Development of Anticancer Peptides Using Artificial Intelligence and Combinational Therapy for Cancer Therapeutics
Abstract
:1. Introduction
2. Development of Therapeutic ACPs
2.1. Classifications of ACPs
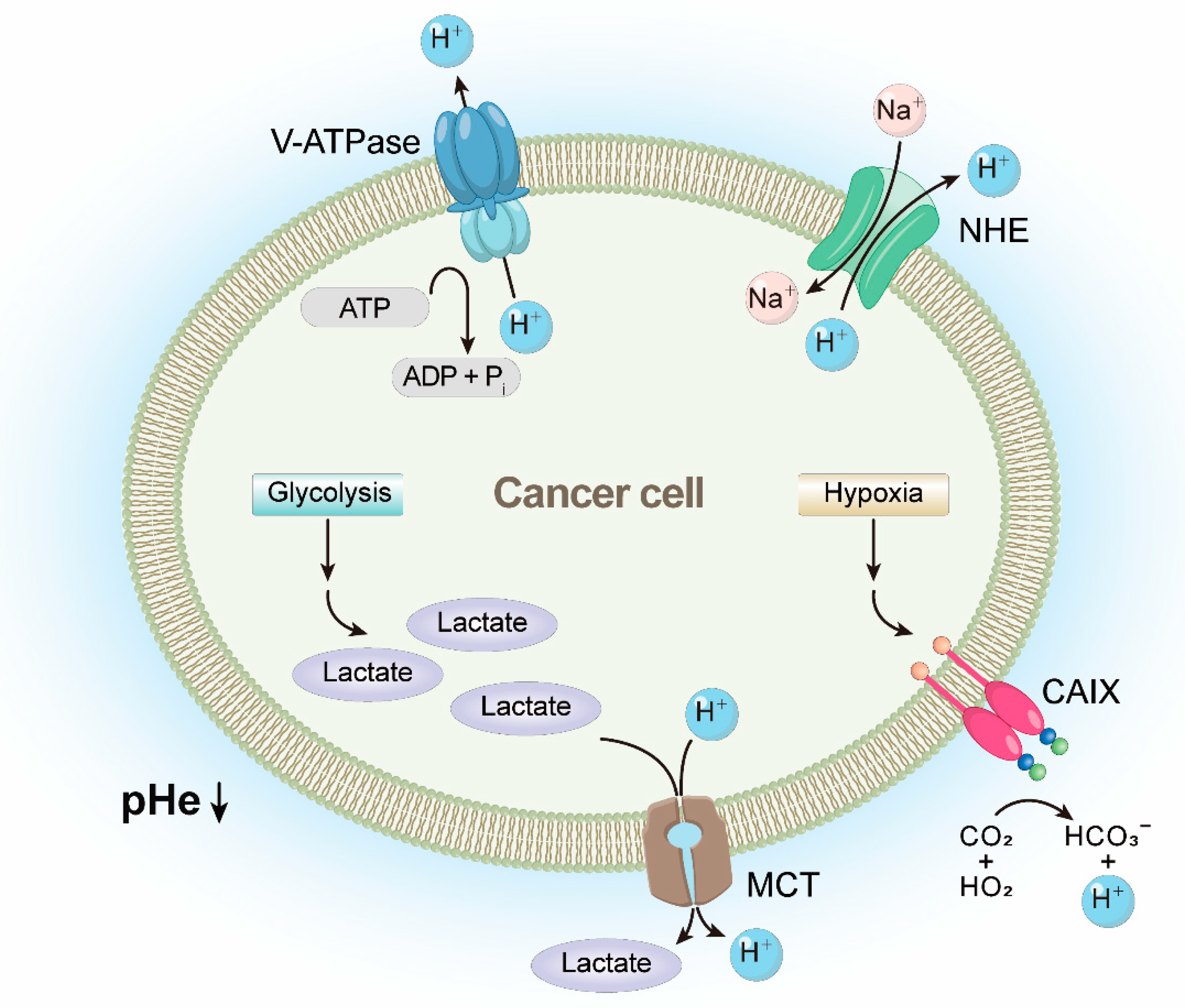
2.2. Characteristics of ACPs and Therapeutic Mechanisms of Cationic ACPs in Cancer Cells
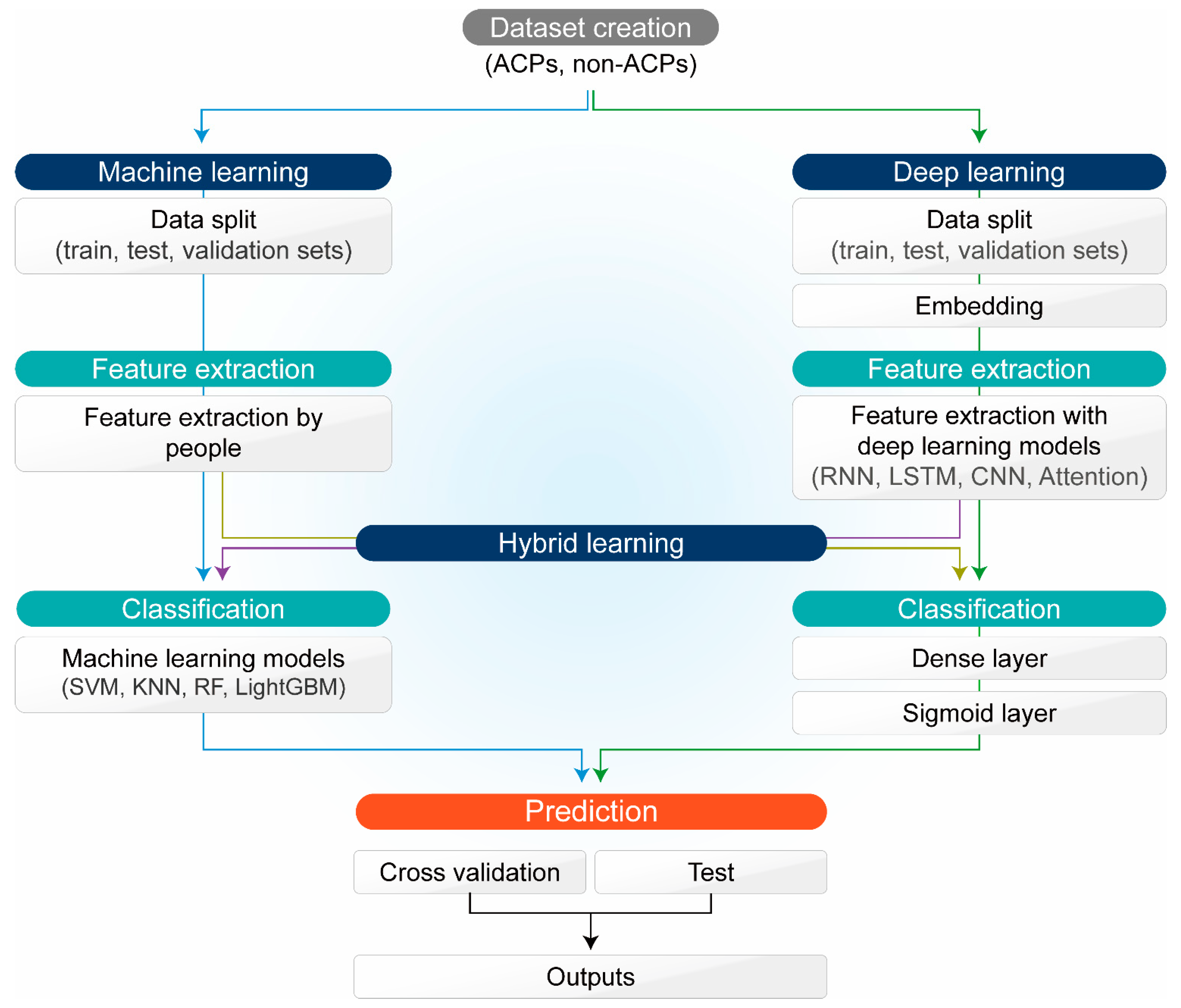
3. Application of ML and DL for ACP Development
4. Future Approaches of Combinational Therapy with ACPs for Cancer Therapeutics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Anand, P.; Kunnumakkara, A.B.; Sundaram, C.; Harikumar, K.B.; Tharakan, S.T.; Lai, O.S.; Sung, B.; Aggarwal, B.B. Cancer is a Preventable Disease that Requires Major Lifestyle Changes. Pharm. Res. 2008, 25, 2097–2116. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [Green Version]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Han, Y.; Xu, Y.; Liu, T.; Cui, M.; Xia, L.; Li, H.; Gu, Y.; Wang, P. Diversified strategies based on nanoscale metal-organic frameworks for cancer therapy: The leap from monofunctional to versatile. Coord. Chem. Rev. 2021, 431, 213676. [Google Scholar] [CrossRef]
- Mansoori, B.; Mohammadi, A.; Davudian, S.; Shirjang, S.; Baradaran, B. The Different Mechanisms of Cancer Drug Resistance: A Brief Review. Adv. Pharm. Bull. 2017, 7, 339–348. [Google Scholar] [CrossRef]
- Berkey, F.J. Managing the adverse effects of radiation therapy. Am. Fam. Physician 2010, 82, 381–388. [Google Scholar]
- Stubblefield, M.D. Clinical Evaluation and Management of Radiation Fibrosis Syndrome. Phys. Med. Rehabil. Clin. N. Am. 2017, 28, 89–100. [Google Scholar] [CrossRef]
- Brook, I. Late side effects of radiation treatment for head and neck cancer. Radiat. Oncol. J. 2020, 38, 84–92. [Google Scholar] [CrossRef]
- Li, S.; Zhang, Z.; Lai, W.-F.; Cui, L.; Zhu, X. How to overcome the side effects of tumor immunotherapy. Biomed. Pharmacother. 2020, 130, 110639. [Google Scholar] [CrossRef]
- Banting, F.G.; Best, C.H.; Collip, J.B.; Campbell, W.R.; Fletcher, A.A. Pancreatic Extracts in the Treatment of Diabetes Mellitus. Can. Med. Assoc. J. 1922, 12, 141–146. [Google Scholar]
- du Vigneaud, V.; Ressler, C.; Trippett, S. The sequence of amino acids in oxytocin, with a proposal for the structure of Oxytocin. J. Biol. Chem. 1953, 205, 949–957. [Google Scholar] [CrossRef]
- Fosgerau, K.; Hoffmann, T. Peptide therapeutics: Current status and future directions. Drug Discov. Today 2015, 20, 122–128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Liu, K.; Xing, R.; Yan, X. Peptide self-assembly: Thermodynamics and kinetics. Chem. Soc. Rev. 2016, 45, 5589–5604. [Google Scholar] [CrossRef] [PubMed]
- Henninot, A.; Collins, J.C.; Nuss, J.M. The Current State of Peptide Drug Discovery: Back to the Future? J. Med. Chem. 2018, 61, 1382–1414. [Google Scholar] [CrossRef]
- Harris, F.; Dennison, S.R.; Singh, J.; Phoenix, D.A. On the selectivity and efficacy of defense peptides with respect to cancer cells. Med. Res. Rev. 2013, 33, 190–234. [Google Scholar] [CrossRef]
- Muttenthaler, M.; King, G.F.; Adams, D.J.; Alewood, P.F. Trends in peptide drug discovery. Nat. Rev. Drug Discov. 2021, 20, 309–325. [Google Scholar] [CrossRef]
- Lau, J.L.; Dunn, M.K. Therapeutic peptides: Historical perspectives, current development trends, and future directions. Bioorganic. Med. Chem. 2018, 26, 2700–2707. [Google Scholar] [CrossRef]
- Chiangjong, W.; Chutipongtanate, S.; Hongeng, S. Anticancer peptide: Physicochemical property, functional aspect and trend in clinical application (Review). Int. J. Oncol. 2020, 57, 678–696. [Google Scholar] [CrossRef]
- Trinidad-Calderón, P.A.; Varela-Chinchilla, C.D.; García-Lara, S. Natural Peptides Inducing Cancer Cell Death: Mechanisms and Properties of Specific Candidates for Cancer Therapeutics. Molecules 2021, 26, 7453. [Google Scholar] [CrossRef]
- Liscano, Y.; Oñate-Garzón, J.; Delgado, J.P. Peptides with Dual Antimicrobial–Anticancer Activity: Strategies to Overcome Peptide Limitations and Rational Design of Anticancer Peptides. Molecules 2020, 25, 4245. [Google Scholar] [CrossRef]
- Basith, S.; Manavalan, B.; Shin, T.H.; Lee, G. Machine intelligence in peptide therapeutics: A next-generation tool for rapid disease screening. Med. Res. Rev. 2020, 40, 1276–1314. [Google Scholar] [CrossRef] [PubMed]
- Boohaker, R.J.; Lee, M.W.; Vishnubhotla, P.; Perez, J.M.; Khaled, A.R. The Use of Therapeutic Peptides to Target and to Kill Cancer Cells. Curr. Med. Chem. 2012, 19, 3794–3804. [Google Scholar] [CrossRef] [PubMed]
- Hoppenz, P.; Els-Heindl, S.; Beck-Sickinger, A.G. Peptide-Drug Conjugates and Their Targets in Advanced Cancer Therapies. Front. Chem. 2020, 8, 571. [Google Scholar] [CrossRef] [PubMed]
- Manavalan, B.; Basith, S.; Shin, T.H.; Choi, S.; Kim, M.O.; Lee, G. MLACP: Machine-learning-based prediction of anticancer peptides. Oncotarget 2017, 8, 77121–77136. [Google Scholar] [CrossRef] [Green Version]
- Lv, Z.; Cui, F.; Zou, Q.; Zhang, L.; Xu, L. Anticancer peptides prediction with deep representation learning features. Briefings Bioinform. 2021, 22, bbab008. [Google Scholar] [CrossRef]
- Bashraheel, S.S.; Domling, A.; Goda, S.K. Update on targeted cancer therapies, single or in combination, and their fine tuning for precision medicine. Biomed. Pharmacother. 2020, 125, 110009. [Google Scholar] [CrossRef]
- Petrou, C.; Sarigiannis, Y. 1-Peptide synthesis: Methods, trends, and challenges. In Peptide Applications in Biomedicine, Biotechnology and Bioengineering; Koutsopoulos, S., Ed.; Woodhead Publishing: Cambridge, UK, 2018; pp. 1–21. [Google Scholar]
- Li, W.; Joshi, M.D.; Singhania, S.; Ramsey, K.H.; Murthy, A.K. Peptide Vaccine: Progress and Challenges. Vaccines 2014, 2, 515–536. [Google Scholar] [CrossRef] [Green Version]
- Verbeke, F.; De Craemer, S.; Debunne, N.; Janssens, Y.; Wynendaele, E.; Van De Wiele, C.; De Spiegeleer, B. Peptides as Quorum Sensing Molecules: Measurement Techniques and Obtained Levels In vitro and In vivo. Front. Neurosci. 2017, 11, 183. [Google Scholar] [CrossRef] [Green Version]
- Basith, S.; Manavalan, B.; Shin, T.H.; Lee, G. iGHBP: Computational identification of growth hormone binding proteins from sequences using extremely randomised tree. Comput. Struct. Biotechnol. J. 2018, 16, 412–420. [Google Scholar] [CrossRef]
- Manavalan, B.; Shin, T.H.; Kim, M.O.; Lee, G. AIPpred: Sequence-Based Prediction of Anti-inflammatory Peptides Using Random Forest. Front. Pharmacol. 2018, 9, 276. [Google Scholar] [CrossRef]
