Pharmacogenetics of Osteoporosis: A Pathway Analysis of the Genetic Influence on the Effects of Antiresorptive Drugs
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Densitometry
2.2. DNA Isolation and Genotyping
2.3. Quality Control and Data Analysis
3. Results
3.1. Patients, BMD Changes and SNPs
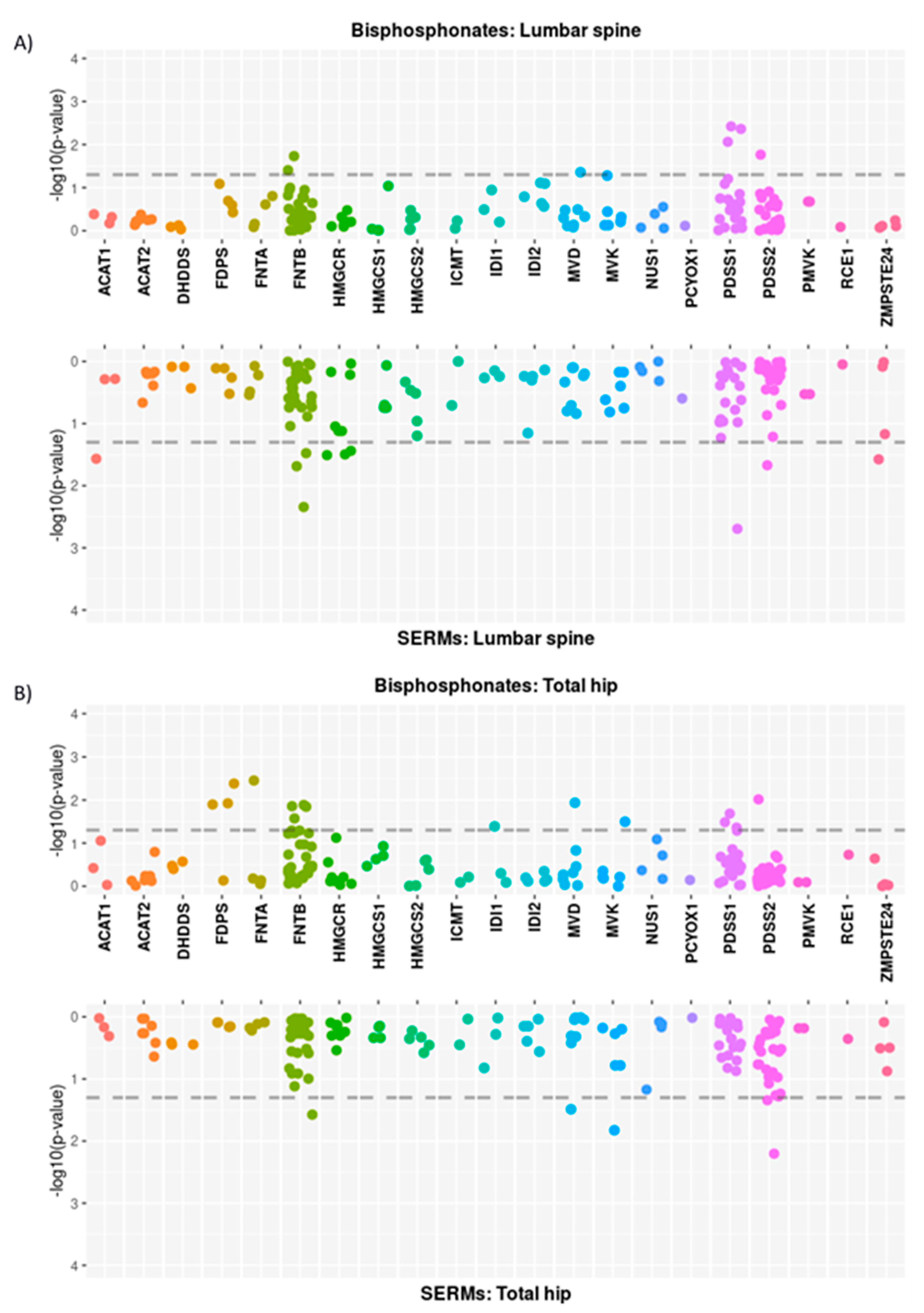
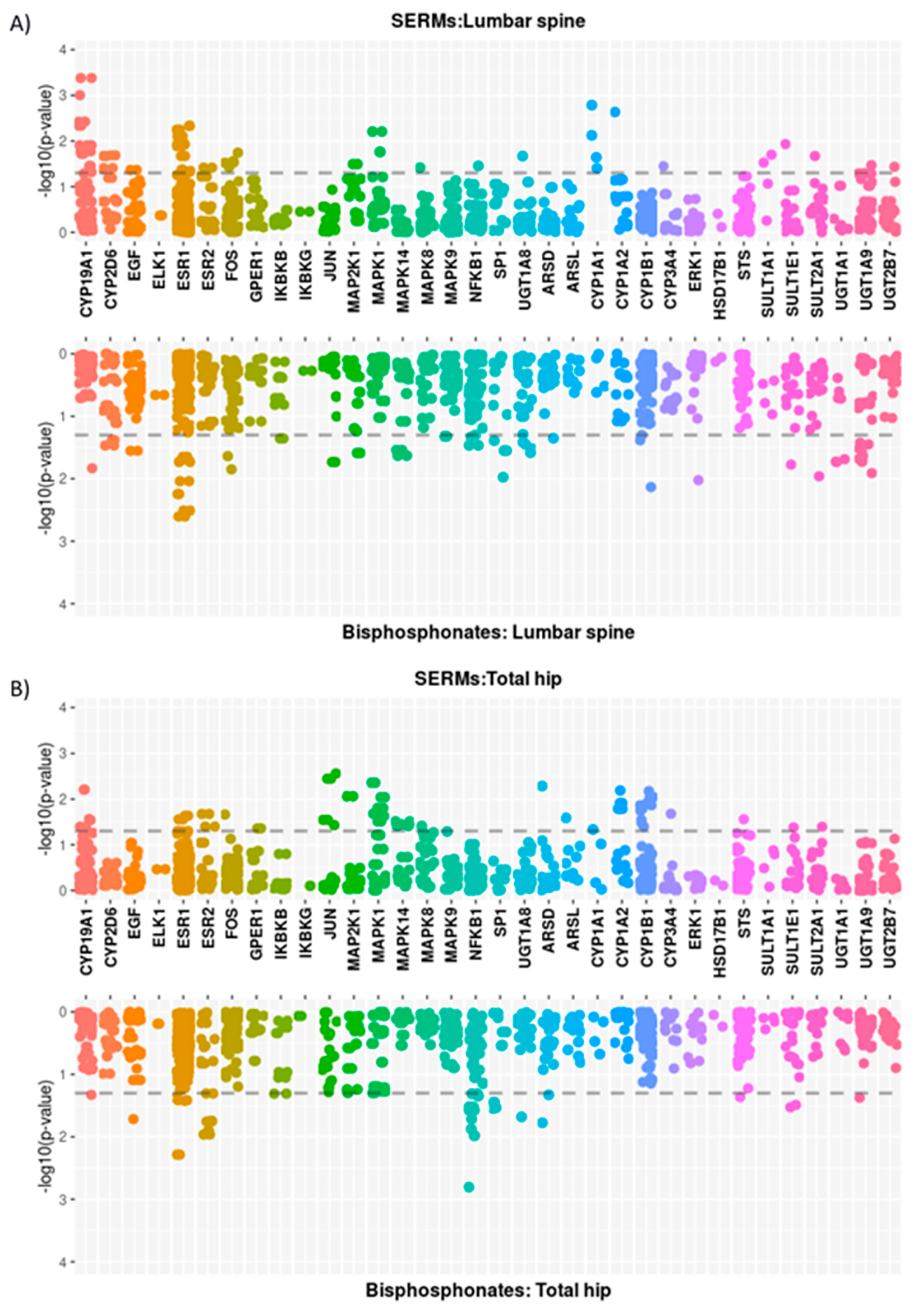
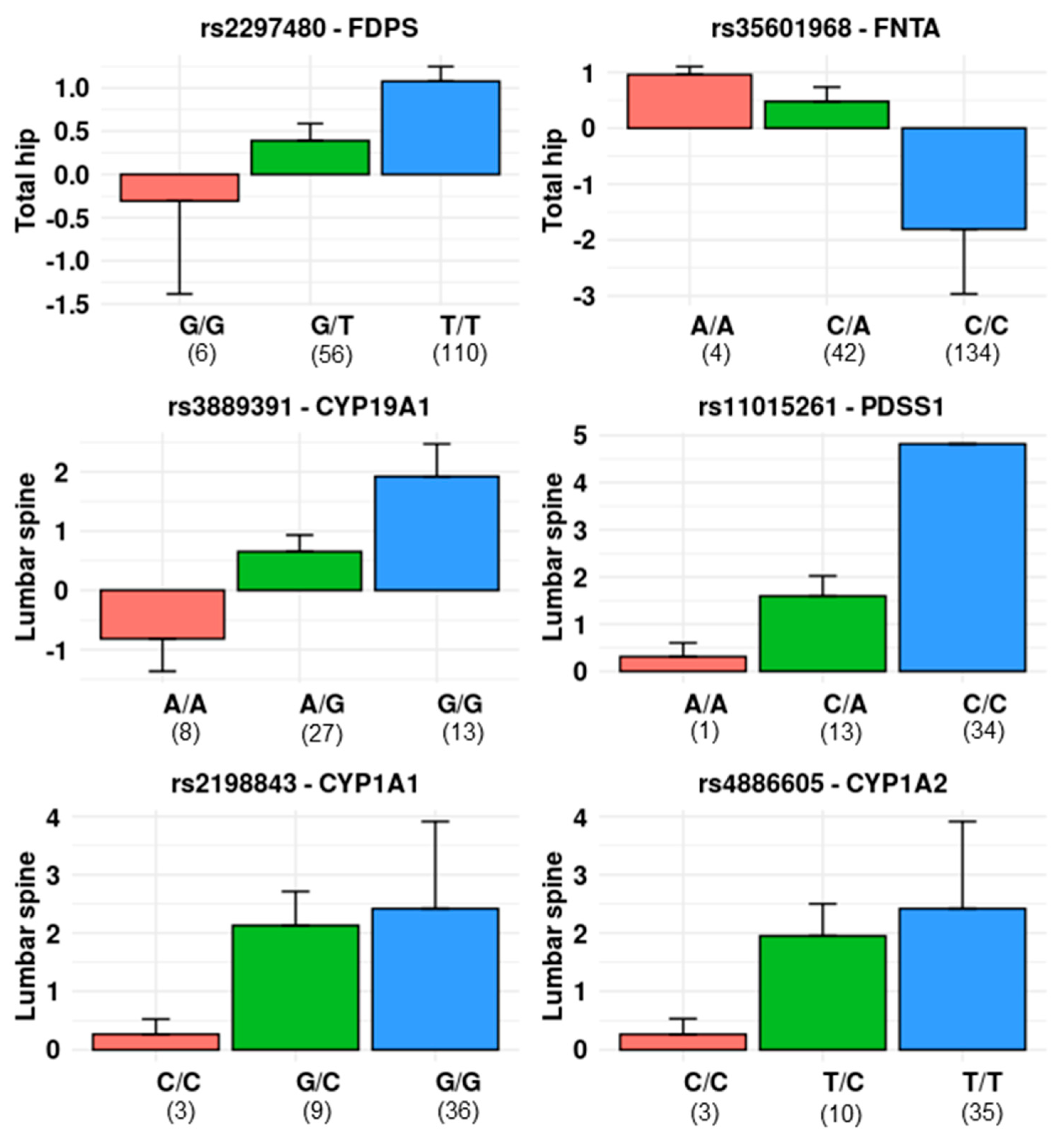
3.2. Single Locus Analysis
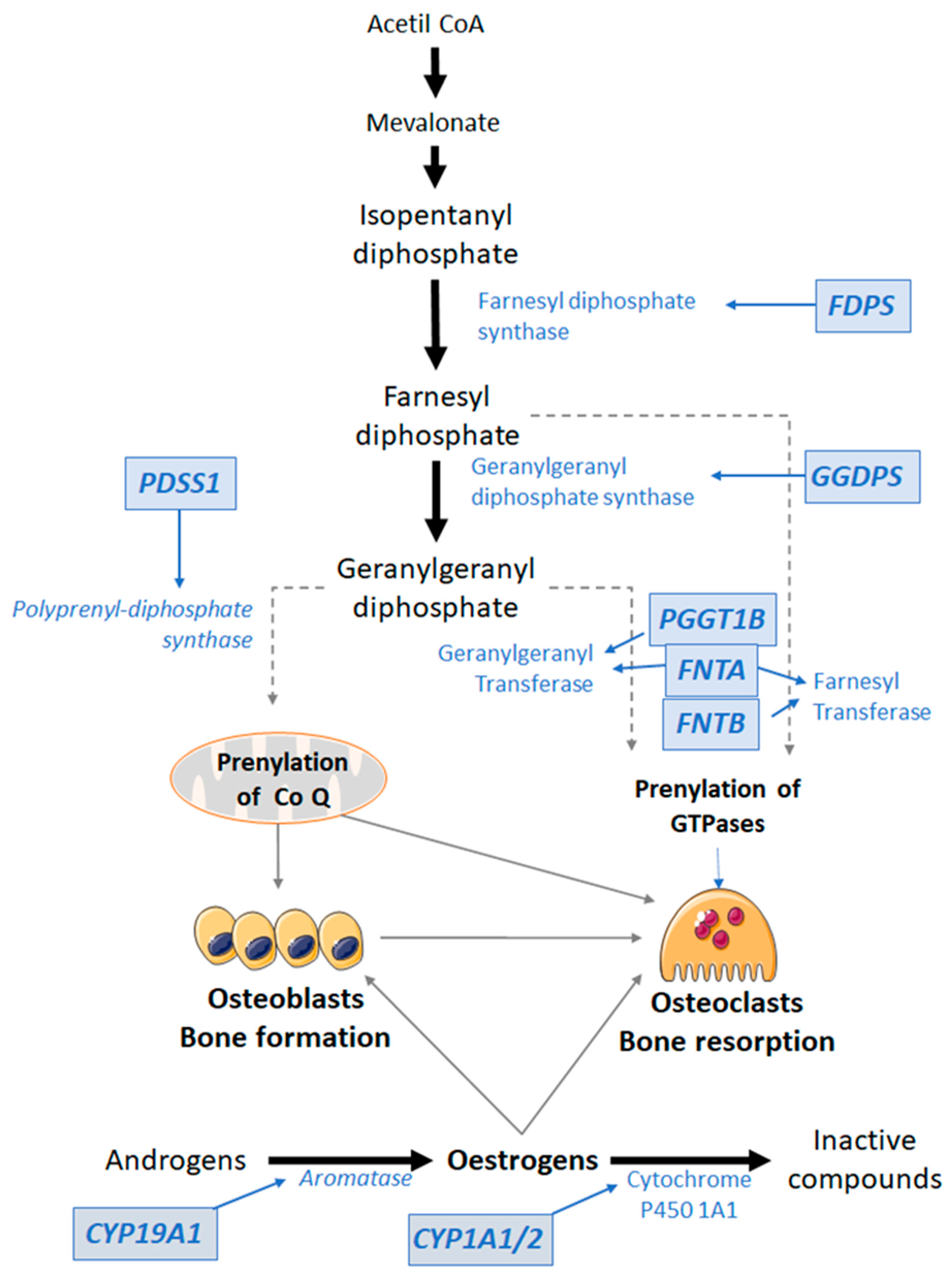
3.2.1. Mevalonate Pathway
3.2.2. Oestrogen-Related Pathway
3.3. Multimarker and Gene-Based Analysis
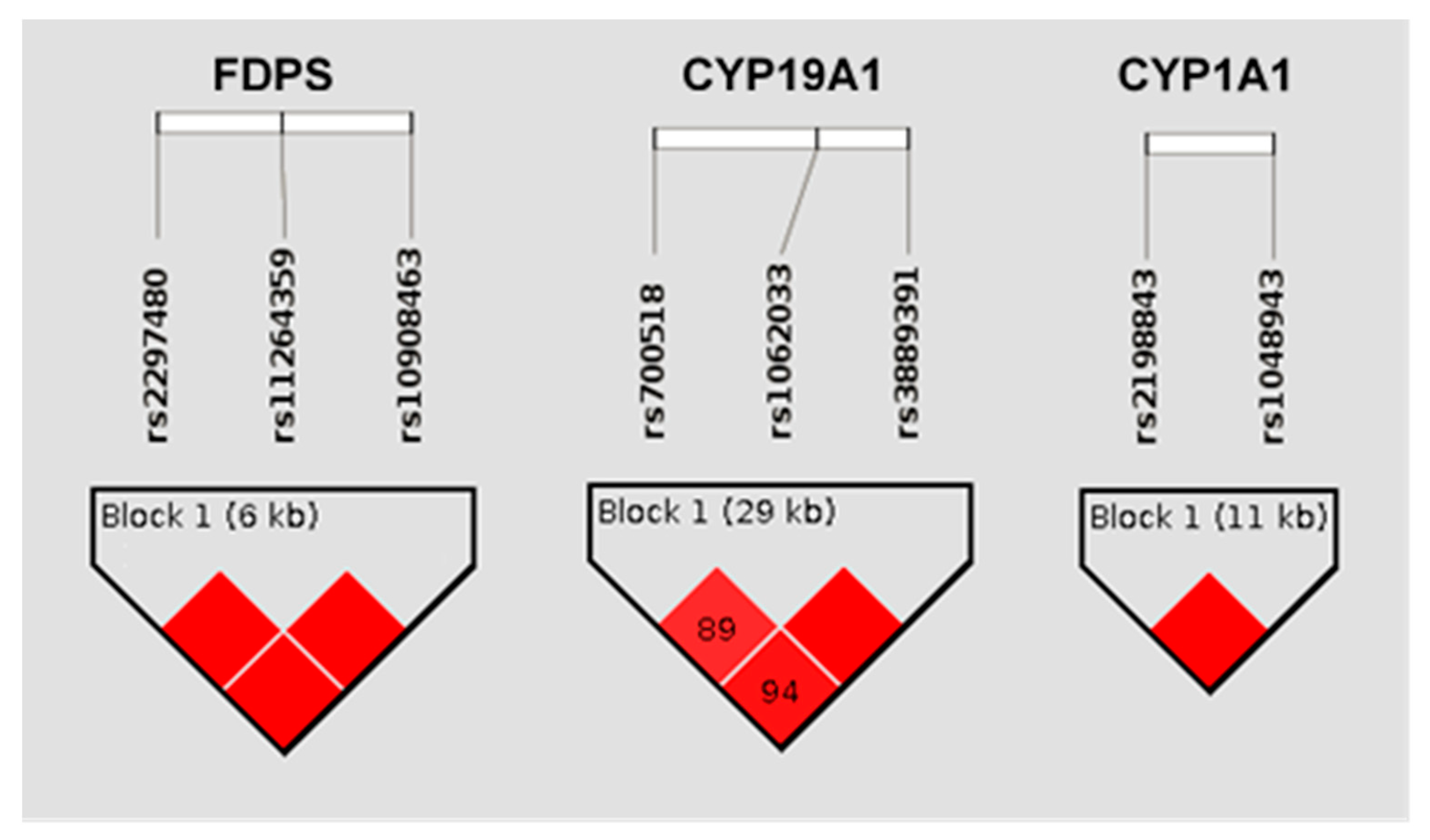
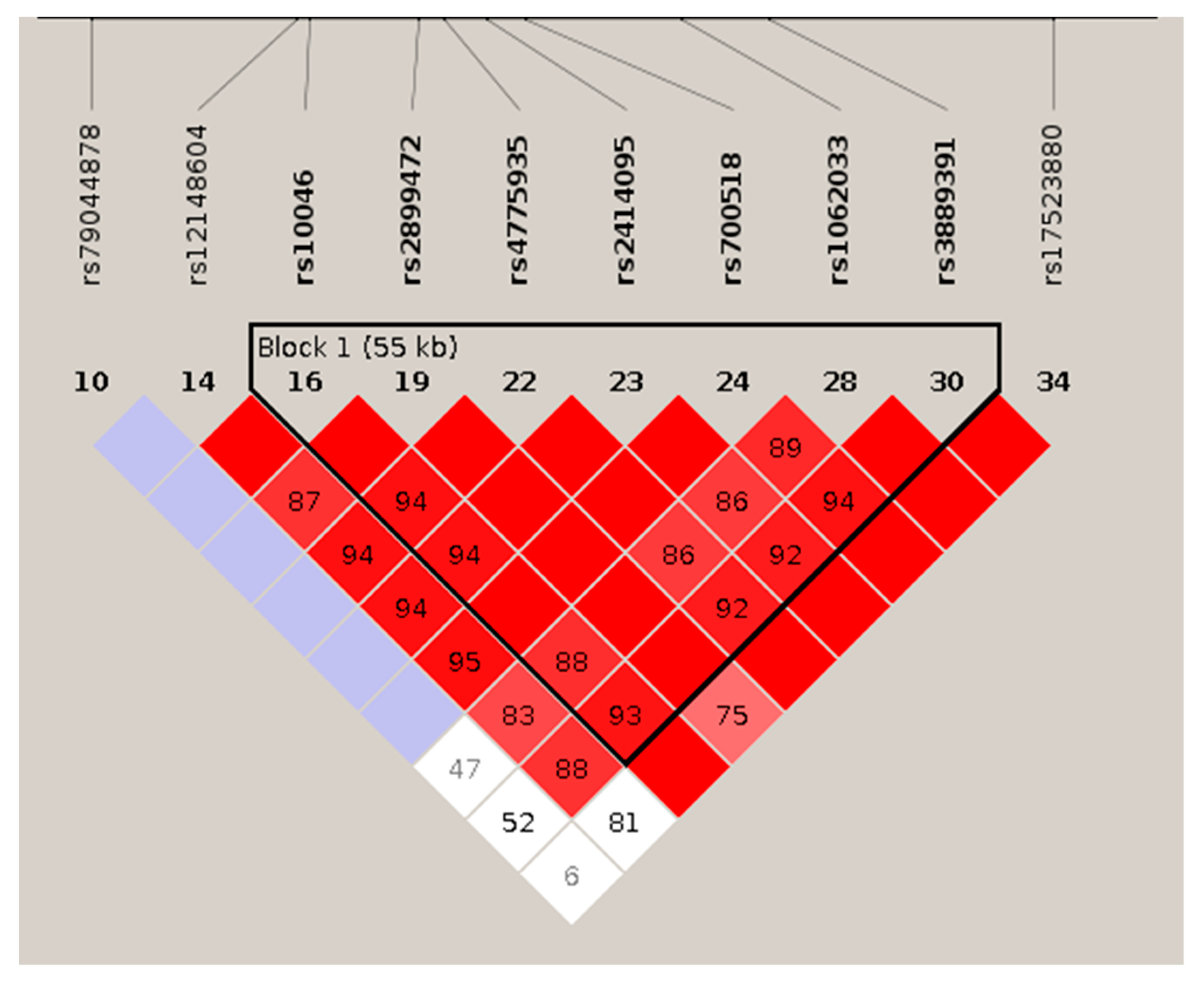
3.3.1. Haplotypic Analysis
3.3.2. Gene-Based Analysis
3.3.3. Multivariate Analysis
4. Discussion
4.1. Pharmacodynamics and Therapeutic Role of Antiresorptive Drugs
4.2. Relationship of Gene Variants with the Effect of Bisphosphonates
4.3. Relationship of Gene Variants with the Effect of SERMs
4.4. Clinical Relevance
4.5. Study Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lorentzon, M.; Cummings, S.R. Osteoporosis: The evolution of a diagnosis. J. Intern. Med. 2015, 277, 650–661. [Google Scholar] [CrossRef] [PubMed]
- Ettinger, B.; Black, D.M.; Mitlak, B.H.; Knickerbocker, R.K.; Nickelsen, T.; Genant, H.K.; Christiansen, C.; Delmas, P.D.; Zanchetta, J.R.; Stakkestad, J.; et al. Reduction of vertebral fracture risk in postmenopausal women with osteoporosis treated with raloxifene: Results from a 3-year randomized clinical trial. Multiple Outcomes of Raloxifene Evaluation (MORE) Investigators. JAMA 1999, 282, 637–645. [Google Scholar] [CrossRef] [PubMed]
- Russell, R.G.G.; Watts, N.B.; Ebetino, F.H.; Rogers, M.J. Mechanisms of action of bisphosphonates: Similarities and differences and their potential influence on clinical efficacy. Osteoporos. Int. 2008, 19, 733–759. [Google Scholar] [CrossRef] [PubMed]
