A Bioluminescence-Based Ex Vivo Burn Wound Model for Real-Time Assessment of Novel Phage-Inspired Enzybiotics
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Growth, and Media
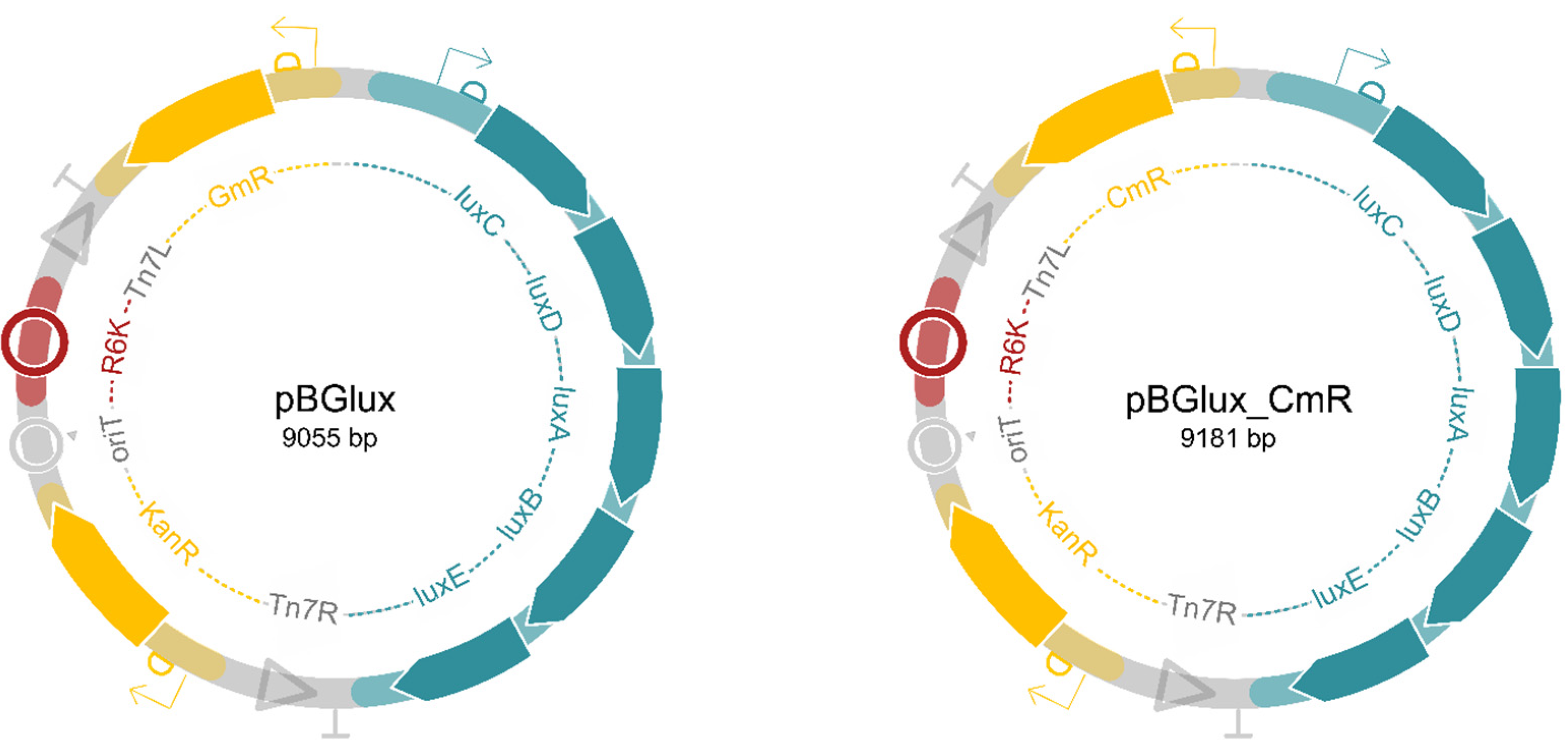
2.2. Construction of Versatile Tagging Plasmids, pBGlux and pBGlux_CmR
2.3. Tagging A Clinical Isolate with the Newly Created Tagging Plasmid, pBGlux_CmR
2.4. Preparation of Pig Skin Explants for Use in the Ex Vivo Model
2.5. Establishing A Correlation between the RLU on the Explant and the CFUs Recovered from the Explant
2.6. Testing Antibacterial Activity in the Bioluminescent Ex Vivo Model
2.7. Data Analysis
3. Results
3.1. Establishing A Flexible Plasmid Collection to Tag Non-Model Organisms
3.2. Integration of the Bioluminescent Reporter Strain in the Ex Vivo Pig Skin Model
3.3. Proof-of-Concept with A Bacteriophage-Derived Engineered Lysin, 1D10
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Centers for Disease Prevention and Control. Antibiotic Resistance Threats in the United States, 2019; U.S. Department of Health and Human Services, CDC: Atlanta, GA, USA, 2019. Available online: https://www.cdc.gov/drugresistance/biggest-threats.html (accessed on 28 March 2020).