- Tesauro, D.; Accardo, A.; Diaferia, C.; Milano, V.; Guillon, J.; Ronga, L.; Rossi, F. Peptide-Based Drug-Delivery Systems in Biotechnological Applications: Recent Advances and Perspectives. Molecules 2019, 24, 351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guidotti, G.; Brambilla, L.; Rossi, D. Cell-Penetrating Peptides: From Basic Research to Clinics. Trends Pharmacol. Sci. 2017, 38, 406–424. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Kang, J.; Jiang, L.; He, B.; Lin, H.; Huang, J. PSBinder: A Web Service for Predicting Polystyrene Surface-Binding Peptides. BioMed Res. Int. 2017, 2017, 5761517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manavalan, B.; Govindaraj, R.G.; Shin, T.H.; Kim, M.O.; Lee, G. iBCE-EL: A New Ensemble Learning Framework for Improved Linear B-Cell Epitope Prediction. Front. Immunol. 2018, 9, 1695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fuchs, J.-A.; Grisoni, F.; Kossenjans, M.; Hiss, J.A.; Schneider, G. Lipophilicity prediction of peptides and peptide derivatives by consensus machine learning. MedChemComm 2018, 9, 1538–1546. [Google Scholar] [CrossRef]
- De la Torre, B.G.; Albericio, F. Peptide Therapeutics 2.0. Molecules 2020, 25, 2293. [Google Scholar] [CrossRef]
- D’Hondt, M.; Bracke, N.; Taevernier, L.; Gevaert, B.; Verbeke, F.; Wynendaele, E.; De Spiegeleer, B. Related impurities in peptide medicines. J. Pharm. Biomed. Anal. 2014, 101, 2–30. [Google Scholar] [CrossRef]
- Robbins, P.F.; Lu, Y.C.; El-Gamil, M.; Li, Y.F.; Gross, C.; Gartner, J.; Lin, J.C.; Teer, J.K.; Cliften, P.; Tycksen, E.; et al. Mining exomic sequencing data to identify mutated antigens recognized by adoptively transferred tumor-reactive T cells. Nat. Med. 2013, 19, 747–752. [Google Scholar] [CrossRef]
- Lee, A.C.-L.; Harris, J.L.; Khanna, K.K.; Hong, J.-H. A Comprehensive Review on Current Advances in Peptide Drug Development and Design. Int. J. Mol. Sci. 2019, 20, 2383. [Google Scholar] [CrossRef] [Green Version]
- Fusaro, V.A.; Mani, D.R.; Mesirov, J.P.; Carr, S.A. Prediction of high-responding peptides for targeted protein assays by mass spectrometry. Nat. Biotechnol. 2009, 27, 190–198. [Google Scholar] [CrossRef] [Green Version]
- Ballester, P.J.; Mitchell, J. A machine learning approach to predicting protein–ligand binding affinity with applications to molecular docking. Bioinformatics 2010, 26, 1169–1175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manavalan, B.; Shin, T.H.; Kim, M.O.; Lee, G. PIP-EL: A New Ensemble Learning Method for Improved Proinflammatory Peptide Predictions. Front. Immunol. 2018, 9, 1783. [Google Scholar] [CrossRef]
- Tallorin, L.; Wang, J.; Kim, W.E.; Sahu, S.; Kosa, N.M.; Yang, P.; Thompson, M.; Gilson, M.K.; Frazier, P.I.; Burkart, M.D.; et al. Discovering de novo peptide substrates for enzymes using machine learning. Nat. Commun. 2018, 9, 5253. [Google Scholar] [CrossRef] [PubMed]
- Yi, H.-C.; You, Z.-H.; Zhou, X.; Cheng, L.; Li, X.; Jiang, T.-H.; Chen, Z.-H. ACP-DL: A Deep Learning Long Short-Term Memory Model to Predict Anticancer Peptides Using High-Efficiency Feature Representation. Mol. Ther.-Nucleic Acids 2019, 17, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agrawal, P.; Bhagat, D.; Mahalwal, M.; Sharma, N.; Raghava, G.P.S. AntiCP 2.0: An updated model for predicting anticancer peptides. Briefings Bioinform. 2020, 22, bbaa153. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, A.; Kapoor, P.; Kumar, R.; Chaudhary, K.; Gautam, A.; Raghava, G.P.S. In Silico Models for Designing and Discovering Novel Anticancer Peptides. Sci. Rep. 2013, 3, srep02984. [Google Scholar] [CrossRef] [PubMed]
- Hajisharifi, Z.; Piryaiee, M.; Beigi, M.M.; Behbahani, M.; Mohabatkar, H. Predicting anticancer peptides with Chou′s pseudo amino acid composition and investigating their mutagenicity via Ames test. J. Theor. Biol. 2014, 341, 34–40. [Google Scholar] [CrossRef]
- Vijayakumar, S.; Lakshmi, P. ACPP: A web server for prediction and design of anti-cancer peptides. Int. J. Pept. Res. Ther. 2015, 21, 99–106. [Google Scholar] [CrossRef]
- Chen, W.; Ding, H.; Feng, P.; Lin, H.; Chou, K.-C. iACP: A sequence-based tool for identifying anticancer peptides. Oncotarget 2016, 7, 16895–16909. [Google Scholar] [CrossRef] [Green Version]
- Akbar, S.; Hayat, M.; Iqbal, M.; Jan, M.A. iACP-GAEnsC: Evolutionary genetic algorithm based ensemble classification of anticancer peptides by utilizing hybrid feature space. Artif. Intell. Med. 2017, 79, 62–70. [Google Scholar] [CrossRef]
- Xu, L.; Liang, G.; Wang, L.; Liao, C. A Novel Hybrid Sequence-Based Model for Identifying Anticancer Peptides. Genes 2018, 9, 158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, L.; Zhou, C.; Chen, H.; Song, J.; Su, R. ACPred-FL: A sequence-based predictor using effective feature representation to improve the prediction of anti-cancer peptides. Bioinformatics 2018, 34, 4007–4016. [Google Scholar] [CrossRef] [PubMed]
- Boopathi, V.; Subramaniyam, S.; Malik, A.; Lee, G.; Manavalan, B.; Yang, D.-C. mACPpred: A Support Vector Machine-Based Meta-Predictor for Identification of Anticancer Peptides. Int. J. Mol. Sci. 2019, 20, 1964. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schaduangrat, N.; Nantasenamat, C.; Prachayasittikul, V.; Shoombuatong, W. ACPred: A Computational Tool for the Prediction and Analysis of Anticancer Peptides. Molecules 2019, 24, 1973. [Google Scholar] [CrossRef] [Green Version]
- Wei, L.; Zhou, C.; Su, R.; Zou, Q. PEPred-Suite: Improved and robust prediction of therapeutic peptides using adaptive feature representation learning. Bioinformatics 2019, 35, 4272–4280. [Google Scholar] [CrossRef]
- Zhao, T.; Hu, Y.; Zang, T. DRACP: A novel method for identification of anticancer peptides. BMC Bioinform. 2020, 21, 559. [Google Scholar] [CrossRef]
- Deo, R.C. Machine Learning in Medicine. Circulation 2015, 132, 1920–1930. [Google Scholar] [CrossRef] [Green Version]
- Rossell, D. Big Data and Statistics: A Statistician’s Perspective. Metode Sci. Stud. J. 2015, 5, 143–149. [Google Scholar] [CrossRef]
- Lee, E.Y.; Fulan, B.M.; Wong, G.C.L.; Ferguson, A.L. Mapping membrane activity in undiscovered peptide sequence space using machine learning. Proc. Natl. Acad. Sci. USA 2016, 113, 13588–13593. [Google Scholar] [CrossRef] [Green Version]
- Manavalan, B.; Subramaniyam, S.; Shin, T.H.; Kim, M.O.; Lee, G. Machine-Learning-Based Prediction of Cell-Penetrating Peptides and Their Uptake Efficiency with Improved Accuracy. J. Proteome Res. 2018, 17, 2715–2726. [Google Scholar] [CrossRef]