- Barrionuevo, P.; Kapoor, E.; Asi, N.; Alahdab, F.; Mohammed, K.; Benkhadra, K.; Almasri, J.; Farah, W.; Sarigianni, M.; Muthusamy, K.; et al. Efficacy of Pharmacological Therapies for the Prevention of Fractures in Postmenopausal Women: A Network Meta-Analysis. J. Clin. Endocrinol. Metab. 2019, 104, 1623–1630. [Google Scholar] [CrossRef]
- Chen, L.X.; Zhou, Z.R.; Li, Y.L.; Ning, G.Z.; Zhang, T.S.; Zhang, D.; Feng, S.Q. Comparison of Bone Mineral Density in Lumbar Spine and Fracture Rate among Eight Drugs in Treatments of Osteoporosis in Men: A Network Meta-Analysis. PLoS ONE 2015, 10, e0128032. [Google Scholar] [CrossRef]
- Freemantle, N.; Cooper, C.; Diez-Perez, A.; Gitlin, M.; Radcliffe, H.; Shepherd, S.; Roux, C. Results of indirect and mixed treatment comparison of fracture efficacy for osteoporosis treatments: A meta-analysis. Osteoporos. Int. 2013, 24, 209–217. [Google Scholar] [CrossRef]
- Ralston, S.H.; Uitterlinden, A.G. Genetics of osteoporosis. Endocr. Rev. 2010, 31, 629–662. [Google Scholar] [CrossRef]
- López-Delgado, L.; Riancho-Zarrabeitia, L.; Riancho, J.A. Genetic and acquired factors influencing the effectiveness and toxicity of drug therapy in osteoporosis. Expert Opin. Drug Metab. Toxicol. 2016, 12, 389–398. [Google Scholar] [CrossRef]
- Riancho, J.A.; Hernández, J.L. Pharmacogenomics of osteoporosis: A pathway approach. Pharmacogenomics 2012, 13, 815–829. [Google Scholar] [CrossRef] [PubMed]
- Morris, J.A.; Kemp, J.P.; Youlten, S.E.; Laurent, L.; Logan, J.G.; Chai, R.C.; Vulpescu, N.A.; Forgetta, V.; Kleinman, A.; Mohanty, S.T.; et al. An atlas of genetic influences on osteoporosis in humans and mice. Nat. Genet. 2019, 51, 258–266. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Bai, W.; Zheng, H. Twelve years of GWAS discoveries for osteoporosis and related traits: Advances, challenges and applications. Bone Res. 2021, 9, 23. [Google Scholar] [CrossRef] [PubMed]
