Pre-Clinical In-Vitro Studies on Parameters Governing Immune Complex Formation
Abstract
1. Introduction
2. Materials and Methods
2.1. Material
2.2. Sample Preparation
2.3. Preparation of Antigen-Drug and Immune Complexes (ICs)
2.4. (Differential) Size Exclusion Chromatography (dSEC)
2.5. Size Exclusion Chromatography with Multi Angle Light Scattering Assessment (SEC-MALS)
2.6. Sedimentation Velocity-Analytical Ultracentrifugation (SV-AUC)
2.7. Mass Photometry
2.8. Dynamic Light Scattering (DLS)
3. Results
3.1. Setup of the Analytical Study
3.2. Assessment of Basic Interactions Relevant for Initiation of Immune Complex Formation
3.3. Assessment of Immune Complexes in Presence of Polyclonal Anti-Biologic Antibodies
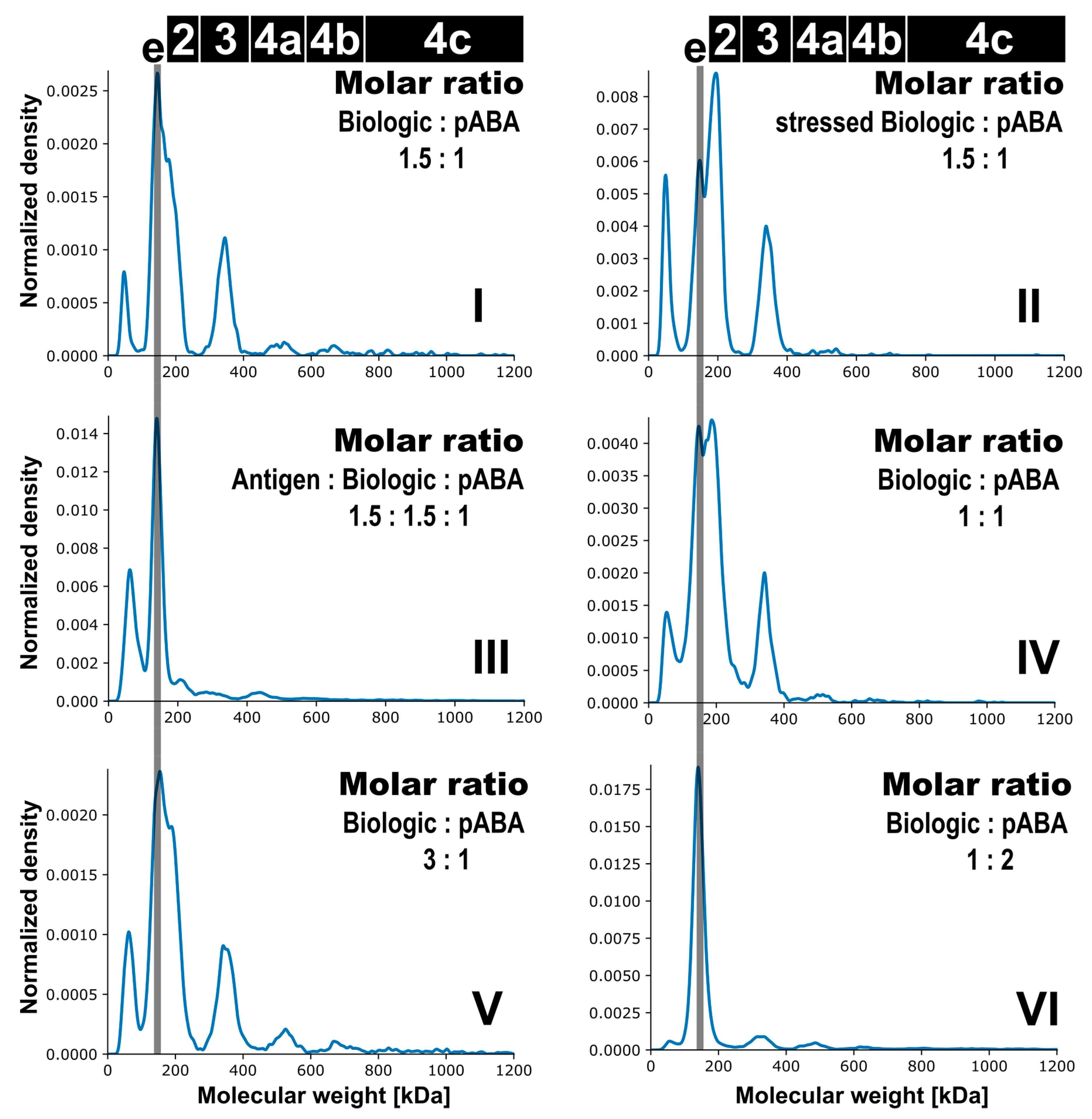
3.4. Assessment of Immune Complexes in Presence of Stressed Biologic Material
3.5. Assessment of Immune Complexes at Low and Physiological Concentrations
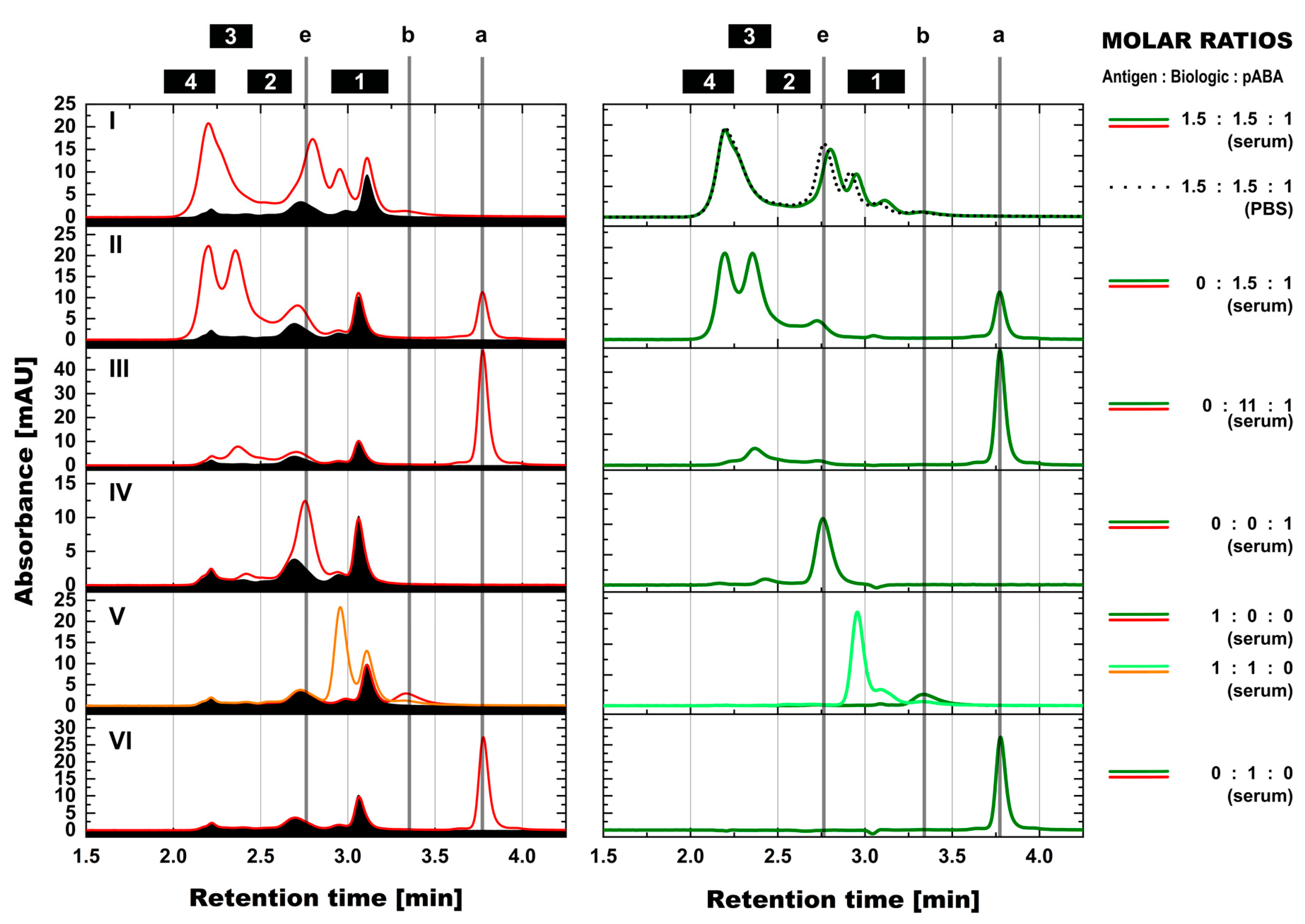
3.6. Assessment of Immune Complexes in Serum Background
4. Discussion
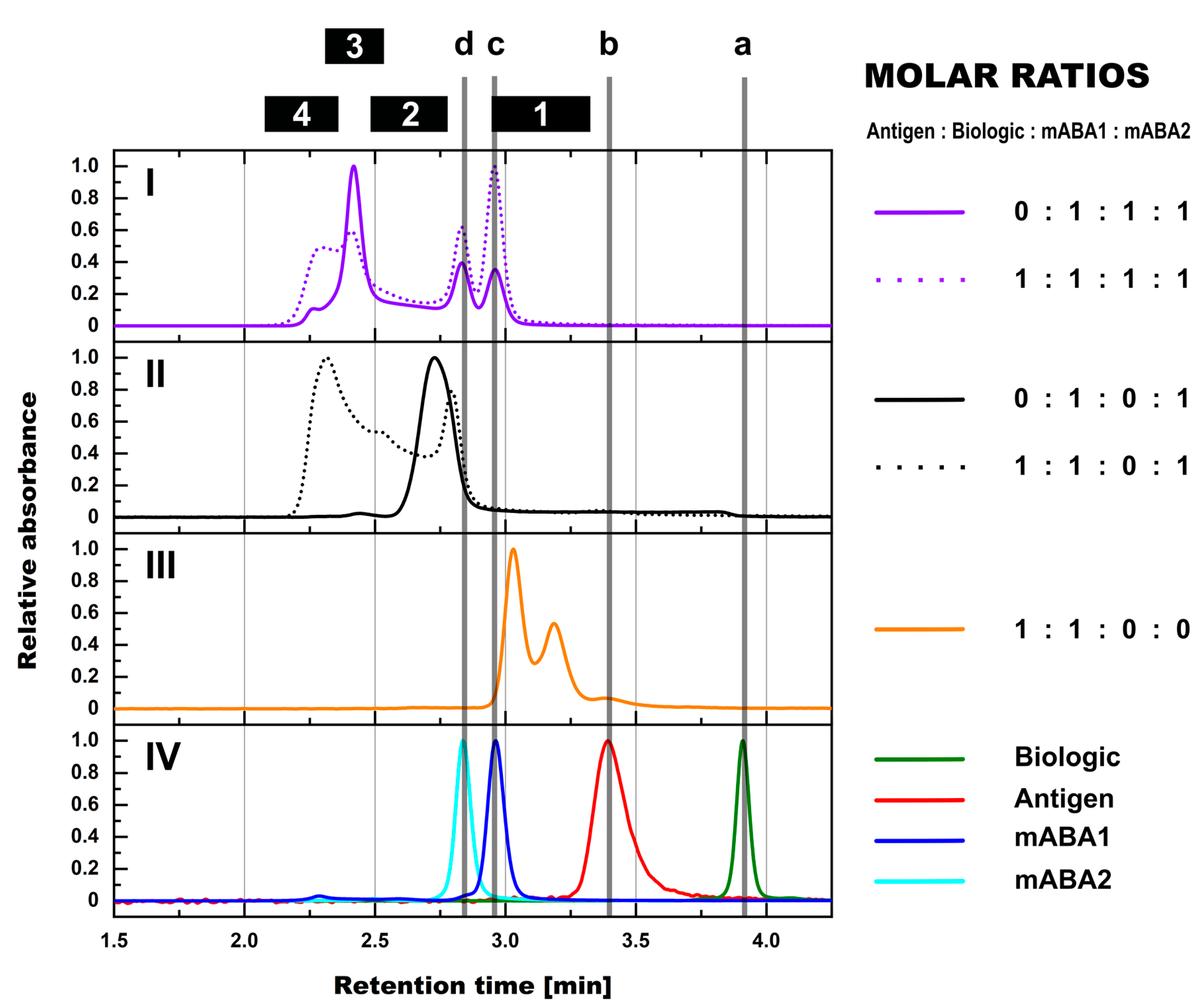
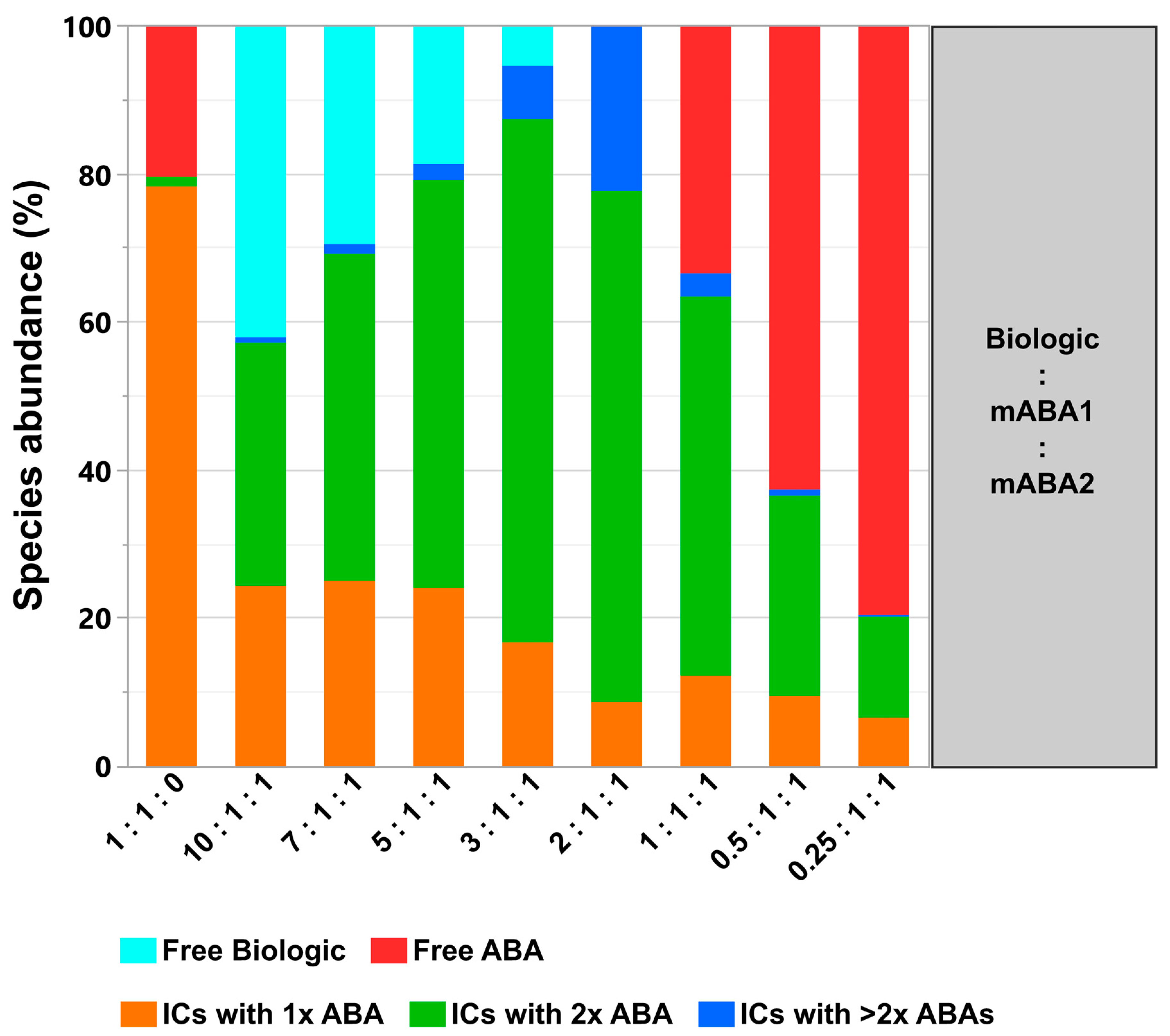
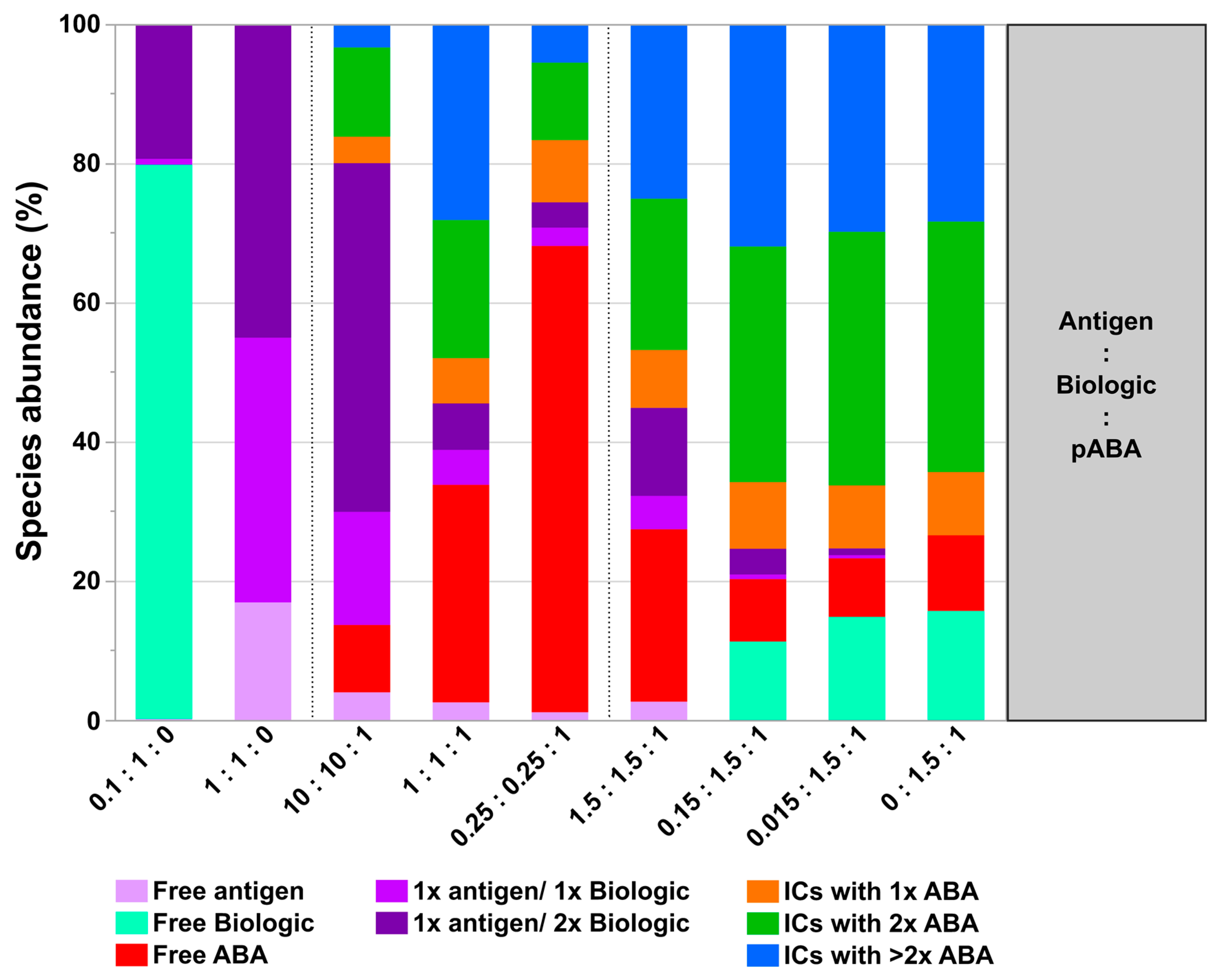
- The most simple test concept between a monomeric biotherapeutic and anti-biotherapeutic mAbs highlights the necessity for having at least two anti-biotherapeutic antibodies, which recognize different and non-overlapping epitopes, to generate ICs larger than a 1:1 biotherapeutic:Ab complex. Alternatively, for multimeric biotherapeutics like therapeutic mAbs, an ADA against a single epitope on the therapeutic mAb is sufficient for the formation of high MW ICs by cross-linking of multiple therapeutic Abs and multiple ADAs. As expected, for the monomeric biologic used in this study, in the presence of two mABAs, which recognize non-overlapping epitopes, the highest amount of ICs (=full conversion of single components) was observed for the equimolar 2:1:1 molar ratio (Figure 10). Under these conditions, also the amount of ICs with higher MW was largest, but decreased dramatically with a molar excess of either the biologic or the mABAs.
