Design and Functional Characterization of HIV-1 Envelope Protein-Coupled T Helper Liposomes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Env Trimer Purification
2.3. Nanoparticle Preparation
2.3.1. First-Generation Liposomes
2.3.2. Uncoupled Anionic T Helper Liposomes
2.3.3. Second-Generation Anionic T Helper Liposomes
2.3.4. Peptide Quantification via HPLC
2.3.5. T Helper VLPs
2.4. Isolation of Murine Primary Cells
2.4.1. B Cells
2.4.2. T Cells
2.4.3. Dendritic Cells (DCs)
2.4.4. Lymphocytes
2.5. Analysis of B Cell Activation and Proliferation
2.6. T Cell Proliferation by T Cell/Dendritic Cell Co-Culture
2.7. Ex Vivo DC Transfer
2.8. In Vitro Intrastructural Help
2.9. Antibody Staining of Liposomes
2.9.1. Immunogold EM Imaging of T Helper Liposomes
2.9.2. Liposomal Surface FACS
2.10. Statistical Analysis
3. Results and Discussion
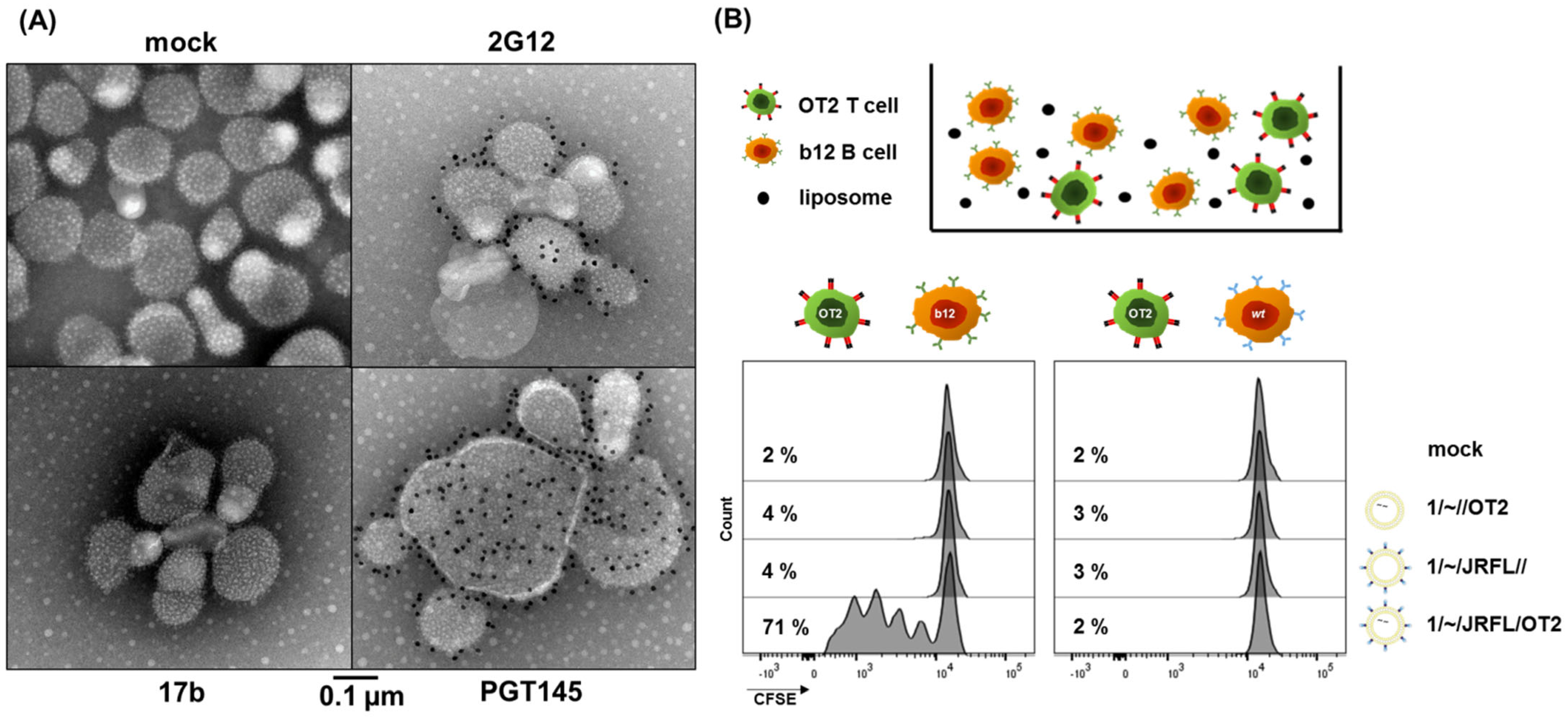
3.1. First-Generation T Helper Liposomes Produced by Passive Peptide Entrapment
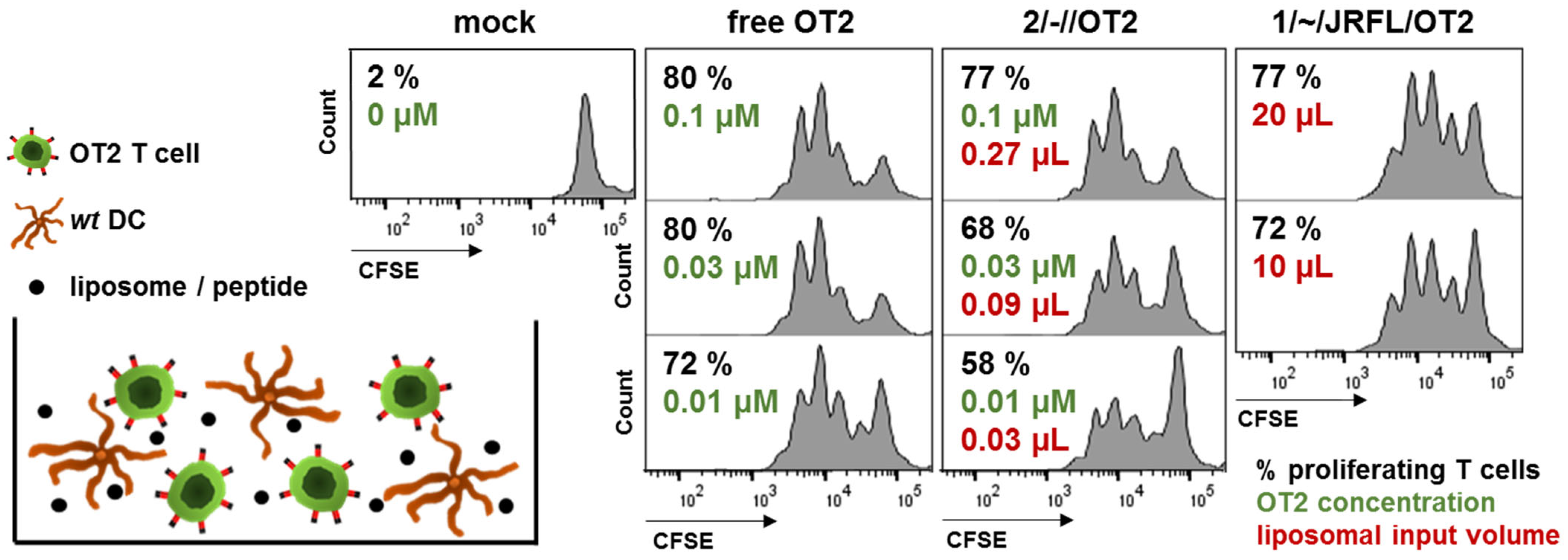
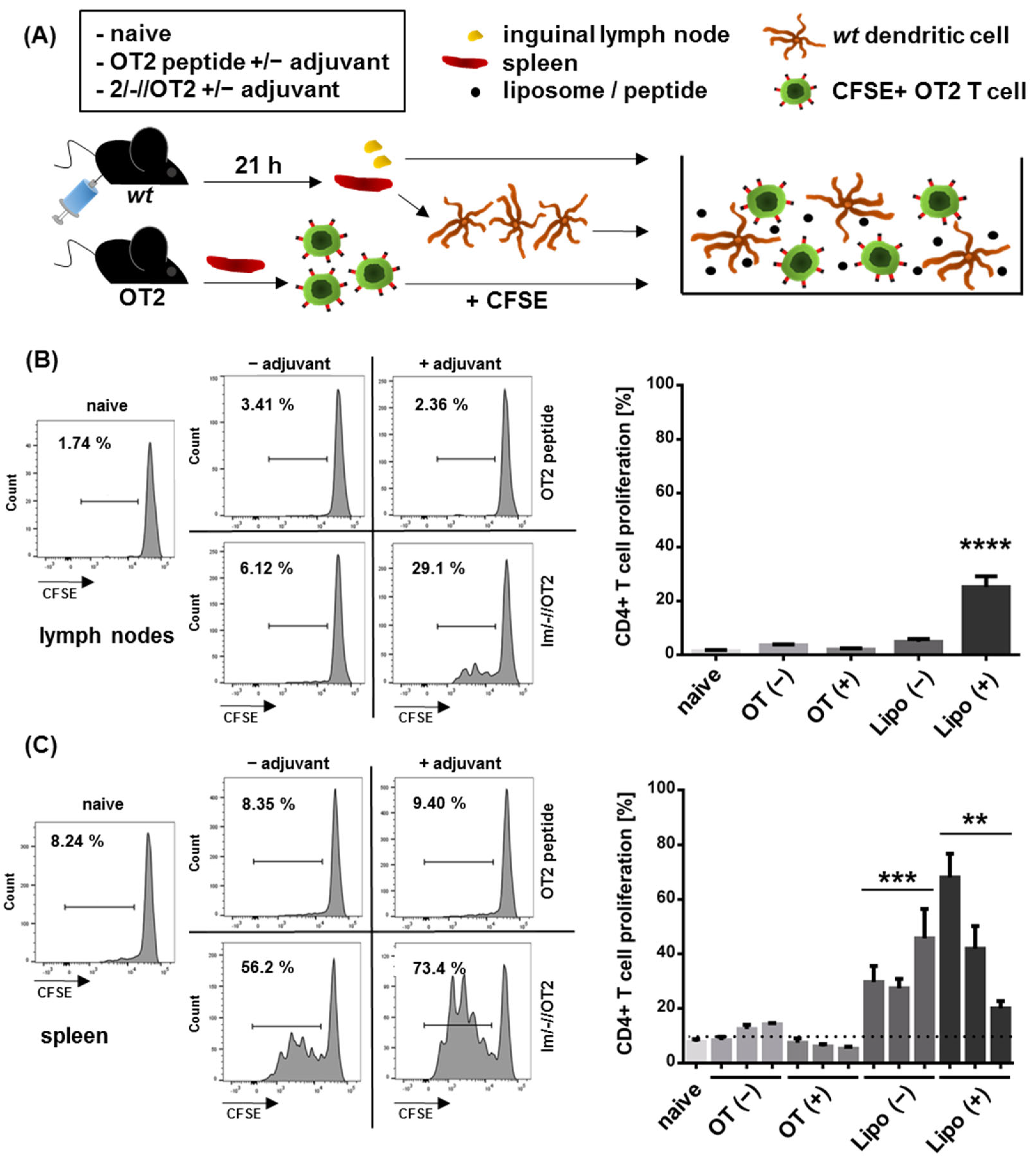
3.2. Functional Characterization of Uncoupled, Anionic Liposomes with Improved Peptide Encapsulation
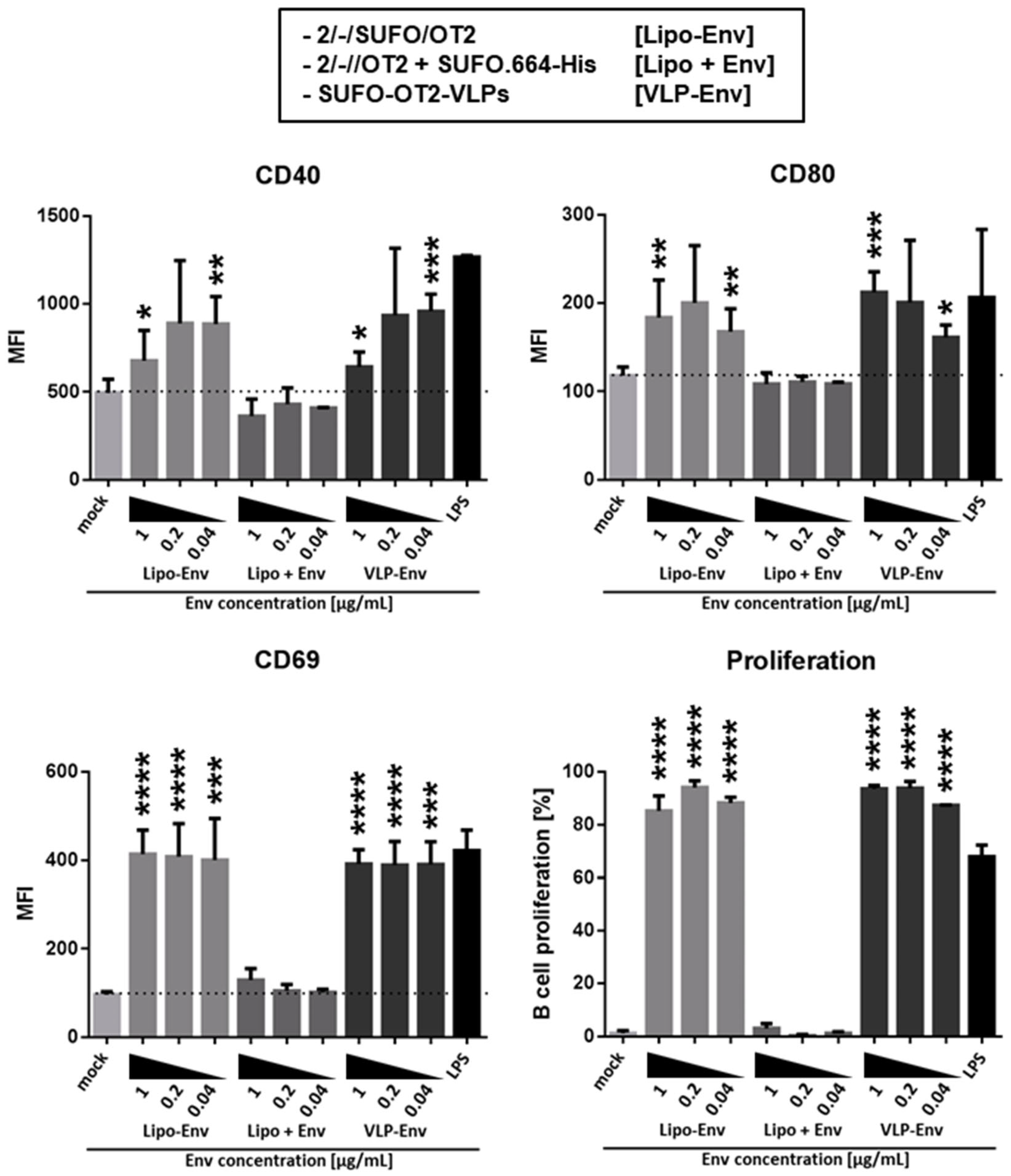
3.3. Production of Second-Generation T Helper Liposomes by Covalent Coupling of Native-Like Env Trimers
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brinkkemper, M.; Sliepen, K. Nanoparticle Vaccines for Inducing HIV-1 Neutralizing Antibodies. Vaccines 2019, 7, 76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Irvine, D.J.; Read, B.J. Shaping humoral immunity to vaccines through antigen-displaying nanoparticles. Curr. Opin. Immunol. 2020, 65, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Nabi, G.; Temchura, V.; Großmann, C.; Kuate, S.; Tenbusch, M.; Überla, K. T cell independent secondary antibody responses to the envelope protein of simian immunodeficiency virus. Retrovirology 2012, 9, 42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fouts, T.R.; Bagley, K.; Prado, I.J.; Bobb, K.L.; Schwartz, J.A.; Xu, R.; Zagursky, R.J.; Egan, M.A.; Eldridge, J.H.; LaBranche, C.C.; et al. Balance of cellular and humoral immunity determines the level of protection by HIV vaccines in rhesus macaque models of HIV infection. Proc. Natl. Acad. Sci. USA 2015, 112, E992–E999. [Google Scholar] [CrossRef] [Green Version]
