DNA Binding and Anticancer Properties of New Pd(II)-Phosphorus Schiff Base Metal Complexes
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Molecular Docking Analysis
2.3. Synthesis of Phosphorus Schiff Base Ligands (L1 and L2)
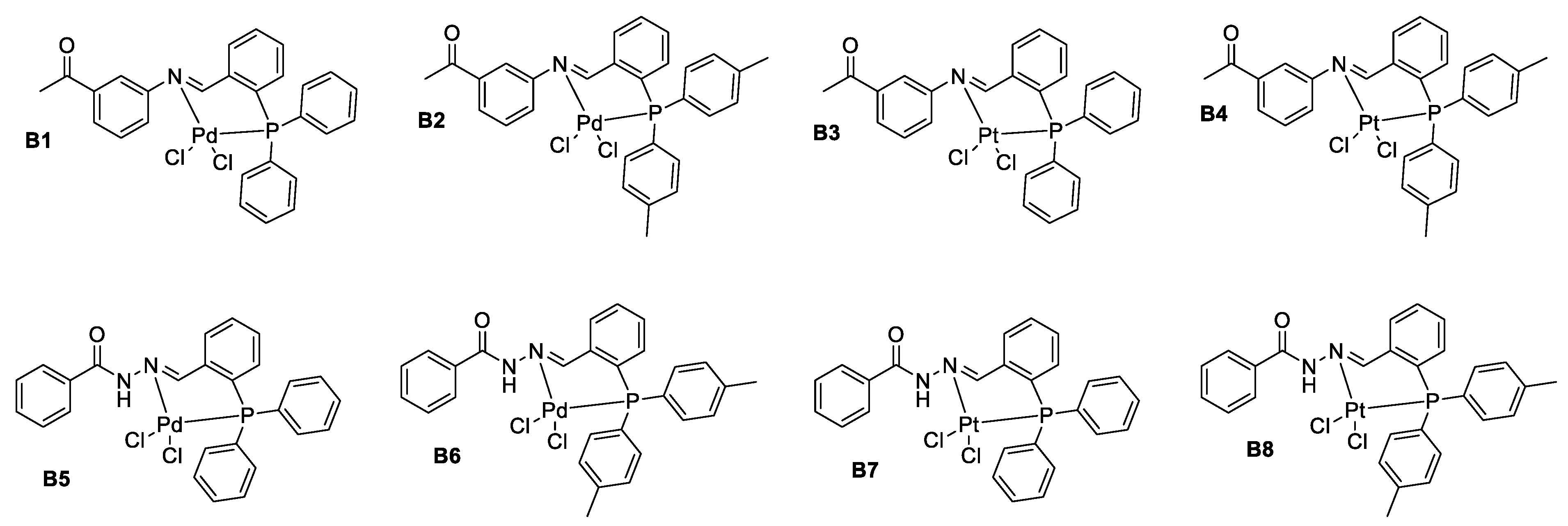
2.4. Synthesis of Pd(II) Complexes of Phosphorus Schiff Base Ligands (B1 and B2)
2.5. Cytotoxicity Assay
2.6. Apoptosis Assay
2.7. Colony Forming Efficiency Assay
2.8. BSA Binding Assay
2.9. DNA Binding Assay
2.10. Statistical Analysis
3. Results and Discussion
3.1. Palladium Complexes B1 and B2 Had the Most Potent Interaction with DNA According to In-Silico Analysis
3.2. Phosphorus Schiff Base Ligands and Palladium(II) Complexes Were Successfully Synthesized and Characterized
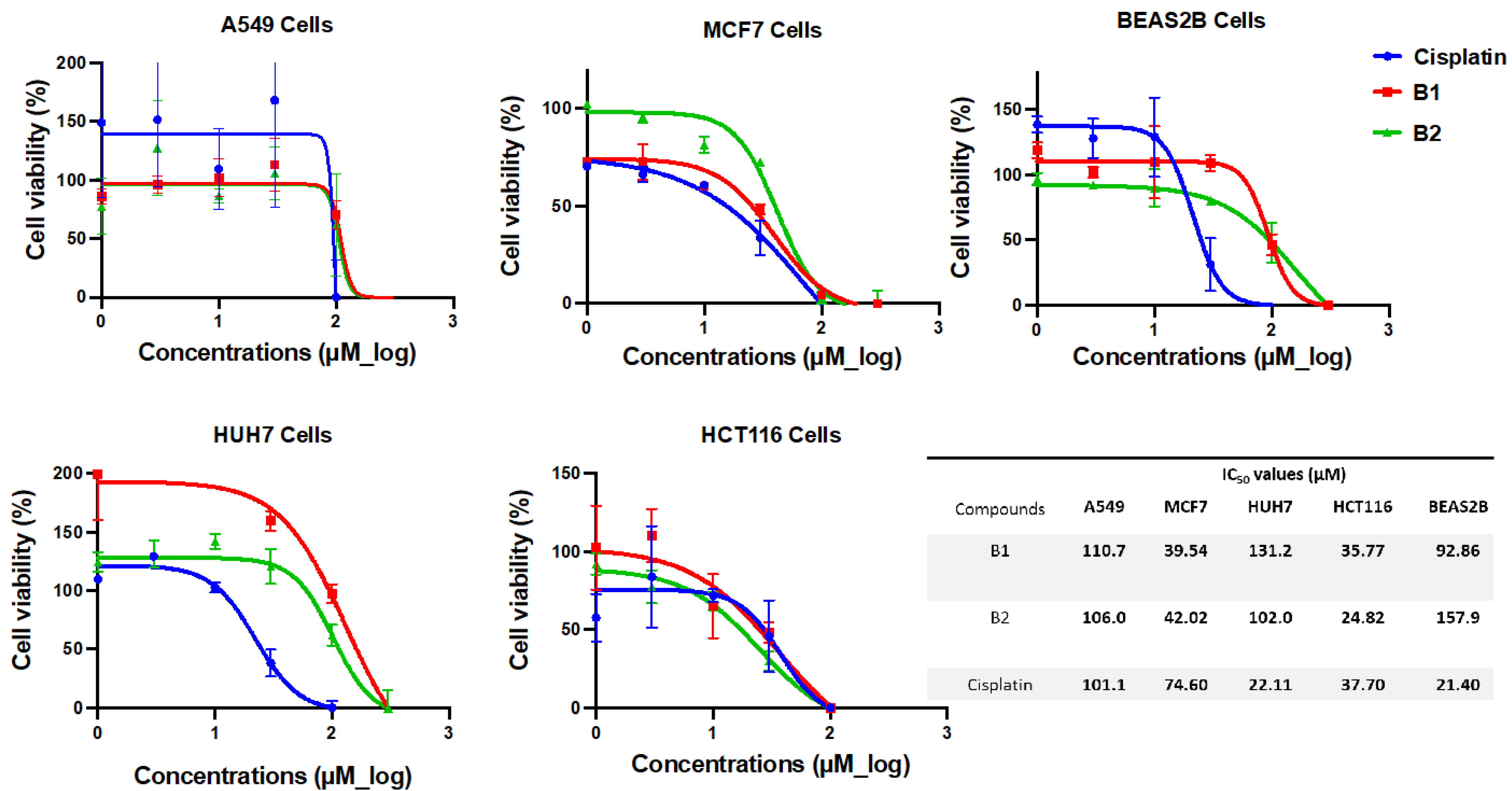
3.3. B1 and B2 Showed Cytotoxic Activity on Cancer Cells
3.4. B1 and B2 Induced Apoptosis in Cancer Cells
3.5. B1 and B2 Suppressed Colony Formation of Cancer Cells
3.6. B1 and B2 Had Strong DNA Intercalating Activity
3.7. B1 and B2 Showed Static BSA Binding
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kumar, P.; Narasimhan, B.; Ramasamy, K.; Mani, V.; Mishra, R.K.; Majeed, A.B.A. Synthesis, antimicrobial, anticancer evaluation and QSAR studies of 3/4-Bromo benzohydrazide derivatives. Curr. Top. Med. Chem. 2015, 15, 1050–1066. [Google Scholar] [CrossRef] [PubMed]
- Allardyce, C.S.; Dorcier, A.; Scolaro, C.; Dyson, P.J. Development of orga-nometallic (organo-transition metal) pharmaceuticals. Appl. Organometal. Chem. 2005, 19, 1–10. [Google Scholar] [CrossRef]
- Bruijnincx, P.C.A.; Sadler, P.J. New trends for metal complexes with anticancer activity. Curr. Opin. Chem. Biol. 2008, 12, 197–206. [Google Scholar] [CrossRef] [PubMed]
- Rau, T.; Eldik, R.V. Mechanistic insight from kinetic studies on the interaction of model palladium(II) complexes with nucleic acid components. Met. Ions. Biol. Syst. 1996, 31, 339–378. [Google Scholar]