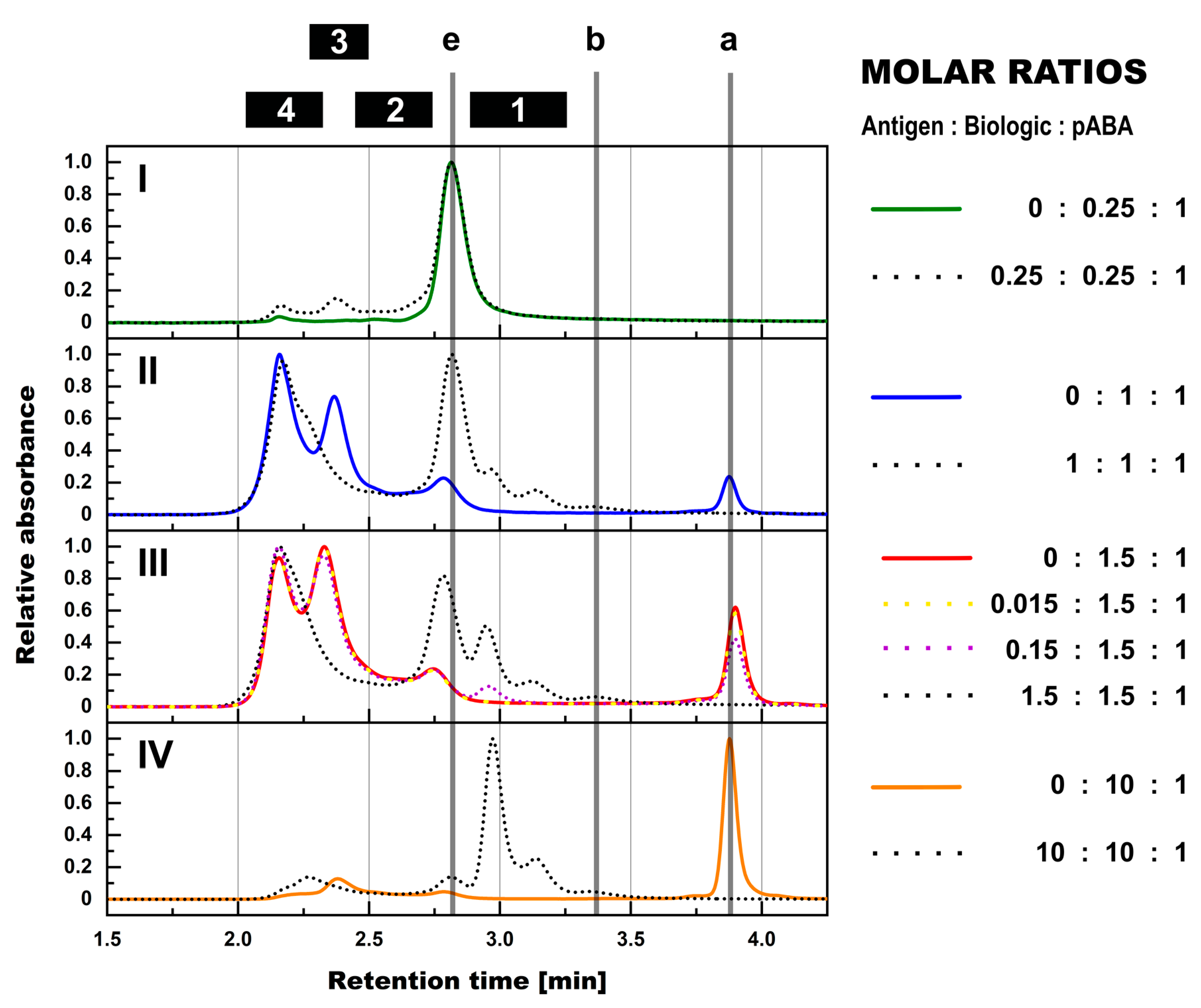
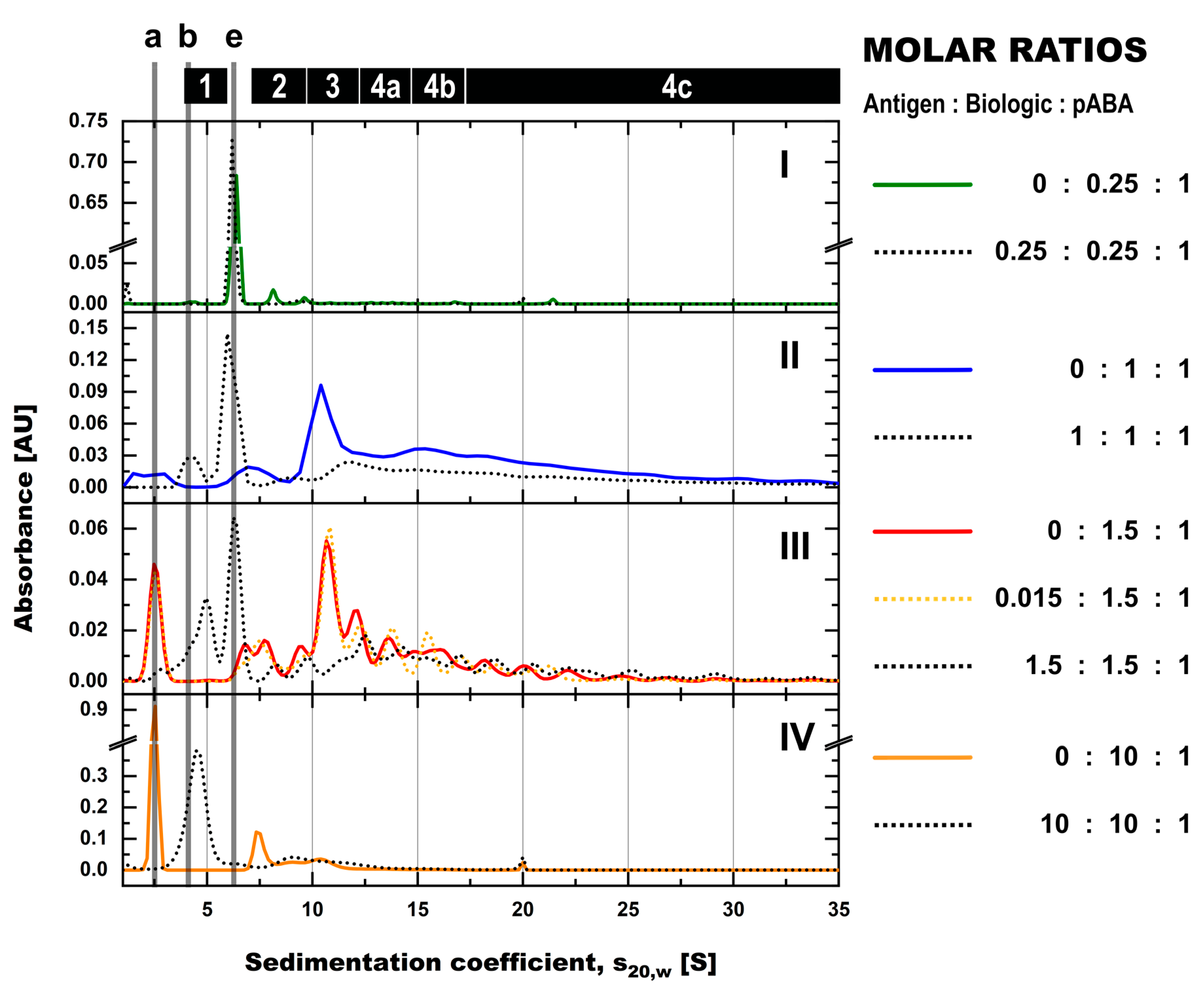
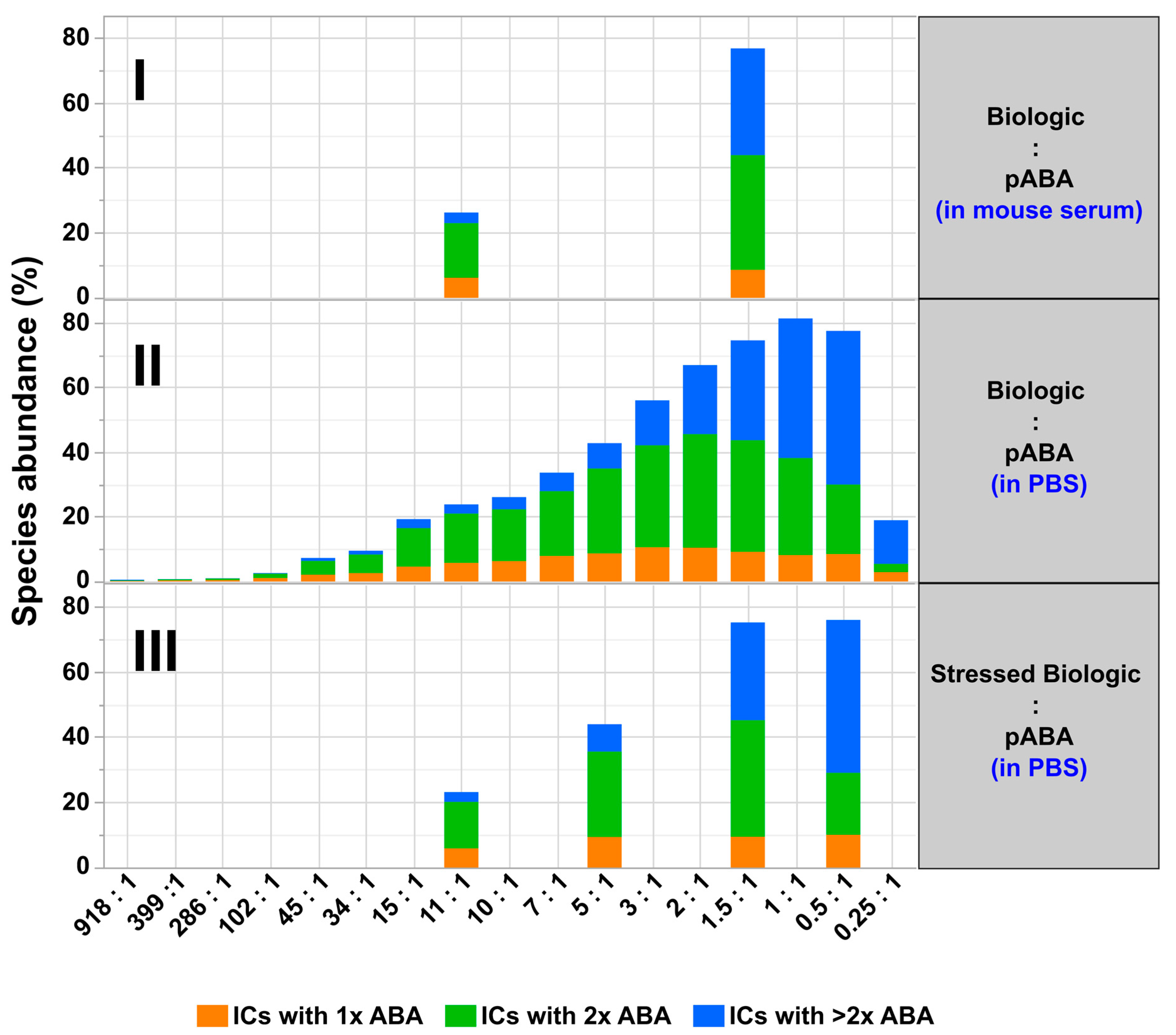
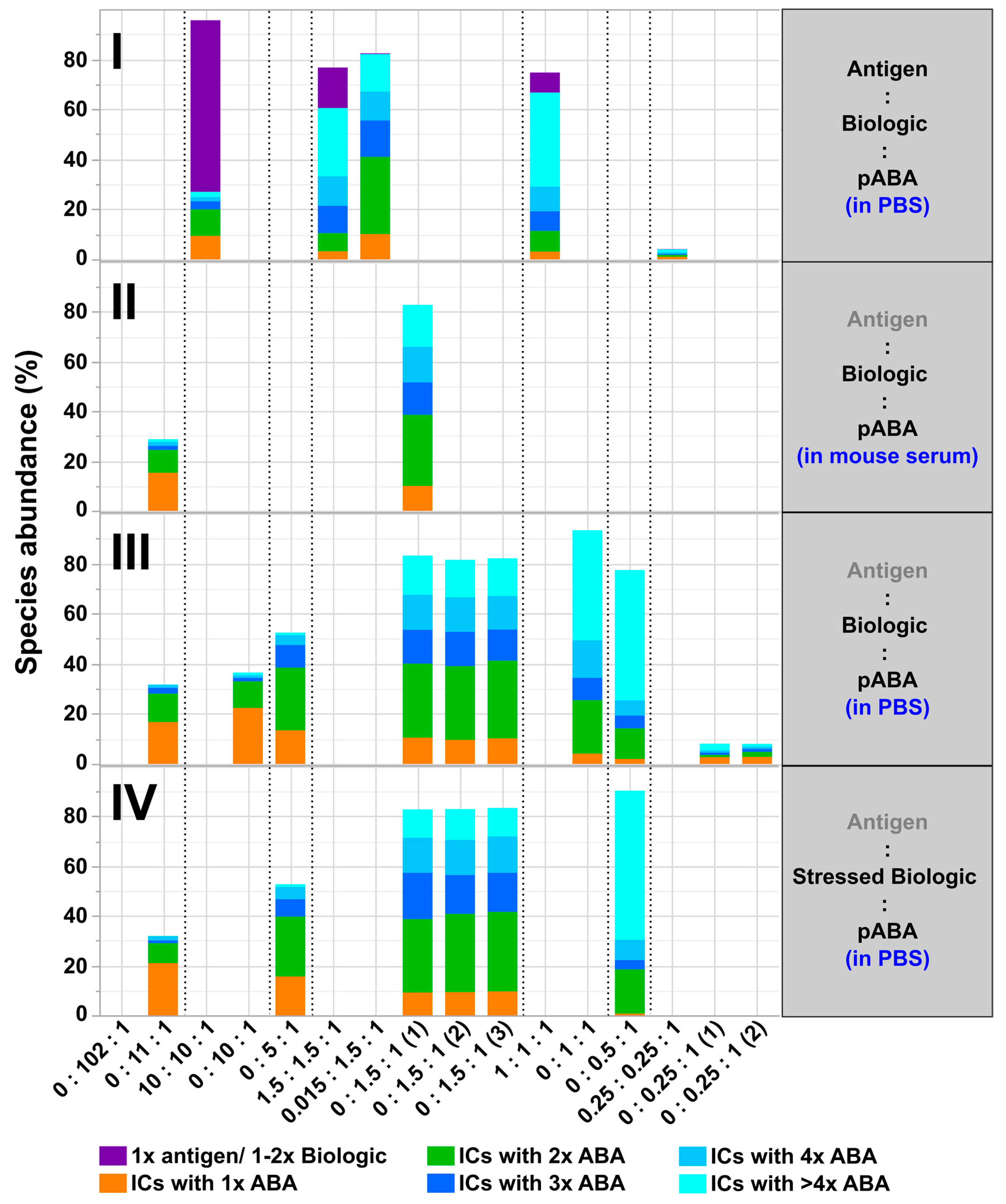
- The expansion of the simplistic model through the exchange of mABAs by surrogate pABAs, which have the potential to cover a broader epitope space, increases the number of options to generate new and more complex forms of ICs. In our hands, the highest levels of ICs were observed at or near to equimolarity between the biologic and the pABA (Figure 11 and Figure 12) in agreement with published data on other IC systems [23]. Our experiments indicate that the most stable complex is formed by cross-stabilization of IC species containing 2 ABAs, presumably avidity driven by all four Fab-arms being involved in the interaction with the biologic molecules. Surprisingly, a non-Gaussian distribution of IC content was observed measuring the abundance of ICs at various molar ratios between the biologic and pABA (Figure 11). The presence of ICs could be detected even at a ~1000-fold molar excess of the biologic over pABA. In contrast, the content of ICs decreased rapidly with molar excess of pABA over the biologic and could not be detected below an excess of pABA of 5-fold (Figure 11 and Figure 12). The ratio between the components was not only dictating the conversion rate of free components to ICs, but had a significant impact on the dynamic distribution between the individual IC species. The amount of smaller ICs, which contain one to two pABA molecules, was highest at a slight molar excess of the biologic (2:1 ratio), and were generally the dominating species at molar excess of the biologic. In contrast, the larger ICs (≥3 pABAs) became the dominant species with molar excess of pABA over the Biologic. The highest amount of larger ICs was observed for the 0.5:1 ratio at non-physiological concentrations.
