila, M.; Lopes-Ramos, C.M.; Weighill, D.; Sonawane, A.R.; Burkholz, R.; Shamsaei, B.; Platig, J.; Glass, K.; Kuijjer, M.L.; Quackenbush, J. GRAND: A database of gene regulatory network models across human conditions. Nucleic Acids Res. 2022, 50, D610–D621. [Google Scholar] [CrossRef] [PubMed]
- Sadegh, S.; Skelton, J.; Anastasi, E.; Bernett, J.; Blumenthal, D.B.; Galindez, G.; Salgado-Albarrán, M.; Lazareva, O.; Flanagan, K.; Cockell, S.; et al. Network medicine for disease module identification and drug repurposing with the NeDRex platform. Nat. Commun. 2021, 12, 6848. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Elenee Argentinis, J.D.; Weber, G. IBM Watson: How Cognitive Computing Can Be Applied to Big Data Challenges in Life Sciences Research. Clin. Ther. 2016, 38, 688–701. [Google Scholar] [CrossRef] [PubMed]
- Karunakaran, K.B.; Chaparala, S.; Ganapathiraju, M.K. Potentially repurposable drugs for schizophrenia identified from its interactome. Sci. Rep. 2019, 9, 12682. [Google Scholar] [CrossRef] [PubMed]
- Batool, M.; Ahmad, B.; Choi, S. A Structure-Based Drug Discovery Paradigm. Int. J. Mol. Sci. 2019, 20, 2783. [Google Scholar] [CrossRef]
- Kulkarni, J. Selective Estrogen Receptor Modulators-A Potential Treatment for Psychotic Symptoms of Schizophrenia? NCT00361543. Available online: https://clinicaltrials.gov/ct2/show/NCT00361543 (accessed on 28 January 2015).
- Kulkarni, J.; Gavrilidis, E.; Gwini, S.M.; Worsley, R.; Grigg, J.; Warren, A.; Gurvich, C.; Gilbert, H.; Berk, M.; Davis, S.R. Effect of Adjunctive Raloxifene Therapy on Severity of Refractory Schizophrenia in Women: A Randomized Clinical Trial. JAMA Psychiatry 2016, 73, 947–954. [Google Scholar] [CrossRef]
- Henry, L. UMCC 2013.051: Prospective Pilot Study Evaluating the Use of Cyclobenzaprine for Treatment of Sleep Disturbance, Fatigue, and Musculoskeletal Symptoms in Aromatase Inhibitor-Treated Breast Cancer Patients. NCT01921296. Available online: https://clinicaltrials.gov/ct2/show/NCT01921296 (accessed on 21 March 2016).
- Hebbring, S.J. The challenges, advantages and future of phenome-wide association studies. Immunology 2014, 141, 157–165. [Google Scholar] [CrossRef]
- Senthil, G.; Dutka, T.; Bingaman, L.; Lehner, T. Genomic resources for the study of neuropsychiatric disorders. Mol. Psychiatry 2017, 22, 1659–1663. [Google Scholar] [CrossRef]
- Shukla, R.; Henkel, N.D.; Alganem, K.; Hamoud, A.-R.; Reigle, J.; Alnafisah, R.S.; Eby, H.M.; Imami, A.S.; Creeden, J.F.; Miruzzi, S.A.; et al. Signature-based approaches for informed drug repurposing: Targeting CNS disorders. Neuropsychopharmacology 2020, 46, 116–130. [Google Scholar] [CrossRef]
- Iorio, F.; Rittman, T.; Ge, H.; Menden, M.; Saez-Rodriguez, J. Transcriptional data: A new gateway to drug repositioning? Drug Discov. Today 2013, 18, 350–357. [Google Scholar] [CrossRef]
- Gaiteri, C.; Ding, Y.; French, B.; Tseng, G.C.; Sibille, E. Beyond modules and hubs: The potential of gene coexpression networks for investigating molecular mechanisms of complex brain disorders. Genes Brain Behav. 2014, 13, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Truong, T.T.; Bortolasci, C.C.; Spolding, B.; Panizzutti, B.; Liu, Z.S.; Kidnapillai, S.; Richardson, M.; Gray, L.; Smith, C.M.; Dean, O.M.; et al. Co-Expression Networks Unveiled Long Non-Coding RNAs as Molecular Targets of Drugs Used to Treat Bipolar Disorder. Front. Pharmacol. 2022, 13, 873271. [Google Scholar] [CrossRef] [PubMed]
- Lamb, J.; Crawford, E.D.; Peck, D.; Modell, J.W.; Blat, I.C.; Wrobel, M.J.; Lerner, J.; Brunet, J.-P.; Subramanian, A.; Ross, K.N.; et al. The Connectivity Map: Using Gene-Expression Signatures to Connect Small Molecules, Genes, and Disease. Science 2006, 313, 1929–1935. [Google Scholar] [CrossRef] [PubMed]
