Green Synthesis of Hexagonal Silver Nanoparticles Using a Novel Microalgae Coelastrella aeroterrestrica Strain BA_Chlo4 and Resulting Anticancer, Antibacterial, and Antioxidant Activities
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Methods
2.2.1. Microalgae Isolation
2.2.2. Morphological and Molecular Identification of Microalgae
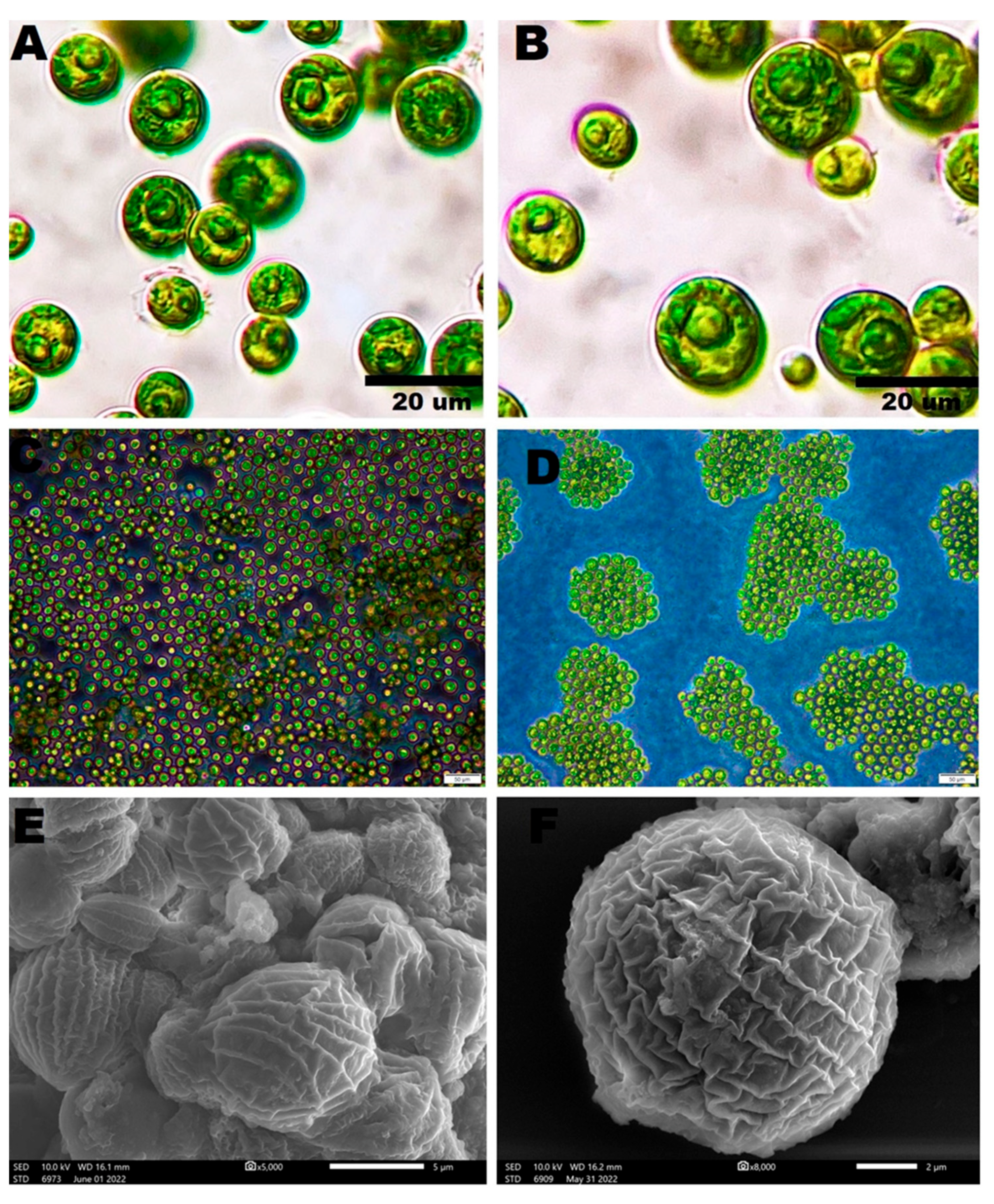
Light and Inverted Light Microscopy
Scanning Electron Microscopy
Molecular Identification
2.2.3. Preparation of Microalgae Extract
2.2.4. Gas Chromatography–Mass Spectrometry (GC-MS) Analysis
2.2.5. Synthesis of Ag-NPs Using Algal Extract
2.2.6. Characterization of Ag-NPs Synthesized by C. aeroterrestrica BA_Chlo4 (C@Ag-NPs)
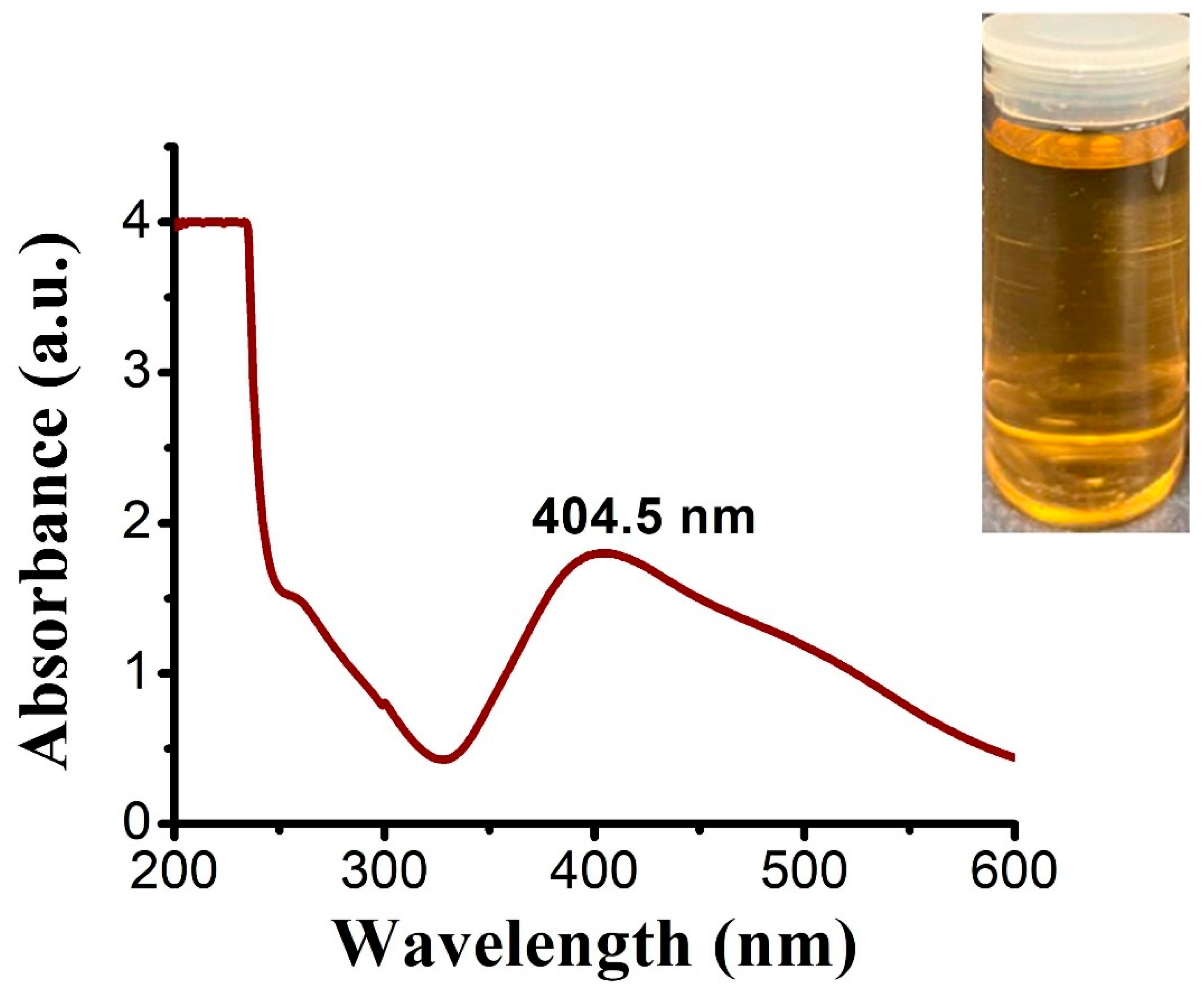
UV-Spectrophotometry
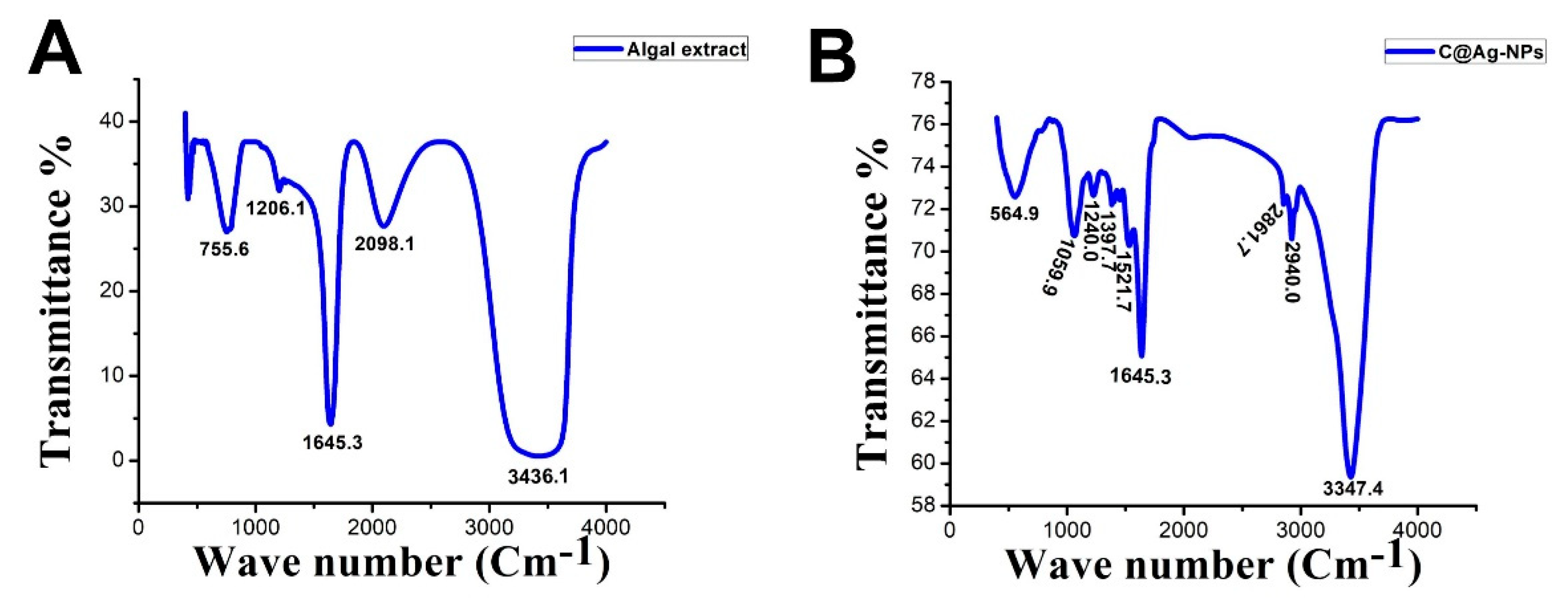
FTIR Spectroscopy
X-ray Diffraction Analysis (XRD)
EDX and Mapping Analyses
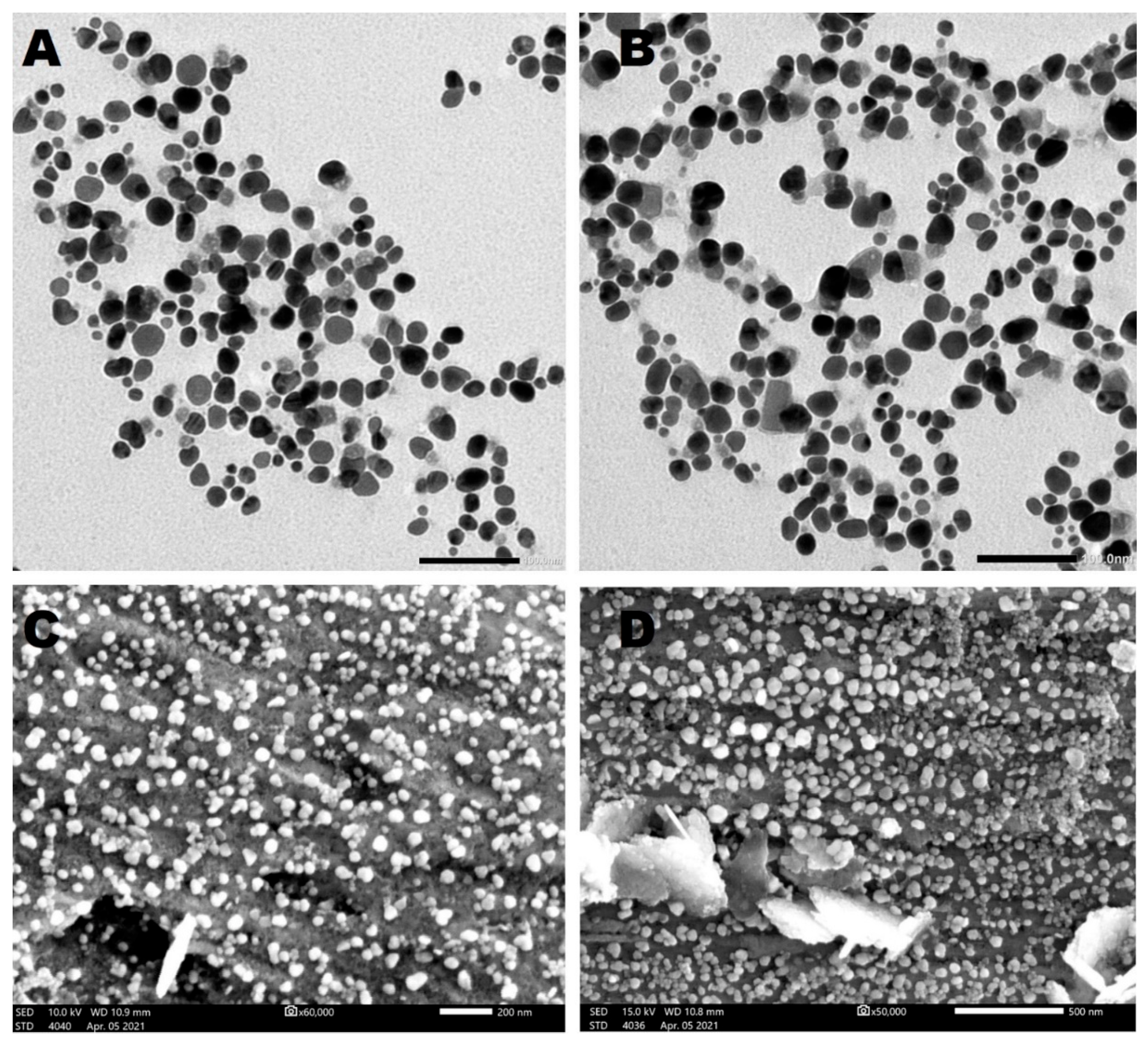
Scanning and Transmission Electron Microscopy (SEM and TEM)
DLS and Zeta Potential
2.2.7. Anticancer Activities of C@Ag-NPs
Cell Culture
MTT Assay
Inverted Light Microscope
2.2.8. Antioxidant Activity
2.2.9. Antimicrobial Activity of C@Ag-NPs
Microbial Culture
Agar Well Diffusion Method
Minimum Inhibition and Biocidal Concentrations (MIC and MBC)
2.2.10. Statistical Analysis
3. Results and Discussion
3.1. Morphological Appearance of Coelastrella aeroterrestrica strain BA_Chlo4
3.2. Phylogenetic Analysis
3.3. GC-MS Analysis
3.4. Characterization of C@Ag-NPs
3.4.1. UV-Spectroscopy
3.4.2. FTIR Spectroscopy
3.4.3. XRD
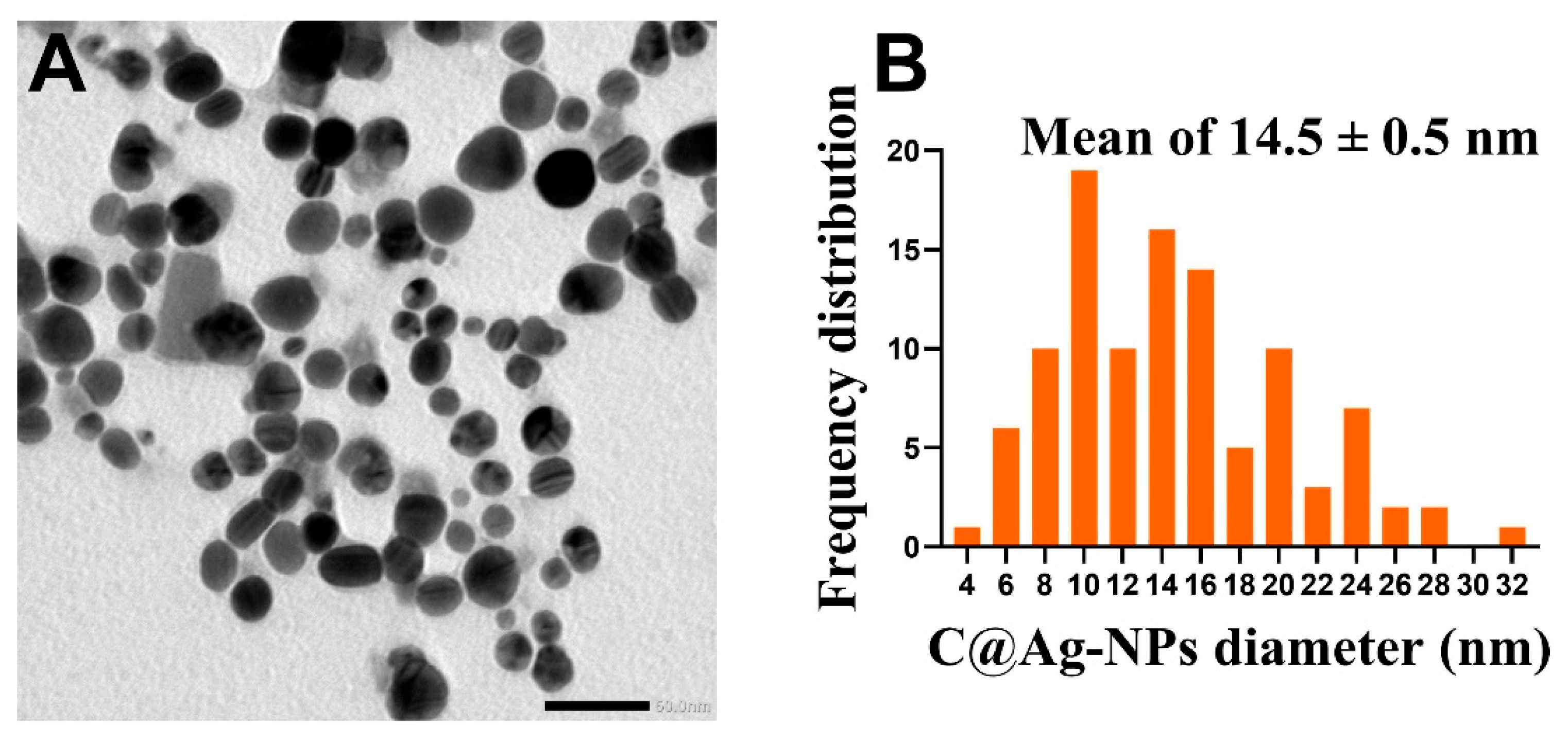
3.4.4. TEM and SEM
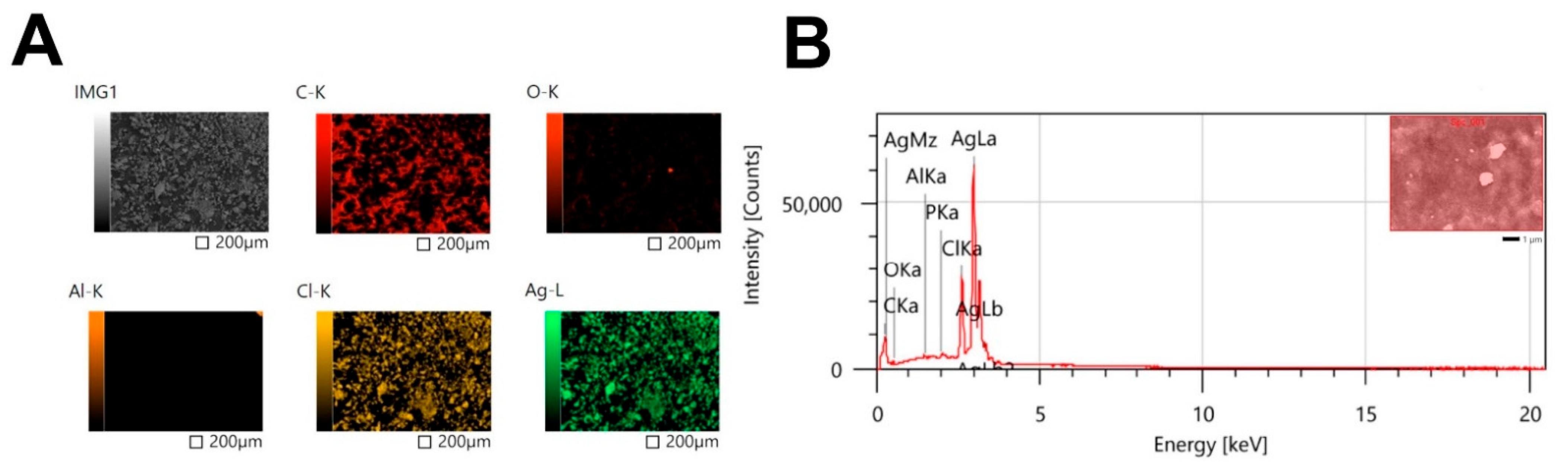
3.4.5. Mapping and EDX
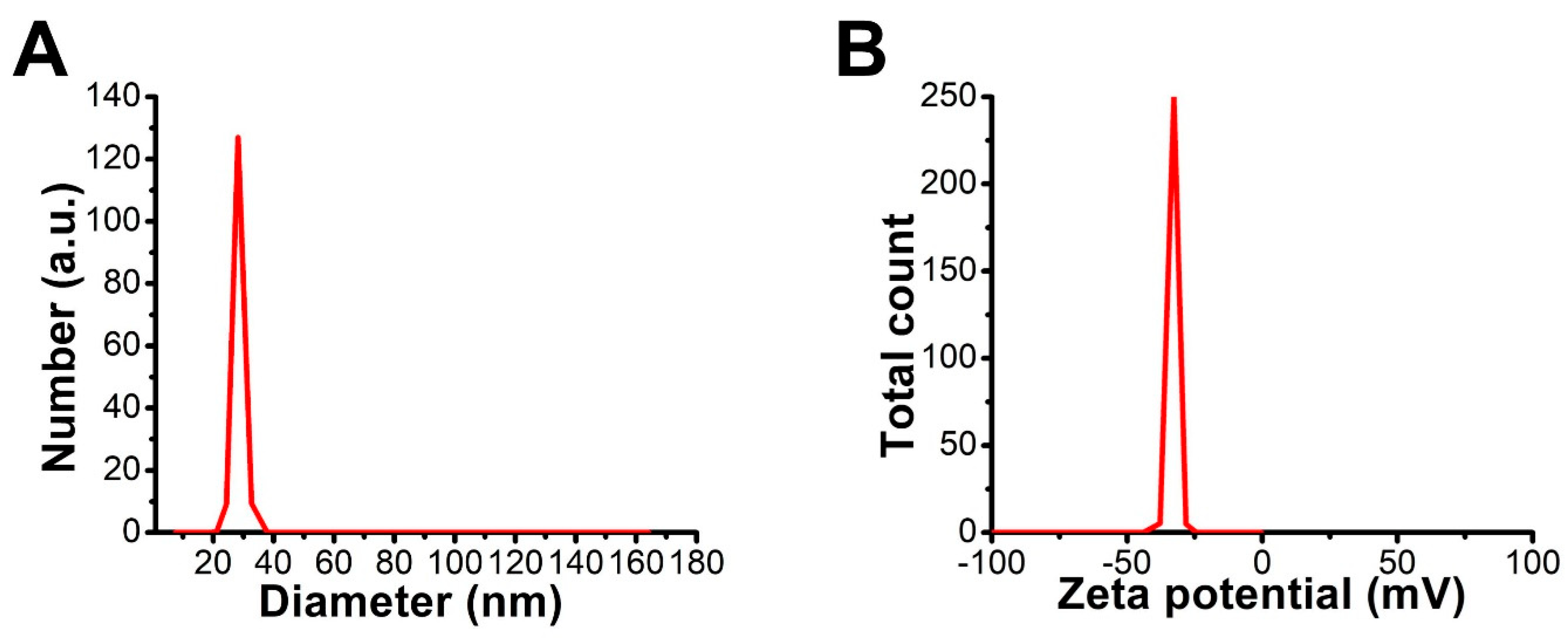
3.4.6. DLS and Zeta Potential
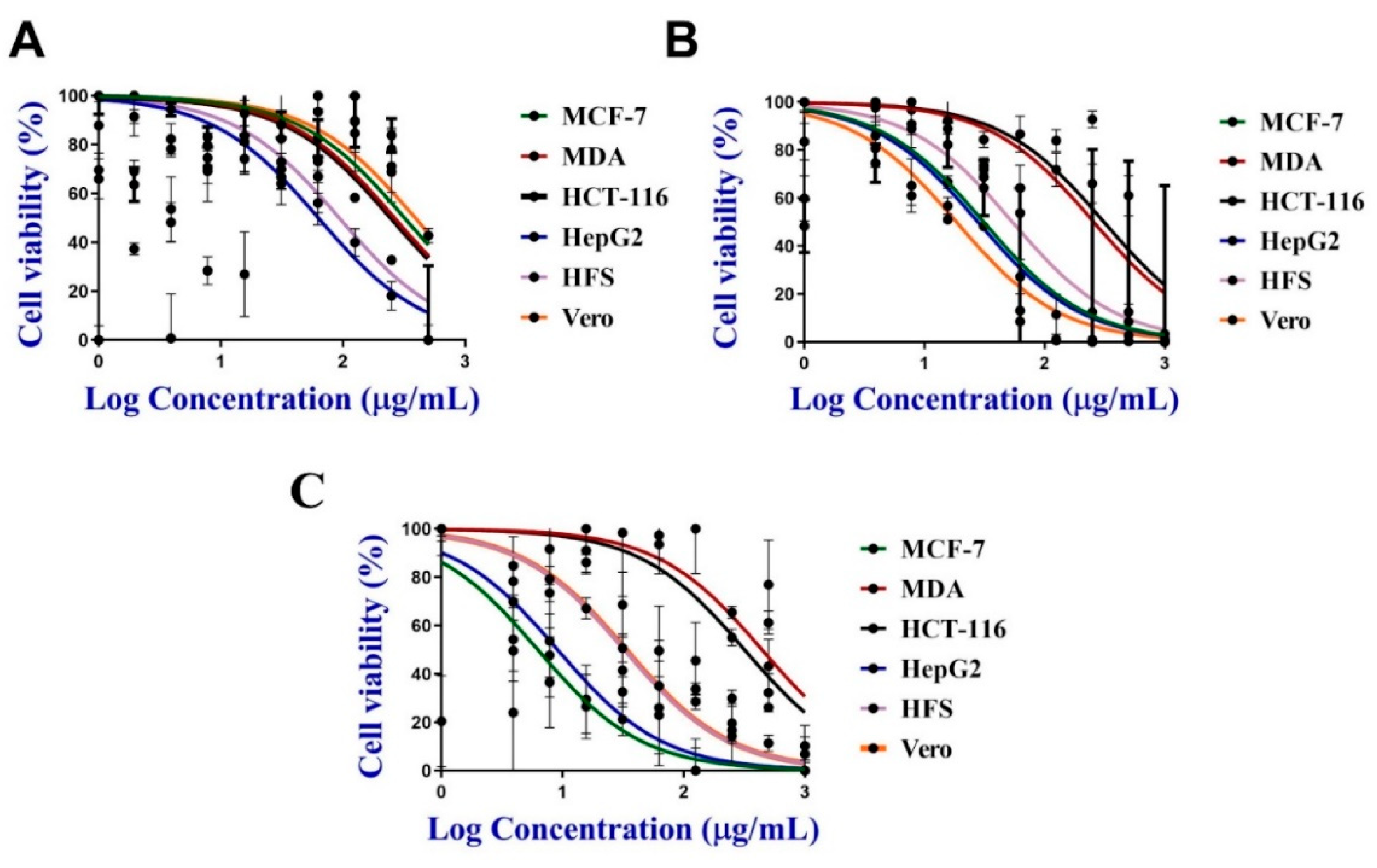
3.5. Toxicity of C@Ag-NPs and Algal Extract against Cancer Cells
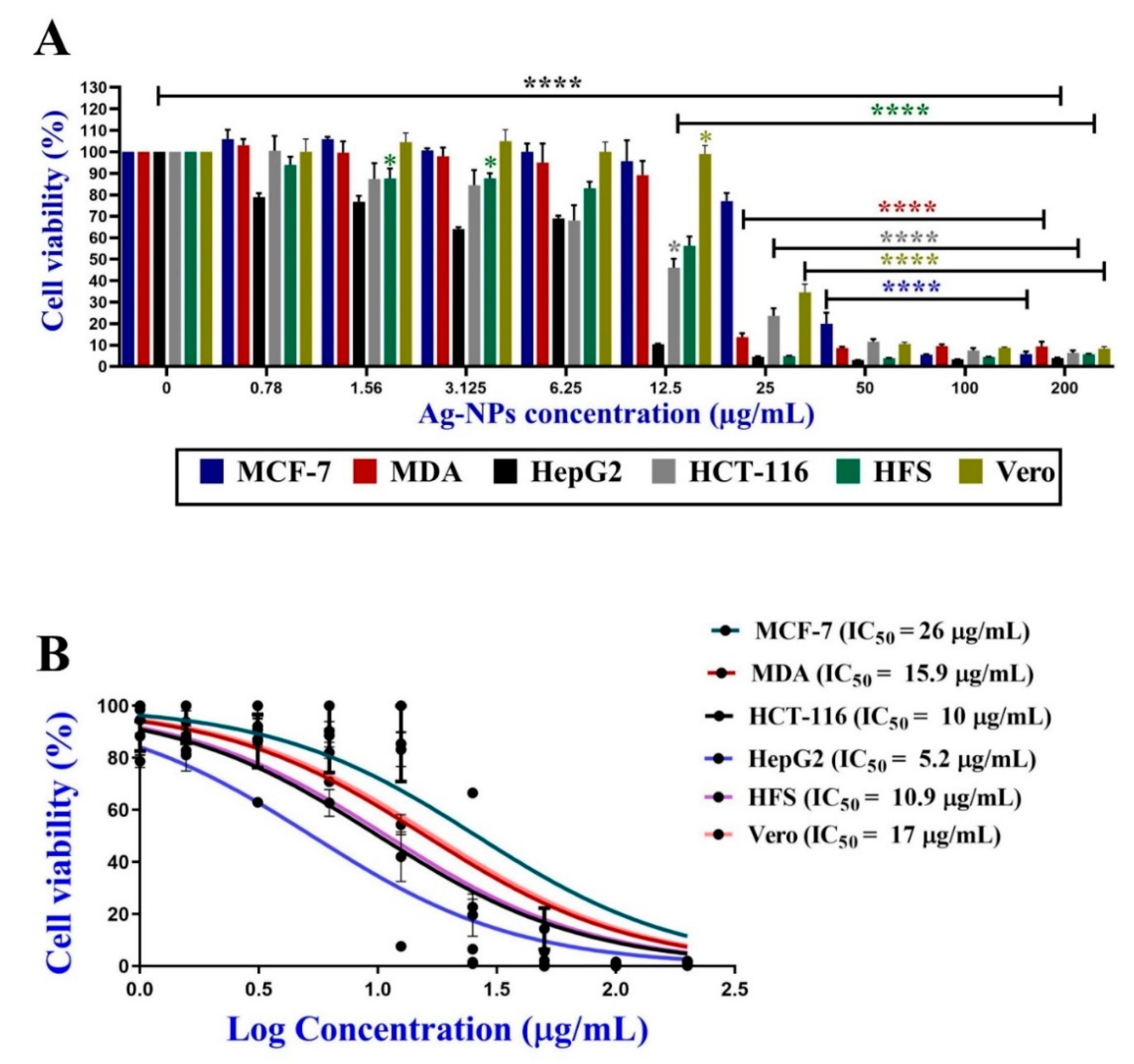
3.5.1. Antiproliferative Activity of C@Ag-NPs
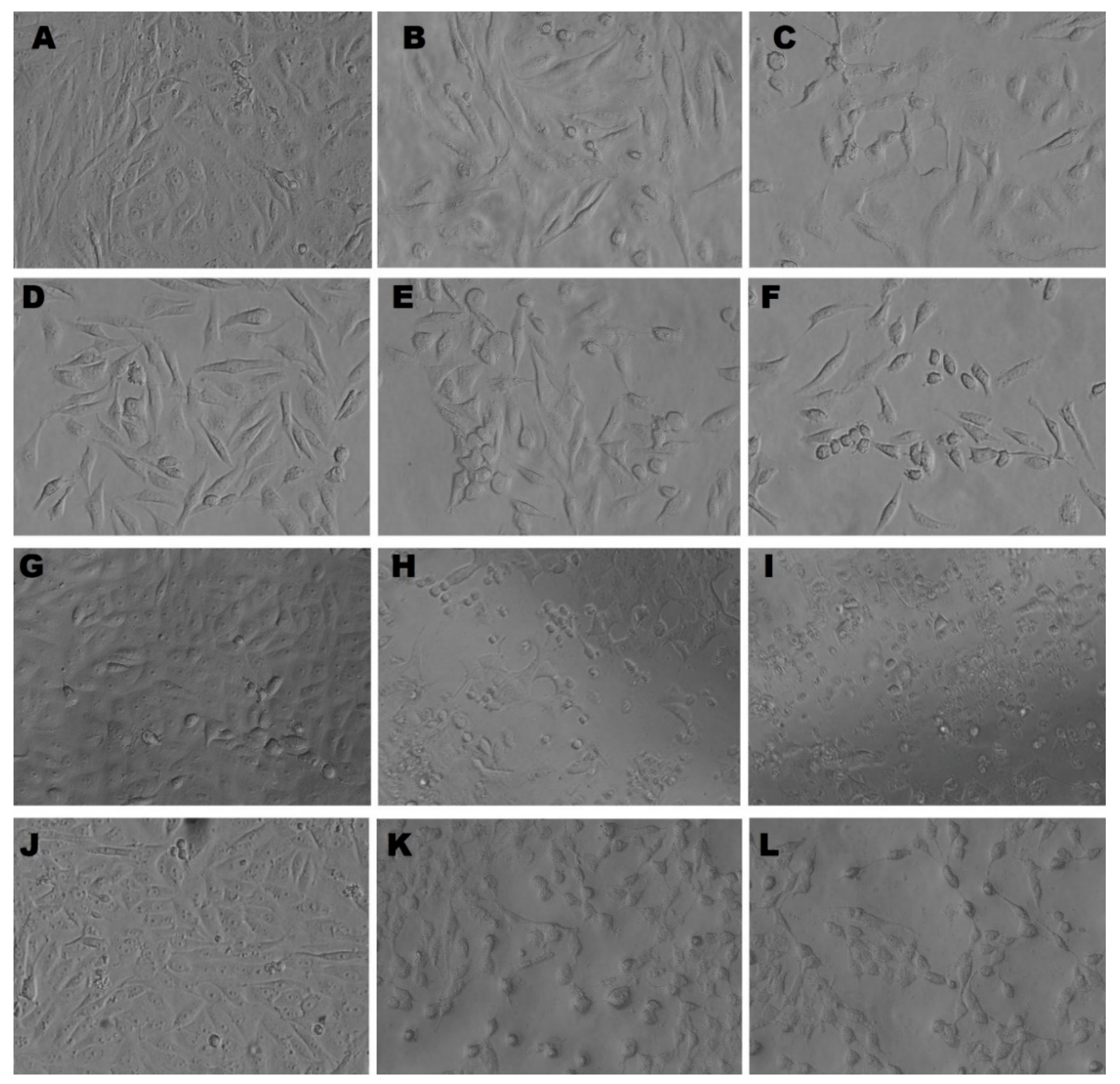
3.5.2. Morphological Appearance of MDA, MCF-7, HCT-116, and HepG2 Cells Treated with C@Ag-NPs
3.6. Scavenging Activity of C@Ag-NPs
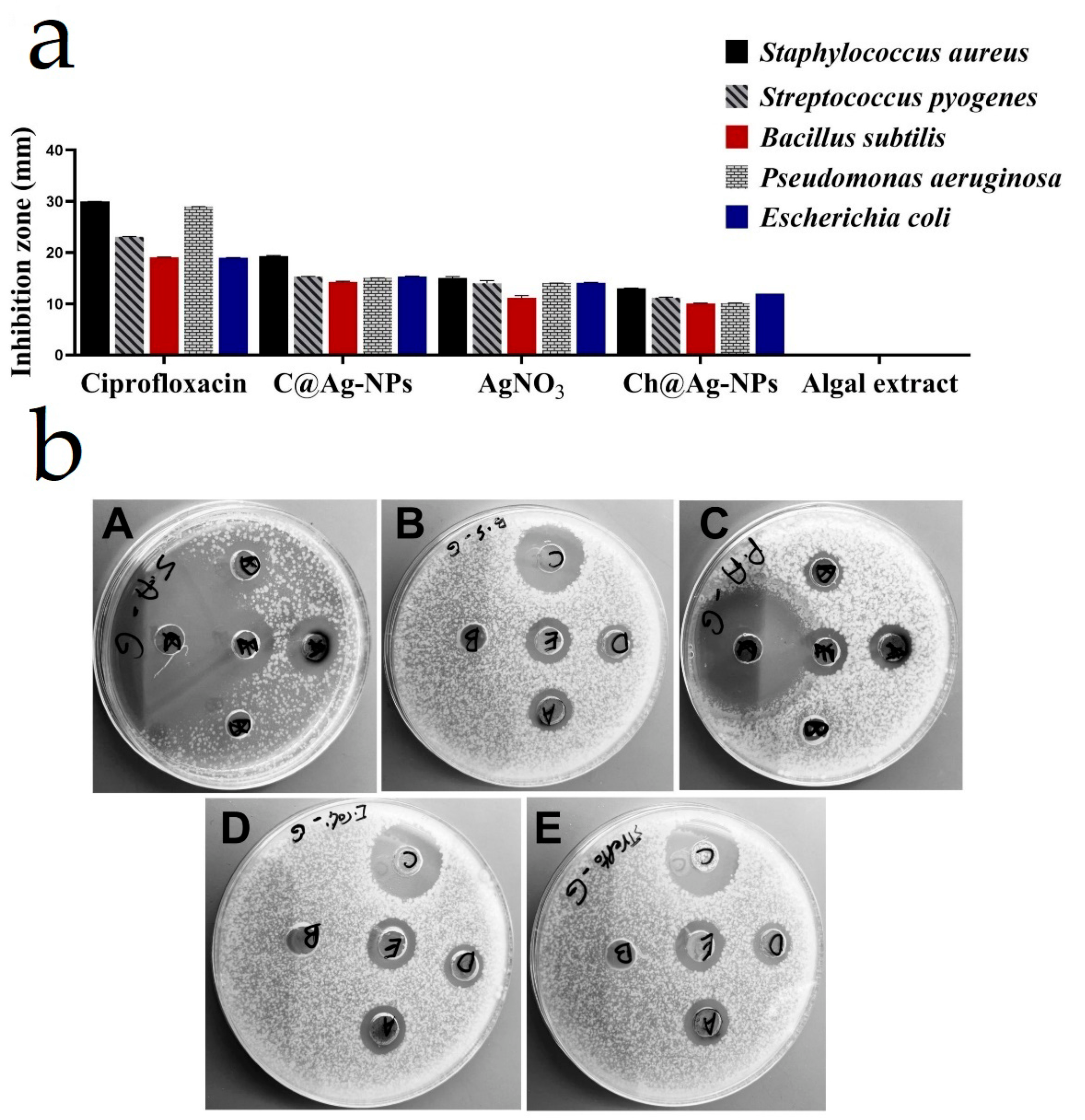
3.7. Antimicrobial Activity
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kirtane, A.R.; Verma, M.; Karandikar, P.; Furin, J.; Langer, R.; Traverso, G. Nanotechnology approaches for global infectious diseases. Nat. Nanotechnol. 2021, 16, 369–384. [Google Scholar] [CrossRef] [PubMed]
- Anselmo, A.C.; Mitragotri, S. Nanoparticles in the clinic: An update. Bioeng. Transl. Med. 2019, 4, e10143. [Google Scholar] [CrossRef] [PubMed]
- Zingale, E.; Romeo, A.; Rizzo, S.; Cimino, C.; Bonaccorso, A.; Carbone, C.; Musumeci, T.; Pignatello, R. Fluorescent nanosystems for drug tracking and theranostics: Recent applications in the ocular field. Pharmaceutics 2022, 14, 955. [Google Scholar] [CrossRef]
- Yan, L.; Shen, J.; Wang, J.; Yang, X.; Dong, S.; Lu, S. Nanoparticle-based drug delivery system: A patient-friendly chemotherapy for oncology. Dose-Response 2020, 18, 1559325820936161. [Google Scholar] [CrossRef] [PubMed]
- Ijaz, I.; Gilani, E.; Nazir, A.; Bukhari, A. Detail review on chemical, physical and green synthesis, classification, characterizations and applications of nanoparticles. Green Chem. Lett. Rev. 2020, 13, 223–245. [Google Scholar] [CrossRef]