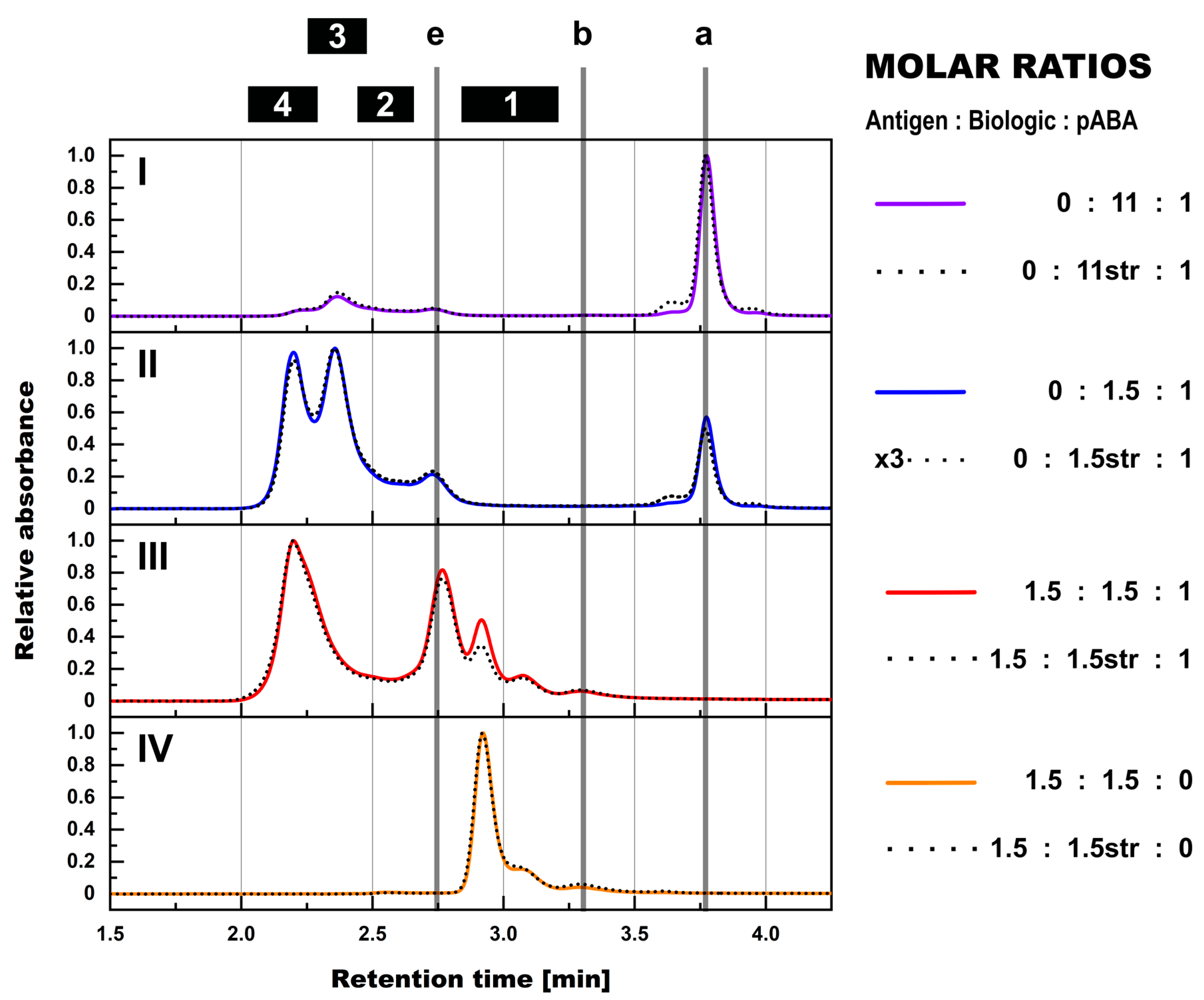
- Expanding the biologic:pABA system by the inclusion of a dimeric antigen provided insights into additional factors contributing to IC formation (Figure 12 and Figure 13). Our experiments demonstrate that the antigen was competing with sub-populations of pABA for free and complexed biologic molecules. In the presence of free biologic material (pABA being the limiting factor), antigen:biologic complexes were formed. In parallel, antigen molecules were getting incorporated into larger biologic:pABA complexes (ICs containing >2 pABA molecules). This led to an increased amount of these species at the expense of smaller ICs (containing ≤2 pABA molecules). Disintegration of the latter with subsequent freeing-up of non-incorporated pABAs was the observed result (Figure 3 and Figure 4). The preferable disintegration of smaller ICs by the antigen indicates an involvement of the biologic’s CDRs in the competitive binding between the antigen and the pABA species, which were involved in the smaller ICs, and/or conformational restrictions imposed by bound antigen. On the other hand, the incorporation of antigen molecules into larger ICs suggests mainly a non-idiotype-driven interaction of pABA species with the biologic in such complexes. Additionally, cross-linking of two biologic molecules by the dimeric antigen provided a two-fold increase in binding sites for pABAs (with exclusion of the CDR region), thereby allowing a higher degree of complexation and new conformational orientations between the components. These larger complexes must be of random conformation and not well structured. The absence of preferred and conformationally stable high MW IC species was detected by strong variability in the amount of individual IC species containing three and more pABAs (Figure 6, triplicate analysis of same composition by SV-AUC). The significance of the antigen for the formation of ICs should not be overestimated. For most biotherapeutics with the aim to neutralize a pharmacological target, the concentration of the administered biologic in the patient is often orders of magnitude higher as compared to the concentration of the endogenous target. The in-vitro experiments show that a reduction in the molar concentration of the antigen in the antigen:biologic:pABA compositions from an equimolar ratio to 10% (=0.1:1 antigen:biologic ratio) strongly reduced the impact of the antigen on the IC composition (Figure 13). A reduction to 1% led to a negligible impact of the antigen on the IC formation.
- In addition to the molar ratio, the absolute concentration of the components is a critical factor for IC formation. The analysis of ICs at low µmolar to high nanomolar (SEC and SV-AUC results) and at low nanomolar (mass photometry) concentrations showed a direct correlation between the concentration and the amount of formed ICs. Hereby, the reduction was relatively more pronounced for the amount of larger ICs (>2 pABAs incorporated) compared to the amount of smaller ICs. The analysis of ICs at low nanomolar concentrations is generally preferable due to a higher relevance for physiological conditions. A significant factor with the potential to impact IC formation and its composition is the KD of ADA molecules to the corresponding target biotherapeutic. At least for low-affinity ADAs (early during the immune response), the KD is reported to be in the medium to low nanomolar range [32]. Not being in the scope for this study, it can be envisioned that fitting of such experimental IC formation data will be of benefit for setting up adverse events evading drug dosing regimens. A direct analysis of the ICs at physiological concentrations without modification of the involved components is a breakthrough and although requiring further refinement, i.e., increasing the precision for the quantification of individual IC species, the new approach we used in this work will significantly change analytics in the field of (immune) complex formation.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- De la Torre, B.G.; Albericio, F. The pharmaceutical industry in 2020. An analysis of fda drug approvals from the perspective of molecules. Molecules 2021, 26, 627. [Google Scholar] [CrossRef] [PubMed]
- Garcês, S.; Demengeot, J. The Immunogenicity of Biologic Therapies. Curr. Probl. Dermatol. 2017, 53, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Khoja, L.; Day, D.; Wei-Wu Chen, T.; Siu, L.L.; Hansen, A.R. Tumour- and class-specific patterns of immune-related adverse events of immune checkpoint inhibitors: A systematic review. Ann. Oncol. 2017, 28, 2377–2385. [Google Scholar] [CrossRef] [PubMed]
- U.S. Food and Drug Administration. FDA Guidance for Industry: Immunogenicity Assessment for Therapeutic Protein Products; U.S. Food and Drug Administration: Silver Spring, MD, USA, 2014.