- Wu, C.; Gao, R.; Zhang, Y.; De Marinis, Y. PTPD: Predicting therapeutic peptides by deep learning and word2vec. BMC Bioinform. 2019, 20, 456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, L.; Jing, R.; Liu, F.; Luo, J.; Li, Y. DeepACP: A Novel Computational Approach for Accurate Identification of Anticancer Peptides by Deep Learning Algorithm. Mol. Ther.-Nucleic Acids 2020, 22, 862–870. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Wang, Y.; Cui, L.; Su, R.; Wei, L. Learning embedding features based on multisense-scaled attention architecture to improve the predictive performance of anticancer peptides. Bioinformatics 2021, 37, 4684–4693. [Google Scholar] [CrossRef]
- Chen, X.-G.; Zhang, W.; Yang, X.; Li, C.; Chen, H. ACP-DA: Improving the Prediction of Anticancer Peptides Using Data Augmentation. Front. Genet. 2021, 12, 698477. [Google Scholar] [CrossRef] [PubMed]
- Xie, M.; Liu, D.; Yang, Y. Anti-cancer peptides: Classification, mechanism of action, reconstruction and modification. Open Biol. 2020, 10, 200004. [Google Scholar] [CrossRef]
- Roudi, R.; Syn, N.; Roudbary, M. Antimicrobial Peptides as Biologic and Immunotherapeutic Agents against Cancer: A Comprehensive Overview. Front. Immunol. 2017, 8, 1320. [Google Scholar] [CrossRef] [Green Version]
- Asadzadeh, Z.; Mohammadi, H.; Safarzadeh, E.; Hemmatzadeh, M.; Mahdian-Shakib, A.; Jadidi-Niaragh, F.; Azizi, G.; Baradaran, B. The paradox of Th17 cell functions in tumor immunity. Cell. Immunol. 2017, 322, 15–25. [Google Scholar] [CrossRef]
- Darabi, A.; Thuring, C.; Paulsson, K.M. HLA-I antigen presentation and tapasin influence immune responses against malignant brain tumors—Considerations for successful immunotherapy. Anti-Cancer Agents Med. Chem. 2014, 14, 1094–1100. [Google Scholar] [CrossRef]
- Dutoit, V.; Migliorini, D.; Ranzanici, G.; Marinari, E.; Widmer, V.; Lobrinus, J.A.; Momjian, S.; Costello, J.; Walker, P.R.; Okada, H.; et al. Antigenic expression and spontaneous immune responses support the use of a selected peptide set from the IMA950 glioblastoma vaccine for immunotherapy of grade II and III glioma. Oncoimmunology 2018, 7, e1391972. [Google Scholar] [CrossRef]
- Jin, H.; Wan, C.; Zou, Z.; Zhao, G.; Zhang, L.; Geng, Y.; Chen, T.; Huang, A.; Jiang, F.; Feng, J.-P.; et al. Tumor Ablation and Therapeutic Immunity Induction by an Injectable Peptide Hydrogel. ACS Nano 2018, 12, 3295–3310. [Google Scholar] [CrossRef]
- Fisher, E.; Pavlenko, K.; Vlasov, A.; Ramenskaya, G. Peptide-Based Therapeutics for Oncology. Pharm. Med. 2019, 33, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Sato, A.K.; Viswanathan, M.; Kent, R.B.; Wood, C.R. Therapeutic peptides: Technological advances driving peptides into development. Curr. Opin. Biotechnol. 2006, 17, 638–642. [Google Scholar] [CrossRef] [PubMed]
- Tornesello, A.L.; Borrelli, A.; Buonaguro, L.; Buonaguro, F.M.; Tornesello, M.L. Antimicrobial Peptides as Anticancer Agents: Functional Properties and Biological Activities. Molecules 2020, 25, 2850. [Google Scholar] [CrossRef] [PubMed]
- Piktel, E.; Niemirowicz, K.; Wnorowska, U.; Wątek, M.; Wollny, T.; Głuszek, K.; Góźdź, S.; Levental, I.; Bucki, R. The Role of Cathelicidin LL-37 in Cancer Development. Arch. Immunol. Ther. Exp. 2016, 64, 33–46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, W.K.; Wang, G.; Coffelt, S.; Betancourt, A.M.; Lee, C.W.; Fan, D.; Wu, K.; Yu, J.; Sung, J.J.; Cho, C.H. Emerging roles of the host defense peptide LL-37 in human cancer and its potential therapeutic applications. Int. J. Cancer 2010, 127, 1741–1747. [Google Scholar] [CrossRef] [PubMed]
- Droin, N.; Hendra, J.-B.; Ducoroy, P.; Solary, E. Human defensins as cancer biomarkers and antitumour molecules. J. Proteom. 2009, 72, 918–927. [Google Scholar] [CrossRef] [PubMed]
- Phan, T.K.; Lay, F.T.; Poon, I.K.; Hinds, M.G.; Kvansakul, M.; Hulett, M.D. Human β-defensin 3 contains an oncolytic motif that binds PI (4, 5) P2 to mediate tumour cell permeabilisation. Oncotarget 2016, 7, 2054–2069. [Google Scholar] [CrossRef] [Green Version]
- Felício, M.R.; Silva, O.N.; Gonçalves, S.; Santos, N.C.; Franco, O.L. Peptides with Dual Antimicrobial and Anticancer Activities. Front. Chem. 2017, 5, 5. [Google Scholar] [CrossRef] [Green Version]
- Tørfoss, V.; Ausbacher, D.; Cavalcanti-Jacobsen, C.d.A.; Hansen, T.; Brandsdal, B.O.; Havelkova, M.; Strøm, M.B. Synthesis of anticancer heptapeptides containing a unique lipophilic β2, 2-amino acid building block. J. Pept. Sci. 2012, 18, 170–176. [Google Scholar] [CrossRef]
- Ke, M.; Dong, J.; Wang, Y.; Zhang, J.; Zhang, M.; Wu, Z.; Lv, Y.; Wu, R. MEL-pep, an analog of melittin, disrupts cell membranes and reverses 5-fluorouracil resistance in human hepatocellular carcinoma cells. Int. J. Biochem. Cell Biol. 2018, 101, 39–48. [Google Scholar] [CrossRef]
- Feng, Z.; Xu, B. Inspiration from the mirror: D-amino acid containing peptides in biomedical approaches. Biomol. Concepts 2016, 7, 179–187. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Du, X.; Yuan, D.; Zhou, J.; Zhou, N.; Huang, Y.; Xu, B. d-Amino Acids Modulate the Cellular Response of Enzymatic-Instructed Supramolecular Nanofibers of Small Peptides. Biomacromolecules 2014, 15, 3559–3568. [Google Scholar] [CrossRef] [Green Version]
- Bastings, J.J.A.J.; Van Eijk, H.M.; Olde Damink, S.W.; Rensen, S.S. d-amino Acids in Health and Disease: A Focus on Cancer. Nutrients 2019, 11, 2205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hilchie, A.L.; Haney, E.F.; Pinto, D.M.; Hancock, R.E.; Hoskin, D.W. Enhanced killing of breast cancer cells by a d-amino acid analog of the winter flounder-derived pleurocidin NRC-03. Exp. Mol. Pathol. 2015, 99, 426–434. [Google Scholar] [CrossRef] [PubMed]
- Maxian, T.; Gerlitz, L.; Riedl, S.; Rinner, B.; Zweytick, D. Effect of L- to D-Amino Acid Substitution on Stability and Activity of Antitumor Peptide RDP215 against Human Melanoma and Glioblastoma. Int. J. Mol. Sci. 2021, 22, 8469. [Google Scholar] [CrossRef] [PubMed]
- Cichoń, T.; Smolarczyk, R.; Matuszczak, S.; Barczyk, M.; Jarosz-Biej, M.; Szala, S. D-K6L9 Peptide Combination with IL-12 Inhibits the Recurrence of Tumors in Mice. Arch. Immunol. Ther. Exp. 2014, 62, 341–351. [Google Scholar] [CrossRef] [Green Version]