- Estrada, K.; Styrkarsdottir, U.; Evangelou, E.; Hsu, Y.H.; Duncan, E.L.; Ntzani, E.E.; Oei, L.; Albagha, O.M.E.; Amin, N.; Kemp, J.P.; et al. Genome-wide meta-analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. Nat. Genet. 2012, 44, 491–501. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; De Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Barrett, J.C.; Fry, B.; Maller, J.; Daly, M.J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 2005, 21, 263–265. [Google Scholar] [CrossRef] [PubMed]
- de Leeuw, C.A.; Mooij, J.M.; Heskes, T.; Posthuma, D. MAGMA: Generalized Gene-Set Analysis of GWAS Data. PLoS Comput. Biol. 2015, 11, e1004219. [Google Scholar] [CrossRef] [PubMed]
- Bouxsein, M.L.; Eastell, R.; Lui, L.Y.; Wu, L.A.; de Papp, A.E.; Grauer, A.; Marin, F.; Cauley, J.A.; Bauer, D.C.; Black, D.M. Change in Bone Density and Reduction in Fracture Risk: A Meta-Regression of Published Trials. J. bone Miner. Res. 2019, 34, 632–642. [Google Scholar] [CrossRef]
- Rogers, M.J.; Mönkkönen, J.; Munoz, M.A. Molecular mechanisms of action of bisphosphonates and new insights into their effects outside the skeleton. Bone 2020, 139, 115493. [Google Scholar] [CrossRef]
- Jiang, P.; Zhang, P.; Mukthavaram, R.; Nomura, N.; Pingle, S.C.; Teng, D.; Chien, S.; Guo, F.; Kesari, S. Anti-cancer effects of nitrogen-containing bisphosphonates on human cancer cells. Oncotarget 2016, 7, 57932–57942. [Google Scholar] [CrossRef]
- Ghavami, S.; Mutawe, M.M.; Schaafsma, D.; Yeganeh, B.; Unruh, H.; Klonisch, T.; Halayko, A.J. Geranylgeranyl transferase 1 modulates autophagy and apoptosis in human airway smooth muscle. Am. J. Physiol. Lung Cell. Mol. Physiol. 2012, 302, L420–L428. [Google Scholar] [CrossRef]
- Das, S.; Edwards, P.A.; Crockett, J.C.; Rogers, M.J. Upregulation of endogenous farnesyl diphosphate synthase overcomes the inhibitory effect of bisphosphonate on protein prenylation in Hela cells. Biochim. Biophys. Acta 2014, 1841, 569–573. [Google Scholar] [CrossRef]
