Nobiletin as a Neuroprotectant against NMDA Receptors: An In Silico Approach
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sequence Retrieval and Analysis
2.2. Sequence Alignment and Phylogenetic Analysis
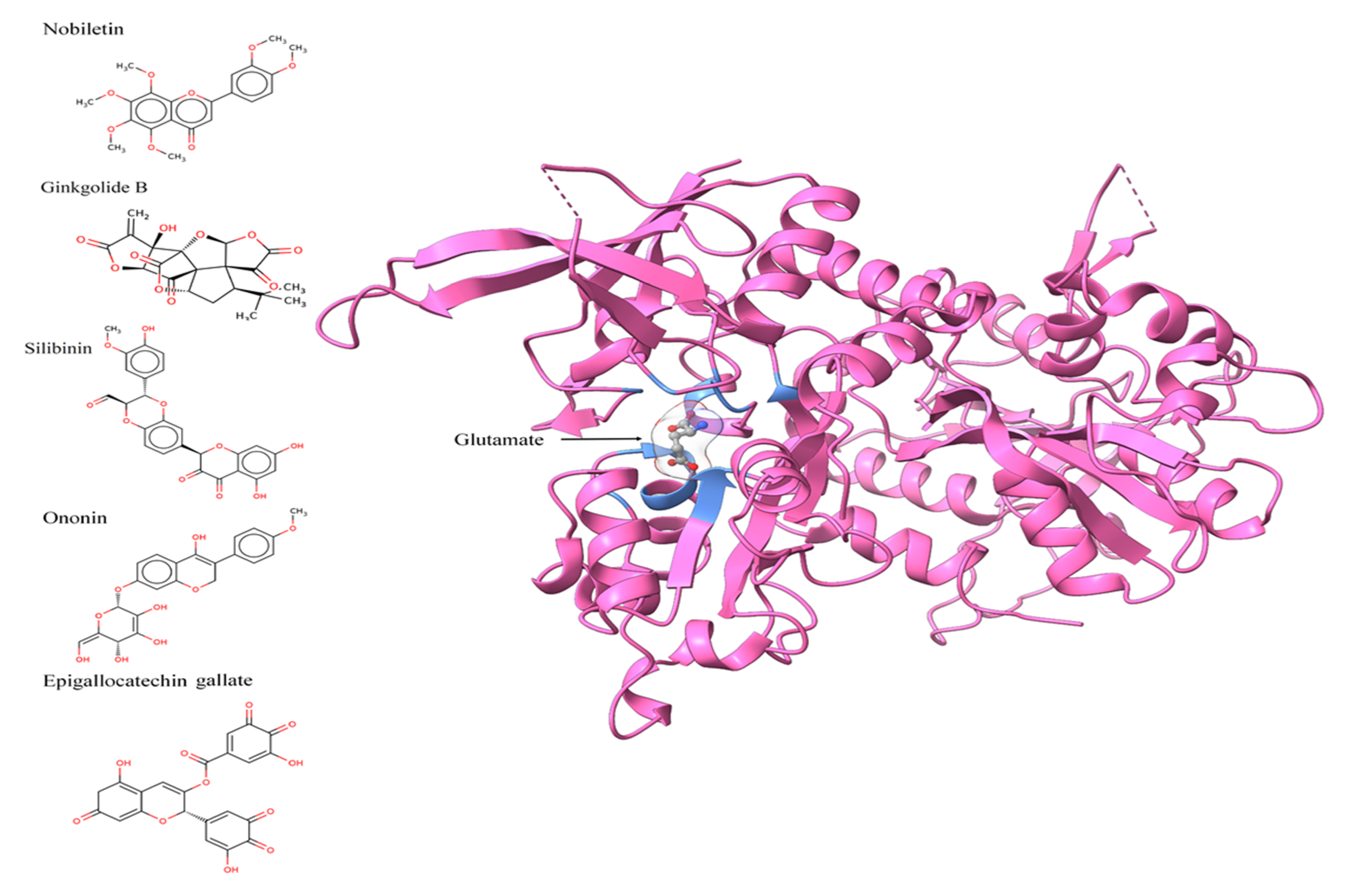
2.3. Protein Structure, Chemical/Ligand Retrieval, and Analysis
2.4. Molecular Docking
2.5. Molecular Dynamic Simulation
2.6. Protein–Ligand Interactions and ADMET Analysis
3. Results
3.1. Sequence and Phylogenetic Analysis
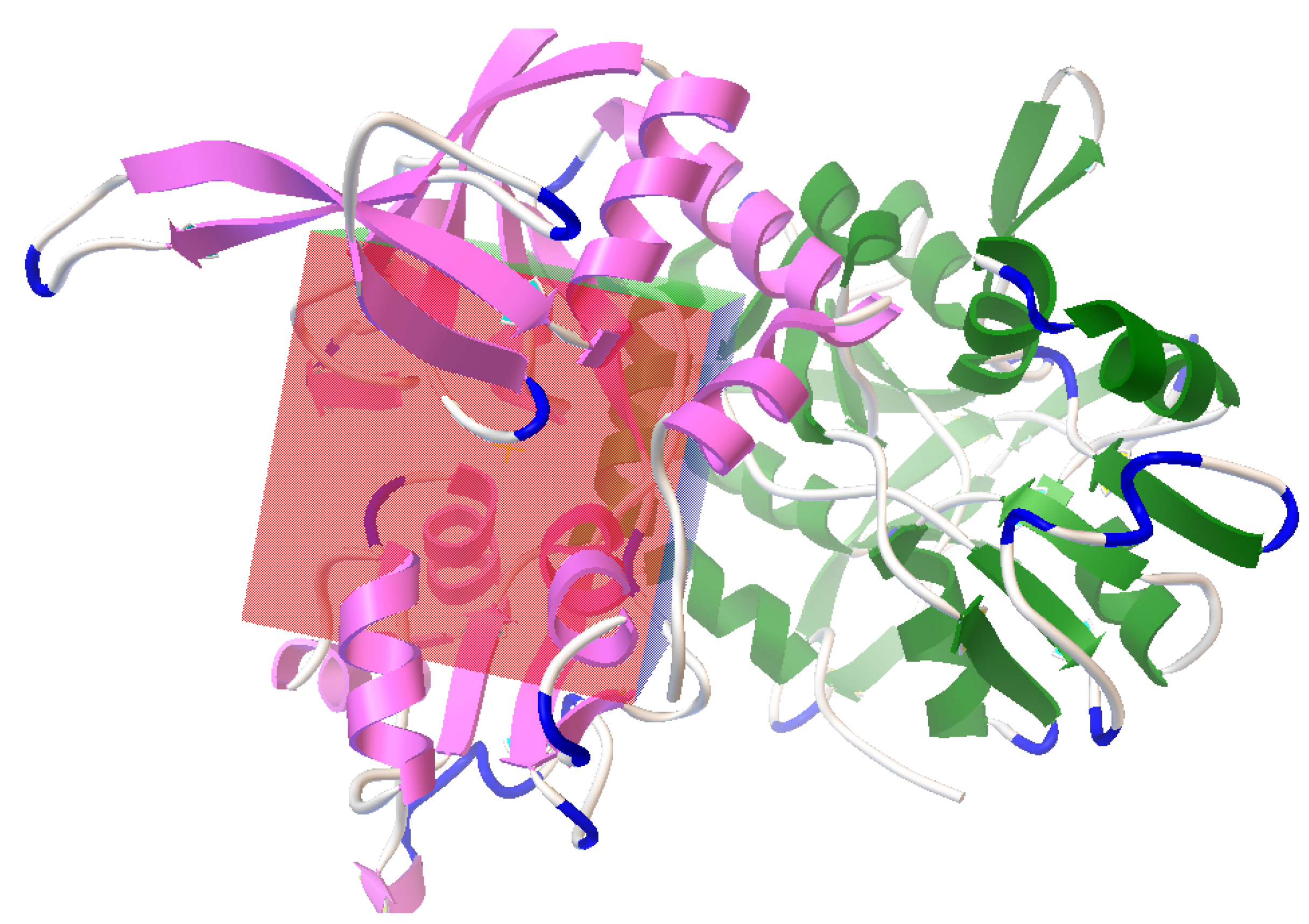
3.2. Active Site Identification and Ligand Preparation
3.3. Molecular Docking
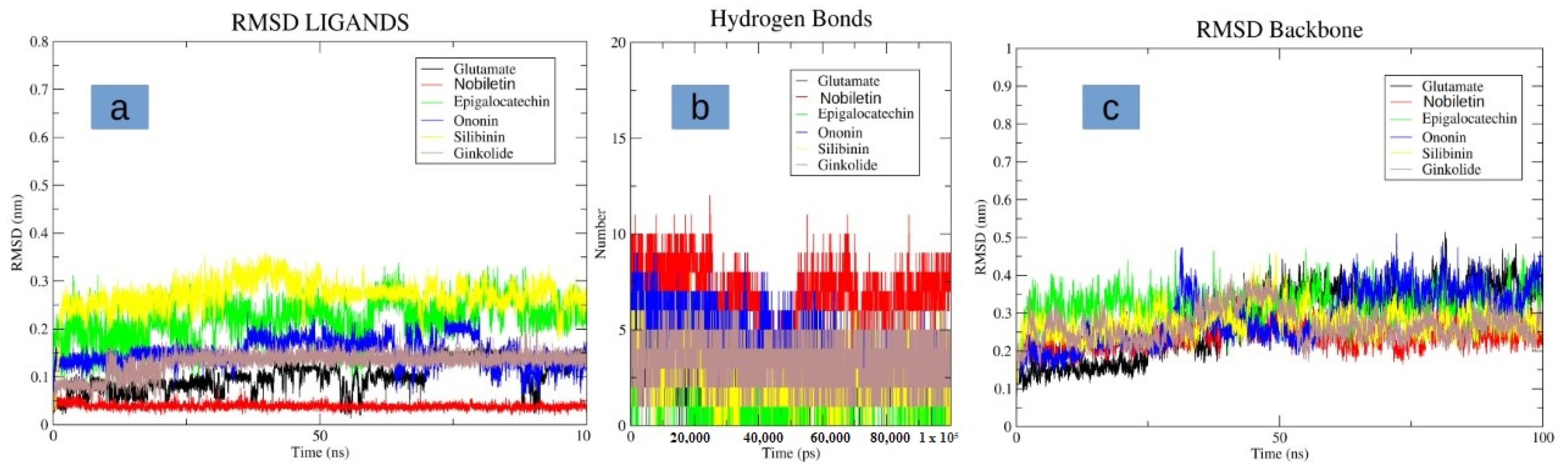
3.4. Molecular Dynamics Simulation
3.5. Protein–Ligand Interaction Analysis
3.6. ADME(T) Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dong, X.X.; Wang, Y.; Qin, Z.H. Molecular Mechanisms of Excitotoxicity and Their Relevance to Pathogenesis of Neurodegenerative Diseases. Acta Pharmacol. Sin. 2009, 30, 379–387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reiner, A.; Levitz, J. Glutamatergic Signaling in the Central Nervous System: Ionotropic and Metabotropic Receptors in Concert. Neuron 2018, 98, 1080–1098. [Google Scholar] [CrossRef] [Green Version]
- Fan, X.; Jin, W.Y.; Wang, Y.T. The NMDA Receptor Complex: A Multifunctional Machine at the Glutamatergic Synapse. Front. Cell. Neurosci. 2014, 8, 160. [Google Scholar] [CrossRef] [Green Version]
- Ince, P.G.; Eggett, C.J.; Shaw, P.J. The Role of Excitotoxicity in Neurological Disease. Rev. Contemp. Pharmacother. 1997, 8, 195–212. [Google Scholar]
- Kirmani, B.F.; Shapiro, L.A.; Shetty, A.K. Neurological and Neurodegenerative Disorders: Novel Concepts and Treatment. Aging Dis. 2021, 12, 950–953. [Google Scholar] [CrossRef] [PubMed]
- Tottori, T.; Fujii, M.; Kuroda, S. NMDAR-Mediated Ca2+ Increase Shows Robust Information Transfer in Dendritic Spines. Biophys. J. 2019, 116, 1748–1758. [Google Scholar] [CrossRef] [PubMed]
- Tachibana, N.; Kinoshita, M.; Saito, Y.; Ikeda, S. Identification of the N-Methyl-D-Aspartate Receptor (NMDAR)- Related Epitope, NR2B, in the Normal Human Ovary: Implication for the Pathogenesis of Anti-NMDAR Encephalitis. Tohoku J. Exp. Med. 2013, 230, 13–16. [Google Scholar] [CrossRef] [Green Version]