- Papo, N.; Seger, D.; Makovitzki, A.; Kalchenko, V.; Eshhar, Z.; Degani, H.; Shai, Y. Inhibition of Tumor Growth and Elimination of Multiple Metastases in Human Prostate and Breast Xenografts by Systemic Inoculation of a Host Defense–Like Lytic Peptide. Cancer Res. 2006, 66, 5371–5378. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Pazgier, M.; Li, C.; Yuan, W.; Liu, M.; Wei, G.; Lu, W.-Y.; Lu, W. Systematic mutational analysis of peptide inhibition of the p53–MDM2/MDMX interactions. J. Mol. Biol. 2010, 398, 200–213. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Liu, C.; Chen, S.; Hu, H.; Su, J.; Zou, Y. d-Amino acid mutation of PMI as potent dual peptide inhibitors of p53-MDM2/MDMX interactions. Bioorg. Med. Chem. Lett. 2017, 27, 4678–4681. [Google Scholar] [CrossRef]
- Fath, M.K.; Babakhaniyan, K.; Zokaei, M.; Yaghoubian, A.; Akbari, S.; Khorsandi, M.; Soofi, A.; Nabi-Afjadi, M.; Zalpoor, H.; Jalalifar, F.; et al. Anti-cancer peptide-based therapeutic strategies in solid tumors. Cell. Mol. Biol. Lett. 2022, 27, 33. [Google Scholar] [CrossRef]
- Sugahara, K.N.; Teesalu, T.; Karmali, P.P.; Kotamraju, V.R.; Agemy, L.; Girard, O.M.; Hanahan, D.; Mattrey, R.F.; Ruoslahti, E. Tissue-Penetrating Delivery of Compounds and Nanoparticles into Tumors. Cancer Cell 2009, 16, 510–520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sugahara, K.N.; Teesalu, T.; Prakash Karmali, P.; Ramana Kotamraju, V.; Agemy, L.; Greenwald, D.R.; Ruoslahti, E. Coadministration of a Tumor-Penetrating Peptide Enhances the Efficacy of Cancer Drugs. Science 2010, 328, 1031–1035. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agemy, L.; Friedmann-Morvinski, D.; Kotamraju, V.R.; Roth, L.; Sugahara, K.N.; Girard, O.M.; Mattrey, R.F.; Verma, I.M.; Ruoslahti, E. Targeted nanoparticle enhanced proapoptotic peptide as potential therapy for glioblastoma. Proc. Natl. Acad. Sci. USA 2011, 108, 17450–17455. [Google Scholar] [CrossRef] [Green Version]
- Teesalu, T.P.; Sugahara, K.N.M.; Ruoslahti, E.M. Tumor-Penetrating Peptides. Front. Oncol. 2013, 3, 216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Danhier, F.; Le Breton, A.; Préat, V. RGD-Based Strategies To Target Alpha(v) Beta(3) Integrin in Cancer Therapy and Diagnosis. Mol. Pharm. 2012, 9, 2961–2973. [Google Scholar] [CrossRef]
- Zuo, H. iRGD: A Promising Peptide for Cancer Imaging and a Potential Therapeutic Agent for Various Cancers. J. Oncol. 2019, 2019, 9367845. [Google Scholar] [CrossRef]
- Tyagi, A.; Tuknait, A.; Anand, P.; Gupta, S.; Sharma, M.; Mathur, D.; Joshi, A.; Singh, S.; Gautam, A.; Raghava, G.P. CancerPPD: A database of anticancer peptides and proteins. Nucleic Acids Res. 2015, 43, D837–D843. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.; Li, X.; Wang, Z. APD3: The antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 2016, 44, D1087–D1093. [Google Scholar] [CrossRef] [Green Version]
- Singh, S.; Chaudhary, K.; Dhanda, S.; Bhalla, S.; Usmani, S.S.; Gautam, A.; Tuknait, A.; Agrawal, P.; Mathur, D.; Raghava, G.P. SATPdb: A database of structurally annotated therapeutic peptides. Nucleic Acids Res. 2016, 44, D1119–D1126. [Google Scholar] [CrossRef] [Green Version]
- Huang, K.-Y.; Tseng, Y.-J.; Kao, H.-J.; Chen, C.-H.; Yang, H.-H.; Weng, S.-L. Identification of subtypes of anticancer peptides based on sequential features and physicochemical properties. Sci. Rep. 2021, 11, 13594. [Google Scholar] [CrossRef]
- Schweizer, F. Cationic amphiphilic peptides with cancer-selective toxicity. Eur. J. Pharmacol. 2009, 625, 190–194. [Google Scholar] [CrossRef] [PubMed]
- Alves, A.C.R.; Ribeiro, D.; Nunes, C.; Reis, S. Biophysics in cancer: The relevance of drug-membrane interaction studies. Biochim. Biophys. Acta (BBA)-Biomembr. 2016, 1858, 2231–2244. [Google Scholar] [CrossRef]
- Kato, Y.; Ozawa, S.; Miyamoto, C.; Maehata, Y.; Suzuki, A.; Maeda, T.; Baba, Y. Acidic extracellular microenvironment and cancer. Cancer Cell Int. 2013, 13, 89. [Google Scholar] [CrossRef] [Green Version]
- Gatenby, R.A.; Gillies, R.J. Why do cancers have high aerobic glycolysis? Nat. Rev. Cancer. 2004, 4, 891–899. [Google Scholar] [CrossRef] [PubMed]
- Muz, B.; de la Puente, P.; Azab, F.; Azab, A.K. The role of hypoxia in cancer progression, angiogenesis, metastasis, and resistance to therapy. Hypoxia 2015, 3, 83–92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petrova, V.; Annicchiarico-Petruzzelli, M.; Melino, G.; Amelio, I. The hypoxic tumour microenvironment. Oncogenesis 2018, 7, 10. [Google Scholar] [CrossRef]
- Švastová, E.; Hulíková, A.; Rafajová, M.; Zat’Ovičová, M.; Gibadulinová, A.; Casini, A.; Cecchi, A.; Scozzafava, A.; Supuran, C.T.; Pastorek, J.; et al. Hypoxia activates the capacity of tumor-associated carbonic anhydrase IX to acidify extracellular pH. FEBS Lett. 2004, 577, 439–445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.-H.; Griffiths, J.R. How and Why Are Cancers Acidic? Carbonic Anhydrase IX and the Homeostatic Control of Tumour Extracellular pH. Cancers 2020, 12, 1616. [Google Scholar] [CrossRef]
- Gaspar, D.; Veiga, A.S.; Castanho, M.A.R.B. From antimicrobial to anticancer peptides. A review. Front. Microbiol. 2013, 4, 294. [Google Scholar] [CrossRef] [Green Version]
- Zhang, B.; Shi, W.; Li, J.; Liao, C.; Yang, L.; Huang, W.; Qian, H. Synthesis and biological evaluation of novel peptides based on antimicrobial peptides as potential agents with antitumor and multidrug resistance-reversing activities. Chem. Biol. Drug Des. 2017, 90, 972–980. [Google Scholar] [CrossRef]
- Wu, D.; Gao, Y.; Qi, Y.; Chen, L.; Ma, Y.; Li, Y. Peptide-based cancer therapy: Opportunity and challenge. Cancer Lett. 2014, 351, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Xu, J.; Jia, X. Research Progress Evaluating the Function and Mechanism of Anti-Tumor Peptides. Cancer Manag. Res. 2020, 12, 397–409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jagot-Lacoussiere, L.; Kotula, E.; Villoutreix, B.O.; Bruzzoni-Giovanelli, H.; Poyet, J.-L. A Cell-Penetrating Peptide Targeting AAC-11 Specifically Induces Cancer Cells Death. Cancer Res. 2016, 76, 5479–5490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Dong, C.; Li, X.; Han, W.; Su, X. Anticancer potential of bioactive peptides from animal sources (Review). Oncol. Rep. 2017, 38, 637–651. [Google Scholar] [CrossRef] [Green Version]
- Hilchie, A.L.; Hoskin, D.W.; Coombs, M.R.P. Anticancer Activities of Natural and Synthetic Peptides. Adv. Exp. Med. Biol. 2019, 1117, 131–147. [Google Scholar] [CrossRef]
- Méndez, N.D.J.H.; Casanova, Y.V.; Chimbi, A.K.G.; Hernández, E.; Castro, A.L.L.; Diaz, J.M.M.; Monroy, Z.J.R.; Castañeda, J.E.G. Synthetic Peptides Derived from Bovine Lactoferricin Exhibit Antimicrobial Activity against E. coli ATCC 11775, S. maltophilia ATCC 13636 and S. enteritidis ATCC 13076. Molecules 2017, 22, 452. [Google Scholar] [CrossRef] [Green Version]