- Olmos, J.M.; Zarrabeitia, M.T.; Hernández, J.L.; Sãudo, C.; González-Macías, J.; Riancho, J.A. Common allelic variants of the farnesyl diphosphate synthase gene influence the response of osteoporotic women to bisphosphonates. Pharm. J. 2012, 12, 227–232. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Castrillón, J.L.; Zarrabeitia, M.T.; Abad, L.; Vega, G.; Ruiz-Mambrilla, M.; Gonzalez-Sagredo, M.; Dueñas-Laita, A.; Riancho, J.A. Polymorphisms of the farnesyl diphosphate synthase gene modulate bone changes in response to atorvastatin. Rheumatol. Int. 2014, 34, 1073–1077. [Google Scholar] [CrossRef] [PubMed]
- Marozik, P.; Alekna, V.; Rudenko, E.; Tamulaitiene, M.; Rudenka, A.; Mastaviciute, A.; Samokhovec, V.; Cernovas, A.; Kobets, K.; Mosse, I. Bone metabolism genes variation and response to bisphosphonate treatment in women with postmenopausal osteoporosis. PLoS ONE 2019, 14, e0221511. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, H.; Li, M.; Zhou, P.; Xing, X.; Xia, W.; Zhang, Z.; Liao, E.; Chen, D.; Liu, J.; et al. Association of farnesyl diphosphate synthase polymorphisms and response to alendronate treatment in Chinese postmenopausal women with osteoporosis. Chin. Med. J. 2014, 127, 662–668. [Google Scholar]
- Marini, F.; Falchetti, A.; Silvestri, S.; Bagger, Y.; Luzi, E.; Tanini, A.; Christiansen, C.; Brandi, M.L. Modulatory effect of farnesyl pyrophosphate synthase (FDPS) rs2297480 polymorphism on the response to long-term amino-bisphosphonate treatment in postmenopausal osteoporosis. Curr. Med. Res. Opin. 2008, 24, 2609–2615. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Wu, T.Y.; Zhao, M.F.; Li, C.J. The balance of protein farnesylation and geranylgeranylation during the progression of nonalcoholic fatty liver disease. J. Biol. Chem. 2020, 295, 5152–5162. [Google Scholar] [CrossRef] [PubMed]