- Centers for Disease Prevention and Control. Antibiotic Resistance Threats in the United States, 2013. Current. 114; U.S. Department of Health and Human Services, CDC: Atlanta, GA, USA, 2013. Available online: https://www.cdc.gov/drugresistance/threat-report-2013/pdf/ar-threats-2013-508.pdf (accessed on 15 October 2018).
- O’Neill, J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations. In The Review on Antimicrobial Resistance; HM Government: London, UK, 2016. [Google Scholar]
- Theuretzbacher, U.; Outterson, K.; Engel, A.; Karlén, A. The global preclinical antibacterial pipeline. Nat. Rev. Microbiol. 2020, 18, 275–285. [Google Scholar] [CrossRef] [PubMed]
- WHO. Prioritization of Pathogens to Guide Discorvery, Research and Development of New Antibiotics for Drug-Resistant Bacterial Infections, Including Tuberculosis; World Health Organization: Geneva, Switzerland, 2017. [Google Scholar]
- Theuretzbacher, U.; Gottwalt, S.; Beyer, P.; Butler, M.; Czaplewski, L.; Lienhardt, C.; Moja, L.; Paul, M.; Paulin, S.; Rex, J.H.; et al. Analysis of the clinical antibacterial and antituberculosis pipeline. Lancet. Infect. Dis. 2019, 19, e40–e50. [Google Scholar] [CrossRef]
- Duarte, A.C.; Fernández, L.; De Maesschalck, V.; Gutiérrez, D.; Campelo, A.B.; Briers, Y.; Lavigne, R.; Rodríguez, A.; García, P. Synergistic action of phage phiIPLA-RODI and lytic protein CHAPSH3b: A combination strategy to target Staphylococcus aureus biofilms. NPJ Biofilms Microbiomes 2021, 7, 39. [Google Scholar] [CrossRef] [PubMed]
- Lewis, K. Platforms for antibiotic discovery. Nat. Rev. Drug Discov. 2013, 12, 371–387. [Google Scholar] [CrossRef]
- Bertesteanu, S.; Triaridis, S.; Stankovic, M.; Lazar, V.; Chifiriuc, M.C.; Vlad, M.; Grigore, R. Polymicrobial wound infections: Pathophysiology and current therapeutic approaches. Int. J. Pharm. 2014, 463, 119–126. [Google Scholar] [CrossRef]
- Clinton, A.; Carter, T. Chronic Wound Biofilms: Pathogenesis and Potential Therapies. Lab. Med. 2015, 46, 277–284. [Google Scholar] [CrossRef]
- Brackman, G.; Coenye, T. In Vitro and In Vivo Biofilm Wound Models and Their Application. Adv. Exp. Med. Biol. 2016, 897, 15–32. [Google Scholar] [CrossRef]
- Parnell, L.; Volk, S.W. The Evolution of Animal Models in Wound Healing Research: 1993–2017. Adv. Wound Care 2019, 8, 692–702. [Google Scholar] [CrossRef]
- Abdullahi, A.; Amini-Nik, S.; Jeschke, M.G. Animal models in burn research. Cell. Mol. Life Sci. CMLS 2014, 71, 3241–3255. [Google Scholar] [CrossRef]