- Hamida, R.S.; Ali, M.A.; Abdelmeguid, N.E.; Al-Zaban, M.I.; Baz, L.; Bin-Meferij, M.M. Lichens—A potential source for nanoparticles fabrication: A review on nanoparticles biosynthesis and their prospective applications. J. Fungi 2021, 7, 291. [Google Scholar] [CrossRef]
- Khanna, P.; Kaur, A.; Goyal, D. Algae-based metallic nanoparticles: Synthesis, characterization and applications. J. Microbiol. Methods 2019, 163, 105656. [Google Scholar] [CrossRef]
- Baker, S.; Harini, B.; Rakshith, D.; Satish, S. Marine microbes: Invisible nanofactories. J. Pharm. Res. 2013, 6, 383–388. [Google Scholar] [CrossRef]
- Vanlalveni, C.; Lallianrawna, S.; Biswas, A.; Selvaraj, M.; Changmai, B.; Rokhum, S.L. Green synthesis of silver nanoparticles using plant extracts and their antimicrobial activities: A review of recent literature. RSC Adv. 2021, 11, 2804–2837. [Google Scholar] [CrossRef]
- Hamida, R.S.; Ali, M.A.; Redhwan, A.; Bin-Meferij, M.M. Cyanobacteria—A promising platform in green nanotechnology: A review on nanoparticles fabrication and their prospective applications. Int. J. Nanomed. 2020, 15, 6033–6066. [Google Scholar] [CrossRef]
- Asmathunisha, N.; Kathiresan, K. A review on biosynthesis of nanoparticles by marine organisms. Colloids Surf. B Biointerfaces 2013, 103, 283–287. [Google Scholar] [CrossRef]
- Aboyewa, J.A.; Sibuyi, N.R.; Meyer, M.; Oguntibeju, O.O. Green synthesis of metallic nanoparticles using some selected medicinal plants from southern africa and their biological applications. Plants 2021, 10, 1929. [Google Scholar] [CrossRef]
- Kamran, U.; Bhatti, H.N.; Iqbal, M.; Nazir, A. Green synthesis of metal nanoparticles and their applications in different fields: A review. Zeitschrift für Physikalische Chemie 2019, 233, 1325–1349. [Google Scholar] [CrossRef]
- Patra, J.K.; Das, G.; Fraceto, L.F.; Campos, E.V.R.; del Pilar Rodriguez-Torres, M.; Acosta-Torres, L.S.; Diaz-Torres, L.A.; Grillo, R.; Swamy, M.K.; Sharma, S. Nano based drug delivery systems: Recent developments and future prospects. J. Nanobiotechnology 2018, 16, 71. [Google Scholar] [CrossRef] [PubMed]
- Hamida, R.S.; Albasher, G.; Bin-Meferij, M.M. Oxidative stress and apoptotic responses elicited by nostoc-synthesized silver nanoparticles against different cancer cell lines. Cancers 2020, 12, 2099. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Chen, Z.; Pan, Y.; Dai, R.; Wu, Y.; Zhuang, Z.; Wang, D.; Peng, Q.; Chen, C.; Li, Y. Functionalization of hollow nanomaterials for catalytic applications: Nanoreactor construction. Adv. Mater. 2019, 31, 1800426. [Google Scholar] [CrossRef]
- Malekzad, H.; Zangabad, P.S.; Mirshekari, H.; Karimi, M.; Hamblin, M.R. Noble metal nanoparticles in biosensors: Recent studies and applications. Nanotechnol. Rev. 2017, 6, 301–329. [Google Scholar] [CrossRef] [PubMed]
- Jacob, J.M.; Ravindran, R.; Narayanan, M.; Samuel, S.M.; Pugazhendhi, A.; Kumar, G. Microalgae: A prospective low cost green alternative for nanoparticle synthesis. Curr. Opin. Environ. Sci. Health 2021, 20, 100163. [Google Scholar] [CrossRef]
- Bulgariu, L.; Bulgariu, D. Bioremediation of toxic heavy metals using marine algae biomass. In Green Materials for Wastewater Treatment; Springer: Berlin, Germany, 2020; pp. 69–98. [Google Scholar]
- Bao, Z.; Lan, C.Q. Mechanism of light-dependent biosynthesis of silver nanoparticles mediated by cell extract of Neochloris oleoabundans. Colloids Surf. B Biointerfaces 2018, 170, 251–257. [Google Scholar] [CrossRef]