- Aloisi, E.; le Corf, K.; Dupuis, J.; Zhang, P.; Ginger, M.; Labrousse, V.; Spatuzza, M.; Georg Haberl, M.; Costa, L.; Shigemoto, R.; et al. Altered Surface MGluR5 Dynamics Provoke Synaptic NMDAR Dysfunction and Cognitive Defects in Fmr1 Knockout Mice. Nat. Commun. 2017, 8, 1103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bacq, A.; Astori, S.; Gebara, E.; Tang, W.; Silva, B.A.; Sanchez-Mut, J.; Grosse, J.; Guillot de Suduiraut, I.; Zanoletti, O.; Maclachlan, C.; et al. Amygdala GluN2B-NMDAR Dysfunction Is Critical in Abnormal Aggression of Neurodevelopmental Origin Induced by St8sia2 Deficiency. Mol. Psychiatry 2020, 25, 2144–2161. [Google Scholar] [CrossRef]
- Haroon, E.; Miller, A.H.; Sanacora, G. Inflammation, Glutamate, and Glia: A Trio of Trouble in Mood Disorders. Neuropsychopharmacology 2017, 42, 193–215. [Google Scholar] [CrossRef]
- Tikhonova, I.G.; Baskin, I.I.; Palyulin, V.A.; Zefirov, N.S.; Bachurin, S.O. Structural basis for understanding structure-activity relationships for the glutamate binding site of the NMDA receptor. J. Med. Chem. 2002, 18, 3836–3843. [Google Scholar] [CrossRef]
- Breijyeh, Z.; Karaman, R. Comprehensive Review on Alzheimer’s Disease: Causes and Treatment. Molecules 2020, 24, 5789. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, S.; Fu, P.; Zhang, Z.; Lin, K.; Ko, J.K.; Yung, K.K. Roles of Glutamate Receptors in Parkinson’s Disease. Int. J. Mol. Sci. 2019, 18, 4391. [Google Scholar] [CrossRef] [Green Version]
- Bardaweel, S.K.; Alzweiri, M.; Ishaqat, A.A. D-Serine in Neurobiology: CNS Neurotransmission and Neuromodulation. Can. J. Neurol. Sci. 2014, 41, 164–176. [Google Scholar] [CrossRef] [Green Version]
- Parsons, M.P.; Raymond, L.A. Extrasynaptic NMDA Receptor Involvement in Central Nervous System Disorders. Neuron 2014, 82, 279–293. [Google Scholar] [CrossRef] [Green Version]
- Franchini, L.; Carrano, N.; di Luca, M.; Gardoni, F. Synaptic GluN2A-Containing NMDA Receptors: From Physiology to Pathological Synaptic Plasticity. Int. J. Mol. Sci. 2020, 21, 1538. [Google Scholar] [CrossRef] [Green Version]
- Barnes, J.R.; Mukherjee, B.; Rogers, B.C.; Nafar, F.; Gosse, M.; Parsons, M.P. The Relationship between Glutamate Dynamics and Activity-Dependent Synaptic Plasticity. J. Neurosci. 2020, 40, 2793–2807. [Google Scholar] [CrossRef]
- Bozic, M.; Valdivielso, J.M. The Potential of Targeting NMDA Receptors Outside the CNS. Expert Opin. Ther. Targets 2015, 19, 399–413. [Google Scholar] [CrossRef]