- Wolf, J.S.; Li, G.; Varadhachary, A.; Petrak, K.; Schneyer, M.; Li, D.; Ongkasuwan, J.; Zhang, X.; Taylor, R.J.; Strome, S.E.; et al. Oral Lactoferrin Results in T Cell–Dependent Tumor Inhibition of Head and Neck Squamous Cell Carcinoma In vivo. Clin. Cancer Res. 2007, 13, 1601–1610. [Google Scholar] [CrossRef] [Green Version]
- Chernysh, S.; Kozuharova, I. Anti-tumor activity of a peptide combining patterns of insect alloferons and mammalian immunoglobulins in naïve and tumor antigen vaccinated mice. Int. Immunopharmacol. 2013, 17, 1090–1093. [Google Scholar] [CrossRef]
- Cui, W.; Aouidate, A.; Wang, S.; Yu, Q.; Li, Y.; Yuan, S. Discovering Anti-Cancer Drugs via Computational Methods. Front. Pharmacol. 2020, 11, 733. [Google Scholar] [CrossRef]
- Chan, H.C.S.; Shan, H.; Dahoun, T.; Vogel, H.; Yuan, S. Advancing Drug Discovery via Artificial Intelligence. Trends Pharmacol. Sci. 2019, 40, 592–604. [Google Scholar] [CrossRef]
- Mohs, R.C.; Greig, N.H. Drug discovery and development: Role of basic biological research. Alzheimers Dement. Transl. Res. Clin. Interv. 2017, 3, 651–657. [Google Scholar] [CrossRef]
- Paul, D.; Sanap, G.; Shenoy, S.; Kalyane, D.; Kalia, K.; Tekade, R.K. Artificial intelligence in drug discovery and development. Drug Discov. Today 2021, 26, 80–93. [Google Scholar] [CrossRef] [PubMed]
- Nascimento, A.C.A.; Prudêncio, R.B.C.; Costa, I.G. A multiple kernel learning algorithm for drug-target interaction prediction. BMC Bioinform. 2016, 17, 46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klopman, G.; Chakravarti, S.K.; Zhu, H.; Ivanov, J.M.; Saiakhov, R.D. ESP: A Method to Predict Toxicity and Pharmacological Properties of Chemicals Using Multiple MCASE Databases. J. Chem. Inf. Comput. Sci. 2004, 44, 704–715. [Google Scholar] [CrossRef]
- Menden, M.P.; Iorio, F.; Garnett, M.; McDermott, U.; Benes, C.H.; Ballester, P.J.; Saez-Rodriguez, J. Machine Learning Prediction of Cancer Cell Sensitivity to Drugs Based on Genomic and Chemical Properties. PLoS ONE 2013, 8, e61318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mak, K.-K.; Pichika, M.R. Artificial intelligence in drug development: Present status and future prospects. Drug Discov. Today 2019, 24, 773–780. [Google Scholar] [CrossRef] [PubMed]
- Refaeilzadeh, P.; Tang, L.; Liu, H. Cross-Validation. In Encyclopedia of Database Systems; Liu, L., Özsu, M.T., Eds.; Springer: New York, NY, USA, 2009; pp. 532–538. [Google Scholar]
- Deisy, C.; Subbulakshmi, B.; Baskar, S.; Ramaraj, N. Efficient dimensionality reduction approaches for feature selection. In Proceedings of the International Conference on Computational Intelligence and Multimedia Applications (ICCIMA 2007), Sivakasi, India, 13–15 December 2007; pp. 121–127. [Google Scholar]
- Hsu, H.-H.; Hsieh, C.-W. Feature Selection via Correlation Coefficient Clustering. J. Softw. 2010, 5, 1371–1377. [Google Scholar] [CrossRef]
- Altmann, A.; Toloşi, L.; Sander, O.; Lengauer, T. Permutation importance: A corrected feature importance measure. Bioinformatics 2010, 26, 1340–1347. [Google Scholar] [CrossRef]
- Malhi, A.; Gao, R. PCA-Based Feature Selection Scheme for Machine Defect Classification. IEEE Trans. Instrum. Meas. 2004, 53, 1517–1525. [Google Scholar] [CrossRef]
- Noble, W.S. What is a support vector machine? Nat. Biotechnol. 2006, 24, 1565–1567. [Google Scholar] [CrossRef]
- Keller, J.M.; Gray, M.R.; Givens, J.A. A fuzzy K-nearest neighbor algorithm. IEEE Trans. Syst. Man Cybern. 1985, SMC-15, 580–585. [Google Scholar] [CrossRef]
- Pal, M. Random forest classifier for remote sensing classification. Int. J. Remote Sens. 2005, 26, 217–222. [Google Scholar] [CrossRef]
- Lakshminarayanan, B.; Pritzel, A.; Blundell, C. Simple and scalable predictive uncertainty estimation using deep ensembles. In Proceedings of the 31st Conference on Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017; pp. 6405–6416. [Google Scholar]
- Ke, G.; Meng, Q.; Finley, T.; Wang, T.; Chen, W.; Ma, W.; Ye, Q.; Liu, T.-Y. LightGBM: A highly efficient gradient boosting decision tree. In Proceedings of the 31st International Conference on Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017; pp. 3149–3157. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. NIPS 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Sutskever, I.; Martens, J.; Hinton, G. Generating Text with recurrent neural networks. In Proceedings of the 28th International Conference on Machine Learning (ICML-11), Bellevue, WA, USA, 28 June–2 July 2011; pp. 1017–1024. [Google Scholar]
- Abiodun, O.I.; Jantan, A.; Omolara, A.E.; Dada, K.V.; Mohamed, N.A.; Arshad, H. State-of-the-art in artificial neural network applications: A survey. Heliyon 2018, 4, e00938. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tran, N.H.; Zhang, X.; Xin, L.; Shan, B.; Li, M. De novo peptide sequencing by deep learning. Proc. Natl. Acad. Sci. USA 2017, 114, 8247–8252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is all you need. In Proceedings of the 31st Conference on Neural Information Processing Systems (NIPS 2017), Long Beach, CA, USA, 4–9 December 2017; pp. 5998–6008. [Google Scholar]
- Roopak, M.; Tian, G.Y.; Chambers, J. Deep learning models for cyber security in IoT networks. In Proceedings of the 2019 IEEE 9th Annual Computing and Communication Workshop and Conference (CCWC), Nevada, TX, USA, 7–9 January 2019; pp. 0452–0457. [Google Scholar]
- Chiou, J.-M. Dynamical functional prediction and classification, with application to traffic flow prediction. Ann. Appl. Stat. 2012, 6, 1588–1614. [Google Scholar] [CrossRef] [Green Version]
- Arlot, S.; Celisse, A. A survey of cross-validation procedures for model selection. Stat. Surv. 2010, 4, 40–79. [Google Scholar] [CrossRef]
- Simon, R.M.; Subramanian, J.; Li, M.-C.; Menezes, S. Using cross-validation to evaluate predictive accuracy of survival risk classifiers based on high-dimensional data. Brief. Bioinform. 2011, 12, 203–214. [Google Scholar] [CrossRef] [Green Version]
- Lin, Y.-C.; Lim, Y.F.; Russo, E.; Schneider, P.; Bolliger, L.; Edenharter, A.; Altmann, K.-H.; Halin, C.; Hiss, J.A.; Schneider, G. Multidimensional Design of Anticancer Peptides. Angew. Chem. Int. Ed. 2015, 54, 10370–10374. [Google Scholar] [CrossRef]
- Grisoni, F.; Neuhaus, C.S.; Gabernet, G.; Müller, A.T.; Hiss, J.A.; Schneider, G. Designing Anticancer Peptides by Constructive Machine Learning. ChemMedChem 2018, 13, 1300–1302. [Google Scholar] [CrossRef]