- Bin-Meferij, M.M.; Hamida, R.S. Biofabrication and antitumor activity of silver nanoparticles utilizing novel Nostoc sp. Bahar M. Int. J. Nanomed. 2019, 14, 9019–9029. [Google Scholar] [CrossRef]
- Hamida, R.S.; Abdelmeguid, N.E.; Ali, M.A.; Bin-Meferij, M.M.; Khalil, M.I. Synthesis of silver nanoparticles using a novel cyanobacteria Desertifilum sp. extract: Their antibacterial and cytotoxicity effects. Int. J. Nanomed. 2020, 15, 49–63. [Google Scholar] [CrossRef] [PubMed]
- Husain, S.; Sardar, M.; Fatma, T. Screening of cyanobacterial extracts for synthesis of silver nanoparticles. World J. Microbiol. Biotechnol. 2015, 31, 1279–1283. [Google Scholar] [CrossRef] [PubMed]
- Tschaikner, A.G.; Kofler, W. Coelastrella aeroterrestrica sp. nov. (Chlorophyta, Scenedesmoideae) a new, obviously often overlooked aeroterrestrial species. Algol. Stud. 2008, 128, 11–20. [Google Scholar] [CrossRef]
- Abed, A.; Derakhshan, M.; Karimi, M.; Shirazinia, M.; Mahjoubin-Tehran, M.; Homayonfal, M.; Hamblin, M.R.; Mirzaei, S.A.; Soleimanpour, H.; Dehghani, S. Platinum nanoparticles in biomedicine: Preparation, anti-cancer activity, and drug delivery vehicles. Front. Pharmacol. 2022, 13, 797804. [Google Scholar] [CrossRef]
- Xue, C.; Hu, S.; Gao, Z.-H.; Wang, L.; Luo, M.-X.; Yu, X.; Li, B.-F.; Shen, Z.; Wu, Z.-S. Programmably tiling rigidified DNA brick on gold nanoparticle as multi-functional shell for cancer-targeted delivery of siRNAs. Nat. Commun. 2021, 12, 2928. [Google Scholar] [CrossRef]
- Koushki, K.; Keshavarz Shahbaz, S.; Keshavarz, M.; Bezsonov, E.E.; Sathyapalan, T.; Sahebkar, A. Gold nanoparticles: Multifaceted roles in the management of autoimmune disorders. Biomolecules 2021, 11, 1289. [Google Scholar] [CrossRef]
- Lin, W.; Zhang, J.; Xu, J.-F.; Pi, J. The advancing of selenium nanoparticles against infectious diseases. Front. Pharmacol. 2021, 12, 1971. [Google Scholar] [CrossRef]
- Baby, E.K.; Reji, C. Metal-based nanoparticles for infectious diseases and therapeutics. In Nanotechnology for Infectious Diseases; Springer Nature: Berlin, Germany, 2022; pp. 103–124. [Google Scholar]
- Vines, J.B.; Yoon, J.-H.; Ryu, N.-E.; Lim, D.-J.; Park, H. Gold nanoparticles for photothermal cancer therapy. Front. Chem. 2019, 7, 167. [Google Scholar] [CrossRef]
- Nguyen, D.D.; Lue, S.J.; Lai, J.-Y. Tailoring therapeutic properties of silver nanoparticles for effective bacterial keratitis treatment. Colloids Surf. B Biointerfaces 2021, 205, 111856. [Google Scholar] [CrossRef]
- Haque, S.; Norbert, C.C.; Acharyya, R.; Mukherjee, S.; Kathirvel, M.; Patra, C.R. Biosynthesized silver nanoparticles for cancer therapy and in vivo bioimaging. Cancers 2021, 13, 6114. [Google Scholar] [CrossRef]
- Hamida, R.S.; Ali, M.A.; Goda, D.A.; Redhwan, A. Anticandidal potential of two cyanobacteria-synthesized silver nanoparticles: Effects on growth, cell morphology, and key virulence attributes of Candida albicans. Pharmaceutics 2021, 13, 1688. [Google Scholar] [CrossRef] [PubMed]
- Jia, D.; Sun, W. Silver nanoparticles offer a synergistic effect with fluconazole against fluconazole-resistant Candida albicans by abrogating drug efflux pumps and increasing endogenous ROS. Infect. Genet. Evol. 2021, 93, 104937. [Google Scholar] [CrossRef] [PubMed]