- Butour, S.; Wimmer, F.; Wimmer, F.; Castan, P. Palladium (II) compounds with potential antitumour properties and their platinum analogues: A comparative study of the reaction of some orotic acid derivatives with DNA in vitro. Chem. Biol. Interact. 1997, 104, 165–178. [Google Scholar] [CrossRef]
- Wimmer, F.Z.; Wimmer, S.; Castan, P.; Cros, S.; Johnson, N.; Colacio-Rodrigez, E. The antitumor activity of some palladium(II) complexes with chelating ligands. Anticancer Res. 1989, 9, 791–794. [Google Scholar] [PubMed]
- Zhao, G.; Lin, H.; Yu, P.; Sun, H.; Zhu, S.; Su, X.; Chen, Y. Ethylenediamine-palladium(II) complexes with pyridine and its derivatives: Synthesis, molecular structure and initial antitumor studies. J. Inorg. Biochem. 1999, 73, 145–149. [Google Scholar] [CrossRef]
- Mansuri-Torshizi, H.; Srivastava, T.S.; Parekh, H.K.; Chitnis, M.P. Synthesis, spectroscopic, cytotoxic, and DNA binding studies of binuclear 2,2 -bipyridine-platinum(II) and -palladium (II) complexes of meso-α,α,′ -diaminoadipic meso α,α-diaminosuberic acids. J. Inorg. Biochem. 1992, 45, 135–148. [Google Scholar] [CrossRef]
- Shurig, J.E.; Harry, H.A.; Timmer, K.; Long, B.H.; Casazza, A.M. Antitumor activity of bis[bis(diphenylphoshino)alkane and alkene] groupVIII metal complexes. Prog. Clin. Biochem. Med. 1989, 10, 205–216. [Google Scholar]
- Malešević, N.; Srdić, T.; Radulović, S.; Sladić, D.; Radulović, V.; Brčeski, I.; Anđelković, K. Synthesis and characterization of a novel Pd(II) complex with the condensation product of 2-(diphenylphosphino)benzaldehyde and ethyl hydrazinoacetate. Cytotoxic activity of the synthesized complex and related Pd(II) and Pt(II) complexes. J. Inorg. Biochem. 2006, 100, 1811–1818. [Google Scholar] [CrossRef]
- Quiroga, A.G.; Perez, J.M.; Montero, E.I.; Masaguer, J.R.; Alonso, C.; Navarro-Ranninger, C. Palladated and platinated complexes derived from phenylacetaldehyde thiosemicarbazone with cytotoxic activity in cis-DDP resistant tumor cells. Formation of DNA interstrand cross-links by these complexes. J. Inorg. Biochem. 1998, 70, 117–123. [Google Scholar] [CrossRef]