- Vidović, D.; Koleti, A.; Schürer, S.C. Large-scale integration of small molecule-induced genome-wide transcriptional responses, Kinome-wide binding affinities and cell-growth inhibition profiles reveal global trends characterizing systems-level drug action. Front. Genet. 2014, 5, 342. [Google Scholar] [CrossRef]
- Kidnapillai, S.; Bortolasci, C.C.; Udawela, M.; Panizzutti, B.; Spolding, B.; Connor, T.; Sanigorski, A.; Dean, O.M.; Crowley, T.; Jamain, S.; et al. The use of a gene expression signature and connectivity map to repurpose drugs for bipolar disorder. World J. Biol. Psychiatry 2020, 21, 775–783. [Google Scholar] [CrossRef]
- Liu, W.; Tu, W.; Li, L.; Liu, Y.; Wang, S.; Li, L.; Tao, H.; He, H. Revisiting Connectivity Map from a gene co-expression network analysis. Exp. Med. 2018, 16, 493–500. [Google Scholar] [CrossRef]
- Keenan, A.B.; Jenkins, S.L.; Jagodnik, K.M.; Koplev, S.; He, E.; Torre, D.; Wang, Z.; Dohlman, A.B.; Silverstein, M.C.; Lachmann, A.; et al. The Library of Integrated Network-Based Cellular Signatures NIH Program: System-Level Cataloging of Human Cells Response to Perturbations. Cell Syst. 2018, 6, 13–24. [Google Scholar] [CrossRef]
- Dolmetsch, R.; Geschwind, D.H. The human brain in a dish: The promise of iPSC-derived neurons. Cell 2011, 145, 831–834. [Google Scholar] [CrossRef]
- Huang, J.K.; Carlin, D.E.; Yu, M.K.; Zhang, W.; Kreisberg, J.F.; Tamayo, P.; Ideker, T. Systematic Evaluation of Molecular Networks for Discovery of Disease Genes. Cell Syst. 2018, 6, 484–495.e485. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, J. A Big World Inside Small-World Networks. PLoS ONE 2009, 4, e5686. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017: Quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef] [PubMed]
- Goh, K.-I.; Cusick Michael, E.; Valle, D.; Childs, B.; Vidal, M.; Barabási, A.-L. The human disease network. Proc. Natl. Acad. Sci. USA 2007, 104, 8685–8690. [Google Scholar] [CrossRef] [PubMed]
- Langhauser, F.; Casas, A.I.; Dao, V.-T.-V.; Guney, E.; Menche, J.; Geuss, E.; Kleikers, P.W.M.; López, M.G.; Barabási, A.-L.; Kleinschnitz, C.; et al. A diseasome cluster-based drug repurposing of soluble guanylate cyclase activators from smooth muscle relaxation to direct neuroprotection. NPJ Syst. Biol. Appl. 2018, 4, 8. [Google Scholar] [CrossRef]
- Menche, J.; Sharma, A.; Kitsak, M.; Ghiassian, S.D.; Vidal, M.; Loscalzo, J.; Barabási, A.-L. Disease networks. Uncovering disease-disease relationships through the incomplete interactome. Science 2015, 347, 1257601. [Google Scholar] [CrossRef] [PubMed]
- Palsson, B.; Zengler, K. The challenges of integrating multi-omic data sets. Nat. Chem. Biol. 2010, 6, 787–789. [Google Scholar] [CrossRef]
- Guo, M.G.; Sosa, D.N.; Altman, R.B. Challenges and opportunities in network-based solutions for biological questions. Brief. Bioinform. 2022, 23, bbab437. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.-F.; Zhao, X.-M. A Survey on the Computational Approaches to Identify Drug Targets in the Postgenomic Era. BioMed Res. Int. 2015, 2015, 239654. [Google Scholar] [CrossRef]
- Arrell, D.K.; Terzic, A. Network systems biology for drug discovery. Clin. Pharm. 2010, 88, 120–125. [Google Scholar] [CrossRef]
- Jarada, T.N.; Rokne, J.G.; Alhajj, R. A review of computational drug repositioning: Strategies, approaches, opportunities, challenges, and directions. J. Cheminform. 2020, 12, 46. [Google Scholar] [CrossRef]
- List of Databases Converted to the OMOP CDM. Available online: https://www.ohdsi.org/web/wiki/doku.php?id=resources:2020_data_network (accessed on 16 May 2022).