- Wang, H.; Agrawal, A.; Wang, Y.; Crawford, D.W.; Siler, Z.D.; Peterson, M.L.; Woofter, R.T.; Labib, M.; Shin, H.Y.; Baumann, A.P.; et al. An ex vivo model of medical device-mediated bacterial skin translocation. Sci. Rep. 2021, 11, 5746. [Google Scholar] [CrossRef]
- Yang, Q.; Phillips, P.L.; Sampson, E.M.; Progulske-Fox, A.; Jin, S.; Antonelli, P.; Schultz, G.S. Development of a novel ex vivo porcine skin explant model for the assessment of mature bacterial biofilms. Wound Repair Regen. 2013, 21, 704–714. [Google Scholar] [CrossRef] [PubMed]
- Alves, D.R.; Booth, S.P.; Scavone, P.; Schellenberger, P.; Salvage, J.; Dedi, C.; Thet, N.T.; Jenkins, A.; Waters, R.; Ng, K.W.; et al. Development of a High-Throughput ex vivo Burn Wound Model Using Porcine Skin, and Its Application to Evaluate New Approaches to Control Wound Infection. Front. Cell. Infect. Microbiol. 2018, 8, 196. [Google Scholar] [CrossRef] [PubMed]
- Andersson M, Å.; Madsen, L.B.; Schmidtchen, A.; Puthia, M. Development of an Experimental ex vivo Wound Model to Evaluate Antimicrobial Efficacy of Topical Formulations. Int. J. Mol. Sci. 2021, 22, 5045. [Google Scholar] [CrossRef] [PubMed]
- Corzo-León, D.E.; Munro, C.A.; MacCallum, D.M. An ex vivo Human Skin Model to Study Superficial Fungal Infections. Front. Microbiol. 2019, 10, 1172. [Google Scholar] [CrossRef]
- Haddock, S.H.; Moline, M.A.; Case, J.F. Bioluminescence in the sea. Annu. Rev. Mar. Sci. 2010, 2, 443–493. [Google Scholar] [CrossRef]
- Li, Y.; He, X.; Zhu, W.; Li, H.; Wang, W. Bacterial bioluminescence assay for bioanalysis and bioimaging. Anal. Bioanal. Chem. 2022, 414, 75–83. [Google Scholar] [CrossRef]
- Avci, P.; Karimi, M.; Sadasivam, M.; Antunes-Melo, W.C.; Carrasco, E.; Hamblin, M.R. In-vivo monitoring of infectious diseases in living animals using bioluminescence imaging. Virulence 2018, 9, 28–63. [Google Scholar] [CrossRef]
- Loh, J.M.; Proft, T. Comparison of firefly luciferase and NanoLuc luciferase for biophotonic labeling of group A Streptococcus. Biotechnol. Lett. 2014, 36, 829–834. [Google Scholar] [CrossRef]
- Persyn, A.; Rogiers, O.; Brock, M.; Vande Velde, G.; Lamkanfi, M.; Jacobsen, I.D.; Himmelreich, U.; Lagrou, K.; van Dijck, P.; Kucharíková, S. Monitoring of Fluconazole and Caspofungin Activity against In Vivo Candida glabrata Biofilms by Bioluminescence Imaging. Antimicrob. Agents Chemother. 2019, 63, e01555-18. [Google Scholar] [CrossRef]
- Vande Velde, G.; Kucharíková, S.; van Dijck, P.; Himmelreich, U. Bioluminescence imaging increases in vivo screening efficiency for antifungal activity against device-associated Candida Albicans Biofilms. Int. J. Antimicrob. Agents 2018, 52, 42–51. [Google Scholar] [CrossRef]