- Hamida, R.S.; Ali, M.A.; Goda, D.A.; Khalil, M.I.; Redhwan, A. Cytotoxic effect of green silver nanoparticles against ampicillin-resistant Klebsiella pneumoniae. RSC Adv. 2020, 10, 21136–21146. [Google Scholar] [CrossRef] [PubMed]
- Acharya, D.; Satapathy, S.; Yadav, K.K.; Somu, P.; Mishra, G. Systemic evaluation of mechanism of cytotoxicity in human colon cancer HCT-116 cells of silver nanoparticles synthesized using marine algae Ulva lactuca extract. J. Inorg. Organomet. Polym. Mater. 2022, 32, 596–605. [Google Scholar] [CrossRef]
- El-Naggar, N.E.-A.; Hussein, M.H.; El-Sawah, A.A. Phycobiliprotein-mediated synthesis of biogenic silver nanoparticles, characterization, in vitro and in vivo assessment of anticancer activities. Sci. Rep. 2018, 8, 8925. [Google Scholar] [CrossRef]
- Ferdous, Z.; Nemmar, A. Health impact of silver nanoparticles: A review of the biodistribution and toxicity following various routes of exposure. Int. J. Mol. Sci. 2020, 21, 2375. [Google Scholar] [CrossRef]
- Kodiha, M.; Wang, Y.M.; Hutter, E.; Maysinger, D.; Stochaj, U. Off to the organelles-killing cancer cells with targeted gold nanoparticles. Theranostics 2015, 5, 357–370. [Google Scholar] [CrossRef]
- Bolch, C.J.; Orr, P.T.; Jones, G.J.; Blackburn, S.I. Genetic, morphological, and toxicological variation among globally distributed strains of Nodularia (Cyanobacteria). J. Phycol. 1999, 35, 339–355. [Google Scholar] [CrossRef]
- Singh, S.P.; Rastogi, R.P.; Häder, D.-P.; Sinha, R.P. An improved method for genomic DNA extraction from cyanobacteria. World J. Microbiol. Biotechnol. 2011, 27, 1225–1230. [Google Scholar] [CrossRef]
- Abd El-Kareem, M.S.; Rabbih, M.A.E.F.; Selim, E.T.M.; Elsherbiny, E.A.E.-m.; El-Khateeb, A.Y. Application of GC/EIMS in combination with semi-empirical calculations for identification and investigation of some volatile components in basil essential oil. Int. J. Anal. Mass Spectrom. Chromatogr. 2016, 4, 14–25. [Google Scholar] [CrossRef]
- Hanna, A.L.; Hamouda, H.M.; Goda, H.A.; Sadik, M.W.; Moghanm, F.S.; Ghoneim, A.M.; Alenezi, M.A.; Alnomasy, S.F.; Alam, P.; Elsayed, T.R. Biosynthesis and characterization of silver nanoparticles produced by phormidium ambiguum and desertifilum tharense cyanobacteria. Bioinorg. Chem. Appl. 2022, 2022, 9072508. [Google Scholar] [CrossRef] [PubMed]
- Hamida, R.S.; Ali, M.A.; Goda, D.A.; Khalil, M.I.; Al-Zaban, M.I. Novel biogenic silver nanoparticle-induced reactive oxygen species inhibit the biofilm formation and virulence activities of methicillin-resistant staphylococcus aureus (MRSA) strain. Front. Bioeng. Biotechnol. 2020, 8, 433. [Google Scholar] [CrossRef] [PubMed]
- Elshikh, M.; Ahmed, S.; Funston, S.; Dunlop, P.; McGaw, M.; Marchant, R.; Banat, I.M. Resazurin-based 96-well plate microdilution method for the determination of minimum inhibitory concentration of biosurfactants. Biotechnol. Lett. 2016, 38, 1015–1019. [Google Scholar] [CrossRef] [PubMed]
- Ragunathan, V.; Pandurangan, J.; Ramakrishnan, T. Gas chromatography-mass spectrometry analysis of methanol extracts from marine red seaweed Gracilaria corticata. Pharmacogn. J. 2019, 11, 547–554. [Google Scholar] [CrossRef]
- González, A.; Noguez, C.; Beránek, J.; Barnard, A. Size, shape, stability, and color of plasmonic silver nanoparticles. J. Phys. Chem. C 2014, 118, 9128–9136. [Google Scholar]
- Rivero, P.J.; Goicoechea, J.; Urrutia, A.; Arregui, F.J. Effect of both protective and reducing agents in the synthesis of multicolor silver nanoparticles. Nanoscale Res. Lett. 2013, 8, 101. [Google Scholar] [CrossRef]