- The All of Us Research Program Investigators. The “All of Us” Research Program. N. Engl. J. Med. 2019, 381, 668–676. [Google Scholar] [CrossRef]
- Agid, Y.; Buzsáki, G.; Diamond, D.M.; Frackowiak, R.; Giedd, J.; Girault, J.-A.; Grace, A.; Lambert, J.J.; Manji, H.; Mayberg, H.; et al. How can drug discovery for psychiatric disorders be improved? Nat. Rev. Drug Discov. 2007, 6, 189–201. [Google Scholar] [CrossRef] [PubMed]
- Wager, T.D.; Woo, C.W. fMRI in analgesic drug discovery. Sci. Transl. Med. 2015, 7, 274fs276. [Google Scholar] [CrossRef] [PubMed]
- Duff, E.P.; Vennart, W.; Wise, R.G.; Howard, M.A.; Harris, R.E.; Lee, M.; Wartolowska, K.; Wanigasekera, V.; Wilson, F.J.; Whitlock, M.; et al. Learning to identify CNS drug action and efficacy using multistudy fMRI data. Sci. Transl. Med. 2015, 7, 274ra216. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, I.; Verma, S.; Kumar, S.; Jere, A.; Anamika, K. Multi-omics Data Integration, Interpretation, and Its Application. Bioinform. Biol. Insights 2020, 14, 1177932219899051. [Google Scholar] [CrossRef]
- Mirza, B.; Wang, W.; Wang, J.; Choi, H.; Chung, N.C.; Ping, P. Machine Learning and Integrative Analysis of Biomedical Big Data. Genes 2019, 10, 87. [Google Scholar] [CrossRef]
- Kuijjer, M.L.; Tung, M.G.; Yuan, G.; Quackenbush, J.; Glass, K. Estimating Sample-Specific Regulatory Networks. iScience 2019, 14, 226–240. [Google Scholar] [CrossRef]
- Liu, C.; Louhimo, R.; Laakso, M.; Lehtonen, R.; Hautaniemi, S. Identification of sample-specific regulations using integrative network level analysis. BMC Cancer 2015, 15, 319. [Google Scholar] [CrossRef]


| Type of Data | Description and Resource | Examples in Psychiatry |
|---|---|---|
| Structome | Chemical structures: ChemBL [53] ChemSpider [54] DrugBank [55] PubChem [56] Macromolecular structures: Protein Data Bank [57] AlphaFold Protein Structure Database [58] | Schizophrenia, sleep disorder [59] |
| Genome/Transcriptome | GWAS (general): GWAS ATLAS [60] NCBI Database of Genotypes and Phenotypes (dbGaP) [61] GWAS (psychiatry): NIMH Repository and Genomics Resource (NRGR) [62] Psychiatric Genomics Consortium (PGC) [63] Autism Sequencing Consortium (ASC) [64] Whole-Genome Sequencing Consortium for Psychiatric Disorders (WGSPD) [65] Human brain resources: PsychENCODE [66] Brain Somatic Mosaicism Network [67] CommonMind Consortium [68] Allen Brain Atlas [69] Drug response: Connectivity Map (CMap) [70] Library of Integrated Network-Based Cellular Signatures (LINCS) [71] Drug Gene Budger (DGB) [72] | Depression [73] Schizophrenia [74] Substance use disorder [75] Autism spectrum disorder [76] |
| Interactome | Protein–protein interaction: Search tool for retrieval of interacting genes/proteins (STRING) [77] Human Protein Reference Database (HPRD) [78] Pathways: Reactome [79] Kyoto Encyclopedia of Genes and Genomes (KEGG) [80] Regulome: The Human Transcription Factors [81] RegulomeDB [82] Catalog of inferred sequence binding preferences [83] JASPAR [84] UniPROBE [85] TRANSFAC [86] Multiple collections: OmniPath [87] | Schizophrenia [88,89] Bipolar disorder [90,91] |