- Riancho, J.A.; Sañudo, C.; Valero, C.; Pipaón, C.; Olmos, J.M.; Mijares, V.; Fernández-Luna, J.L.; Zarrabeitia, M.T. Association of the aromatase gene alleles with BMD: Epidemiological and functional evidence. J. Bone Miner. Res. 2009, 24, 1709–1718. [Google Scholar] [CrossRef]
- Valero, C.; Pérez-Castrillón, J.L.; Zarrabeitia, M.T.; Hernández, J.L.; Alonso, M.A.; Del Pino-Montes, J.; Olmos, J.M.; González-Macías, J.; Riancho, J.A. Association of aromatase and estrogen receptor gene polymorphisms with hip fractures. Osteoporos. Int. 2008, 19, 787–792. [Google Scholar] [CrossRef][Green Version]
- Riancho, J.A.; Valero, C.; Naranjo, A.; Morales, D.J.; Sañudo, C.; Zarrabeitia, M.T. Identification of an aromatase haplotype that is associated with gene expression and postmenopausal osteoporosis. J. Clin. Endocrinol. Metab. 2007, 92, 660–665. [Google Scholar] [CrossRef][Green Version]
- Guan, Q.; Song, X.; Zhang, Z.; Zhang, Y.; Chen, Y.; Li, J. Identification of Tamoxifen-Resistant Breast Cancer Cell Lines and Drug Response Signature. Front. Mol. Biosci. 2020, 7, 564005. [Google Scholar] [CrossRef]
- Zheng, D.; Cui, C.; Shao, C.; Wang, Y.; Ye, C.; Lv, G. Coenzyme Q10 inhibits RANKL-induced osteoclastogenesis by regulation of mitochondrial apoptosis and oxidative stress in RAW264.7 cells. J. Biochem. Mol. Toxicol. 2021, 35, e22778. [Google Scholar] [CrossRef] [PubMed]
- Ling, W.; Krager, K.; Richardson, K.K.; Warren, A.D.; Ponte, F.; Aykin-Burns, N.; Manolagas, S.C.; Almeida, M.; Kim, H.N. Mitochondrial Sirt3 contributes to the bone loss caused by aging or estrogen deficiency. JCI Insight 2021, 6, e146728. [Google Scholar] [CrossRef] [PubMed]
- Morse, L.R.; Troy, K.L.; Fang, Y.; Nguyen, N.; Battaglino, R.; Goldstein, R.F.; Gupta, R.; Taylor, J.A. Combination Therapy With Zoledronic Acid and FES-Row Training Mitigates Bone Loss in Paralyzed Legs: Results of a Randomized Comparative Clinical Trial. JBMR Plus 2019, 3, e101617. [Google Scholar] [CrossRef]