- Matesanz, A.I.; Perez, J.M.; Navarro, P.; Moreno, J.M.; Colacio, E.; Souza, P. Synthesis and characterization of novel palladium(II) complexes of bis(thiosemicarbazone). Structure, cytotoxic activity and DNA binding of Pd(II)-benzyl bis(thiosemicarbazonate). J. Inorg. Biochem. 1999, 76, 29–37. [Google Scholar] [CrossRef]
- Budzisz, E.; Krajewska, U.; Rózalski, M. Cytotoxic and proapoptotic effects of new Pd(II) and Pt(II)-complexes with 2-ethanimidoyl-2-methoxy-2H-1,2-benzoxaphosphinin-4-ol-2-oxide. Pol. J. Pharmacol. 2004, 56, 473–478. [Google Scholar] [PubMed]
- Marchand, N.; Lienard, P.; Siehl, H.U.; Izato, H. Applications of Molecular Simulation Software SCIGRESS in Industry and University. Fujitsu Sci. Tech. J. 2014, 50, 46–51. [Google Scholar]
- Rose, Y.; Duarte, J.M.; Lowe, R.; Segura, J.; Bi, C.; Bhikadiya, C.; Chen, L.; Rose, A.S.; Bittrich, S.; Burley, S.K.; et al. RCSB protein data bank: Architectural advances towards integrated searching and efficient access to macromolecular structure data from the PDB archive. J. Mol. Biol. 2021, 433, 166704. [Google Scholar] [CrossRef]
- Chattaraj, P.K.; Gir, S.; Duley, S. Update 2 of: Electrophilicity index. Chem. Rev. 2011, 111, 43–75. [Google Scholar] [CrossRef]
- Accelrys Software Inc. Discovery Studio Modeling Environment, Release 3.5; Accelrys Software Inc.: San Diego, CA, USA, 2013. [Google Scholar]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Saygideğer-Kont, Y.; Minas, T.Z.; Jones, H.; Hour, S.; Çelik, H.; Temel, I.; Han, J.; Atabey, N.; Erkizan, H.V.; Toretsky, J.A.; et al. Ezrin Enhances EGFR Signaling and Modulates Erlotinib Sensitivity in Non-Small Cell Lung Cancer Cells. Neoplasia 2016, 18, 111–120. [Google Scholar] [CrossRef]
- APC Annexin V Apoptosis Detection Kit with PI. Available online: https://www.biolegend.com/nl-be/products/apc-annexin-v-apoptosis-detection-kit-with-pi-9788 (accessed on 25 January 2022).
- Marmur, J.; Doty, P. Determination of the base composition of deoxyribonucleic acid from its thermal denaturation temperature. J. Mol. Biol. 1962, 5, 109–118. [Google Scholar] [CrossRef]