| Phenome | Side effects: SIDER [92] Drug targets: DrugBank [55] PharmGKB [93] Drug–Gene Interaction Database (DGIdb) [94] DrugCentral [95] canSARblack [96] KEGG DRUG [97] IUPHAR/BPS Guide to PHARMACOLOGY (GtoPdb) [98] Search Tool for Interacting Chemicals (STITCH) [99,100] Therapeutic Target Database (TTD) [101] Drug Signatures Database (DSigDB) [102] Pharos [103] Binding assay profiles: Psychoactive Drug Screening Program (PDSP) [104] BindingDB [105] Disease-associated targets: Online Mendelian Inheritance in Man (OMIM) [106] ClinVar [107] MalaCards [108] DisGeNET [109] Human Phenotype Ontology (HPO) [110] Monarch [111] GPCards [112] Disease symptoms: Human symptoms–disease network [113] Human Phenotype Ontology (HPO) [110] DMPatternUMLS [114] Clinical trials: ClinicalTrials.gov [115] | Opioid use disorders [116] Schizophrenia [117] Schizophrenia, bipolar disorder, autism spectrum disorder [118] |
| Network-based drug discovery platforms | GRAND [119] PharmOmics [100] NeDRex [120] IBM Watson for Drug Discovery [121] |
| Studies | Diseases | Databases Used | Inference Model and Network Type | Key Finding (Original Indication/ Mechanism–Repurposed Indications) | Validation |
|---|---|---|---|---|---|
| [59] | Schizophrenia Sleep disorder | DrugBank PubChem | GBA: Drug–drug similarity | Raloxifene (estrogen receptor modulator → SCZ) Cyclobenzaprine (muscle relaxant → sleep disorder) | Literature-based (clinical trials, research articles), expert consultation |
| [73] | Depression | DGIdb ChEMBL PDSP Pharos PubChem DSigDB | ABC: Phenotype-informed drug-target network (http://drugtargetor.com/, accessed on 2 June 2022), i.e., an integration of drug-disease associations (GWAS pathway analysis p-values), target-disease associations (GWAS gene-wise analysis p-values, genetically predicted expression z-scores), and drug-target connections | Verapamil (calcium channel blocker → MDD) Pregabalin, Gabapentin and Nitrendipine (calcium channel modulators → MDD) Brompheniramine and Chlorphenamine (antihistamines → MDD) Lasofoxifene (estrogen receptor modulator → MDD) Levonorgestrel (sex hormones → MDD) Alizapride and Mesoridazine (D2 antagonists → MDD) Quinagolide (D2 agonist → MDD) | Literature-based (clinical trials, research articles) |
| [118] | Schizophrenia Bipolar Disorder Autism Spectrum Disorder | PubMed DrugBank Open Targets | ABC: Literature-mined disease–gene–drug association | AC-480, Mubritinib, CP724714, Trastuzumab, Ertumaxomab, and MM-302 (Target ERBB2 gene → SZ) SLC6A9 (glycine transporter → SZ) Bitopertin and PF-03463275 (? → SZ) Levetiracetan and Brivaracetam (anticonvulsant → SZ) CEACAM5 (? → BD) Lebrikizumab and Tralokinumab (act on IL3 →ASD) | Literature-based (clinical trials and research articles) |
| [74] | Schizophrenia | DGIdb | ABC: Brain co-expression network + TWAS predicted expression polygenic risk scores + drug-target interactions | Zonisamide (antiepileptic/ antiparkinsonian → SZ) Bevacizumab (antineoplastic agent → SZ) Fluticasone (cortisone analogue → SZ) | Literature-based (research articles) |
| [75] | Substance Use Disorder | DGIdb | ABC: Disease-related co-expression networks + drug-target interactions | MAOA inhibitors (antidepressants → SUD) Dextromethorphan (cough suppressant → SUB with suicide) Eglumegad and loxapine (? → non-suicidal SUD) Clozapine and olanzapine (antypsychotics SZ → non-suicidal SUD) Modafinil (sleep disorder → SUD) | Literature-based (research articles) |