- Ho-Le, T.P.; Tran, H.T.T.; Center, J.R.; Eisman, J.A.; Nguyen, H.T.; Nguyen, T.V. Assessing the clinical utility of genetic profiling in fracture risk prediction: A decision curve analysis. Osteoporos. Int. 2021, 32, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Forgetta, V.; Keller-Baruch, J.; Forest, M.; Durand, A.; Bhatnagar, S.; Kemp, J.P.; Nethander, M.; Evans, D.; Morris, J.A.; Kiel, D.P.; et al. Development of a polygenic risk score to improve screening for fracture risk: A genetic risk prediction study. PLoS Med. 2020, 17, e1003152. [Google Scholar] [CrossRef]
- Bauer, D.C.; Black, D.M.; Bouxsein, M.L.; Lui, L.Y.; Cauley, J.A.; de Papp, A.E.; Grauer, A.; Khosla, S.; McCulloch, C.E.; Eastell, R. Treatment-Related Changes in Bone Turnover and Fracture Risk Reduction in Clinical Trials of Anti-Resorptive Drugs: A Meta-Regression. J. Bone Miner. Res. 2018, 33, 634–642. [Google Scholar] [CrossRef]
- Eastell, R.; Black, D.M.; Lui, L.Y.; Chines, A.; Marin, F.; Khosla, S.; de Papp, A.E.; Cauley, J.A.; Mitlak, B.; McCulloch, C.E.; et al. Treatment-Related Changes in Bone Turnover and Fracture Risk Reduction in Clinical Trials of Antiresorptive Drugs: Proportion of Treatment Effect Explained. J. Bone Miner. Res. 2021, 36, 236–243. [Google Scholar] [CrossRef]
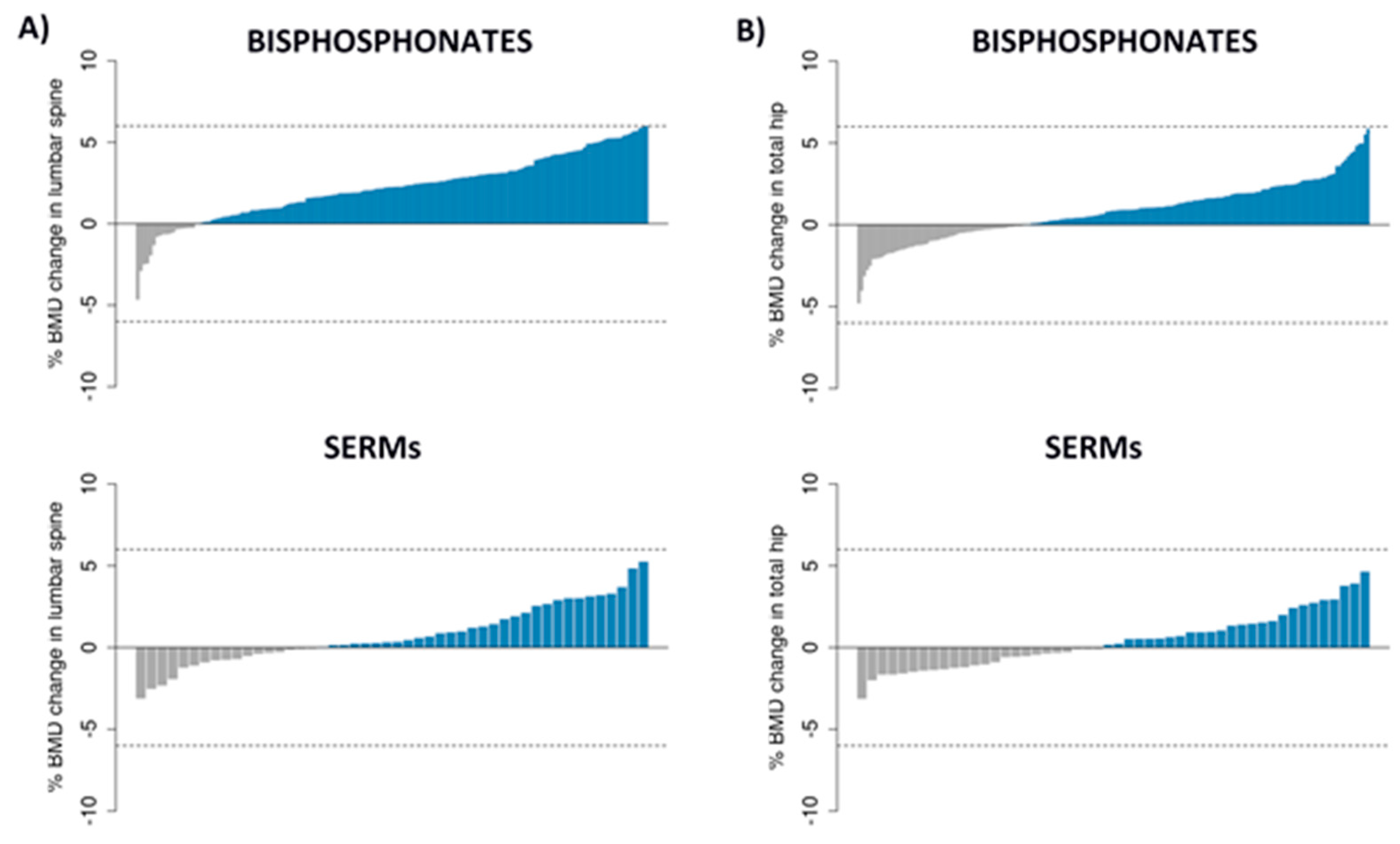


| Patient Characteristics | Bisphosphonate Group (n = 192; LS = 154; TH = 180) | SERM Group (n = 51; LS = 48; TH = 50) |
|---|---|---|
| Age, yr | 67 ± 9 | 59 ± 7 |
| BMI, Kg/m2 | 25.7 ± 4 | 25.9 ± 3.8 |
| Baseline Spine DXA, g/cm2 | 0.745 ± 0.098 | 0.757 ± 0.093 |
| Baseline Hip DXA, g/cm2 | 0.733 ± 0.095 | 0.781 ± 0.085 |
| Alendronate, n | 143 | - |
| Risedronate, n | 49 | - |
| Raloxifene, n | - | 43 |
| Bazedoxifene, n | - | 8 |
| Between-DXA interval, yr | 2.6 ± 0.6 | 2.6 ± 0.7 |
| Probes Group | Region | Treatment | SNPs with p < 0.05 (n) | Genes with Significantly- Associated SNPs |
|---|---|---|---|---|
| Mevalonate genes | Lumbar spine | Bisphosphonates | 7 | FNTB; MVD; PDSS1; PDSS2 |