- Reichmann, M.E.; Rice, A.S.; Thomas, C.A.; Doty, P. A further examination of the molecular weight and size of desoxypentose nucleic acid. J. Am. Chem. Soc. 1954, 76, 3047–3053. [Google Scholar] [CrossRef]
- Wolfe, A.; Shimer, G.H., Jr.; Meehan, T. Polycyclic aromatic hydrocarbons physically intercalate into duplex regions of denatured DNA. Biochemistry 1987, 26, 6392–6396. [Google Scholar] [CrossRef]
- Yilmaz, M.K. Palladium(II) complexes with new bidentate phosphine-imine ligands for the Suzuki C-C coupling reactions in supercritical carbon dioxide. J. Supercrit. Fluids 2018, 138, 221–227. [Google Scholar] [CrossRef]
- Yılmaz, M.K.; İnce, S.; Yılmaz, S.; Keleş, M. Palladium(II) catalyzed Suzuki C-C coupling reactions with imino- and amino-phosphine ligands. Inorg. Chim. Acta 2018, 482, 252–258. [Google Scholar] [CrossRef]
- Suárez-Meneses, J.V.; Oukhrib, A.; Gouygou, M.; Urrutigoïty, M.; Daran, J.C.; Cordero-Vargas, A.; Ortega-Alfaro, M.C.; López-Cortés, J.G. [N,P]-pyrrole PdCl2 complexes catalyzed the formation of dibenzo-α-pyrone and lactam analogues. Dalton Trans. 2016, 45, 9621–9630. [Google Scholar] [CrossRef] [PubMed]
- Hejchman, E.; Kruszewska, H.; Maciejewska, D.; Sowirka-Taciak, B.; Tomczyk, M.; Sztoksz-Ignasiak, A.; Jankowski, J.; Młynarczuk-Biały, I. Design, synthesis, and biological activity of Schiff bases bearing salicyl and 7-hydroxycoumarinyl moieties. Mon. Chem. 2019, 155, 255–266. [Google Scholar] [CrossRef]
- Arafath, M.A.; Adam, F.; Razali, M.R.; Hassan, L.E.A.; Khadeer Ahamed, M.B.; Majid, A.M.S.A. Synthesis, characterization and anticancer studies of Ni(II), Pd(II) and Pt(II) complexes with Schiff base derived from N-methylhydrazinecarbothioamide and 2-hydroxy-5-methoxy-3-nitrobenzaldehyde. J. Mol. Struct. 2017, 1130, 791–798. [Google Scholar] [CrossRef]
- Frezza, M.; Hindo, S.; Chen, D.; Davenport, A.; Schmitt, S.; Tomco, D.; Dou, Q.P. Novel metals and metal complexes as platforms for cancer therapy. Curr. Pharm. Des. 2010, 16, 1813–1825. [Google Scholar] [CrossRef]
- Ndagi, U.; Mhlongo, N.; Soliman, M.E. Metal complexes in cancer therapy—An update from drug design perspective. Drug Des. Devel. Ther. 2017, 11, 599–616. [Google Scholar] [CrossRef]
