Mono-Rhamnolipid Biosurfactants Synthesized by Pseudomonas aeruginosa Detrimentally Affect Colorectal Cancer Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strain, Cell Lines and Culture Conditions
2.2. Polymerase Chain Reaction (PCR) and DNA Sequencing
2.3. Mono-RL Biosynthesis, Extraction, and Purification
2.4. Critical Micelle Determination
2.5. HPLC-MS Analysis of RLs
2.6. Cytotoxicity Assessment
2.7. Morphological Examination
2.8. Acridine Orange and Propidium Iodine Staining
2.9. Statistical Analysis
3. Results
3.1. PCR Confirmation of P. aeruginosa PAO1 ∆rhlC Strain Identity and Mutation
3.2. Molecular Profile and Physical Analysis of RLs Synthesised by P. aeruginosa PAO1 ∆rhlC
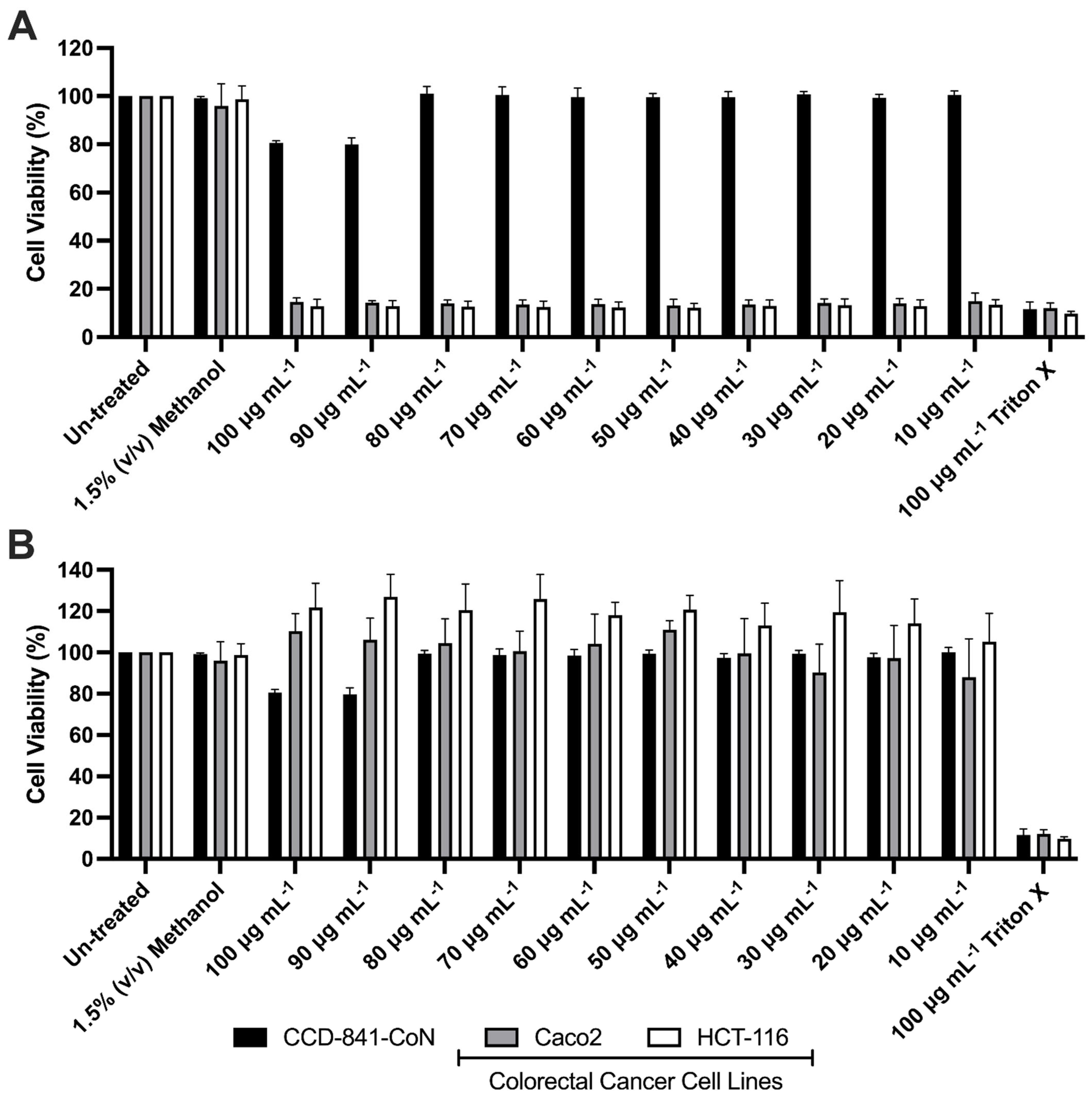
3.3. Effects of Mono-RL on the Viability of Colorectal Cancer Cell Lines & Healthy Gut Epithelia
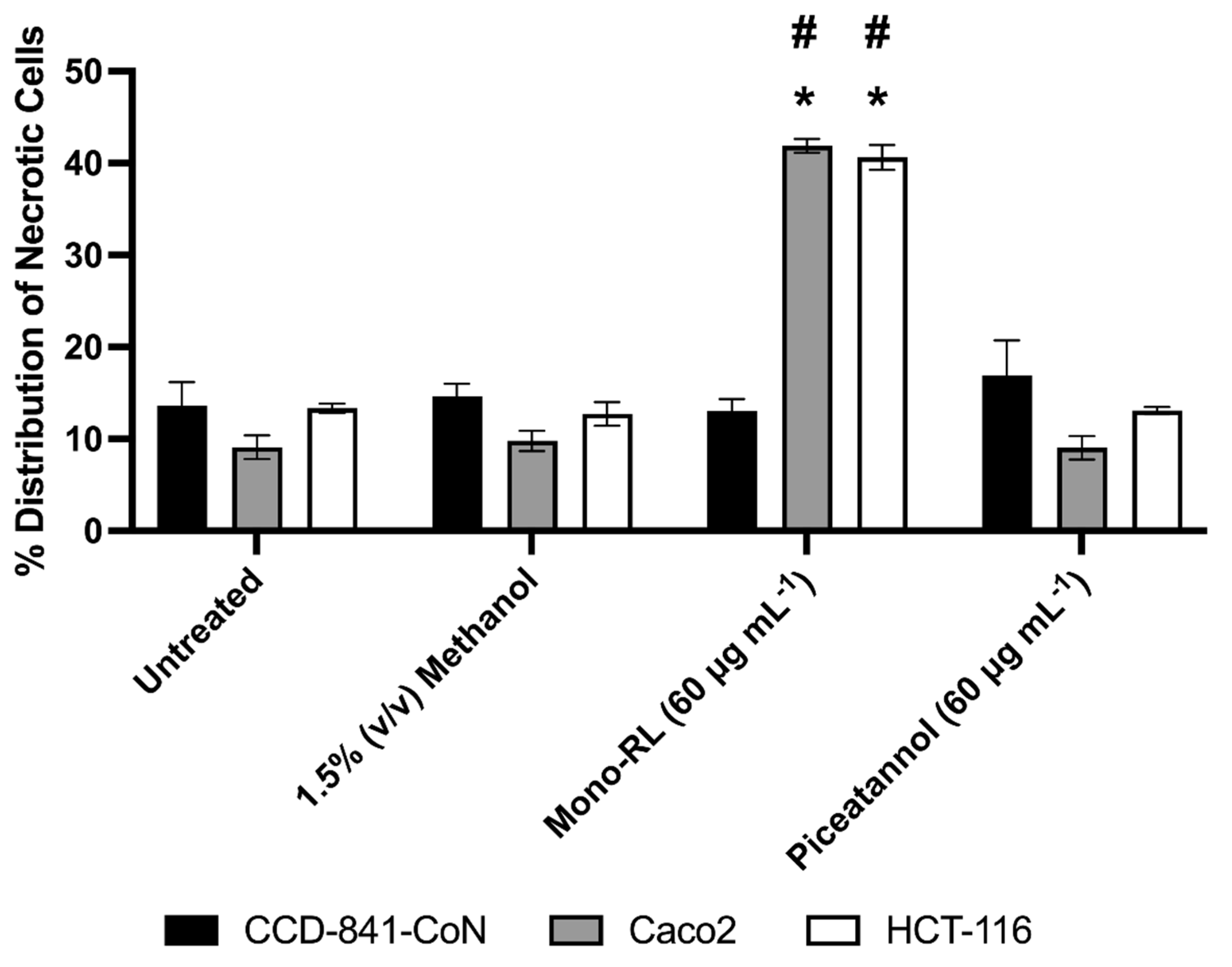
3.4. Morphological Assessment of Colorectal Cancer Cell Lines & Healthy Gut Epithelial Cells Treated with Mono-RL and Determination of Cell Death Mechanisms
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Center for Disease Control. Available online: https://www.cdc.gov/tobacco/basic_information/health_effects/cancer/index.htm (accessed on 30 August 2022).