| SERMs | 10 | ACAT1; FNTB; HMGCR; PDSS1; PDSS2; ZMPSTE24 | ||
| Total hip | Bisphosphonates | 15 | FDPS; FNTA; FNTB; IDI1; MVD; MVK; PDSS1; PDSS2 | |
| SERMs | 5 | FNTB; MVD; MVK; PDSS2 | ||
| Oestrogen-related genes | Lumbar spine | Bisphosphonates | 50 | CYP19A1; CYP2D6; EGF; ESR1; FOS; IKBKB; JUN; MAP2K1; MAPK14; MAPK9; NFKB1; SP1; UGT1A8; ARSD; CYP1B1 |
| SERMs | 49 | CYP19A1; CYP2D6; EGF; ESR1; ESR2; FOS; MAP2K1; MAPK1; MAPK8; NFKB1; UGT1A8; CYP1A1; CYP1A2; CYP3A4; SULT1A1 | ||
| Total hip | Bisphosphonates | 31 | CYP19A1; EGF; ESR1; ESR2; IKBKB; NFKB1; SP1; UGT1A8; ARSD; STS; SULT1E1; UGT1A9 | |
| SERMs | 50 | CYP19A1; ESR1; ESR2; FOS; GPER1; JUN; MAP2K1; MAPK1; MAPK14; MAPK8; ARSD; ARSL; CYP1A1; CYP1A2; CYP1B1 |
| Gene | Number of SNPs | Adjusted p Threshold | Bisphosphonates | SERMs | Significant SNPs (n) | |
|---|---|---|---|---|---|---|
| Lowest p-Value; Spine | Lowest p-Value; Hip | Lowest p-Value; Spine | ||||
| FDPS | 4 | 0.0125 | 0.0816 | 0.0041 | 0.3010 | 2 |
| FNTA | 4 | 0.0125 | 0.1570 | 0.0035 | 0.2889 | 1 |
| PDSS1 | 21 | 0.0024 | 0.0038 | 0.0206 | 0.0020 | 1 |
| CYP19A1 | 42 | 0.0012 | 0.0147 | 0.0469 | 0.0004 | 2 |
| CYP1A1 | 4 | 0.0125 | 0.2408 | 0.2104 | 0.0016 | 2 |
| CYP1A2 | 16 | 0.0031 | 0.0835 | 0.1776 | 0.0023 | 1 |
| Gene | Haplotype | β | r2 | p | Frequency | SNPs |
|---|---|---|---|---|---|---|
| CYP19A1 | GA | −1.35 | 0.2393 | 0.0004 | 0.447 | rs1062033; rs3889391 |
| CYP19A1 | GG | 0.74 | 0.0067 | 0.5801 | 0.020 | rs1062033; rs3889391 |
| CYP19A1 | CG | 1.26 | 0.2120 | 0.0009 | 0.531 | rs1062033; rs3889391 |
| CYP1A1 | GC | 2.27 | 0.1483 | 0.0075 | 0.053 | rs2198843; rs1048943 |
| CYP1A1 | GT | 0.97 | 0.0637 | 0.0869 | 0.085 | rs2198843; rs1048943 |
| CYP1A1 | CT | −1.52 | 0.2000 | 0.0016 | 0.861 | rs2198843; rs1048943 |
| FDPS | GT | −0.69 | 0.0473 | 0.0041 | 0.197 | rs2297480; rs10908463 |
| FDPS | TC | 0.63 | 0.0418 | 0.0071 | 0.799 | rs2297480; rs10908463 |
| Gene | Bisphosphonates | SERMs | |
