Abstract
The blood–brain barrier is a major obstacle in treating brain-related disorders, as it does not allow the delivery of drugs into the brain. We developed a method for predicting blood–brain barrier penetrating peptides to facilitate drug delivery into the brain. These blood–brain barrier penetrating peptides (B3PPs) can act as therapeutics, as well as drug delivery agents. We trained, tested, and evaluated our models on blood–brain barrier peptides obtained from the B3Pdb database. First, we computed a wide range of peptide features. Then, we selected relevant peptide features. Finally, we developed numerous machine-learning-based models for predicting blood–brain barrier peptides using the selected features. The random-forest-based model performed the best with respect to the top 80 selected features and achieved a maximal 85.08% accuracy with an AUROC of 0.93. We also developed a webserver, B3pred, that implements our best models. It has three major modules that allow users to predict/design B3PPs and scan B3PPs in a protein sequence.
1. Introduction
The blood–brain barrier (BBB) is the primary barrier between the brain’s interstitial fluid and the blood. It is the connection between the central nervous system (CNS) and the peripheral nervous system (PNS) [1,2,3,4]. The neurovascular unit (NVU) is the structural and functional unit of the BBB, formed by neurons, macrophages, endothelial cells, astrocytes, and pericytes [5] (Figure 1). The NVU regulates the biochemical environment between the blood and the brain, which is essential for neural function. The endothelial cells of the NVU allow the entry or exit of molecules, such as glucose, amino acids, and proteins/peptides, into or from the CNS [6,7,8]. In the last few decades, researchers have made many attempts to develop drug delivery systems that can deliver drugs into the brain. Despite advances made by the scientific community in developing drug delivery systems, it is still challenging to penetrate the BBB [9].
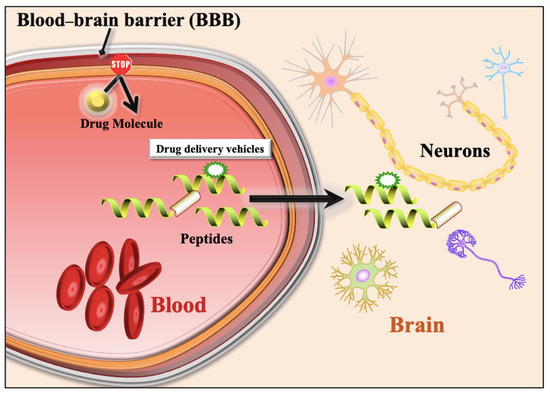
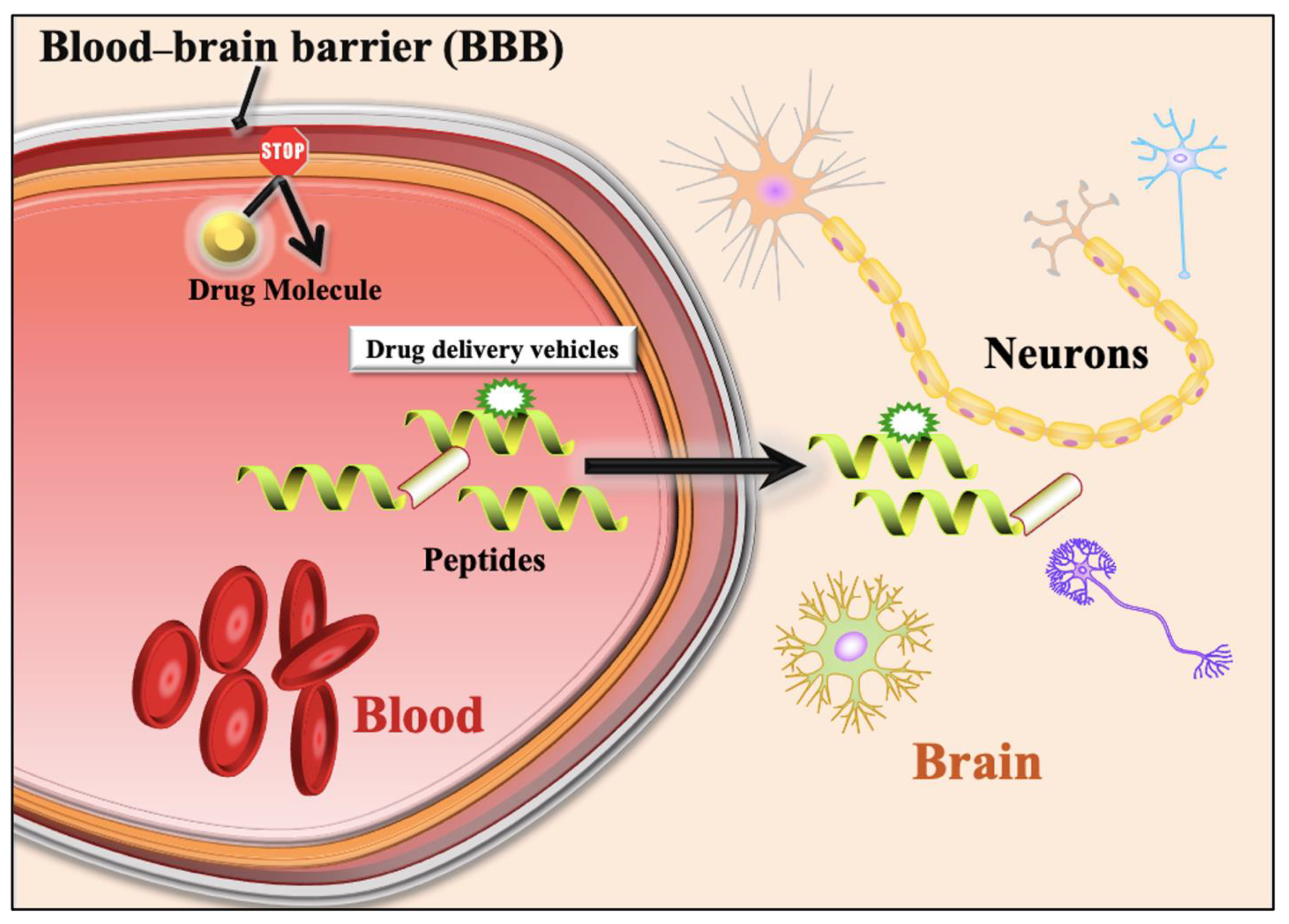
Figure 1.
A schematic diagram shows inhibition of drug molecules entry from blood to brain due to Blood–brain barrier. It also shows entry of drug molecules from blood to brain with the support of Blood–brain barrier penetrating peptides.