- Ferlay, J.; Ervik, M.; Lam, F.; Colombet, M.; Mery, L.; Piñeros, M.; Znaor, A.; Soerjomataram, I.; Bray, F. Global Cancer Observatory: Cancer Today. Available online: https://gco.iarc.fr/today (accessed on 30 August 2022).
- Cancer Research UK. Available online: https://www.cancerresearchuk.org/health-professional/cancer-statistics/statistics-by-cancer-type/bowel-cancer (accessed on 30 August 2022).
- National Cancer Registry Ireland. Available online: https://www.ncri.ie/sites/ncri/files/factsheets/Factsheetcervix.pdf (accessed on 25 October 2022).
- Tremblay, J.; Richardson, A.P.; Lépine, F.; Déziel, E. Self-produced extracellular stimuli modulate the Pseudomonas aeruginosa swarming motility behaviour. Environ. Microbiol. 2007, 9, 2622–2630. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, M.J.; Lavery, I.; Yothers, G.; Paik, S.; Clark-Langone, K.M.; Lopatin, M.; Watson, D.; Baehner, F.L.; Shak, S.; Baker, J.; et al. Relationship Between Tumor Gene Expression and Recurrence in Four Independent Studies of Patients With Stage II/III Colon Cancer Treated With Surgery Alone or Surgery Plus Adjuvant Fluorouracil Plus Leucovorin. J. Clin. Oncol. 2010, 28, 3937–3944. [Google Scholar] [CrossRef] [PubMed]
- Mazevet, M.; Moulin, M.; Llach-Martinez, A.; Chargari, C.; Deutsch, É.; Gomez, A.-M.; Morel, É. Complications of chemotherapy, a basic science update. Presse Med. 2013, 42, e352–e361. [Google Scholar] [CrossRef] [PubMed]
- Farinha, P.; Pinho, J.O.; Matias, M.; Gaspar, M.M. Nanomedicines in the treatment of colon cancer: A focus on metallodrugs. Drug Deliv. Transl. Res. 2022, 12, 49–66. [Google Scholar] [CrossRef]
- Kelloff, G.J.; Boone, C.W.; Crowell, J.A.; Steele, V.E.; Lubet, R.A.; Doody, L.A.; Malone, W.F.; Hawk, E.T.; Sigman, C.C. New agents for cancer chemoprevention. J. Cell. Biochem. 1996, 63, 1–28. [Google Scholar] [CrossRef]
- D’Incalci, M.; Steward, W.P.; Gescher, A.J. Use of cancer chemopreventive phytochemicals as antineoplastic agents. Lancet. Oncol. 2005, 6, 899–904. [Google Scholar] [CrossRef]
- Adu, S.A.; Twigg, M.S.; Naughton, P.J.; Marchant, R.; Banat, I.M. Biosurfactants as Anticancer Agents: Glycolipids Affect Skin Cells in a Differential Manner Dependent on Chemical Structure. Pharmaceutics 2022, 14, 360. [Google Scholar] [CrossRef]
- Callaghan, B.; Twigg, M.S.; Baccile, N.; Van Bogaer, I.N.A.; Marchant, R.; Mitchell, C.A.; Banat, I.M. Microbial sophorolipids inhibit colorectal tumour cell growth in vitro and restore haematocrit in Apcmin+/− mice. Appl. Microbiol. Biotechnol. 2022, 106, 6003–6016. [Google Scholar] [CrossRef]