- Tenbusch, M.; Ignatius, R.; Temchura, V.; Nabi, G.; Tippler, B.; Stewart-Jones, G.; Salazar, A.M.; Sauermann, U.; Stahl-Hennig, C.; Überla, K. Risk of immunodeficiency virus infection may increase with vaccine-induced immune response. J. Virol. 2012, 86, 10533–10539. [Google Scholar] [CrossRef] [Green Version]
- Temchura, V.; Tenbusch, M. The two faces of vaccine-induced immune response: Protection or increased risk of HIV infection?! Virol. Sin. 2014, 29, 7–9. [Google Scholar] [CrossRef]
- Russell, S.M.; Liew, F.Y. T cells primed by influenza virion internal components can cooperate in the antibody response to haemagglutinin. Nature 1979, 280, 147–148. [Google Scholar] [CrossRef]
- Milich, D.R.; McLachlan, A.; Thornton, G.B.; Hughes, J.L. Antibody production to the nucleocapsid and envelope of the hepatitis B virus primed by a single synthetic T cell site. Nature 1987, 329, 547–549. [Google Scholar] [CrossRef]
- Elsayed, H.; Nabi, G.; McKinstry, W.J.; Khoo, K.K.; Mak, J.; Salazar, A.M.; Tenbusch, M.; Temchura, V.; Überla, K. Intrastructural Help: Harnessing T Helper Cells Induced by Licensed Vaccines for Improvement of HIV Env Antibody Responses to Virus-Like Particle Vaccines. J. Virol. 2018, 92, e00141-18. [Google Scholar] [CrossRef] [Green Version]
- Klessing, S.; Temchura, V.; Tannig, P.; Peter, A.S.; Christensen, D.; Lang, R.; Überla, K. Cd4+ T cells induced by tuberculosis subunit vaccine h1 can improve the hiv-1 env humoral response by intrastructural help. Vaccines 2020, 8, 604. [Google Scholar] [CrossRef]
- Damm, D.; Rojas-Sánchez, L.; Theobald, H.; Sokolova, V.; Wyatt, R.T.; Überla, K.; Epple, M.; Temchura, V. Calcium phosphate nanoparticle-based vaccines as a platform for improvement of HIV-1 env antibody responses by intrastructural help. Nanomaterials 2019, 9, 1389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Temchura, V.; Überla, K. Intrastructural help. Curr. Opin. HIV AIDS 2017, 12, 272–277. [Google Scholar] [CrossRef] [PubMed]