- Grisoni, F.; Neuhaus, C.S.; Hishinuma, M.; Gabernet, G.; Hiss, J.A.; Kotera, M.; Schneider, G. De novo design of anticancer peptides by ensemble artificial neural networks. J. Mol. Model. 2019, 25, 112. [Google Scholar] [CrossRef]
- Zhao, Y.; Wang, S.; Fei, W.; Feng, Y.; Shen, L.; Yang, X.; Wang, M.; Wu, M. Prediction of Anticancer Peptides with High Efficacy and Low Toxicity by Hybrid Model Based on 3D Structure of Peptides. Int. J. Mol. Sci. 2021, 22, 5630. [Google Scholar] [CrossRef]
- Dhanda, S.K.; Mahajan, S.; Paul, S.; Yan, Z.; Kim, H.; Jespersen, M.C.; Jurtz, V.; Andreatta, M.; Greenbaum, J.A.; Marcatili, P.; et al. IEDB-AR: Immune epitope database—analysis resource in 2019. Nucleic Acids Res. 2019, 47, W502–W506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vita, R.; Mahajan, S.; Overton, J.A.; Dhanda, S.K.; Martini, S.; Cantrell, J.R.; Wheeler, D.K.; Sette, A.; Peters, B. The Immune Epitope Database (IEDB): 2018 update. Nucleic Acids Res. 2019, 47, D339–D343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vita, R.; Overton, J.A.; Greenbaum, J.A.; Ponomarenko, J.; Clark, J.D.; Cantrell, J.R.; Wheeler, D.K.; Gabbard, J.L.; Hix, D.; Sette, A.; et al. The immune epitope database (IEDB) 3.0. Nucleic Acids Res. 2015, 43, D405–D412. [Google Scholar] [CrossRef]
- Gautam, A.; Singh, H.; Tyagi, A.; Chaudhary, K.; Kumar, R.; Kapoor, P.; Raghava, G.P.S. CPPsite: A curated database of cell penetrating peptides. Database 2012, 2012, bas015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agrawal, P.; Bhalla, S.; Usmani, S.S.; Singh, S.; Chaudhary, K.; Raghava, G.P.S.; Gautam, A. CPPsite 2.0: A repository of experimentally validated cell-penetrating peptides. Nucleic Acids Res. 2016, 44, D1098–D1103. [Google Scholar] [CrossRef]
- Kumar, R.; Chaudhary, K.; Sharma, M.; Nagpal, G.; Chauhan, J.S.; Singh, S.; Gautam, A.; Raghava, G.P. AHTPDB: A comprehensive platform for analysis and presentation of antihypertensive peptides. Nucleic Acids Res. 2015, 43, D956–D962. [Google Scholar] [CrossRef] [Green Version]
- Manavalan, B.; Basith, S.; Shin, T.H.; Wei, L.; Lee, G. mAHTPred: A sequence-based meta-predictor for improving the prediction of anti-hypertensive peptides using effective feature representation. Bioinformatics 2019, 35, 2757–2765. [Google Scholar] [CrossRef]
- Minkiewicz, P.; Dziuba, J.; Iwaniak, A.; Dziuba, M.; Darewicz, M. BIOPEP Database and Other Programs for Processing Bioactive Peptide Sequences. J. AOAC Int. 2008, 91, 965–980. [Google Scholar] [CrossRef] [Green Version]
- Hasan, M.; Schaduangrat, N.; Basith, S.; Lee, G.; Shoombuatong, W.; Manavalan, B. HLPpred-Fuse: Improved and robust prediction of hemolytic peptide and its activity by fusing multiple feature representation. Bioinformatics 2020, 36, 3350–3356. [Google Scholar] [CrossRef]
- Sarosiek, K.A.; Ni Chonghaile, T.; Letai, A. Mitochondria: Gatekeepers of response to chemotherapy. Trends Cell Biol. 2013, 23, 612–619. [Google Scholar] [CrossRef] [Green Version]
- Pérez-Herrero, E.; Fernández-Medarde, A. Advanced targeted therapies in cancer: Drug nanocarriers, the future of chemotherapy. Eur. J. Pharm. Biopharm. 2015, 93, 52–79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Girard, N.; Mornex, F.; Van Houtte, P.; Cordier, J.-F.; van Schil, P. Thymoma: A Focus on Current Therapeutic Management. J. Thorac. Oncol. 2009, 4, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Pearce, A.; Haas, M.; Viney, R.; Pearson, S.-A.; Haywood, P.; Brown, C.; Ward, R. Incidence and severity of self-reported chemotherapy side effects in routine care: A prospective cohort study. PLoS ONE 2017, 12, e0184360. [Google Scholar] [CrossRef]
- Zakrzewski, W.; Dobrzyński, M.; Szymonowicz, M.; Rybak, Z. Stem cells: Past, present, and future. Stem Cell Res. Ther. 2019, 5, 170–182. [Google Scholar] [CrossRef]
- Petrelli, F.; Zaniboni, A.; Coinu, A.; Cabiddu, M.; Ghilardi, M.; Sgroi, G.; Barni, S. Cisplatin or Not in Advanced Gastric Cancer: A Systematic Review and Meta-Analysis. PLoS ONE 2013, 8, e83022. [Google Scholar] [CrossRef] [Green Version]
- Su, X.; Dong, C.; Zhang, J.; Su, L.; Wang, X.; Cui, H.; Chen, Z. Combination therapy of anti-cancer bioactive peptide with Cisplatin decreases chemotherapy dosing and toxicity to improve the quality of life in xenograft nude mice bearing human gastric cancer. Cell Biosci. 2014, 4, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Porporato, P.E.; Filigheddu, N.; Pedro, J.M.B.-S.; Kroemer, G.; Galluzzi, L. Mitochondrial metabolism and cancer. Cell Res. 2018, 28, 265–280. [Google Scholar] [CrossRef]
- Moreno-Sánchez, R.; Marín-Hernández, Á.; Gallardo-Pérez, J.C.; Pacheco-Velázquez, S.C.; Robledo-Cadena, D.X.; Padilla-Flores, J.A.; Saavedra, E.; Rodríguez-Enríquez, S. Physiological Role of Glutamate Dehydrogenase in Cancer Cells. Front. Oncol. 2020, 10, 429. [Google Scholar] [CrossRef]
- Farsinejad, S.; Gheisary, Z.; Samani, S.E.; Alizadeh, A.M. Mitochondrial targeted peptides for cancer therapy. Tumor Biol. 2015, 36, 5715–5725. [Google Scholar] [CrossRef]
- Hsu, P.P.; Sabatini, D.M. Cancer Cell Metabolism: Warburg and Beyond. Cell 2008, 134, 703–707. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Chen, S.; Yu, D. Metabolic Reprogramming of Chemoresistant Cancer Cells and the Potential Significance of Metabolic Regulation in the Reversal of Cancer Chemoresistance. Metabolites 2020, 10, 289. [Google Scholar] [CrossRef] [PubMed]
- Pranzini, E.; Pardella, E.; Paoli, P.; Fendt, S.-M.; Taddei, M.L. Metabolic Reprogramming in Anticancer Drug Resistance: A Focus on Amino Acids. Trends Cancer 2021, 7, 682–699. [Google Scholar] [CrossRef] [PubMed]
- Wise, D.R.; Thompson, C.B. Glutamine addiction: A new therapeutic target in cancer. Trends Biochem. Sci. 2010, 35, 427–433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, W.; Pelicano, H.; Huang, P. Cancer Metabolism: Is Glutamine Sweeter than Glucose? Cancer Cell. 2010, 18, 199–200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, L.; Alesi, G.N.; Kang, S. Glutaminolysis as a target for cancer therapy. Oncogene 2016, 35, 3619–3625. [Google Scholar] [CrossRef] [Green Version]
- Gao, P.; Tchernyshyov, I.; Chang, T.-C.; Lee, Y.-S.; Kita, K.; Ochi, T.; Zeller, K.I.; De Marzo, A.M.; Van Eyk, J.E.; Mendell, J.T.; et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature 2009, 458, 762–765. [Google Scholar] [CrossRef] [Green Version]
- Reynolds, M.R.; Lane, A.N.; Robertson, B.; Kemp, S.; Liu, Y.; Hill, B.G.; Dean, D.C.; Clem, B.F. Control of glutamine metabolism by the tumor suppressor Rb. Oncogene 2014, 33, 556–566. [Google Scholar] [CrossRef] [Green Version]