| [76] | Autism Spectrum Disorder | STRING DrugBank Drug Targetor CMap | ABC: Disease-related co-expression networks + drug–gene interactome Mental disease and compounds knowledge graph (MCKG) based on literature mining for validation | Baclofen (GABA agonist for pain and muscle spasms → ASD) Sulpiride (D2 receptor antagonist, for SZ and ASD, confirmatory) Estradiol (steroid sex hormone → ASD) Entinostat (HDAC inhibitor → ASD) Everolimus (seizures → ASD) Fluvoxamine, Curcumin, Calcitriol, Metronidazole, and zinc (diverse mechanisms and uses → ASD) | Literature-based (research articles) |
| [116] | Opioid Use Disorders | STITCH SIDER STRING DrugBank | ABC: Drug side effect + protein interactome | Tramadol (pain → OUD) Olanzapine (SZ → OUD) Mirtazapine and Bupropion (MDD→OUD) Atomoxetine (ADHD → OUD) | Literature-based (clinical trials and research articles), clinical corroboration (retrospective case-control study of top candidates in population-level EHR data) |
| [117] | Schizophrenia | DrugBank MATADOR PDSP Ki Database BindingDB | GBA: SZ drug target–non-SZ drug interactome | 264 SZ related drugs, 39 being investigated in clinical trials (Listed in Figure 3 of the corresponding publication) | Literature-based (clinical trials and research articles) |
| [122] repurposing based on network built by [88] | Schizophrenia | Psychiatric Genomics Consortium (PGC) HPRD Ensembl DrugBank | ABC: Disease risk gene–drug interactome | Sargramostin, Regorafenib, Theophylline (cancer and respiratory drugs → SZ) Cromoglicic acid (asthma prophylaxis → SZ) Acetazolamide (glaucoma, mountain sickness → SZ) Cinnarizine (Motion sickness, vertigo → SZ) Alfacalcidol (targets the VDR protein → SZ) Amiloride (on clinical trial for ADHD → SZ) Antazoline (targets ubiquitination and proteasome degradation → SZ) Danazol and Miconazole (target ESR1 and NOS3 associated with Alzheimer’s Disease → SZ) | Literature-based (clinical trials and research articles) |
| [89] | Schizophrenia | Psychiatric Genomics Consortium (PGC) STRING DGIdb | ABC: Disease risk gene–untargeted neighbor gene interactome | 19 drugs to repurpose, one major example: Galantamine (Alzheimer’s disease → SZ) | Literature-based (research articles) |
| [91] | Bipolar Disorder | GEO CMap (via PharmacoGx package) | GBA: Transcription factor-target association | Chlorpromazine, Lavomepromazine, Perphenazine, Zuclopenthixol, Haloperidol, Promazine (antipsychotics → BD) Maprotiline, Desipramine, Mianserin (antidepressants → BD) Diflorasone (corticosteroid → BD) Meclofenamic acid, Ketorolac, Trolox c, and Acetylsalicylsalicylic acid (antiinflamatory/antirheumatic → BD) | Literature-based (research articles) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Truong, T.T.T.; Panizzutti, B.; Kim, J.H.; Walder, K. Repurposing Drugs via Network Analysis: Opportunities for Psychiatric Disorders. Pharmaceutics 2022, 14, 1464. https://doi.org/10.3390/pharmaceutics14071464
Truong TTT, Panizzutti B, Kim JH, Walder K. Repurposing Drugs via Network Analysis: Opportunities for Psychiatric Disorders. Pharmaceutics. 2022; 14(7):1464. https://doi.org/10.3390/pharmaceutics14071464
Chicago/Turabian StyleTruong, Trang T. T., Bruna Panizzutti, Jee Hyun Kim, and Ken Walder. 2022. "Repurposing Drugs via Network Analysis: Opportunities for Psychiatric Disorders" Pharmaceutics 14, no. 7: 1464. https://doi.org/10.3390/pharmaceutics14071464
APA StyleTruong, T. T. T., Panizzutti, B., Kim, J. H., & Walder, K. (2022). Repurposing Drugs via Network Analysis: Opportunities for Psychiatric Disorders. Pharmaceutics, 14(7), 1464. https://doi.org/10.3390/pharmaceutics14071464