- Deml, L.; Speth, C.; Dierich, M.P.; Wolf, H.; Wagner, R. Recombinant HIV-1 Pr55gag virus-like particles: Potent stimulators of innate and acquired immune responses. Mol. Immunol. 2005, 42, 259–277. [Google Scholar] [CrossRef] [PubMed]
- Thompson, C.M.; Petiot, E.; Mullick, A.; Aucoin, M.G.; Henry, O.; Kamen, A.A. Critical assessment of influenza VLP production in Sf9 and HEK293 expression systems. BMC Biotechnol. 2015, 15, 31. [Google Scholar] [CrossRef] [Green Version]
- Bulbake, U.; Doppalapudi, S.; Kommineni, N.; Khan, W. Liposomal Formulations in Clinical Use: An Updated Review. Pharmaceutics 2017, 9, 12. [Google Scholar] [CrossRef]
- Semple, S.C.; Leone, R.; Barbosa, C.J.; Tam, Y.K.; Lin, P.J.C. Lipid Nanoparticle Delivery Systems to Enable mRNA-Based Therapeutics. Pharmaceutics 2022, 14, 398. [Google Scholar] [CrossRef]
- Glück, R.; Metcalfe, I.C. New technology platforms in the development of vaccines for the future. Vaccine 2002, 20 (Suppl. S5), B10–B16. [Google Scholar] [CrossRef]
- Sanders, R.W.; Vesanen, M.; Schuelke, N.; Master, A.; Schiffner, L.; Kalyanaraman, R.; Paluch, M.; Berkhout, B.; Maddon, P.J.; Olson, W.C.; et al. Stabilization of the soluble, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J. Virol. 2002, 76, 8875–8889. [Google Scholar] [CrossRef] [Green Version]
- Sharma, S.K.; de Val, N.; Bale, S.; Guenaga, J.; Tran, K.; Feng, Y.; Dubrovskaya, V.; Ward, A.B.; Wyatt, R.T. Cleavage-Independent HIV-1 Env Trimers Engineered as Soluble Native Spike Mimetics for Vaccine Design. Cell Rep. 2015, 11, 539–550. [Google Scholar] [CrossRef] [Green Version]
- Ingale, J.; Stano, A.; Guenaga, J.; Sharma, S.K.; Nemazee, D.; Zwick, M.B.; Wyatt, R.T. High-Density Array of Well-Ordered HIV-1 Spikes on Synthetic Liposomal Nanoparticles Efficiently Activate B Cells. Cell Rep. 2016, 15, 1986–1999. [Google Scholar] [CrossRef] [Green Version]