|---|---|---|---|
| Spine | Hip | Spine | |
| FDPS | 0.1804 | 0.0111 | 0.6361 |
| FNTA | 0.4662 | 0.0665 | 0.6553 |
| PDSS1 | 0.0277 | 0.1269 | 0.1195 |
| CYP19A1 | 0.9562 | 0.8625 | 0.0086 |
| CYP1A1 | 0.5439 | 0.6565 | 0.0055 |
| CYP1A2 | 0.1211 | 0.8432 | 0.1692 |
| Gene | β | r | p |
|---|---|---|---|
| FDPS | −0.68 (−1.17 to −0.18) | 0.21 | 0.008 |
| FNTA | −0.93 (−1.48 to −0.38) | 0.26 | 0.001 |
| FDPS & FNTA | - | 0.34 | 0.00009 |
| Gene | β | r | p |
|---|---|---|---|
| CYP19A1 | −1.32 (−2.06 to −0.583) | 0.48 | 0.001 |
| PDSS1 | 1.65 (0.69–2.61) | 0.47 | 0.001 |
| CYP1A1 | 1.35 (0.49 to 2.02) | 0.44 | 0.003 |
| CYP1A2 | 1.29 (0.44–2.14) | 0.43 | 0.004 |
| CYP19A1,PDSS1,CYP1A1,CYP1A2 | - | 0.63 | 0.0004 |
| CYP19A1 & PDSS1 | - | 0.62 | 0.0003 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
del Real, Á.; Valero, C.; Olmos, J.M.; Hernández, J.L.; Riancho, J.A. Pharmacogenetics of Osteoporosis: A Pathway Analysis of the Genetic Influence on the Effects of Antiresorptive Drugs. Pharmaceutics 2022, 14, 776. https://doi.org/10.3390/pharmaceutics14040776
del Real Á, Valero C, Olmos JM, Hernández JL, Riancho JA. Pharmacogenetics of Osteoporosis: A Pathway Analysis of the Genetic Influence on the Effects of Antiresorptive Drugs. Pharmaceutics. 2022; 14(4):776. https://doi.org/10.3390/pharmaceutics14040776
Chicago/Turabian Styledel Real, Álvaro, Carmen Valero, José M. Olmos, Jose L. Hernández, and José A. Riancho. 2022. "Pharmacogenetics of Osteoporosis: A Pathway Analysis of the Genetic Influence on the Effects of Antiresorptive Drugs" Pharmaceutics 14, no. 4: 776. https://doi.org/10.3390/pharmaceutics14040776
APA Styledel Real, Á., Valero, C., Olmos, J. M., Hernández, J. L., & Riancho, J. A. (2022). Pharmacogenetics of Osteoporosis: A Pathway Analysis of the Genetic Influence on the Effects of Antiresorptive Drugs. Pharmaceutics, 14(4), 776. https://doi.org/10.3390/pharmaceutics14040776