- Twigg, M.S.; Baccile, N.; Banat, I.M.; Déziel, E.; Marchant, R.; Roelants, S.; Van Bogaert, I.N.A. Microbial biosurfactant research: Time to improve the rigour in the reporting of synthesis, functional characterization and process development. Microb. Biotechnol. 2021, 14, 147–170. [Google Scholar] [CrossRef]
- Banat, I.M.; Franzetti, A.; Gandolfi, I.; Bestetti, G.; Martinotti, M.G.; Fracchia, L.; Smyth, T.J.; Marchant, R. Microbial biosurfactants production, applications and future potential. Appl. Microbiol. Biotechnol. 2010, 87, 427–444. [Google Scholar] [CrossRef]
- Marchant, R.; Banat, I.M. Biosurfactants: A sustainable replacement for chemical surfactants? Biotechnol. Lett. 2012, 34, 1597–1605. [Google Scholar] [CrossRef] [PubMed]
- Sarubbo, L.A.; Silva, M.d.G.C.; Durval, I.J.B.; Bezerra, K.G.O.; Ribeiro, B.G.; Silva, I.A.; Twigg, M.S.; Banat, I.M. Biosurfactants: Production, Properties, Applications, Trends, and General Perspectives. Biochem. Eng. J. 2022, 181, 108377. [Google Scholar] [CrossRef]
- Fracchia, L.; Ceresa, C.; Banat, I.M. Biosurfactants in Cosmetic, Biomedical and Pharmaceutical Industry. In Microbial Biosurfactants and their Environmental and Industrial Applications; Thavasi, R., Banat, I.M., Eds.; CRC Press: Boca Raton, FL, USA, 2019; pp. 258–287. ISBN 9781315271767. [Google Scholar]
- Ceresa, C.; Fracchia, L.; Fedeli, E.; Porta, C.; Banat, I.M. Recent advances in biomedical, therapeutic and pharmaceutical applications of microbial surfactants. Pharmaceutics 2021, 13, 466. [Google Scholar] [CrossRef] [PubMed]
- Ohadi, M.; Shahravan, A.; Dehghannoudeh, N.; Eslaminejad, T.; Banat, I.M.; Dehghannoudeh, G. Potential use of microbial surfactant in microemulsion drug delivery system: A systematic review. Drug Des. Devel. Ther. 2020, 14, 541–550. [Google Scholar] [CrossRef] [PubMed]
- Adu, S.A.; Twigg, M.S.; Naughton, P.J.; Marchant, R.; Banat, I.M. Characterisation of cytotoxicity and immunomodulatory effects of glycolipid biosurfactants on human keratinocytes. Appl. Microbiol. Biotechnol. 2022. [Google Scholar] [CrossRef]
- Ochsner, U.A.; Fiechter, A.; Reiser, J. Isolation, characterization, and expression in Escherichia coli of the Pseudomonas aeruginosa rhlAB genes encoding a rhamnosyltransferase involved in rhamnolipid biosurfactant synthesis. J. Biol. Chem. 1994, 269, 19787–19795. [Google Scholar] [CrossRef] [PubMed]
- Funston, S.J.; Tsaousi, K.; Rudden, M.; Smyth, T.J.; Stevenson, P.S.; Marchant, R.; Banat, I.M. Characterising rhamnolipid production in Burkholderia thailandensis E264, a non-pathogenic producer. Appl. Microbiol. Biotechnol. 2016, 100, 7945–7956. [Google Scholar] [CrossRef]