- Mock, J.; Barbic, M.; Smith, D.; Schultz, D.; Schultz, S. Shape effects in plasmon resonance of individual colloidal silver nanoparticles. J. Chem. Phys. 2002, 116, 6755–6759. [Google Scholar] [CrossRef]
- Kashyap, M.; Samadhiya, K.; Ghosh, A.; Anand, V.; Shirage, P.M.; Bala, K. Screening of microalgae for biosynthesis and optimization of Ag/AgCl nano hybrids having antibacterial effect. RSC Adv. 2019, 9, 25583–25591. [Google Scholar] [CrossRef]
- Soleimani, M.; Habibi-Pirkoohi, M. Biosynthesis of silver nanoparticles using Chlorella vulgaris and evaluation of the antibacterial efficacy against Staphylococcus aureus. Avicenna J. Med. Biotechnol. 2017, 9, 120–125. [Google Scholar]
- Mora-Godínez, S.; Abril-Martínez, F.; Pacheco, A. Green synthesis of silver nanoparticles using microalgae acclimated to high CO2. Mater. Today Proc. 2020, 48, 5–9. [Google Scholar] [CrossRef]
- Öztürk, B.Y. Intracellular and extracellular green synthesis of silver nanoparticles using Desmodesmus sp.: Their antibacterial and antifungal effects. Caryologia 2019, 72, 29–43. [Google Scholar]
- Jeon, M.S.; Han, S.-I.; Park, Y.H.; Kim, H.S.; Choi, Y.-E. Rapid green synthesis of silver nanoparticles using sulfated polysaccharides originating from Porphyridium cruentum UTEX 161: Evaluation of antibacterial and catalytic activities. J. Appl. Phycol. 2021, 33, 3091–3101. [Google Scholar] [CrossRef]
- Karthik, L.; Kumar, G.; Kirthi, A.V.; Rahuman, A.; Bhaskara Rao, K. Streptomyces sp. LK3 mediated synthesis of silver nanoparticles and its biomedical application. Bioprocess Biosyst. Eng. 2014, 37, 261–267. [Google Scholar] [CrossRef] [PubMed]
- Roy, K.; Sarkar, C.; Ghosh, C. Plant-mediated synthesis of silver nanoparticles using parsley (Petroselinum crispum) leaf extract: Spectral analysis of the particles and antibacterial study. Appl. Nanosci. 2015, 5, 945–951. [Google Scholar] [CrossRef]
- Ssekatawa, K.; Byarugaba, D.K.; Kato, C.D.; Wampande, E.M.; Ejobi, F.; Nakavuma, J.L.; Maaza, M.; Sackey, J.; Nxumalo, E.; Kirabira, J.B. Green strategy-based synthesis of silver nanoparticles for antibacterial applications. Front. Nanotechnol. 2021, 3, 59. [Google Scholar] [CrossRef]
- Rajeshkumar, S. Synthesis of silver nanoparticles using fresh bark of Pongamia pinnata and characterization of its antibacterial activity against gram positive and gram negative pathogens. Resour.-Effic. Technol. 2016, 2, 30–35. [Google Scholar] [CrossRef]
- Arokiyaraj, S.; Arasu, M.V.; Vincent, S.; Prakash, N.U.; Choi, S.H.; Oh, Y.-K.; Choi, K.C.; Kim, K.H. Rapid green synthesis of silver nanoparticles from Chrysanthemum indicum L and its antibacterial and cytotoxic effects: An in vitro study. Int. J. Nanomed. 2014, 9, 379–388. [Google Scholar] [CrossRef]
- Meng, Y. A sustainable approach to fabricating Ag nanoparticles/PVA hybrid nanofiber and its catalytic activity. Nanomaterials 2015, 5, 1124–1135. [Google Scholar] [CrossRef]
- Kemper, F.; Beckert, E.; Eberhardt, R.; Tünnermann, A. Light filter tailoring—The impact of light emitting diode irradiation on the morphology and optical properties of silver nanoparticles within polyethylenimine thin films. RSC Adv. 2017, 7, 41603–41609. [Google Scholar] [CrossRef]
- Lengke, M.F.; Fleet, M.E.; Southam, G. Biosynthesis of silver nanoparticles by filamentous cyanobacteria from a silver (I) nitrate complex. Langmuir 2007, 23, 2694–2699. [Google Scholar] [CrossRef]