- Pérez-Otaño, I.; Larsen, R.S.; Wesseling, J.F. Emerging Roles of GluN3-Containing NMDA Receptors in the CNS. Nat. Rev. Neurosci. 2016, 17, 623–635. [Google Scholar] [CrossRef]
- Zhou, Q.; Sheng, M. NMDA Receptors in Nervous System Diseases. Neuropharmacology 2013, 74, 69–75. [Google Scholar] [CrossRef] [Green Version]
- Salussolia, C.L.; Prodromou, M.L.; Borker, P.; Wollmuth, L.P. Arrangement of Subunits in Functional NMDA Receptors. J. Neurosci. 2011, 31, 11295–11304. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Stein, R.A.; Yoshioka, C.; Lee, C.H.; Goehring, A.; McHaourab, H.S.; Gouaux, E. Mechanism of NMDA Receptor Inhibition and Activation. Cell 2016, 165, 704–714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hansen, K.B.; Yi, F.; Perszyk, R.E.; Furukawa, H.; Wollmuth, L.P.; Gibb, A.J.; Traynelis, S.F. Structure, Function, and Allosteric Modulation of NMDA Receptors. J. Gen. Physiol. 2018, 150, 1081–1105. [Google Scholar] [CrossRef] [PubMed]
- Kalia, L.V.; Kalia, S.K.; Salter, M.W. NMDA Receptors in Clinical Neurology: Excitatory Times Ahead. Lancet Neurol. 2008, 7, 742–755. [Google Scholar] [CrossRef] [Green Version]
- Salter, M.W.; Kalia, L.V. SRC Kinases: A Hub for NMDA Receptor Regulation. Nat. Rev. Neurosci. 2004, 5, 317–328. [Google Scholar] [CrossRef]
- Fan, M.M.Y.; Raymond, L.A. N-Methyl-d-Aspartate (NMDA) Receptor Function and Excitotoxicity in Huntington’s Disease. Prog. Neurobiol. 2007, 81, 272–293. [Google Scholar] [CrossRef]
- Wesseling, J.F.; Pérez-Otaño, I. Modulation of GluN3A Expression in Huntington Disease a New N-Methyl-D-Aspartate Receptor-Based Therapeutic Approach? JAMA Neurol. 2015, 72, 468–473. [Google Scholar] [CrossRef] [Green Version]
- Danysz, W.; Parsons, C.G. Alzheimer’s Disease, β-Amyloid, Glutamate, NMDA Receptors and Memantine-Searching for the Connections. Br. J. Pharmacol. 2012, 167, 324–352. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Chang, L.; Song, Y.; Li, H.; Wu, Y. The Role of NMDA Receptors in Alzheimer’s Disease. Front. Neurosci. 2019, 13, 43. [Google Scholar] [CrossRef] [Green Version]
- Wang, R.; Reddy, P.H. Role of Glutamate and NMDA Receptors in Alzheimer’s Disease. J. Alzheimer’s Dis. 2017, 57, 1041–1048. [Google Scholar] [CrossRef] [Green Version]