In the past, researchers have attempted to develop peptide/protein-based drug delivery vehicles. In this approach, a major challenge is to identify peptides that can penetrate the BBB [10]. In addition, researchers are exploring peptide-based therapeutics to treat CNS-associated diseases, including neurodegenerative disorders such as Parkinson’s disease, Alzheimer’s disease [11,12], and glioblastoma [13]. This means that peptides can be used as therapeutic agents as well as drug delivery vehicles. In recent studies, numerous peptides, such as shuttle peptides [14], self-assembled peptides [15], and peptide-decorated nanoparticles [16], have been used for efficient drug delivery into the brain. Some neuropeptides are utilized as potential therapeutic targets against many neurological diseases, such as epilepsy [17,18], depression [19,20], and neuroimmune disorders [21]. Due to the low toxicity of these peptides, they may act as potential peptide-based drug candidates against neurological diseases. The major limitation of these peptide-based drugs is low bioavailability, short half-life [22], and weak penetration of the BBB [23]. For example, tumor homing peptides (THPs) [24] and cell-penetrating peptides (CPPs) [25] can be used as drug delivery vehicles [26,27]. The tumor homing peptides need a carrier to cross the BBB, while selected CPPs can directly pass through the BBB [28].
The cell-penetrating peptides are short peptides which act as molecular delivery vehicles, and are able to deliver various therapeutic molecules inside a cell [29,30]. There are CPPs that can even cross the blood–brain barrier, which are called blood–brain barrier penetrating peptides (B3PPs). These B3PPs can be used to deliver several cargo molecules (e.g., peptides/proteins, siRNA, plasmid DNA) into the brain [31,32,33,34]. Mainly, these peptides are obtained from naturally occurring proteins/peptides such as signal peptides, RNA/DNA-binding proteins, viral proteins, and antimicrobial peptides [35]. Several studies have shown that B3PPs may be synthesized chemically or designed with rDNA technology [36,37,38] to enhance the stability and half-life of the B3PPs [39]. In the past, several methods have been developed for predicting cell-penetrating peptides, such as cellPPD, SkipCPP-Pred, CPPred-RF, KELM-CPPpred, CellPPDMod, and CPPred-FL [40,41,42,43,44,45]. In addition, various methods have been developed for predicting chemical-based drug delivery vehicles to cross the blood–brain barrier [46,47,48]. In contrast, a limited attempt has been made to develop methods to predict B3PPs. Recently, Dai et al. developed an in silico method, BBPpred, to identify B3PPs [49].