- Committee for Medicinal Products for Human Use. EMEA Guidance Document CHMP Guideline on Immunogenicity Assessment of Biotechnology-Derived Therapeutic Proteins; European Medicines Agency: London, UK, 2008. [Google Scholar]
- Warncke, M.; Calzascia, T.; Coulot, M.; Balke, N.; Touil, R.; Kolbinger, F.; Heusser, C. Different Adaptations of IgG Effector Function in Human and Nonhuman Primates and Implications for Therapeutic Antibody Treatment. J. Immunol. 2012, 188, 4405–4411. [Google Scholar] [CrossRef]
- Van Norman, G.A. Drugs, Devices, and the FDA: Part 1: An Overview of Approval Processes for Drugs. JACC Basic Transl. Sci. 2016, 1, 170–179. [Google Scholar] [CrossRef]
- Brown, D.G.; Wobst, H.J.; Kapoor, A.; Kenna, L.A.; Southall, N. Clinical development times for innovative drugs. Nat. Rev. Drug Discov. 2021. ahead of print. [Google Scholar] [CrossRef]
- Lorenz, T.; Fiaux, J.; Heitmann, D.; Gupta, K.; Kocher, H.P.; Knopf, H.P.; Hartmann, S. Developability assessment of biologics by integrated biologics profiling. Am. Pharm. Rev. 2014, 17. [Google Scholar]
- Xu, Y.; Wang, D.; Mason, B.; Rossomando, T.; Li, N.; Liu, D.; Cheung, J.K.; Xu, W.; Raghava, S.; Katiyar, A.; et al. Structure, heterogeneity and developability assessment of therapeutic antibodies. In MAbs; Taylor & Francis: Abingdon, UK, 2019; Volume 11, pp. 239–264. [Google Scholar] [CrossRef]
- Doneva, N.; Doytchinova, I.; Dimitrov, I. Predicting immunogenicity risk in biopharmaceuticals. Symmetry 2021, 13, 388. [Google Scholar] [CrossRef]
- Karle, A.C. Applying MAPPs Assays to Assess Drug Immunogenicity. Front. Immunol. 2020, 11, 698. [Google Scholar] [CrossRef]
- Rombach-Riegraf, V.; Karle, A.C.; Wolf, B.; Sordé, L.; Koepke, S.; Gottlieb, S.; Krieg, J.; Djidja, M.C.; Baban, A.; Spindeldreher, S.; et al. Aggregation of human recombinant monoclonal antibodies influences the capacity of dendritic cells to stimulate adaptive T-cell responses in vitro. PLoS ONE 2014, 9, e86322. [Google Scholar] [CrossRef]
- Kroenke, M.A.; Milton, M.N.; Kumar, S.; Bame, E.; White, J.T. Immunogenicity Risk Assessment for Multi-specific Therapeutics. AAPS J. 2021, 23, 115. [Google Scholar] [CrossRef] [PubMed]
- Schuster, J.; Koulov, A.; Mahler, H.C.; Detampel, P.; Huwyler, J.; Singh, S.; Mathaes, R. In Vivo Stability of Therapeutic Proteins. Pharm. Res. 2020, 37, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Krishna, M.; Nadler, S.G. Immunogenicity to biotherapeutics—The role of anti-drug immune complexes. Front. Immunol. 2016, 7, 21. [Google Scholar] [CrossRef] [PubMed]
- Hock, B.D.; McKenzie, J.L.; Strother, M.; Goddard, L.; Butt, L.; Currie, M.J. Functional effects of immune complexes formed between pembrolizumab and patient-generated anti-drug antibodies. Cancer Immunol. Immunother. 2020, 69, 2453–2464. [Google Scholar] [CrossRef] [PubMed]
- Chirmule, N.; Jawa, V.; Meibohm, B. Immunogenicity to therapeutic proteins: Impact on PK/PD and efficacy. AAPS J. 2012, 14, 296–302. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Piszczek, G. Standard protocol for mass photometry experiments. Eur. Biophys. J. 2021, 50, 403–409. [Google Scholar] [CrossRef]
- Lobo, S.A.; Bączyk, P.; Wyss, B.; Widmer, J.C.; Jesus, L.P.; Gomes, J.; Batista, A.P.; Hartmann, S.; Wassmann, P. Stability liabilities of biotherapeutic proteins: Early assessment as mitigation strategy. J. Pharm. Biomed. Anal. 2021, 192, 113650. [Google Scholar] [CrossRef]
- Harding, F.A.; Stickler, M.M.; Razo, J.; DuBridge, R.B. The immunogenicity of humanized and fully human antibodies: Residual immunogenicity resides in the CDR regions. MAbs 2010, 2, 256–265. [Google Scholar] [CrossRef]
- Lundahl, M.L.E.; Fogli, S.; Colavita, P.E.; Scanlan, E.M. Aggregation of protein therapeutics enhances their immunogenicity: Causes and mitigation strategies. RSC Chem. Biol. 2021, 2, 1004–1020. [Google Scholar] [CrossRef]
- Van Schie, K.A.; Kruithof, S.; De Heer, P.O.; Derksen, N.I.L.; Van De Bovenkamp, F.S.; Saris, A.; Vidarsson, G.; Bentlage, A.E.H.; Jiskoot, W.; Romeijn, S.; et al. Restricted immune activation and internalisation of anti-idiotype complexes between drug and antidrug antibodies. Ann. Rheum. Dis. 2018, 77, 1471–1479. [Google Scholar] [CrossRef]
- Lebedeva, M.A.; Palmieri, E.; Kukura, P.; Fletcher, S.P. Emergence and Rearrangement of Dynamic Supramolecular Aggregates Visualized by Interferometric Scattering Microscopy. ACS Nano 2020, 14, 11160–11168. [Google Scholar] [CrossRef] [PubMed]
- Demeule, B.; Shire, S.J.; Liu, J. A therapeutic antibody and its antigen form different complexes in serum than in phosphate-buffered saline: A study by analytical ultracentrifugation. Anal. Biochem. 2009, 388, 279–287. [Google Scholar] [CrossRef] [PubMed]
- Baarsma, G.S.; Luyendijk, L.; Kijlstra, A.; De Vries, J.; Peperkamp, E.; Mertens, D.A.E.; Van Meurs, J.C. Analysis of local antibody production in the vitreous humor of patients with severe uveitis. Am. J. Ophthalmol. 1991, 112, 147–150. [Google Scholar] [CrossRef]
- Schuster, J.; Mahler, H.C.; Joerg, S.; Huwyler, J.; Mathaes, R. Analytical Challenges Assessing Protein Aggregation and Fragmentation Under Physiologic Conditions. J. Pharm. Sci. 2021, 110, 3103–3110. [Google Scholar] [CrossRef] [PubMed]
- Krieckaert, C.; Rispens, T.; Wolbink, G. Immunogenicity of biological therapeutics: From assay to patient. Curr. Opin. Rheumatol. 2012, 24, 306–311. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; de Jong, R.N.; van den Bremer, E.T.J.; Beurskens, F.J.; Labrijn, A.F.; Ugurlar, D.; Gros, P.; Schuurman, J.; Parren, P.W.H.I.; Heck, A.J.R. Molecular Basis of Assembly and Activation of Complement Component C1 in Complex with Immunoglobulin G1 and Antigen. Mol. Cell 2016, 63, 135–145. [Google Scholar] [CrossRef]