- Meighen, E.A. Bacterial bioluminescence: Organization, regulation, and application of the lux genes. FASEB J. 1993, 7, 1016–1022. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, A.; Lebeaux, D.; Decante, B.; Kriegel, I.; Escande, M.C.; Ghigo, J.M.; Beloin, C. A rat model of central venous catheter to study establishment of long-term bacterial biofilm and related acute and chronic infections. PLoS ONE 2012, 7, e37281. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, A.; Schlüter, T.; Melkonian, K.; Takeda, A.; Nakagami, H.; Mine, A. A versatile Tn7 transposon-based bioluminescence tagging tool for quantitative and spatial detection of bacteria in plants. Plant Commun. 2021, 3, 100227. [Google Scholar] [CrossRef] [PubMed]
- Gerstmans, H.; Grimon, D.; Gutiérrez, D.; Lood, C.; Rodríguez, A.; van Noort, V.; Lammertyn, J.; Lavigne, R.; Briers, Y. A VersaTile-driven platform for rapid hit-to-lead development of engineered lysins. Sci. Adv. 2020, 6, eaaz1136. [Google Scholar] [CrossRef] [PubMed]
- Figurski, D.H.; Helinski, D.R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 1979, 76, 1648–1652. [Google Scholar] [CrossRef]
- Michiels, J.E.; van den Bergh, B.; Fauvart, M.; Michiels, J. Draft genome sequence of Acinetobacter baumannii strain NCTC 13423, a multidrug-resistant clinical isolate. Stand. Genom. Sci. 2016, 11, 57. [Google Scholar] [CrossRef]
- Turton, J.F.; Kaufmann, M.E.; Gill, M.J.; Pike, R.; Scott, P.T.; Fishbain, J.; Craft, D.; Deye, G.; Riddell, S.; Lindler, L.E.; et al. Comparison of Acinetobacter baumannii isolates from the United Kingdom and the United States that were associated with repatriated casualties of the Iraq conflict. J. Clin. Microbiol. 2006, 44, 2630–2634. [Google Scholar] [CrossRef]
- Green, R.; Rogers, E.J. Transformation of chemically competent E. coli. Methods Enzymol. 2013, 529, 329–336. [Google Scholar] [CrossRef]
- Gibson, D.G.; Young, L.; Chuang, R.Y.; Venter, J.C.; Hutchison, C.A., 3rd; Smith, H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 2009, 6, 343–345. [Google Scholar] [CrossRef]
- Choi, K.H.; Gaynor, J.B.; White, K.G.; Lopez, C.; Bosio, C.M.; Karkhoff-Schweizer, R.R.; Schweizer, H.P. A Tn7-based broad-range bacterial cloning and expression system. Nat. Methods 2005, 2, 443–448. [Google Scholar] [CrossRef]
- Zobel, S.; Benedetti, I.; Eisenbach, L.; de Lorenzo, V.; Wierckx, N.; Blank, L.M. Tn7-Based Device for Calibrated Heterologous Gene Expression in Pseudomonas putida. ACS Synth. Biol. 2015, 4, 1341–1351. [Google Scholar] [CrossRef] [PubMed]