- Jin, G.; Bai, D.; Yin, S.; Yang, Z.; Zou, D.; Zhang, Z.; Li, X.; Sun, Y.; Zhu, Q. Silibinin Rescues Learning and Memory Deficits by Attenuating Microglia Activation and Preventing Neuroinflammatory Reactions in SAMP8 Mice. Neurosci. Lett. 2016, 629, 256–261. [Google Scholar] [CrossRef]
- Bai, S.; Zhang, X.; Chen, Z.; Wang, W.; Hu, Q.; Liang, Z.; Shen, P.; Gui, S.; Zeng, L.; Liu, Z.; et al. Insight into the Metabolic Mechanism of Diterpene Ginkgolides on Antidepressant Effects for Attenuating Behavioural Deficits Compared with Venlafaxine. Sci. Rep. 2017, 7, 9591. [Google Scholar] [CrossRef] [Green Version]
- Spanaki, C.; Zaganas, I.; Kounoupa, Z.; Plaitakis, A. The Complex Regulation of Human Glud1 and Glud2 Glutamate Dehydrogenases and Its Implications in Nerve Tissue Biology. Neurochem. Int. 2012, 61, 470–481. [Google Scholar] [CrossRef]
- Huang, H.; Li, L.; Shi, W.; Liu, H.; Yang, J.; Yuan, X.; Wu, L. The Multifunctional Effects of Nobiletin and Its Metabolites In Vivo and In Vitro. Evid.-Based Complement. Altern. Med. 2016, 2016, 2918796. [Google Scholar] [CrossRef] [Green Version]
- Jahan, S.; Ansari, U.A.; Siddiqui, A.J.; Iqbal, D.; Khan, J.; Banawas, S.; Alshehri, B.; Alshahrani, M.M.; Alsagaby, S.A.; Redhu, N.S. Nobiletin Ameliorates Cellular Damage and Stress Response and Restores Neuronal Identity Altered by Sodium Arsenate Exposure in Human iPSCs-Derived hNPCs. Pharmaceuticals 2022, 15, 593. [Google Scholar] [CrossRef]
- Liu, Z.; Zhao, W.; Xu, T.; Pei, D.; Peng, Y. Alterations of NMDA Receptor Subunits NR1, NR2A and NR2B MRNA Expression and Their Relationship to Apoptosis following Transient Forebrain Ischemia. Brain Res. 2010, 1361, 133–139. [Google Scholar] [CrossRef]
- Arnemann, J. NCBI. In Lexikon der Medizinischen Laboratoriumsdiagnostik; Springer: Berlin/Heidelberg, Germany, 2018. [Google Scholar]
- Apweiler, R.; Bairoch, A.; Wu, C.H.; Barker, W.C.; Boeckmann, B.; Ferro, S.; Gasteiger, E.; Huang, H.; Lopez, R.; Magrane, M.; et al. UniProt: The Universal Protein Knowledgebase. Nucleic Acids Res. 2004, 32, D115–D119. [Google Scholar] [CrossRef]
- Garg, V.K.; Avashthi, H.; Tiwari, A.; Jain, A.; Ramkete, P.W.; Kayastha, A.M.; Singh, K. MFPPI-Multi FASTA ProtParam Interface. Bioinformation 2016, 12, 74. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [Green Version]