- Opolka-Hoffmann, E.; Jordan, G.; Otteneder, M.; Kieferle, R.; Lechmann, M.; Winter, G.; Staack, R.F. The impact of immunogenicity on therapeutic antibody pharmacokinetics: A preclinical evaluation of the effect of immune complex formation and antibody effector function on clearance. In MAbs; Taylor & Francis: Abingdon, UK, 2021; Volume 13. [Google Scholar] [CrossRef]
- Vaisman-Mentesh, A.; Gutierrez-Gonzalez, M.; DeKosky, B.J.; Wine, Y. The Molecular Mechanisms That Underlie the Immune Biology of Anti-drug Antibody Formation Following Treatment With Monoclonal Antibodies. Front. Immunol. 2020, 11, 1951. [Google Scholar] [CrossRef]
- Li, J.; Schantz, A.; Schwegler, M.; Shankar, G. Detection of low-affinity anti-drug antibodies and improved drug tolerance in immunogenicity testing by Octet® biolayer interferometry. J. Pharm. Biomed. Anal. 2011, 54, 286–294. [Google Scholar] [CrossRef]
- Gandhi, A.V.; Pothecary, M.R.; Bain, D.L.; Carpenter, J.F. Some Lessons Learned from a Comparison Between Sedimentation Velocity Analytical Ultracentrifugation and Size Exclusion Chromatography to Characterize and Quantify Protein Aggregates. J. Pharm. Sci. 2017, 106, 2178–2186. [Google Scholar] [CrossRef]
- Gorbet, G.E.; Mohapatra, S.; Demeler, B. Multi-speed sedimentation velocity implementation in UltraScan-III. Eur. Biophys. J. 2018, 47, 825–835. [Google Scholar] [CrossRef]
- Stafford, W.F.; Braswell, E.H. Sedimentation velocity, multi-speed method for analyzing polydisperse solutions. Biophys. Chem. 2004, 108, 273–279. [Google Scholar] [CrossRef] [PubMed]
- Gorbet, G.E.; Pearson, J.Z.; Demeler, A.K.; Cölfen, H.; Demeler, B. Next-Generation AUC: Analysis of Multiwavelength Analytical Ultracentrifugation Data. Methods Enzymol. 2015, 562, 27–47. [Google Scholar] [CrossRef] [PubMed]
- Walter, J.; Sherwood, P.J.; Lin, W.; Segets, D.; Stafford, W.F.; Peukert, W. Simultaneous Analysis of Hydrodynamic and Optical Properties Using Analytical Ultracentrifugation Equipped with Multiwavelength Detection. Anal. Chem. 2015, 87, 3396–3403. [Google Scholar] [CrossRef] [PubMed]
- Padrick, S.B.; Deka, R.K.; Chuang, J.L.; Wynn, R.M.; Chuang, D.T.; Norgard, M.V.; Rosen, M.K.; Brautigam, C.A. Determination of protein complex stoichiometry through multisignal sedimentation velocity experiments. Anal. Biochem. 2010, 407, 89–103. [Google Scholar] [CrossRef][Green Version]
- Padrick, S.B.; Brautigam, C.A. Evaluating the stoichiometry of macromolecular complexes using multisignal sedimentation velocity. Methods 2011, 54, 39–55. [Google Scholar] [CrossRef][Green Version]
- Brautigam, C.A.; Padrick, S.B.; Schuck, P. Multi-Signal Sedimentation Velocity Analysis with Mass Conservation for Determining the Stoichiometry of Protein Complexes. PLoS ONE 2013, 8, e62694. [Google Scholar] [CrossRef]
- den Boer, M.A.; Lai, S.-H.; Xue, X.; van Kampen, M.D.; Bleijlevens, B.; Heck, A.J.R. Comparative Analysis of Antibodies and Heavily Glycosylated Macromolecular Immune Complexes by Size-Exclusion Chromatography Multi-Angle Light Scattering, Native Charge Detection Mass Spectrometry, and Mass Photometry. Anal. Chem. 2022, 94, 892–900. [Google Scholar] [CrossRef]
- Fischer, K.; Schmidt, M. Pitfalls and novel applications of particle sizing by dynamic light scattering. Biomaterials 2016, 98, 79–91. [Google Scholar] [CrossRef]
- Velichko, E.; Makarov, S.; Nepomnyashchaya, E.; Dong, G. Molecular Aggregation in Immune System Activation Studied by Dynamic Light Scattering. Biology 2020, 9, 123. [Google Scholar] [CrossRef]
- Wright, R.T.; Hayes, D.B.; Stafford, W.F.; Sherwood, P.J.; Correia, J.J. Characterization of therapeutic antibodies in the presence of human serum proteins by AU-FDS analytical ultracentrifugation. Anal. Biochem. 2018, 550, 72–83. [Google Scholar] [CrossRef]
- Ducret, A.; Ackaert, C.; Bessa, J.; Bunce, C.; Hickling, T.; Jawa, V.; Kroenke, M.A.; Lamberth, K.; Manin, A.; Penny, H.L.; et al. Assay format diversity in pre-clinical immunogenicity risk assessment: Toward a possible harmonization of antigenicity assays. In MAbs; Taylor & Francis: Abingdon, UK, 2022; Volume 14. [Google Scholar] [CrossRef]
- Schuster, J.; Kamuju, V.; Mathaes, R. Assessment of antibody stability in a novel protein-free serum model. Pharmaceutics 2021, 13, 774. [Google Scholar] [CrossRef] [PubMed]
- Pierog, P.; Krishna, M.; Yamniuk, A.; Chauhan, A.; DeSilva, B. Detection of drug specific circulating immune complexes from in vivo cynomolgus monkey serum samples. J. Immunol. Methods 2015, 416, 124–136. [Google Scholar] [CrossRef] [PubMed]
- Boysen, M.; Schlicksupp, L.; Dreher, I.; Loebbert, R.; Richter, M. SEC Based Method for Size Determination of Immune Complexes of Therapeutic Antibodies in Animal Matrix. J. Immunol. Res. 2016, 2016, 9096059. [Google Scholar] [CrossRef]
- Collins, P.W.; Mathias, M.; Hanley, J.; Keeling, D.; Keenan, R.; Laffan, M.; Perry, D.; Liesner, R. Rituximab and immune tolerance in severe hemophilia A: A consecutive national cohort. J. Thromb. Haemost. 2009, 7, 787–794. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fichter, M.; Richter, G.; Bepperling, A.; Wassmann, P. Pre-Clinical In-Vitro Studies on Parameters Governing Immune Complex Formation. Pharmaceutics 2022, 14, 1254. https://doi.org/10.3390/pharmaceutics14061254
Fichter M, Richter G, Bepperling A, Wassmann P. Pre-Clinical In-Vitro Studies on Parameters Governing Immune Complex Formation. Pharmaceutics. 2022; 14(6):1254. https://doi.org/10.3390/pharmaceutics14061254
Chicago/Turabian StyleFichter, Marie, Gesa Richter, Alexander Bepperling, and Paul Wassmann. 2022. "Pre-Clinical In-Vitro Studies on Parameters Governing Immune Complex Formation" Pharmaceutics 14, no. 6: 1254. https://doi.org/10.3390/pharmaceutics14061254
APA StyleFichter, M., Richter, G., Bepperling, A., & Wassmann, P. (2022). Pre-Clinical In-Vitro Studies on Parameters Governing Immune Complex Formation. Pharmaceutics, 14(6), 1254. https://doi.org/10.3390/pharmaceutics14061254