- Haas, K.L.; Franz, K.J. Application of metal coordination chemistry to explore and manipulate cell biology. Chem. Rev. 2009, 109, 4921–4960. [Google Scholar] [CrossRef]
- Kelly, J.M.; Tossi, A.B.; McConnell, D.J.; OhUigin, C. A study of the interactions of some polypyridylruthenium(II) complexes with DNA using fluorescence spectroscopy, topoisomerisation and thermal denaturation. Nucl. Acids Res. 1985, 13, 6017–6034. [Google Scholar] [CrossRef]
- Abdel-Rahman, L.H.; El-Khatib, R.M.; Nassr, L.A.E.; AbuDief, A.M.; Ismail, M.; Seleem, A.A. Metal based pharmacologically active agents: Synthesis, structural characterization, molecular modeling, CT-DNA binding studies and in vitro antimicrobial screening of iron(II) bromosalicylidene amino acid chelates. Spectrochim. Acta 2014, 117, 366–378. [Google Scholar] [CrossRef] [PubMed]
- Rahban, M.; Divsalar, A.; Saboury, A.A.; Golestani, A. Nanotoxicity and spectroscopy studies of silver nanoparticle: Calf Thymus DNA and K562 as targets. J. Phys. Chem. C 2010, 114, 5798–5803. [Google Scholar] [CrossRef]
- Li, W.; Ji, Y.Y.; Wang, J.W.; Zhu, Y.M. Cytotoxic activities and DNA binding properties of 1-methyl-7H-indeno [1,2-b]quinolinium-7-(4-dimethylamino) benzylidene triflate. DNA Cell Biol. 2012, 31, 1046–1053. [Google Scholar] [CrossRef] [PubMed]
- Abu-Dief, A.M.; El-khatib, R.M.; Aljohani, F.S.; Alzahrani, S.O.; Mahran, A.; Khalifa, M.E.; El-Metwaly, N.M. Synthesis and intensive characterization for novel Zn(II), Pd(II), Cr(III) and VO(II)-Schiff base complexes; DNA-interaction, DFT, drug-likeness and molecular docking studies. J. Mol. Struct. 2021, 1242, 130693. [Google Scholar] [CrossRef]
- Raja, D.S.; Bhuvanesh, N.S.P.; Natarajan, K. Biological evaluation of a novel water soluble sulphur bridged binuclear copper(II) thiosemicarbazone complex. Eur. J. Med. Chem. 2011, 46, 4584–4594. [Google Scholar] [CrossRef]