- Hu, J.K.; Crampton, J.C.; Cupo, A.; Ketas, T.; van Gils, M.J.; Sliepen, K.; de Taeye, S.W.; Sok, D.; Ozorowski, G.; Deresa, I.; et al. Murine Antibody Responses to Cleaved Soluble HIV-1 Envelope Trimers Are Highly Restricted in Specificity. J. Virol. 2015, 89, 10383–10398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dubrovskaya, V.; Tran, K.; Ozorowski, G.; Guenaga, J.; Wilson, R.; Bale, S.; Cottrell, C.A.; Turner, H.L.; Seabright, G.; O’Dell, S.; et al. Vaccination with Glycan-Modified HIV NFL Envelope Trimer-Liposomes Elicits Broadly Neutralizing Antibodies to Multiple Sites of Vulnerability. Immunity 2019, 51, 915–929.e7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martinez-Murillo, P.; Tran, K.; Guenaga, J.; Lindgren, G.; Àdori, M.; Feng, Y.; Phad, G.E.; Vázquez Bernat, N.; Bale, S.; Ingale, J.; et al. Particulate Array of Well-Ordered HIV Clade C Env Trimers Elicits Neutralizing Antibodies that Display a Unique V2 Cap Approach. Immunity 2017, 46, 804–817.e7. [Google Scholar] [CrossRef] [Green Version]
- Suleiman, E.; Damm, D.; Batzoni, M.; Temchura, V.; Wagner, A.; Überla, K.; Vorauer-Uhl, K. Electrostatically Driven Encapsulation of Hydrophilic, Non-Conformational Peptide Epitopes into Liposomes. Pharmaceutics 2019, 11, 619. [Google Scholar] [CrossRef] [Green Version]
- Forbes, N.; Hussain, M.T.; Briuglia, M.L.; Edwards, D.P.; Ter Horst, J.H.; Szita, N.; Perrie, Y. Rapid and scale-independent microfluidic manufacture of liposomes entrapping protein incorporating in-line purification and at-line size monitoring. Int. J. Pharm. 2019, 556, 68–81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giraldo, K.A.; Bermudez, J.S.; Torres, C.E.; Reyes, L.H.; Osma, J.F.; Cruz, J.C. Microfluidics for Multiphase Mixing and Liposomal Encapsulation of Nanobioconjugates: Passive vs. Acoustic Systems. Fluids 2021, 6, 309. [Google Scholar] [CrossRef]
- Barnden, M.J.; Allison, J.; Heath, W.R.; Carbone, F.R. Defective TCR expression in transgenic mice constructed using cDNA-based α- and β-chain genes under the control of heterologous regulatory elements. Immunol. Cell Biol. 1998, 76, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Escolano, A.; Steichen, J.M.; Dosenovic, P.; Kulp, D.W.; Golijanin, J.; Sok, D.; Freund, N.T.; Gitlin, A.D.; Oliveira, T.; Araki, T.; et