- Husain, S.; Verma, S.K.; Azam, M.; Sardar, M.; Haq, Q.; Fatma, T. Antibacterial efficacy of facile cyanobacterial silver nanoparticles inferred by antioxidant mechanism. Mater. Sci. Eng. C 2021, 122, 111888. [Google Scholar] [CrossRef] [PubMed]
- Singh, H.; Du, J.; Yi, T.-H. Kinneretia THG-SQI4 mediated biosynthesis of silver nanoparticles and its antimicrobial efficacy. Artif. Cells Nanomed. Biotechnol. 2017, 45, 602–608. [Google Scholar] [CrossRef] [PubMed]
- Cepoi, L.; Rudi, L.; Zinicovscaia, I.; Chiriac, T.; Miscu, V.; Rudic, V. Biochemical changes in microalga Porphyridium cruentum associated with silver nanoparticles biosynthesis. Arch. Microbiol. 2021, 203, 1547–1554. [Google Scholar] [CrossRef] [PubMed]
- Honary, S.; Zahir, F. Effect of zeta potential on the properties of nano-drug delivery systems-a review (Part 1). Trop. J. Pharm. Res. 2013, 12, 255–264. [Google Scholar]
- Honary, S.; Zahir, F. Effect of zeta potential on the properties of nano-drug delivery systems-a review (Part 2). Trop. J. Pharm. Res. 2013, 12, 265–273. [Google Scholar]
- Pearson, R.M.; Juettner, V.V.; Hong, S. Biomolecular corona on nanoparticles: A survey of recent literature and its implications in targeted drug delivery. Front. Chem. 2014, 2, 108. [Google Scholar] [CrossRef]
- Nel, A.E.; Mädler, L.; Velegol, D.; Xia, T.; Hoek, E.; Somasundaran, P.; Klaessig, F.; Castranova, V.; Thompson, M. Understanding biophysicochemical interactions at the nano-bio interface. Nat. Mater. 2009, 8, 543–557. [Google Scholar] [CrossRef]
- Farheen, S.; Oanz, A.M.; Khan, N.; Umar, M.S.; Jamal, F.; Altaf, I.; Kashif, M.; Alshameri, A.W.; Somavarapu, S.; Wani, I. Fabrication of microbicidal silver nanoparticles: Green synthesis and implications in the containment of bacterial biofilm on orthodontal appliances. Front. Nanotechnol. 2022, 4, 780783. [Google Scholar] [CrossRef]
- Bélteky, P.; Rónavári, A.; Zakupszky, D.; Boka, E.; Igaz, N.; Szerencsés, B.; Pfeiffer, I.; Vágvölgyi, C.; Kiricsi, M.; Kónya, Z. Are smaller nanoparticles always better? Understanding the biological effect of size-dependent silver nanoparticle aggregation under biorelevant conditions. Int. J. Nanomed. 2021, 16, 3021–3040. [Google Scholar] [CrossRef]
- Jeong, Y.; Lim, D.W.; Choi, J. Assessment of size-dependent antimicrobial and cytotoxic properties of silver nanoparticles. Adv. Mater. Sci. Eng. 2014, 2014, 763807. [Google Scholar] [CrossRef]
- Yang, B.; Shi, J. Chemistry of advanced nanomedicines in cancer cell metabolism regulation. Adv. Sci. 2020, 7, 2001388. [Google Scholar] [CrossRef] [PubMed]
- Durán, N.; Durán, M.; De Jesus, M.B.; Seabra, A.B.; Fávaro, W.J.; Nakazato, G. Silver nanoparticles: A new view on mechanistic aspects on antimicrobial activity. Nanomed. Nanotechnol. Biol. Med. 2016, 12, 789–799. [Google Scholar] [CrossRef] [PubMed]
- Abbaszadegan, A.; Ghahramani, Y.; Gholami, A.; Hemmateenejad, B.; Dorostkar, S.; Nabavizadeh, M.; Sharghi, H. The effect of charge at the surface of silver nanoparticles on antimicrobial activity against gram-positive and gram-negative bacteria: A preliminary study. J. Nanomater. 2015, 2015, 53. [Google Scholar] [CrossRef]
- Cheon, J.Y.; Kim, S.J.; Rhee, Y.H.; Kwon, O.H.; Park, W.H. Shape-dependent antimicrobial activities of silver nanoparticles. Int. J. Nanomed. 2019, 14, 2773–2780. [Google Scholar] [CrossRef]