- Degtyarenko, K.; de matos, P.; Ennis, M.; Hastings, J.; Zbinden, M.; Mcnaught, A.; Alcántara, R.; Darsow, M.; Guedj, M.; Ashburner, M. ChEBI: A Database and Ontology for Chemical Entities of Biological Interest. Nucleic Acids Res. 2008, 36, D344–D350. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Meng, E.C.; Couch, G.S.; Croll, T.I.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Structure Visualization for Researchers, Educators, and Developers. Protein Sci. 2021, 30, 70–82. [Google Scholar] [CrossRef]
- Barret, R. Lipinski’s Rule of Five. In Medicinal Chemistry; Elsevier: Amsterdam, The Netherlands, 2018. [Google Scholar]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A Free Web Tool to Evaluate Pharmacokinetics, Drug-Likeness and Medicinal Chemistry Friendliness of Small Molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef] [Green Version]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [Green Version]
- Seeliger, D.; de Groot, B.L. Ligand Docking and Binding Site Analysis with PyMOL and Autodock/Vina. J. Comput.-Aided Mol. Des. 2010, 24, 417–422. [Google Scholar] [CrossRef] [Green Version]
- Cosconati, S.; Forli, S.; Perryman, A.L.; Harris, R.; Goodsell, D.S.; Olson, A.J. Virtual Screening with AutoDock: Theory and Practice. Expert Opin. Drug Discov. 2010, 5, 597–607. [Google Scholar] [CrossRef] [Green Version]
- Sharma, T.; Abohashrh, M.; Baig, M.H.; Baig, M.H.; Dong, J.-J.; Alam, M.M.; Ahmad, I.; Irfan, S. Screening of drug databank against WT and mutant main protease of SARS-CoV-2: Towards finding potential compound for repurposing against COVID-19. Saudi J. Biol. Sci. 2021, 28, 3152–3159. [Google Scholar] [CrossRef]
- Sharma, T.; Siddiqi, M.I. In silico identification and design of potent peptide inhibitors against PDZ-3 domain of Postsynaptic Density Protein (PSD-95). J. Biomol. Struct. Dyn. 2019, 37, 1241–1253. [Google Scholar] [CrossRef]
- Sharma, T.; Harioudh, M.K.; Kuldeep, J.; Kumar, S.; Banerjee, D.; Ghosh, J.K.; Siddiqi, M.I. Identification of Potential Inhibitors of Cathepsin-B using Shape & Pharmacophore-based Virtual Screening, Molecular Docking and Explicit Water Thermodynamics. Mol. Inform. 2020, 39, e1900023. [Google Scholar]
- Mothay, D.; Ramesh, K.V. Binding Site Analysis of Potential Protease Inhibitors of COVID-19 Using AutoDock. VirusDisease 2020, 31, 194–199. [Google Scholar] [CrossRef]