In this study, we have developed a computational tool named “B3Pred” for predicting B3PPs with high reliability and precision. This method has the ability to classify BBPs vs. non-BBPs and CPPs vs. BBPs; it uses a large dataset for training and validation. We used three datasets, i.e., Dataset_1 (269 B3PPs and 269 CPPs), Dataset_2 (269 B3PPs, and 269 non-B3PPs), and Dataset_3 (269 B3PPs and 2690 non-B3PPs), for training and validation. We have used more than 9000 descriptors/features for the generation of the prediction models using several machine learning techniques, such as RF, DT, LR, XGB, SVM, and GBM.
2. Materials and Methods
2.1. Dataset Collection
In this study, we collected 465 blood–brain barrier penetrating peptides (B3PPs) from the B3Pdb database (https://webs.iiitd.edu.in/raghava/b3pdb/, accessed on 22 July 2020) [50]. We considered B3PPs having a length between 6 and 30 amino acid (AA) residues, inclusive. For the positive dataset, we collected 269 unique B3PPs. The major challenge of this type of study is to generate an authenticated negative dataset. We used three negative datasets in this study. Firstly, we collected unique 269 cell-penetrating peptides (CPPs) [51], other than B3PPs, and called them non-B3PPs or negative Dataset_1. In negative Dataset_2, we randomly generated 269 non-B3PPs from the Swiss-Prot database [52]. Our third negative dataset is ten times larger than the positive dataset, i.e., 2690 unique non-B3PPs randomly generated using the Swiss-Prot database. Finally, we combined the three datasets, i.e., Dataset_1 (269 B3PPs and 269 CPPs), Dataset_2 (269 B3PPs, and 269 non-B3PPs), and Dataset_3 (269 B3PPs and 2690 non-B3PPs).
2.2. Amino Acid Composition
Amino acid composition (AAC) analysis of peptides helped us to find out whether there were any amino acid compositional similarities/differences in different types of peptides. We compared the amino acid composition of B3PPs, CPPs, and randomly generated peptides. The following equation is used to calculate AAC:
where AACi and AARi are the percentage composition and number of residues of type i in a peptide, respectively. TNR is the total number of residues in a peptide [53].
2.3. Two Sample Logo
The Two Sample Logo (TSL) tool was used to identify the amino acid preference at a specific position in the peptide sequences [54]. This tool needed an input amino acid sequence vector of fixed length, since the minimum size of peptides in all datasets was five residues; hence, we selected five residues from the N-terminal, and five amino acids from the C-terminal, of the peptide sequences. To create a fixed input vector, the N-terminus side residues and C-terminus residues were grouped together to generate a sequence of 10 amino acid residues. We used the 10-residue sequences generated from our dataset peptides to develop TSLs. To build these Two Sample Logos, we used all B3PPs and all non-B3PPs from the three different negative datasets.
2.4. Generation of Peptide Features
In order to calculate a wide range of features from the protein or peptide sequences, we used the Pfeature package [55]. Pfeature is used to generate thousands of features/descriptors. We computed the composition-based module of Pfeature to calculate >9000 descriptors of peptide sequences for positive and negative datasets. This module calculated fifteen types of features (AAC, DPC, RRI, DDOR, SE, SER, SEP, CTD, CeTD, PAAC, APAAC, QSO, TPC, ABC, and SOCN). The input vector of 9189 descriptors was used further for feature selection and machine learning purposes (Supplementary Table S1).
2.5. Feature Selection
This study used the SVC-L1 feature selection technique to extract an essential set of features from all the datasets. We chose the SVC-L1 method because it is much faster than other feature selection methods [56]. This method applies the L1 penalty to select a relevant set of features, after selecting the non-zero coefficients. SVC-L1 mainly considers regularization and the loss function. During the optimization process, the L1 regularization generates a sparse matrix by choosing some model features. The other important parameter used in this technique is the “C” parameter; its value is directly proportional to the selected features. The smaller the value of “C”, the fewer the number of features determined by the method. We chose the default value (i.e., 0.01) of the “C” parameter [57]. Using SVC-L1, 73 important features were identified from the 9189 features for Dataset_1 (B3PPs and CPPs peptides) and Dataset_2 (B3PPs and balanced non-B3PPs). Similarly, 145 features were selected for Dataset_3 (i.e., B3PPs and random non-B3PPs).