al. Sequential Immunization Elicits Broadly Neutralizing Anti-HIV-1 Antibodies in Ig Knockin Mice. Cell 2016, 166, 1445–1458.e12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ota, T.; Doyle-Cooper, C.; Cooper, A.B.; Doores, K.J.; Aoki-Ota, M.; Le, K.; Schief, W.R.; Wyatt, R.T.; Burton, D.R.; Nemazee, D. B cells from knock-in mice expressing broadly neutralizing HIV antibody b12 carry an innocuous B cell receptor responsive to HIV vaccine candidates. J. Immunol. 2013, 191, 3179–3185. [Google Scholar] [CrossRef] [Green Version]
- Aldon, Y.; McKay, P.F.; Allen, J.; Ozorowski, G.; Felfödiné Lévai, R.; Tolazzi, M.; Rogers, P.; He, L.; de Val, N.; Fábián, K.; et al. Rational Design of DNA-Expressed Stabilized Native-Like HIV-1 Envelope Trimers. Cell Rep. 2018, 24, 3324–3338.e5. [Google Scholar] [CrossRef] [Green Version]
- Bale, S.; Goebrecht, G.; Stano, A.; Wilson, R.; Ota, T.; Tran, K.; Ingale, J.; Zwick, M.B.; Wyatt, R.T. Covalent Linkage of HIV-1 Trimers to Synthetic Liposomes Elicits Improved B Cell and Antibody Responses. J. Virol. 2017, 91, e00443-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suleiman, E.; Mayer, J.; Lehner, E.; Kohlhauser, B.; Katholnig, A.; Batzoni, M.; Damm, D.; Temchura, V.; Wagner, A.; Überla, K.; et al. Conjugation of native-like hiv-1 envelope trimers onto liposomes using edc/sulfo-nhs chemistry: Requirements and limitations. Pharmaceutics 2020, 12, 979. [Google Scholar] [CrossRef] [PubMed]
- Nabi, G.; Genannt Bonsmann, M.; Tenbusch, M.; Gardt, O.; Barouch, D.H.; Temchura, V.; Überla, K. GagPol-specific CD4+ T-cells increase the antibody response to Env by intrastructural help. Retrovirology 2013, 10, 117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kolenbrander, A.; Grewe, B.; Nemazee, D.; Überla, K.; Temchura, V. Generation of T follicular helper cells in vitro: Requirement for B-cell receptor cross-linking and cognate B- and T-cell interaction. Immunology 2018, 153, 214–224. [Google Scholar] [CrossRef] [Green Version]