- Forli, S.; Huey, R.; Pique, M.E.; Sanner, M.F.; Goodsell, D.S.; Olson, A.J. Computational Protein-Ligand Docking and Virtual Drug Screening with the AutoDock Suite. Nat. Protoc. 2016, 11, 905–919. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goga, N.; Marin, I.; VasilǍţeanu, A.; PǍvǍloiu, I.B.; Kadiri, K.O.; Awodele, O. Improved GROMACS Algorithms Using the MPI Parallelization. In Proceedings of the 2015 E-Health and Bioengineering Conference (EHB), Iasi, Romania, 19–21 November 2015. [Google Scholar]
- MacKerell, A.D.; Banavali, N.; Foloppe, N. Development and Current Status of the CHARMM Force Field for Nucleic Acids. Biopolymers 2000, 56, 257–265. [Google Scholar] [CrossRef]
- Boonstra, S.; Onck, P.R.; van der Giessen, E. CHARMM TIP3P Water Model Suppresses Peptide Folding by Solvating the Unfolded State. J. Phys. Chem. B 2016, 120, 3692–3698. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; Swindells, M.B. LigPlot+: Multiple Ligand-Protein Interaction Diagrams for Drug Discovery. J. Chem. Inf. Model. 2011, 51, 2778–2786. [Google Scholar] [CrossRef]
- Hubbard, S.J.; Thornton, J.M. NACCESS; Department of Biochemistry and Molecular Biology, University College London: London, UK, 1993. [Google Scholar]
- Schyman, P.; Liu, R.; Desai, V.; Wallqvist, A. VNN Web Server for ADMET Predictions. Front. Pharmacol. 2017, 8, 889. [Google Scholar] [CrossRef] [Green Version]
- Delaney, J.S. ESOL: Estimating Aqueous Solubility Directly from Molecular Structure. J. Chem. Inf. Comput. Sci. 2004, 44, 1000–1005. [Google Scholar] [CrossRef]
- Ali, J.; Camilleri, P.; Brown, M.B.; Hutt, A.J.; Kirton, S.B. Revisiting the General Solubility Equation: In Silico Prediction of Aqueous Solubility Incorporating the Effect of Topographical Polar Surface Area. J. Chem. Inf. Model. 2012, 52, 420–428. [Google Scholar] [CrossRef]
- Baell, J.B.; Holloway, G.A. New Substructure Filters for Removal of Pan Assay Interference Compounds (PAINS) from Screening Libraries and for Their Exclusion in Bioassays. J. Med. Chem. 2010, 53, 2719–2740. [Google Scholar] [CrossRef] [Green Version]



| Name of Gene | Refseq Nucleotide Accession No. | Isoforms | Protein Encoded | UniProt Accession No. | Isoforms (at SwissProt) |
|---|---|---|---|---|---|
| GRIN2A | BC117131.1 | 3 | GLUN2A | Q12879 | 3 |
| GRIN2B | BC113620.1 | 1 | GLUN2B | Q13224 | 1 |
| GRIN2C | BC140801.1 | 2 | GLUN2C | Q14957 | 2 |
| GRIN2D | U77783.1 | 1 | GLUN2D | O15399 | 1 |