- Silva-Rocha, R.; Martínez-García, E.; Calles, B.; Chavarría, M.; Arce-Rodríguez, A.; de Las Heras, A.; Páez-Espino, A.D.; Durante-Rodríguez, G.; Kim, J.; Nikel, P.I.; et al. The Standard European Vector Architecture (SEVA): A coherent platform for the analysis and deployment of complex prokaryotic phenotypes. Nucleic Acids Res. 2013, 41, D666–D675. [Google Scholar] [CrossRef] [PubMed]
- Gregor, C.; Gwosch, K.C.; Sahl, S.J.; Hell, S.W. Strongly enhanced bacterial bioluminescence with the ilux operon for single-cell imaging. Proc. Natl. Acad. Sci. USA 2018, 115, 962–967. [Google Scholar] [CrossRef] [PubMed]
- Bowler, P.G. The 10(5) bacterial growth guideline: Reassessing its clinical relevance in wound healing. Ostomy/Wound Manag. 2003, 49, 44–53. [Google Scholar]
- Czaplewski, L.; Bax, R.; Clokie, M.; Dawson, M.; Fairhead, H.; Fischetti, V.A.; Foster, S.; Gilmore, B.F.; Hancock, R.E.; Harper, D.; et al. Alternatives to antibiotics-a pipeline portfolio review. Lancet Infect. Dis. 2016, 16, 239–251. [Google Scholar] [CrossRef]
- De Maesschalck, V.; Gutiérrez, D.; Paeshuyse, J.; Lavigne, R.; Briers, Y. Advanced engineering of third-generation lysins and formulation strategies for clinical applications. Crit. Rev. Microbiol. 2020, 46, 548–564. [Google Scholar] [CrossRef]
- Jun, S.Y.; Jang, I.J.; Yoon, S.; Jang, K.; Yu, K.S.; Cho, J.Y.; Seong, M.W.; Jung, G.M.; Yoon, S.J.; Kang, S.H. Pharmacokinetics and Tolerance of the Phage Endolysin-Based Candidate Drug SAL200 after a Single Intravenous Administration among Healthy Volunteers. Antimicrob. Agents Chemother. 2017, 61, e02629-16. [Google Scholar] [CrossRef]
- Totté, J.; van Doorn, M.B.; Pasmans, S. Successful Treatment of Chronic Staphylococcus aureus-Related Dermatoses with the Topical Endolysin Staphefekt SA.100: A Report of 3 Cases. Case Rep. Dermatol. 2017, 9, 19–25. [Google Scholar] [CrossRef]
- Shivak, D.J.; MacKenzie, K.D.; Watson, N.L.; Pasternak, J.A.; Jones, B.D.; Wang, Y.; DeVinney, R.; Wilson, H.L.; Surette, M.G.; White, A.P. A Modular, Tn7-Based System for Making Bioluminescent or Fluorescent Salmonella and Escherichia coli Strains. Appl. Environ. Microbiol. 2016, 82, 4931–4943. [Google Scholar] [CrossRef]
- Bachman, N.; Biery, M.C.; Boeke, J.D.; Craig, N.L. Tn7-mediated mutagenesis of Saccharomyces cerevisiae genomic DNA in vitro. Methods Enzymol. 2002, 350, 230–247. [Google Scholar] [CrossRef]
- Yeh, H.W.; Karmach, O.; Ji, A.; Carter, D.; Martins-Green, M.M.; Ai, H.W. Red-shifted luciferase-luciferin pairs for enhanced bioluminescence imaging. Nat. Methods 2017, 14, 971–974. [Google Scholar] [CrossRef] [PubMed]