- Twigg, M.S.; Tripathi, L.; Zompra, A.; Salek, K.; Irorere, V.U.; Gutierrez, T.; Spyroulias, G.A.; Marchant, R.; Banat, I.M. Identification and characterisation of short chain rhamnolipid production in a previously uninvestigated, non-pathogenic marine pseudomonad. Appl. Microbiol. Biotechnol. 2018, 102, 8537–8549. [Google Scholar] [CrossRef]
- Tripathi, L.; Twigg, M.S.; Zompra, A.; Salek, K.; Irorere, V.U.; Gutierrez, T.; Spyroulias, G.A.; Marchant, R.; Banat, I.M. Biosynthesis of rhamnolipid by a Marinobacter species expands the paradigm of biosurfactant synthesis to a new genus of the marine microflora. Microb. Cell Fact. 2019, 18, 164. [Google Scholar] [CrossRef]
- Déziel, E.; Lépine, F.; Dennie, D.; Boismenu, D.; Mamer, O.A.; Villemur, R. Liquid chromatography/mass spectrometry analysis of mixtures of rhamnolipids produced by Pseudomonas aeruginosa strain 57RP grown on mannitol or naphthalene. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1999, 1440, 244–252. [Google Scholar] [CrossRef]
- Rudden, M.; Tsauosi, K.; Marchant, R.; Banat, I.M.; Smyth, T.J. Development and validation of an ultra-performance liquid chromatography tandem mass spectrometry (UPLC-MS/MS) method for the quantitative determination of rhamnolipid congeners. Appl. Microbiol. Biotechnol. 2015, 99, 9177–9187. [Google Scholar] [CrossRef] [PubMed]
- Zhu, K.; Rock, C.O. RhlA converts β-hydroxyacyl-acyl carrier protein intermediates in fatty acid synthesis to the β-hydroxydecanoyl-β-hydroxydecanoate component of rhamnolipids in Pseudomonas aeruginosa. J. Bacteriol. 2008, 190, 3147–3154. [Google Scholar] [CrossRef] [PubMed]
- Rahim, R.; Ochsner, U.A.; Olvera, C.; Graninger, M.; Messner, P.; Lam, J.S.; Soberón-Chávez, G. Cloning and functional characterization of the Pseudomonas aeruginosa rhlC gene that encodes rhamnosyltransferase 2, an enzyme responsible for di-rhamnolipid biosynthesis. Mol. Microbiol. 2001, 40, 708–718. [Google Scholar] [CrossRef] [PubMed]
- Dubeau, D.; Déziel, E.; Woods, D.E.; Lépine, F. Burkholderia thailandensis harbors two identical rhl gene clusters responsible for the biosynthesis of rhamnolipids. BMC Microbiol. 2009, 9, 263. [Google Scholar] [CrossRef] [PubMed]
- Nakata, K.; Yoshimoto, A.; Yamada, Y. Correlation between autoinducers and rhamnolipids production by Pseudomonas aeruginosa IFO 3924. J. Ferment. Bioeng. 1998, 86, 608–610. [Google Scholar] [CrossRef]
- Thanomsub, B.; Pumeechockchai, W.; Limtrakul, A.; Arunrattiyakorn, P.; Petchleelaha, W.; Nitoda, T.; Kanzaki, H. Chemical structures and biological activities of rhamnolipids produced by Pseudomonas aeruginosa B189 isolated from milk factory waste. Bioresour. Technol. 2006, 97, 2457–2461. [Google Scholar] [CrossRef] [PubMed]