- Elorza, A.; Soro-Arnáiz, I.; Meléndez-Rodríguez, F.; Rodríguez-Vaello, V.; Marsboom, G.; de Cárcer, G.; Acosta-Iborra, B.; Albacete-Albacete, L.; Ordóñez, A.; Oviedo, L.S.; et al. HIF2α Acts as an mTORC1 Activator through the Amino Acid Carrier SLC7A5. Mol. Cell 2012, 48, 681–691. [Google Scholar] [CrossRef] [Green Version]
- Nicklin, P.; Bergman, P.; Zhang, B.; Triantafellow, E.; Wang, H.; Nyfeler, B.; Yang, H.; Hild, M.; Kung, C.; Wilson, C.; et al. Bidirectional Transport of Amino Acids Regulates mTOR and Autophagy. Cell 2009, 136, 521–534. [Google Scholar] [CrossRef] [Green Version]
- Hassanein, M.; Hoeksema, M.D.; Shiota, M.; Qian, J.; Harris, B.K.; Chen, H.; Clark, J.E.; Alborn, W.E.; Eisenberg, R.; Massion, P.P. SLC1A5 Mediates Glutamine Transport Required for Lung Cancer Cell Growth and Survival. Clin. Cancer Res. 2013, 19, 560–570. [Google Scholar] [CrossRef] [Green Version]
- Hassanein, M.; Qian, J.; Hoeksema, M.D.; Wang, J.; Jacobovitz, M.; Ji, X.; Harris, F.T.; Harris, B.K.; Boyd, K.L.; Chen, H.; et al. Targeting SLC1a5-mediated glutamine dependence in non-small cell lung cancer. Int. J. Cancer 2015, 137, 1587–1597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q.; Beaumont, K.A.; Otte, N.J.; Font, J.; Bailey, C.G.; Van Geldermalsen, M.; Sharp, D.M.; Tiffen, J.C.; Ryan, R.M.; Jormakka, M.; et al. Targeting glutamine transport to suppress melanoma cell growth. Int. J. Cancer 2014, 135, 1060–1071. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.M.; McBryant, S.J.; Tsukamoto, T.; Rojas, C.; Ferraris, D.; Hamilton, S.K.; Hansen, J.C.; Curthoys, N.P. Novel mechanism of inhibition of rat kidney-type glutaminase by bis-2-(5-phenylacetamido-1,2,4-thiadiazol-2-yl)ethyl sulfide (BPTES). Biochem. J. 2007, 406, 407–414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gross, M.I.; Demo, S.D.; Dennison, J.B.; Chen, L.; Chernov-Rogan, T.; Goyal, B.; Janes, J.R.; Laidig, G.J.; Lewis, E.R.; Li, J.; et al. Antitumor Activity of the Glutaminase Inhibitor CB-839 in Triple-Negative Breast Cancer. Mol. Cancer Ther. 2014, 13, 890–901. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.-B.; Erickson, J.W.; Fuji, R.; Ramachandran, S.; Gao, P.; Dinavahi, R.; Wilson, K.F.; Ambrosio, A.L.; Dias, S.M.; Dang, C.V.; et al. Targeting Mitochondrial Glutaminase Activity Inhibits Oncogenic Transformation. Cancer Cell 2010, 18, 207–219. [Google Scholar] [CrossRef] [Green Version]
- Csibi, A.; Fendt, S.-M.; Li, C.; Poulogiannis, G.; Choo, A.Y.; Chapski, D.J.; Jeong, S.M.; Dempsey, J.M.; Parkhitko, A.; Morrison, T.; et al. TThe mTORC1 pathway stimulates glutamine metabolism and cell proliferation by repressing SIRT4. Cell 2013, 153, 840–854. [Google Scholar] [CrossRef] [Green Version]
- Jin, L.; Li, D.; Alesi, G.N.; Fan, J.; Kang, H.-B.; Lu, Z.; Boggon, T.J.; Jin, P.; Yi, H.; Wright, E.R.; et al. Glutamate Dehydrogenase 1 Signals through Antioxidant Glutathione Peroxidase 1 to Regulate Redox Homeostasis and Tumor Growth. Cancer Cell 2015, 27, 257–270. [Google Scholar] [CrossRef] [Green Version]
- Korangath, P.; Teo, W.W.; Sadik, H.; Han, L.; Mori, N.; Huijts, C.M.; Wildes, F.; Bharti, S.; Zhang, Z.; Santa-Maria, C.A.; et al. Targeting Glutamine Metabolism in Breast Cancer with Aminooxyacetate. Clin. Cancer Res. 2015, 21, 3263–3273. [Google Scholar] [CrossRef] [Green Version]
- Thornburg, J.M.; Nelson, K.K.; Clem, B.F.; Lane, A.N.; Arumugam, S.; Simmons, A.; Eaton, J.W.; Telang, S.; Chesney, J. Targeting aspartate aminotransferase in breast cancer. Breast Cancer Res. 2008, 10, R84. [Google Scholar] [CrossRef] [Green Version]
- Tan, S.; Li, D.; Zhu, X. Cancer immunotherapy: Pros, cons and beyond. Biomed. Pharmacother. 2020, 124, 109821. [Google Scholar] [CrossRef]
- Kooshkaki, O.; Derakhshani, A.; Hosseinkhani, N.; Torabi, M.; Safaei, S.; Brunetti, O.; Racanelli, V.; Silvestris, N.; Baradaran, B. Combination of Ipilimumab and Nivolumab in Cancers: From Clinical Practice to Ongoing Clinical Trials. Int. J. Mol. Sci. 2020, 21, 4427. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, L.B.; Salama, A. A review of cancer immunotherapy toxicity. CA A Cancer J. Clin. 2020, 70, 86–104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chuang, C.-M.; Monie, A.; Wu, A.; Mao, C.-P.; Hung, C.-F. Treatment with LL-37 peptide enhances antitumor effects induced by CpG oligodeoxynucleotides against ovarian cancer. Hum. Gene Ther. 2009, 20, 303–313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmad, S.S.; Duke, S.; Jena, R.; Williams, M.V.; Burnet, N.G. Advances in radiotherapy. BMJ 2012, 345, e7765. [Google Scholar] [CrossRef] [PubMed]
- Haymart, M.R.; Banerjee, M.; Stewart, A.K.; Koenig, R.J.; Birkmeyer, J.D.; Griggs, J.J. Use of radioactive iodine for thyroid cancer. JAMA 2011, 306, 721–728. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamazaki, T.; Wennerberg, E.; Hensler, M.; Buqué, A.; Kraynak, J.; Fucikova, J.; Zhou, X.K.; Sveinbjørnsson, B.; Rekdal, Ø.; Demaria, S. LTX-315-enabled, radiotherapy-boosted immunotherapeutic control of breast cancer by NK cells. Oncoimmunology 2021, 10, 1962592. [Google Scholar] [CrossRef]
- Sveinbjørnsson, B.; Camilio, K.A.; Haug, B.E.; Rekdal, O. LTX-315: A first-in-class oncolytic peptide that reprograms the tumor microenvironment. Future Med. Chem. 2017, 9, 1339–1344. [Google Scholar] [CrossRef] [Green Version]
- Jha, S.; Sharma, P.K.; Malviya, R. Hyperthermia: Role and Risk Factor for Cancer Treatment. Achiev. Life Sci. 2016, 10, 161–167. [Google Scholar] [CrossRef] [Green Version]
- Jia, D.; Liu, J. Current devices for high-performance whole-body hyperthermia therapy. Expert Rev. Med. Devices 2010, 7, 407–423. [Google Scholar] [CrossRef]
- Wust, P.; Hildebrandt, B.; Sreenivasa, G.; Rau, B.; Gellermann, J.; Riess, H.; Felix, R.; Schlag, P.M. Hyperthermia in combined treatment of cancer. Lancet Oncol. 2002, 3, 487–497. [Google Scholar] [CrossRef]
- Chang, D.; Lim, M.; Goos, J.; Qiao, R.; Ng, Y.Y.; Mansfeld, F.M.; Jackson, M.; Davis, T.P.; Kavallaris, M. Biologically Targeted Magnetic Hyperthermia: Potential and Limitations. Front. Pharmacol. 2018, 9, 831. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moktan, S.; Raucher, D. Anticancer Activity of Proapoptotic Peptides is Highly Improved by Thermal Targeting using Elastin-like Polypeptides. Int. J. Pept. Res. Ther. 2012, 18, 227–237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kojima, S.; Suzuki, H.; Akakura, K.; Shimbo, M.; Ichikawa, T.; Ito, H. Alternative Antiandrogens to Treat Prostate Cancer Relapse After Initial Hormone Therapy. J. Urol. 2004, 171, 679–683. [Google Scholar] [CrossRef] [PubMed]