- Storcksdieck genannt Bonsmann, M.; Niezold, T.; Temchura, V.; Pissani, F.; Ehrhardt, K.; Brown, E.P.; Osei-Owusu, N.Y.; Hannaman, D.; Hengel, H.; Ackerman, M.E.; et al. Enhancing the Quality of Antibodies to HIV-1 Envelope by GagPol-Specific Th Cells. J. Immunol. 2015, 195, 4861–4872. [Google Scholar] [CrossRef]
- Zilker, C.; Kozlova, D.; Sokolova, V.; Yan, H.; Epple, M.; Überla, K.; Temchura, V. Nanoparticle-based B-cell targeting vaccines: Tailoring of humoral immune responses by functionalization with different TLR-ligands. Nanomed. Nanotechnol. Biol. Med. 2017, 13, 173–182. [Google Scholar] [CrossRef]
- Arnold, P.; Himmels, P.; Weiß, S.; Decker, T.-M.; Markl, J.; Gatterdam, V.; Tampé, R.; Bartholomäus, P.; Dietrich, U.; Dürr, R. Antigenic and 3D structural characterization of soluble X4 and hybrid X4-R5 HIV-1 Env trimers. Retrovirology 2014, 11, 42. [Google Scholar] [CrossRef] [Green Version]
- Brouwer, P.J.M.; Antanasijevic, A.; de Gast, M.; Allen, J.D.; Bijl, T.P.L.; Yasmeen, A.; Ravichandran, R.; Burger, J.A.; Ozorowski, G.; Torres, J.L.; et al. Immunofocusing and enhancing autologous Tier-2 HIV-1 neutralization by displaying Env trimers on two-component protein nanoparticles. NPJ Vaccines 2021, 6, 24. [Google Scholar] [CrossRef]
- Xu, X.; Khan, M.A.; Burgess, D.J. Predicting hydrophilic drug encapsulation inside unilamellar liposomes. Int. J. Pharm. 2012, 423, 410–418. [Google Scholar] [CrossRef]
- Martins, K.A.O.; Steffens, J.T.; van Tongeren, S.A.; Wells, J.B.; Bergeron, A.A.; Dickson, S.P.; Dye, J.M.; Salazar, A.M.; Bavari, S. Toll-Like Receptor Agonist Augments Virus-Like Particle-Mediated Protection from Ebola Virus with Transient Immune Activation. PLoS ONE 2014, 9, e89735. [Google Scholar] [CrossRef] [Green Version]
- Feray, A.; Szely, N.; Guillet, E.; Hullo, M.; Legrand, F.-X.; Brun, E.; Pallardy, M.; Biola-Vidamment, A. How to Address the Adjuvant Effects of Nanoparticles on the Immune System. Nanomaterials 2020, 10, 425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Damm, D.; Kostka, K.; Weingärtner, C.; Wagner, J.T.; Rojas-Sánchez, L.; Gensberger-Reigl, S.; Sokolova, V.; Überla, K.; Epple, M.; Temchura, V. Covalent coupling of HIV-1 glycoprotein trimers to biodegradable calcium phosphate nanoparticles via genetically encoded aldehyde-tags. Acta Biomater. 2022, 140, 586–600. [Google Scholar] [CrossRef] [PubMed]