- Raja, D.S.; Bhuvanesh, N.S.P.; Bhuvanesh, J.H.; Reibenspies, R.; Renganathan, K.N. Effect of terminal N-substitution in 2-oxo-1,2-dihydroquinoline-3-carbaldehyde thiosemicarbazones on the mode of coordination, structure, interaction with protein, radical scavenging and cytotoxic activity of copper(II) complexes. Dalton Trans. 2011, 40, 4548–4559. [Google Scholar] [CrossRef]
- Gupta, R.K.; Sharma, G.; Pandey, R.; Kumar, A.; Koch, B.; Li, P.-Z.; Xu, Q.; Pandey, D.S. DNA/protein binding, molecular docking, and in vitro anticancer activity of some thioether-dipyrrinato complexes. Inorg. Chem. 2013, 52, 13984–13996. [Google Scholar] [CrossRef]






| Name | Binding Energy (kcal/mol) | RMSD (Å) | Inhibition Constant, (Ki, µM) |
|---|---|---|---|
| cisplatin | −2.73 | 2.61 | 9980 |
| triphenylphosphine | −4.02 | 2.24 | 1140 |
| B1 | −4.51 | 2.34 | 4910 |
| B2 | −6.04 | 2.05 | 3730 |
| B3 | −2.30 | 2.42 | 20,620 |
| B4 | −2.38 | 2.36 | 18,040 |
| B5 | 4.56 | 24.01 | - |
| B6 | 17.54 | 24.73 | - |
| B7 | 1.54 | 24.11 | - |
| B8 | 48.85 | 24.77 | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saygıdeğer Demir, B.; İnce, S.; Yilmaz, M.K.; Sezan, A.; Derinöz, E.; Taskin-Tok, T.; Saygideger, Y. DNA Binding and Anticancer Properties of New Pd(II)-Phosphorus Schiff Base Metal Complexes. Pharmaceutics 2022, 14, 2409. https://doi.org/10.3390/pharmaceutics14112409
Saygıdeğer Demir B, İnce S, Yilmaz MK, Sezan A, Derinöz E, Taskin-Tok T, Saygideger Y. DNA Binding and Anticancer Properties of New Pd(II)-Phosphorus Schiff Base Metal Complexes. Pharmaceutics. 2022; 14(11):2409. https://doi.org/10.3390/pharmaceutics14112409
Chicago/Turabian StyleSaygıdeğer Demir, Burcu, Simay İnce, Mustafa Kemal Yilmaz, Aycan Sezan, Ezgi Derinöz, Tugba Taskin-Tok, and Yasemin Saygideger. 2022. "DNA Binding and Anticancer Properties of New Pd(II)-Phosphorus Schiff Base Metal Complexes" Pharmaceutics 14, no. 11: 2409. https://doi.org/10.3390/pharmaceutics14112409
APA StyleSaygıdeğer Demir, B., İnce, S., Yilmaz, M. K., Sezan, A., Derinöz, E., Taskin-Tok, T., & Saygideger, Y. (2022). DNA Binding and Anticancer Properties of New Pd(II)-Phosphorus Schiff Base Metal Complexes. Pharmaceutics, 14(11), 2409. https://doi.org/10.3390/pharmaceutics14112409