2.6. Feature Ranking
After selecting an important set of features, we ranked the features based on their importance in classification. The Feature-selector method is based on a decision-tree-like algorithm and uses the Light Gradient Boosting Machine (LightGBM) method [58]. It computes the rank of each feature based on the feature that is used to split the dataset across all the trees. Further, the top-most ranked features for each dataset were used in different machine learning techniques for the classification of B3PPs and non-B3PPs.
2.7. Machine Learning Techniques
We used several machine learning algorithms to classify B3PPs and non-B3PPs. In this study, we implemented decision tree (DT), random forest (RF), Logistic Regression (LR), k-nearest neighbors (KNN), Gaussian Naive Bayes (GNB), XGBoost (XGB), and Support Vector Classifier (SVC) machine learning classifiers. The different classification methods were implemented with the help of a python-based library known as Scikit-learn [59]. DT algorithms work based on non-parametric supervised learning models. The major aim of the classifier is to identify the output instance by learning various decision rules, provided in the form of input data [60]. The GNB method is a probabilistic classifier and builds on Bayes’ theorem. It is based on the assumption that the consecutive variable of every group follows the Gaussian (or normal) distribution [61]. Random forest is an ensemble-based classifier, which predicts a single tree as a response variable by training the number of decision trees. It also controls the overfitting of the models [62]. The LR technique is used to train the logistic/logit model, which gives the likelihood of an event happening. It applies a logistic function to predict the response variable or occurrence of a class [63]. The KNN method is an instance-based classifier. It usually collects the instances of the training dataset. Its prediction is based on the maximum number of votes given to a particular class which is closest to the nearest neighbor data point [64]. The XGB classifier uses the scalable tree boosting algorithm, in which an iterative approach is used for the prediction of the final output [65]. The SVC is developed on the library of support vector machines. It usually fits the data points provided as input features and provides the most suitable fit of a hyperplane that categorizes the data into two classes [66].
2.8. Cross-Validation Techniques
We used internal and external validation techniques to assess the performance of our classification models. In the past, several methods used 80:20 splitting of the complete dataset for training and validation [67,68]. In the current study, we implemented a similar strategy to evaluate our classification models. For each dataset, 80% of the data were used for training, and the remaining 20% were used for external validation. We applied 5-fold cross-validation techniques on the training dataset; this is called internal validation. In internal validation, training data are equally divided into five sets/folds in which four folds were used for training, and the fifth fold is used for testing the model (Supplementary Table S2–S4). This process is repeated five times so that each set is used once for testing. The final performance is computed by taking the average of the performance on the five sets. In the case of external validation, the performance of the best model on the training dataset was evaluated on a validation, or independent, dataset.
2.9. Performance Evaluation Parameters
We used standard evaluation parameters to compute the performance of the classification models. Threshold-dependent and -independent parameters were used in this study. The performance of the models was calculated using threshold-dependent parameters, such as sensitivity (Sens), accuracy (Acc), and specificity (Spec). Area Under the Receiver Operating Characteristic (AUROC) curve, a threshold-independent parameter, was used to measure the models’ performance. AUROC generates a curve by plotting sensitivity against (1-specificity) on various thresholds. Threshold-dependent parameters were computed using the given equations:
where TP, FP, TN, and FN are true positive, false positive, true negative, and false negative predictions, respectively.
2.10. Webserver Implementation
We developed a webserver named “B3Pred” (https://webs.iiitd.edu.in/raghava/b3pred/, accessed on 22 February 2021) to identify blood–brain barrier penetrating peptides and non-B3PPs. We used HTML5, JAVA, CSS3, and PHP scripts to develop the front-end and back-end of the webserver. The B3Pred server is compatible with all the latest devices, such as mobiles, tablets, iMacs, and desktop computers. It mainly incorporates the predict, design, and protein scan modules.