- Rajamanickam, K.; Sudha, S.; Francis, M.; Sowmya, T.; Rengaramanujam, J.; Sivalingam, P.; Prabakar, K. Microalgae associated Brevundimonas sp. MSK 4 as the nano particle synthesizing unit to produce antimicrobial silver nanoparticles. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2013, 113, 10–14. [Google Scholar] [CrossRef]




| No. | Biomolecule Name | Retention Time | Area % | Mentioned Factor | Molecular Formula | Molecular Weight | Structure |
|---|---|---|---|---|---|---|---|
| 1 | 1-Heptanol | 4.10 | 0.31 | 738 | C7H16O | 117 | 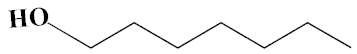 |
| 2 | 2-Ethyl-1-hexanethiol | 4.14 | 0.25 | 648 | C8H18S | 146 | 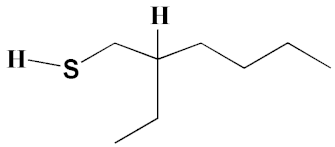 |
| 3 | 1-Octene | 4.35 | 4.58 | 637 | C8H16 | 112 |  |
| 4 | 1-Deoxy-d-mannitol | 5.05 | 0.46 | 635 | C6H14O5 | 166 |  |
| 5 | 1-Chlorotetradecane | 11.65, 14.01 | 0.51, 0.40 | 583, 614 | C14H29Cl | 232 |  |
| 6 | Methyl 10-methylundecanoate | 14.71 | 3.76 | 816 | C13H26O2 | 214 | 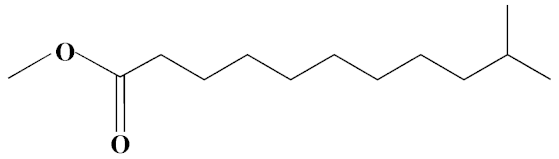 |
| 7 | 1-Nonadecene | 17.95 | 0.52 | 704 | C19H38 | 266 |  |
| 8 | 2-Methylhexadecan-1-ol | 18.46 | 0.50 | 684 | C17H36O | 256 |  |
| 9 | Arachidic acid methyl ester | 19.11 | 3.68 | 906 | C21H42O2 | 326 |  |
| 10 | Oxiraneundecanoic acid, 3-pentyl-, methyl ester, cis- | 20.42 | 0.33 | 651 | C19H36O3 | 312 | 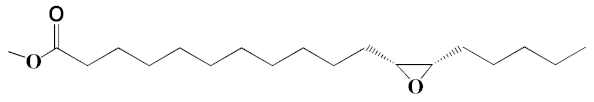 |
| 11 | Methyl 12-methyltetradecanoate | 21.17, 24.48 | 1.10, 1.43 | 685, 671 | C16H32O2 | 256 | 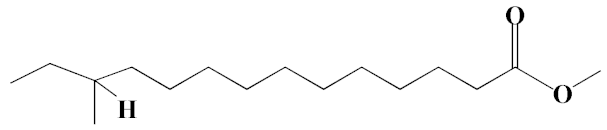 |
| 12 | Caffeine (3,7-Dihydro-1,3,7-trimethyl-1H-purine-2,6-dione) | 22.19, 22.69 | 6.12, 0.97 | 891, 830 | C8H10N4O2 | 194 | 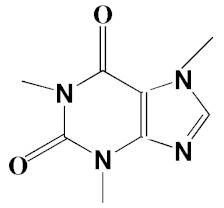 |
| 13 | Methyl palmitate (Hexadecanoic acid, methyl ester) | 23.23 | 34.13 | 930 | C17H34O2 | 270 |  |
| 14 | Methyl juniperate (Methyl 16-hydroxy-hexadecanoate) | 24.33 | 0.64 | 689 | C17H34O3 | 286 | 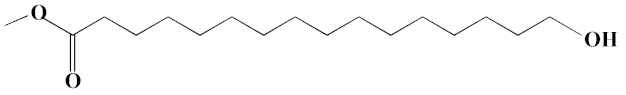 |
| 15 | Cyclopentanetridecanoic acid, methyl ester | 25.02 | 1.27 | 753 | C19H36O2 | 296 |  |
| 16 | E,E-11,14-Octadecadienoic acid, methyl ester | 26.23 | 0.43 | 779 | C19H34O2 | 294 |  |
| 17 | Oleic acid, methyl ester | 26.38, 26.49 | 14.18, 4.61 | 902, 871 | C19H36O2 | 296 | 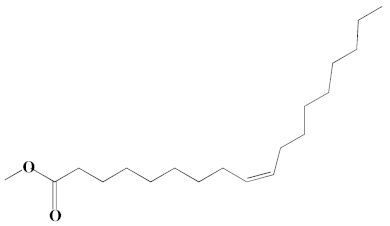 |
| 18 | Methyl stearate | 26.89 | 18.50 | 900 | C19H38O2 | 298 |  |
| 19 | 2E,15Z-14-Methyl-2,15-octadecadien-1-ol | 27.99 | 0.66 | 658 | C19H36O | 280 | 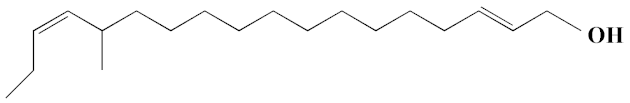 |
| 20 | Dotriacontane | 38.43 | 0.65 | 674 | C32H66 | 450 |  |
| Element | Line | Mass% | Atom% |
|---|---|---|---|
| C | K | 8.01 ± 0.02 | 37.05 ± 0.08 |
| O | K | 1.49 ± 0.02 | 5.17 ± 0.07 |
| Al | K | 0.21 ± 0.01 | 0.44 ± 0.02 |
| P | K | 0.40 ± 0.01 | 0.72 ± 0.02 |
| Cl | K | 9.79 ± 0.02 | 15.34 ± 0.04 |
| Ag | L | 80.10 ± 0.1 | 41.27 ± 0.05 |
| Total | 100.00 | 100.00 |
| Cells | Drugs (µg/mL) | |||||||
|---|---|---|---|---|---|---|---|---|
| C@Ag-NPs | C. aeroterrestrica Aqueous Extract | Ch@Ag-NPs | 5-FU | |||||
| IC25 | IC50 | IC25 | IC50 | IC25 | IC50 | IC25 | IC50 | |
| MCF-7 | 13.015 | 26.03 | 159.55 | 319.10 | 15.59 | 31.18 | 28.24 | 56.48 |
| MDA | 7.96 | 15.92 | 132.15 | 264.3 | 128.45 | 256.9 | 22.13 | 44.26 |
| HCT-116 | 5.04 | 10.08 | 125.75 | 251.5 | 156.25 | 312.50 | 16.07 | 32.14 |
| HepG2 | 2.645 | 5.29 | 31.435 | 62.87 | 13.955 | 27.91 | 42.89 | 85.78 |