- Chlebowski, R.T.; Anderson, G.L. Changing Concepts: Menopausal Hormone Therapy and Breast Cancer. JNCI J. Natl. Cancer Inst. 2012, 104, 517–527. [Google Scholar] [CrossRef] [Green Version]
- Frampton, J.E.; Lyseng-Williamson, K.A. Degarelix. Drugs 2009, 69, 1967–1976. [Google Scholar] [CrossRef]
- Colleoni, M.; Giobbie-Hurder, A. Benefits and adverse effects of endocrine therapy. Ann. Oncol. 2010, 21, vii107–vii111. [Google Scholar] [CrossRef]
- McClements, L.; Annett, S.; Yakkundi, A.; O’Rourke, M.; Valentine, A.; Moustafa, N.; Alqudah, A.; Simões, B.M.; Furlong, F.; Short, A.; et al. FKBPL and its peptide derivatives inhibit endocrine therapy resistant cancer stem cells and breast cancer metastasis by downregulating DLL4 and Notch4. BMC Cancer 2019, 19, 351. [Google Scholar] [CrossRef] [Green Version]
- Dougherty, T.J.; Gomer, C.J.; Henderson, B.W.; Jori, G.; Kessel, D.; Korbelik, M.; Moan, J.; Peng, Q. Photodynamic therapy. J. Natl. Cancer Inst. 1998, 90, 889–905. [Google Scholar] [CrossRef] [Green Version]
- Baskaran, R.; Lee, J.; Yang, S.-G. Clinical development of photodynamic agents and therapeutic applications. Biomater. Res. 2018, 22, 25. [Google Scholar] [CrossRef]
- Calixto, G.M.F.; Bernegossi, J.; De Freitas, L.M.; Fontana, C.R.; Chorilli, M.; Grumezescu, A.M. Nanotechnology-Based Drug Delivery Systems for Photodynamic Therapy of Cancer: A Review. Molecules 2016, 21, 342. [Google Scholar] [CrossRef]
- Ellerby, H.M.; Arap, W.; Ellerby, L.M.; Kain, R.; Andrusiak, R.; Del Rio, G.; Krajewski, S.; Lombardo, C.R.; Rao, R.; Ruoslahti, E.; et al. Anti-cancer activity of targeted pro-apoptotic peptides. Nat. Med. 1999, 5, 1032–1038. [Google Scholar] [CrossRef] [PubMed]
- Han, K.; Lei, Q.; Wang, S.-B.; Hu, J.-J.; Qiu, W.-X.; Zhu, J.-Y.; Yin, W.-N.; Luo, X.; Zhang, X.-Z. Dual-Stage-Light-Guided Tumor Inhibition by Mitochondria-Targeted Photodynamic Therapy. Adv. Funct. Mater. 2015, 25, 2961–2971. [Google Scholar] [CrossRef]



| Peptide Name | Brand Name | Indication | First Approval | Reference |
|---|---|---|---|---|
| Ixazomib | Ninlaro | Multiple myeloma | 2015 (FDA) | [37] |
| Carfilzomib | Kyprolis | Multiple myeloma | 2012 (FDA) | [38] |
| Bortezomib | Velcade | Multiple myeloma | 2003 (FDA) | |
| Goserelin | Zoladex | Prostate cancer | 1989 (FDA) | |
| Histrelin | Vantas | Prostate cancer | 2004 (FDA) | |
| Leuprolide | Lupron | Prostate cancer | 1985 (FDA) | |
| Degarelix | Firmagon | Prostate cancer | 2008 (FDA) | |
| Romidepsin | Istodax | T-cell lymphoma | 2009 (EMA) | |
| Thymalfasin | Zadaxin | Hepatocellular carcinoma | 2002 (EMA) | |
| Triptorelin | Trelstar | Hormone-responsive cancers | 2010 (FDA) | |
| Mifamurtide | Mepact | Osteosarcoma | 2009 (EMA) |
| Name | Datasets Size | Model URL | Method | Accuracy | References |
|---|---|---|---|---|---|
| MLACP | T: 187 ACPs and 398 non ACPs I: 422 ACPs and 422 non ACPs | http://www.thegleelab.org/MLACP/MLACP.html | ML | 88.72% | [24] |
| Lv et al. | T: 861 ACPs and 861 non ACPs I: 970 ACPs and 970 non ACPs | https://github.com/zhibinlv/iACP-DRLF | Hybrid learning | 93.5% | [25] |
| ACP-DL | T: 376 ACPs and 364 non ACPs I: 129 ACPs and 111 non ACPs | https://github.com/haichengyi/ACP-DL | DL | 81.48% and 85.42% | [45] |
| AntiCP 2.0 | T: 861 ACPs and 861 non ACPs I: 970 ACPs and 970 non ACPs | https://webs.iiitd.edu.in/raghava/anticp2/ | ML | 72.81% and 88.81% | [46] |
| Hajisharifi et al. | T: 138 ACPs and 206 non ACPs I: 22 ACPs | NA | ML | 83.82% and 89.7% | [48] |
| ACPP | T: 217 ACPs and 3979 non ACPs I: 40 ACPs and 40 non ACPs | http://acpp.bicpu.edu.in/predict.php | ML | 96% | [49] |
| iACP | T: 138 ACPs and 206 non ACPs I: 150 ACPs and 150 non ACPs | http://lin.uestc.edu.cn/server/iACP | ML | 92.67% | [50] |
| iACP-GAEnsC | T: 138 ACPs and 206 non ACPs I: NA | NA | ML | 96.45% | [51] |
| SAP | T: 138 ACPs and 206 non ACPs I: NA | NA | ML | 91% | [52] |
| ACPred-FL | T: 250 ACPs and 250 non ACPs I: 82 ACPs and 82 non ACPs | http://server.malab.cn/ACPred-FL | ML | 91.4% | [53] |
| mACPpred | T: 266 ACPs and 266 non ACPs I: 157 ACPs and 157 non ACPs | http://thegleelab.org/mACPpred/ | ML | 91.7% | [54] |
| ACPred | T: 138 ACPs and 205 non ACPs I: 250 ACPs and 250 non ACPs | http://codes.bio/acpred/ | ML | 92.87% | [55] |
| PEPred-Suite | T: 250 ACPs and 250 non ACPs I: 82 ACPs and 82 non ACPs | http://server.malab.cn/PEPred-Suite | ML | NA | [56] |
| DRACP | T: 138 ACPs and 206 non ACPs I: 150 ACPs and 150 non ACPs | https://github.com/zty2009/ACP | ML | 96% | [57] |
| PTPD | T: 225 ACPs and 2250 non ACPs I: 138 ACPs and 206 non ACPs | NA | DL | 96% | [62] |
| DeepACP | T: 250 ACPs and 250 non ACPs I: 82 ACPs and 82 non ACPs | https://github.com/jingry/autoBioSeqpy/tree/master/examples/anticancer_peptide_prediction | DL | 84.9% | [63] |
| ACPred-LAF | T: 558 ACPs and 558 non ACPs I: 148 ACPs and 148 non ACPs | https://github.com/TearsWaiting/ACPred-LAF | DL | 81.15% | [64] |
| ACP-DA | T: 376 ACPs and 364 non ACPs I: 129 ACPs and 111 non ACPs | https://github.com/chenxgscuec/ACPDA | Hybrid learning | 82.03% and 88.33% | [65] |
| Database | Total ACPs | Database URL | Reference |
|---|---|---|---|
| CancerPPD | 3491 | http://crdd.osdd.net/raghava/cancerppd/ | [98] |
| APD3 | 185 | http://aps.unmc.edu/AP/ | [99] |
| SATPdb | 1099 | http://crdd.osdd.net/raghava/satpdb/ | [100] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hwang, J.S.; Kim, S.G.; Shin, T.H.; Jang, Y.E.; Kwon, D.H.; Lee, G. Development of Anticancer Peptides Using Artificial Intelligence and Combinational Therapy for Cancer Therapeutics. Pharmaceutics 2022, 14, 997. https://doi.org/10.3390/pharmaceutics14050997
Hwang JS, Kim SG, Shin TH, Jang YE, Kwon DH, Lee G. Development of Anticancer Peptides Using Artificial Intelligence and Combinational Therapy for Cancer Therapeutics. Pharmaceutics. 2022; 14(5):997. https://doi.org/10.3390/pharmaceutics14050997
Chicago/Turabian StyleHwang, Ji Su, Seok Gi Kim, Tae Hwan Shin, Yong Eun Jang, Do Hyeon Kwon, and Gwang Lee. 2022. "Development of Anticancer Peptides Using Artificial Intelligence and Combinational Therapy for Cancer Therapeutics" Pharmaceutics 14, no. 5: 997. https://doi.org/10.3390/pharmaceutics14050997
APA StyleHwang, J. S., Kim, S. G., Shin, T. H., Jang, Y. E., Kwon, D. H., & Lee, G. (2022). Development of Anticancer Peptides Using Artificial Intelligence and Combinational Therapy for Cancer Therapeutics. Pharmaceutics, 14(5), 997. https://doi.org/10.3390/pharmaceutics14050997