- Resendiz-Sharpe, A.; da Silva, R.P.; Geib, E.; Vanderbeke, L.; Seldeslachts, L.; Hupko, C.; Brock, M.; Lagrou, K.; Vande Velde, G. Longitudinal multimodal imaging-compatible mouse model of triazole-sensitive and -resistant invasive pulmonary aspergillosis. Dis. Models Mech. 2022, 15, dmm049165. [Google Scholar] [CrossRef] [PubMed]
- Vanherp, L.; Ristani, A.; Poelmans, J.; Hillen, A.; Lagrou, K.; Janbon, G.; Brock, M.; Himmelreich, U.; Vande Velde, G. Sensitive bioluminescence imaging of fungal dissemination to the brain in mouse models of cryptococcosis. Dis. Models Mech. 2019, 12, dmm039123. [Google Scholar] [CrossRef] [PubMed]


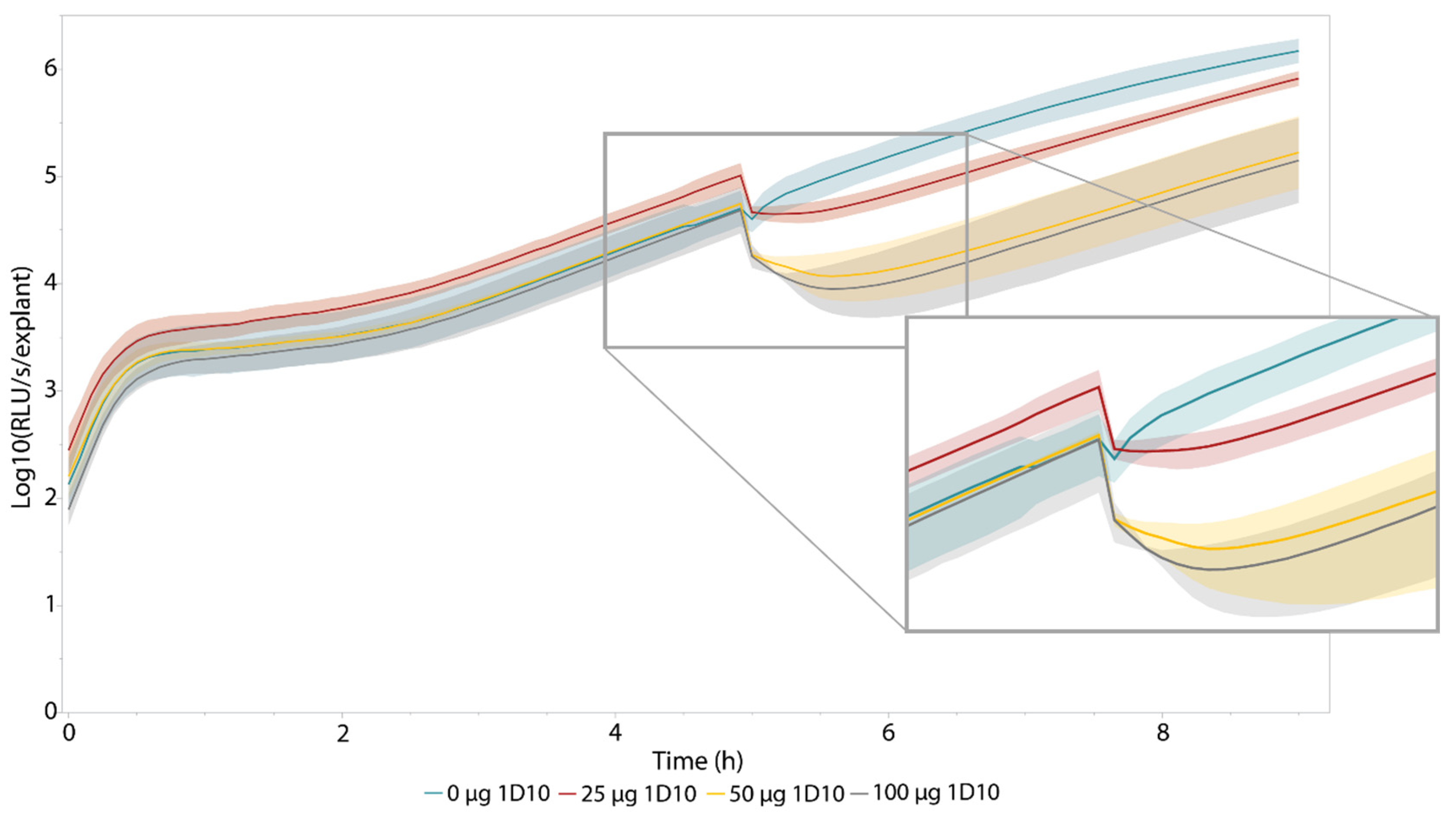

| Treatment | Relative Drop in Luminescence (%) | p-Value | Time to Reach Minimum (h) | p-Value | Area under the Curve (t5–t9) (×102) | p-Value |
|---|---|---|---|---|---|---|
| 0 µg 1D10 | 20.3 ± 9.31 | N/A | 0.0833 ± 0 | N/A | 41.0 ± 4.41 | N/A |
| 25 µg 1D10 | 58.3 ± 17.0 | 0.0405 | 0.278 ± 0.210 | 0.125 | 10.0 ± 3.87 | 0.000858 |
| 50 µg 1D10 | 79.4 ±8.41 | 0.00129 | 0.694 ± 0.337 | 0.0440 | 4.29 ± 3.47 | 0.000346 |
| 100 µg 1D10 | 77.0 ± 16.4 | 0.0121 | 0.639 ± 0.315 | 0.0464 | 4.97 ± 3.69 | 0.000481 |
| Treatment | a | b | f’(x) | f’(9)-f’(5) | R2 of Fit |
|---|---|---|---|---|---|
| 0 µg 1D10 | 950 | 0.845 | 803 e0.845x | 1.56 × 106 | 0.980 |
| 25 µg 1D10 | 638 | 0.794 | 507 e0.794x | 6.17 × 105 | 1.000 |
| 50 µg 1D10 | 215 | 0.745 | 160 e0.745x | 1.24 × 105 | 0.990 |
| 100 µg 1D10 | 150 | 0.770 | 116 e0.770x | 1.13 × 105 | 0.991 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Maesschalck, V.; Gutiérrez, D.; Paeshuyse, J.; Briers, Y.; Vande Velde, G.; Lavigne, R. A Bioluminescence-Based Ex Vivo Burn Wound Model for Real-Time Assessment of Novel Phage-Inspired Enzybiotics. Pharmaceutics 2022, 14, 2553. https://doi.org/10.3390/pharmaceutics14122553
De Maesschalck V, Gutiérrez D, Paeshuyse J, Briers Y, Vande Velde G, Lavigne R. A Bioluminescence-Based Ex Vivo Burn Wound Model for Real-Time Assessment of Novel Phage-Inspired Enzybiotics. Pharmaceutics. 2022; 14(12):2553. https://doi.org/10.3390/pharmaceutics14122553
Chicago/Turabian StyleDe Maesschalck, Vincent, Diana Gutiérrez, Jan Paeshuyse, Yves Briers, Greetje Vande Velde, and Rob Lavigne. 2022. "A Bioluminescence-Based Ex Vivo Burn Wound Model for Real-Time Assessment of Novel Phage-Inspired Enzybiotics" Pharmaceutics 14, no. 12: 2553. https://doi.org/10.3390/pharmaceutics14122553
APA StyleDe Maesschalck, V., Gutiérrez, D., Paeshuyse, J., Briers, Y., Vande Velde, G., & Lavigne, R. (2022). A Bioluminescence-Based Ex Vivo Burn Wound Model for Real-Time Assessment of Novel Phage-Inspired Enzybiotics. Pharmaceutics, 14(12), 2553. https://doi.org/10.3390/pharmaceutics14122553