- Sanjivkumar, M.; Deivakumari, M.; Immanuel, G. Investigation on spectral and biomedical characterization of rhamnolipid from a marine associated bacterium Pseudomonas aeruginosa (DKB1). Arch. Microbiol. 2021, 203, 2297–2314. [Google Scholar] [CrossRef] [PubMed]
- Semkova, S.; Antov, G.; Iliev, I.; Tsoneva, I.; Lefterov, P.; Christova, N.; Nacheva, L.; Stoineva, I.; Kabaivanova, L.; Staneva, G.; et al. Rhamnolipid biosurfactants—Possible natural anticancer agents and autophagy inhibitors. Separations 2021, 8, 92. [Google Scholar] [CrossRef]
- Jiang, L.; Shen, C.; Long, X.; Zhang, G.; Meng, Q. Rhamnolipids elicit the same cytotoxic sensitivity between cancer cell and normal cell by reducing surface tension of culture medium. Appl. Microbiol. Biotechnol. 2014, 98, 10187–10196. [Google Scholar] [CrossRef]
- Callaghan, B.; Lydon, H.; Roelants, S.L.K.W.; Van Bogaert, I.N.A.; Marchant, R.; Banat, I.M.; Mitchell, C.A. Lactonic Sophorolipids Increase Tumor Burden in Apcmin+/− Mice. PLoS ONE 2016, 11, e0156845. [Google Scholar] [CrossRef]
- Du Noüy, P.L. An interfacial tensiometer for universal use. J. Gen. Physiol. 1925, 7, 625–632. [Google Scholar] [CrossRef] [PubMed]
- Smyth, T.J.P.; Rudden, M.; Tsaousi, K.; Marchant, R.; Banat, I.M. Protocols for the Detection and Chemical Characterisation of Microbial Glycolipids. In Hydrocarbon and Lipid Microbiology Protocols; McGenity, T.J., Timmis, K.N., Nogales, B., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 29–60. ISBN 978-3-662-49137-9. [Google Scholar]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Ribble, D.; Goldstein, N.B.; Norris, D.A.; Shellman, Y.G. A simple technique for quantifying apoptosis in 96-well plates. BMC Biotechnol. 2005, 5, 12. [Google Scholar] [CrossRef] [PubMed]
- Naughton, P.J.; Marchant, R.; Naughton, V.; Banat, I.M. Microbial biosurfactants: Current trends and applications in agricultural and biomedical industries. J. Appl. Microbiol. 2019, 127, 12–28. [Google Scholar] [CrossRef]
- McQuade, R.M.; Bornstein, J.C.; Nurgali, K. Anti-Colorectal Cancer Chemotherapy-Induced Diarrhoea: Current Treatments and Side-Effects. Int. J. Clin. Med. 2014, 05, 393–406. [Google Scholar] [CrossRef]
- Zhao, J.; Wu, Y.; Alfred, A.T.; Xin, X.; Yang, S. Chemical structures and biological activities of rhamnolipid biosurfactants produced by Pseudomonas aeruginosa M14808. J. Chem. Pharm. Res. 2013, 5, 177–182. [Google Scholar]