- Sliepen, K.; Han, B.W.; Bontjer, I.; Mooij, P.; Garces, F.; Behrens, A.-J.; Rantalainen, K.; Kumar, S.; Sarkar, A.; Brouwer, P.J.M.; et al. Structure and immunogenicity of a stabilized HIV-1 envelope trimer based on a group-M consensus sequence. Nat. Commun. 2019, 10, 2355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanders, R.W.; Derking, R.; Cupo, A.; Julien, J.-P.; Yasmeen, A.; de Val, N.; Kim, H.J.; Blattner, C.; de la Peña, A.T.; Korzun, J.; et al. A Next-Generation Cleaved, Soluble HIV-1 Env Trimer, BG505 SOSIP.664 gp140, Expresses Multiple Epitopes for Broadly Neutralizing but Not Non-Neutralizing Antibodies. PLoS Pathog. 2013, 9, e1003618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Utz, U.; Britt, W.; Vugler, L.; Mach, M. Identification of a neutralizing epitope on glycoprotein gp58 of human cytomegalovirus. J. Virol. 1989, 63, 1995–2001. [Google Scholar] [CrossRef] [Green Version]

| First-generation T helper liposomes | ||
|---|---|---|
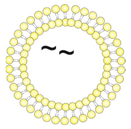 | 1/~//OT2 | Uncoupled liposomes with zeta potential in a neutral range. OT2 peptides (black) were encapsulated by passive inclusion during hydration of the lipid film. |
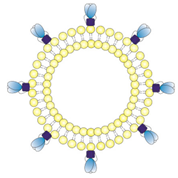 | 1/~/JRFL// | Empty liposomes with zeta potential in a neutral range. JRFL NFL trimers (blue) were coupled to the liposomal surface by interaction of C-terminal His-tags with Ni-NTA lipids. |
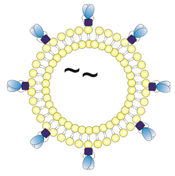 | 1/~/JRFL/OT2 | T helper liposomes with zeta potential in a neutral range. OT2 peptides (black) were encapsulated by passive inclusion during hydration of the lipid film. JRFL NFL trimers (blue) were coupled to the liposomal surface by interaction of C-terminal His-tags with Ni-NTA lipids. |
| Second-generation T helper liposomes | ||