| Rigid Docking | |||||
|---|---|---|---|---|---|
| Epigallocatechin Gallate | Ginkgolide B | Nobiletin | Ononin | Silibinin | |
| Binding energy | −2.87 | 42.73 | −6.66 | −4.37 | 1.67 |
| Ligand efficiency | −0.09 | 1.42 | −0.23 | −0.14 | 0.05 |
| Intermolecular energy | −4.95 | 42.43 | −8.75 | −7.35 | −0.42 |
| Vdw_hb_desolvation energy | −4.73 | 42.7 | −8.39 | −6.95 | 0.21 |
| Electrostatic energy | −0.22 | −0.26 | −0.36 | −0.38 | 0.21 |
| Total internal energy | −2.39 | −0.14 | −1.21 | −2.83 | 1.34 |
| Torsional energy | 2.09 | 0.3 | 2.09 | 2.98 | 2.09 |
| Unbound energy | −2.39 | 0.14 | −1.21 | −2.83 | 1.34 |
| refRMS | 25.37 | 27.76 | 7.4 | 7.43 | 11.15 |
| Name of Compounds | Name of Residues |
|---|---|
| Epigallocatechin gallate | Leu13, Glu14, Glu15, Gly85, Lys86, His87, Gly88, Lys89, Asn96, Ser113, Thr115, Arg120, Val168, Pro169, Asn170, Gly171, Ser172, Thr173, Lys194, Gly195, Val196, Glu197, Tyr213, Asp214, Val217, Tyr244 |
| Ginkgolide B | Leu13, Glu14, Glu15, Lys86, His87, Ser113, Thr115, Arg120, Thr133, Gly134, Ile135, Val168, Pro169, Asn170, Gly171, Ser172, Thr173, Asn176, Gly195, Val196, Tyr213, Asp214, Val217, Thr242, Tyr244 |
| Nobiletin | Glu15, Gly85, Lys86, His87, Gly88, Lys89, Asn96, Ser113, Thr115, Arg120, Thr133, Gly134, Ile135, Val168, Pro169, Asn170, Gly171, Ser172, Thr173, Lys194, Tyr213, Asp214, Val217, Tyr244 |
| Ononin | Glu15 Gly85, Lys86, His87, Gly88, Lys89, Asn96, Ser113, Thr115, Arg120, Thr133, Gly134, Ile135, Val168, Pro169, Asn170, Gly171, Ser172, Thr173, Lys194, Tyr213, Asp214, Tyr244 |
| Silibinin | Leu13, Glu14, Glu15, Val82, Thr83, Gly85, Lys86, His87, Val168, Pro169, Asn170, Asn192, Gln193, Lys194, Gly195, Val196, Glu197, Asp198, Tyr213 |
| Water Solubility (Log S and Class) | Bioavailability Score | Medicinal Chemistry | ||||
|---|---|---|---|---|---|---|
| ESOL Method | Ali Method | Synthetic Accessibility | PAINS | Brenk | ||
| Nobiletin | −4.18 Moderately soluble | −4.47 Moderately soluble | 0.55 | 3.90 | 0 alert | 0 alert |
| Ginkgolide B | −2.22 Soluble | −2.29 Soluble | 0.55 | 6.18 | 0 alert | 3 alerts: diketo_group, michael_acceptor_1, more_than_2_esters |
| Epigallocatechin gallate | −1.55 Very soluble | −1.99 Very soluble | 0.11 | 4.81 | 2 alerts: imine_one_A, quinone_D | 2 alerts: chinone_2, diketo_group |