- Chen, J.; Song, X.; Zhang, H.; Qu, Y.B.; Miao, J.Y. Sophorolipid produced from the new yeast strain Wickerhamiella domercqiae induces apoptosis in H7402 human liver cancer cells. Appl. Microbiol. Biotechnol. 2006, 72, 52–59. [Google Scholar] [CrossRef]
- Fu, S.L.; Wallner, S.R.; Bowne, W.B.; Hagler, M.D.; Zenilman, M.E.; Gross, R.; Bluth, M.H. Sophorolipids and Their Derivatives Are Lethal Against Human Pancreatic Cancer Cells. J. Surg. Res. 2008, 148, 77–82. [Google Scholar] [CrossRef]
- Zompra, A.A.; Chasapi, S.A.; Twigg, M.S.; Salek, K.; Anestopoulos, I.; Galanis, A.; Pappa, A.; Gutierrez, T.; Banat, I.M.; Marchant, R.; et al. Multi-method biophysical analysis in discovery, identification, and in-depth characterization of surface-active compounds. Front. Mar. Sci. 2022, 9, 1023287. [Google Scholar] [CrossRef]
- Zhang, Y.; Jia, D.; Sun, W.; Yang, X.; Zhang, C.; Zhao, F.; Lu, W. Semicontinuous sophorolipid fermentation using a novel bioreactor with dual ventilation pipes and dual sieve-plates coupled with a novel separation system. Microb. Biotechnol. 2018, 11, 455–464. [Google Scholar] [CrossRef]
- Zhao, H.; Yan, L.; Xu, X.; Jiang, C.; Shi, J.; Zhang, Y.; Liu, L.; Lei, S.; Shao, D.; Huang, Q. Potential of Bacillus subtilis lipopeptides in anti-cancer I: Induction of apoptosis and paraptosis and inhibition of autophagy in K562 cells. AMB Express 2018, 8, 78. [Google Scholar] [CrossRef] [PubMed]
- Rahimi, K.; Lotfabad, T.B.; Jabeen, F.; Mohammad Ganji, S. Cytotoxic effects of mono- and di-rhamnolipids from Pseudomonas aeruginosa MR01 on MCF-7 human breast cancer cells. Colloids Surf. B Biointerfaces 2019, 181, 943–952. [Google Scholar] [CrossRef] [PubMed]
- Siedlecka-Kroplewska, K.; Wrońska, A.; Kmieć, Z. Piceatannol, a structural analog of resveratrol, is an apoptosis inducer and a multidrug resistance modulator in hl-60 human acute myeloid leukemia cells. Int. J. Mol. Sci. 2021, 22, 10597. [Google Scholar] [CrossRef] [PubMed]
- Kuo, P.L.; Hsu, Y.L. The grape and wine constituent piceatannol inhibits proliferation of human bladder cancer cells via blocking cell cycle progression and inducing Fas/membrane bound Fas ligand-mediated apoptotic pathway. Mol. Nutr. Food Res. 2008, 52, 408–418. [Google Scholar] [CrossRef]
- Morales, P.; Haza, A.I. Selective apoptotic effects of piceatannol and myricetin in human cancer cells. J. Appl. Toxicol. 2012, 32, 986–993. [Google Scholar] [CrossRef] [PubMed]