 | 2/-//OT2 | Uncoupled liposomes with an anionic zeta potential. OT2 peptides (black) were quantitatively encapsulated by an electrostatically driven approach. |
 | 2/-/SUFO/OT2 | T helper liposomes with an anionic zeta potential. OT2 peptides (black) were encapsulated by an electrostatically driven approach. conSOSL.UFO.664 trimers (red) were covalently coupled to the liposomal surface by His-tag / Ni-NTA interaction followed by EDC / Sulfo-NHS crosslinking. |
| Lentiviral T helper VLPs | ||
 | SUFO-OT2-VLP | T helper VLPs that display conSOSL.UFO.750 trimers (SUFO.750, red) and 293T producer cell-derived proteins (black) on the surface and encapsulate both HIV-1 capsid proteins (grey) and OT2 peptides (black). |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Damm, D.; Suleiman, E.; Theobald, H.; Wagner, J.T.; Batzoni, M.; Ahlfeld, B.; Walkenfort, B.; Albrecht, J.-C.; Ingale, J.; Yang, L.; et al. Design and Functional Characterization of HIV-1 Envelope Protein-Coupled T Helper Liposomes. Pharmaceutics 2022, 14, 1385. https://doi.org/10.3390/pharmaceutics14071385
Damm D, Suleiman E, Theobald H, Wagner JT, Batzoni M, Ahlfeld B, Walkenfort B, Albrecht J-C, Ingale J, Yang L, et al. Design and Functional Characterization of HIV-1 Envelope Protein-Coupled T Helper Liposomes. Pharmaceutics. 2022; 14(7):1385. https://doi.org/10.3390/pharmaceutics14071385
Chicago/Turabian StyleDamm, Dominik, Ehsan Suleiman, Hannah Theobald, Jannik T. Wagner, Mirjam Batzoni, Bianca Ahlfeld (née Kohlhauser), Bernd Walkenfort, Jens-Christian Albrecht, Jidnyasa Ingale, Lifei Yang, and et al. 2022. "Design and Functional Characterization of HIV-1 Envelope Protein-Coupled T Helper Liposomes" Pharmaceutics 14, no. 7: 1385. https://doi.org/10.3390/pharmaceutics14071385
APA StyleDamm, D., Suleiman, E., Theobald, H., Wagner, J. T., Batzoni, M., Ahlfeld, B., Walkenfort, B., Albrecht, J.-C., Ingale, J., Yang, L., Hasenberg, M., Wyatt, R. T., Vorauer-Uhl, K., Überla, K., & Temchura, V. (2022). Design and Functional Characterization of HIV-1 Envelope Protein-Coupled T Helper Liposomes. Pharmaceutics, 14(7), 1385. https://doi.org/10.3390/pharmaceutics14071385