| Silibinin | −4.81 Moderately soluble | −5.78 Moderately soluble | 0.55 | 4.65 | 1 alert: imine_one_A | 2 alerts: aldehyde, diketo_group |
| Ononin | −3.53 Soluble | −4.11 Moderately soluble | 0.55 | 4.80 | 0 alert | 2 alerts: acyclic_C=C-O, stilbene |
| GI Absorption | P-gp Substrate | CYP1A2 Inhibitor | CYP2C19 Inhibitor | CYP2C9 Inhibitor | CYP2D6 Inhibitor | CYP3A4 Inhibitor | Log Kp (Skin Permeation) (cm/s) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Swiss ADME | Swiss ADME | vNN- ADMET | Swiss ADME | vNN- ADMET | Swiss ADME | vNN- ADMET | Swiss ADME | vNN- ADMET | Swiss ADME | vNN- ADMET | Swiss ADME | vNN- ADMET | Swiss ADME | |
| Nobiletin | High | No | Yes | Yes | No | No | Yes | Yes | No | No | No | Yes | No | −6.62 |
| Silibinin | Low | No | No | No | No | No | No | Yes | Yes | No | No | Yes | Yes | −7.10 |
| Ononin | Low | Yes | No | No | No | No | No | No | No | No | No | No | No | −7.78 |
| Ginkgolide B | Low | Yes | Yes | No | No | No | No | No | No | No | No | No | No | −9.02 |
| Epigallocatechin gallate | Low | No | Yes | No | No | No | No | No | No | No | No | No | No | −10.00 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jahan, S.; Redhu, N.S.; Siddiqui, A.J.; Iqbal, D.; Khan, J.; Banawas, S.; Alaidarous, M.; Alshehri, B.; Mir, S.A.; Adnan, M.; et al. Nobiletin as a Neuroprotectant against NMDA Receptors: An In Silico Approach. Pharmaceutics 2022, 14, 1123. https://doi.org/10.3390/pharmaceutics14061123
Jahan S, Redhu NS, Siddiqui AJ, Iqbal D, Khan J, Banawas S, Alaidarous M, Alshehri B, Mir SA, Adnan M, et al. Nobiletin as a Neuroprotectant against NMDA Receptors: An In Silico Approach. Pharmaceutics. 2022; 14(6):1123. https://doi.org/10.3390/pharmaceutics14061123
Chicago/Turabian StyleJahan, Sadaf, Neeru Singh Redhu, Arif Jamal Siddiqui, Danish Iqbal, Johra Khan, Saeed Banawas, Mohammed Alaidarous, Bader Alshehri, Shabir Ahmad Mir, Mohd Adnan, and et al. 2022. "Nobiletin as a Neuroprotectant against NMDA Receptors: An In Silico Approach" Pharmaceutics 14, no. 6: 1123. https://doi.org/10.3390/pharmaceutics14061123
APA StyleJahan, S., Redhu, N. S., Siddiqui, A. J., Iqbal, D., Khan, J., Banawas, S., Alaidarous, M., Alshehri, B., Mir, S. A., Adnan, M., & Pant, A. B. (2022). Nobiletin as a Neuroprotectant against NMDA Receptors: An In Silico Approach. Pharmaceutics, 14(6), 1123. https://doi.org/10.3390/pharmaceutics14061123