- Müller, M.M.; Hörmann, B.; Kugel, M.; Syldatk, C.; Hausmann, R. Evaluation of rhamnolipid production capacity of Pseudomonas aeruginosa PAO1 in comparison to the rhamnolipid over-producer strains DSM 7108 and DSM 2874. Appl. Microbiol. Biotechnol. 2011, 89, 585–592. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Zu, Y.; Li, X.; Meng, Q.; Long, X. Recent progress towards industrial rhamnolipids fermentation: Process optimization and foam control. Bioresour. Technol. 2020, 298, 122394. [Google Scholar] [CrossRef] [PubMed]
- Cabrera-Valladares, N.; Richardson, A.P.; Olvera, C.; Treviño, L.G.; Déziel, E.; Lépine, F.; Soberón-Chávez, G. Monorhamnolipids and 3-(3-hydroxyalkanoyloxy)alkanoic acids (HAAs) production using Escherichia coli as a heterologous host. Appl Microbiol Biotechnol. 2006, 73, 187–194. [Google Scholar] [CrossRef]
- Wittgens, A.; Tiso, T.; Arndt, T.T.; Wenk, P.; Hemmerich, J.; Müller, C.; Wichmann, R.; Küpper, B.; Zwick, M.; Wilhelm, S.; et al. Growth independent rhamnolipid production from glucose using the non-pathogenic Pseudomonas putida KT2440. Microb Cell Fact. 2011, 10, 80. [Google Scholar] [CrossRef]
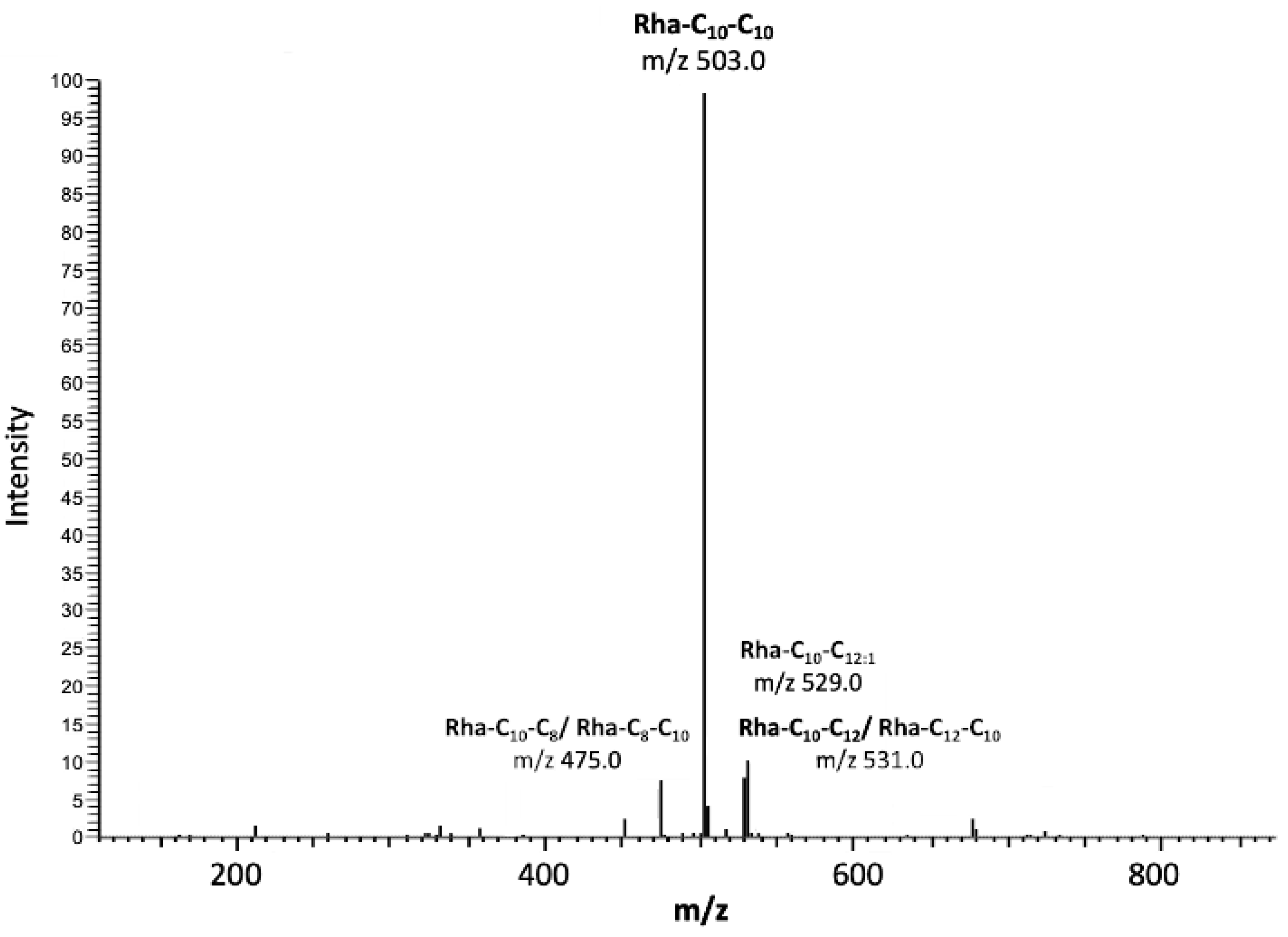

| Retention Time (Min) | m/z | Mw | Relative Abundance (%) | Congener |
|---|---|---|---|---|
| 20.93 | 333.0 | 1.07 | Rha-C10 | |
| 24.37 | 356.8 | 0.76 | Rha-C12:2 | |
| 17.62 | 475.0 | 5.60 | Rha-C10-C8 | |
| 20.78 | 503.0 | 74.59 | Rha-C10-C10 | |
| 22.47 | 529.0 | 5.89 | Rha-C10-C12:1 | |
| 23.87 | 531.0 | 7.62 | Rha-C10-C12 | |
| 24.37 | 557.0 | 0.27 | Rha-C10-C14:1 | |
| 26.09 | 559.1 | 0.22 | Rha-C12-C12 | |
| 21.97 | 517.0 | 0.59 | Rha-C10-C10-CH3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Twigg, M.S.; Adu, S.A.; Sugiyama, S.; Marchant, R.; Banat, I.M. Mono-Rhamnolipid Biosurfactants Synthesized by Pseudomonas aeruginosa Detrimentally Affect Colorectal Cancer Cells. Pharmaceutics 2022, 14, 2799. https://doi.org/10.3390/pharmaceutics14122799
Twigg MS, Adu SA, Sugiyama S, Marchant R, Banat IM. Mono-Rhamnolipid Biosurfactants Synthesized by Pseudomonas aeruginosa Detrimentally Affect Colorectal Cancer Cells. Pharmaceutics. 2022; 14(12):2799. https://doi.org/10.3390/pharmaceutics14122799
Chicago/Turabian StyleTwigg, Matthew S., Simms A. Adu, Suguru Sugiyama, Roger Marchant, and Ibrahim M. Banat. 2022. "Mono-Rhamnolipid Biosurfactants Synthesized by Pseudomonas aeruginosa Detrimentally Affect Colorectal Cancer Cells" Pharmaceutics 14, no. 12: 2799. https://doi.org/10.3390/pharmaceutics14122799
APA StyleTwigg, M. S., Adu, S. A., Sugiyama, S., Marchant, R., & Banat, I. M. (2022). Mono-Rhamnolipid Biosurfactants Synthesized by Pseudomonas aeruginosa Detrimentally Affect Colorectal Cancer Cells. Pharmaceutics, 14(12), 2799. https://doi.org/10.3390/pharmaceutics14122799