| HFS | 5.485 | 10.97 | 46.63 | 93.26 | 27.03 | 54.06 | 16.205 | 32.41 |
| Vero | 8.56 | 17.12 | 195.00 | 390.00 | 9.255 | 18.51 | 16.555 | 33.11 |
| Concentrations (µg/mL) | Ascorbic Acid | C@Ag-NPs | C. aeroterrestrica Extract |
|---|---|---|---|
| 1000 | 82.7 ± 0.8 | 54.2 ± 1.0 | 38.2 ± 0.2 |
| 500 | 72.2 ± 0.7 | 40.0 ± 2.5 | 26.7 ± 0.9 |
| 250 | 53.3 ± 1.6 | 33.4 ± 1.4 | 20.7 ± 1.4 |
| 125 | 30.6 ± 4.2 | 25.4 ± 1.8 | 19.2 ± 0.9 |
| 62.5 | 31.9 ± 3.5 | 23.9 ± 0.4 | 16.7 ± 0.4 |
| 31.25 | 10.1 ± 0.7 | 22.8 ± 1.2 | 15.4 ± 0.2 |
| 15.6 | 8.4 ± 1.2 | 22.6 ± 1.3 | 15.4 ± 0.1 |
| 7.8 | 5.4 ± 0.9 | 22.9 ± 1.8 | 15.2 ± 0.2 |
| 3.9 | 2.9 ± 0.7 | 22.5 ± 0.9 | 12. 6 ± 0.9 |
| 1.95 | 2.5 ± 0.8 | 21.7 ± 3.1 | 12.7 ± 3.1 |
| Microbes | Treatments | |||||
|---|---|---|---|---|---|---|
| C. aeroterrestrica Extract (µg/mL) | C@Ag-NPs (µg/mL) | |||||
| MIC | MBC | MIC/MBC | MIC | MBC | MIC/MBC | |
| Staphylococcus aureus | >500 | >500 | 1.0 | <0.98 | <0.98 | 1.0 |
| Escherichia coli | >500 | >500 | 1.0 | 0.98 | 1.95 | 0.5 |
| Pseudomonas aeruginosa | >500 | >500 | 1.0 | 0.98 | 1.95 | 0.5 |
| Streptococcus pyogenes | >500 | >500 | 1.0 | 0.98 | 1.95 | 0.5 |
| Bacillus subtilis | >500 | >500 | 1.0 | 1.95 | 3.9 | 0.5 |
| Microbes | Inhibition Zone Diameter (mm) | ||||
|---|---|---|---|---|---|
| C@Ag-NPs | C. aeroterrestrica Extract | AgNO3 | Ch@Ag-NPs | Ciprofloxacin | |
| Staphylococcus aureus | 19.3 ± 0.15 | 0.0 ± 0.0 | 15.0 ± 0.33 | 13.0 ± 0.06 | 30.0 ± 0.03 |
| Escherichia coli | 15.3 ± 0.08 | 0.0 ± 0.0 | 14.1 ± 0.11 | 12.0 ± 0.0 | 19.03 ± 0.03 |
| Pseudomonas aeruginosa | 15.0 ± 0.04 | 0.0 ± 0.0 | 14.0 ± 0.04 | 10.1 ± 0.06 | 29.0 ± 0.03 |
| Streptococcus pyogenes | 15.3 ± 0.05 | 0.0 ± 0.0 | 14.03 ± 0.51 | 11.2 ± 0.11 | 23.1 ± 0.05 |
| Bacillus subtilis | 14.27 ± 0.15 | 0.0 ± 0.0 | 11.17 ± 0.44 | 10.07 ± 0.06 | 19.07 ± 0.06 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hamida, R.S.; Ali, M.A.; Almohawes, Z.N.; Alahdal, H.; Momenah, M.A.; Bin-Meferij, M.M. Green Synthesis of Hexagonal Silver Nanoparticles Using a Novel Microalgae Coelastrella aeroterrestrica Strain BA_Chlo4 and Resulting Anticancer, Antibacterial, and Antioxidant Activities. Pharmaceutics 2022, 14, 2002. https://doi.org/10.3390/pharmaceutics14102002
Hamida RS, Ali MA, Almohawes ZN, Alahdal H, Momenah MA, Bin-Meferij MM. Green Synthesis of Hexagonal Silver Nanoparticles Using a Novel Microalgae Coelastrella aeroterrestrica Strain BA_Chlo4 and Resulting Anticancer, Antibacterial, and Antioxidant Activities. Pharmaceutics. 2022; 14(10):2002. https://doi.org/10.3390/pharmaceutics14102002
Chicago/Turabian StyleHamida, Reham Samir, Mohamed Abdelaal Ali, Zakiah Nasser Almohawes, Hadil Alahdal, Maha Abdullah Momenah, and Mashael Mohammed Bin-Meferij. 2022. "Green Synthesis of Hexagonal Silver Nanoparticles Using a Novel Microalgae Coelastrella aeroterrestrica Strain BA_Chlo4 and Resulting Anticancer, Antibacterial, and Antioxidant Activities" Pharmaceutics 14, no. 10: 2002. https://doi.org/10.3390/pharmaceutics14102002
APA StyleHamida, R. S., Ali, M. A., Almohawes, Z. N., Alahdal, H., Momenah, M. A., & Bin-Meferij, M. M. (2022). Green Synthesis of Hexagonal Silver Nanoparticles Using a Novel Microalgae Coelastrella aeroterrestrica Strain BA_Chlo4 and Resulting Anticancer, Antibacterial, and Antioxidant Activities. Pharmaceutics, 14(10), 2002. https://doi.org/10.3390/pharmaceutics14102002