3. Results
3.1. Amino Acid Composition Analysis
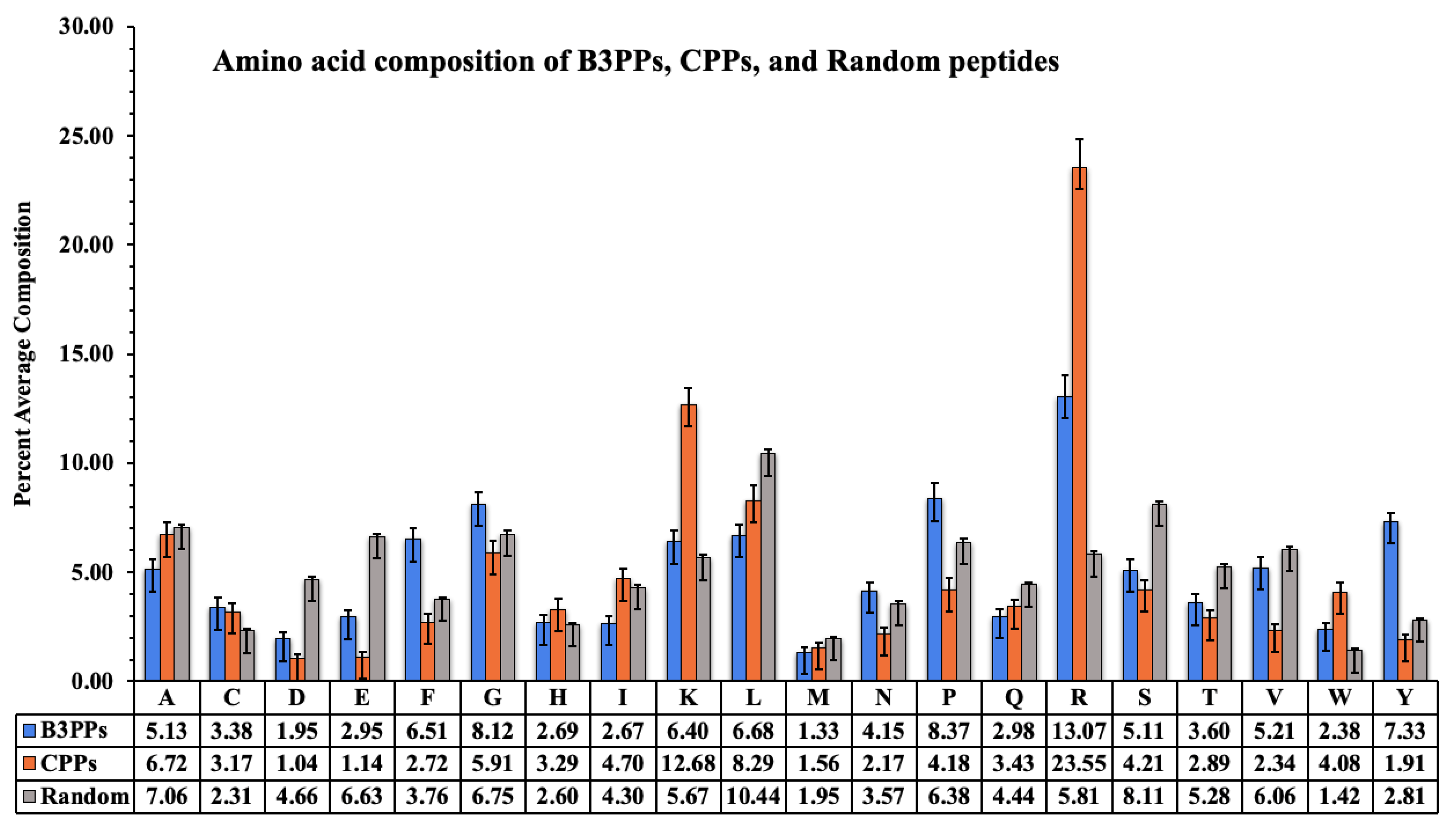
The acid composition of B3PPs, CPPs, and random peptides is shown by a graph (Figure 2); the compositional difference is clearly visible. Arginine is highest in CPPs and B3PPs, which shows that it plays a crucial role in the penetration of peptides into cells. Tyrosine, an aromatic amino acid, is high in B3PPs as compared to other types of peptides. The unique amino acids proline and glycine are prevalent in B3PPs, which contrasts with other types of peptides.
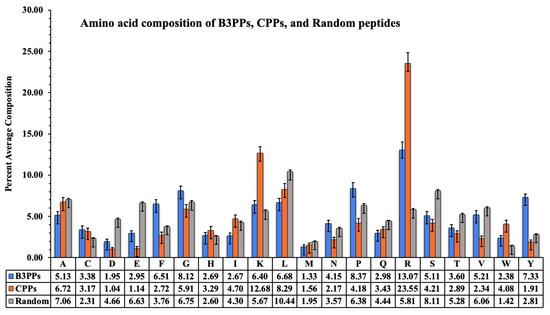
Figure 2.
A bar graph to represent percentage amino acid composition of blood–brain barrier penetrating peptides (B3PPs), cell-penetrating peptides (CPPs), and random peptides.
3.2. Amino Acid Position Analysis
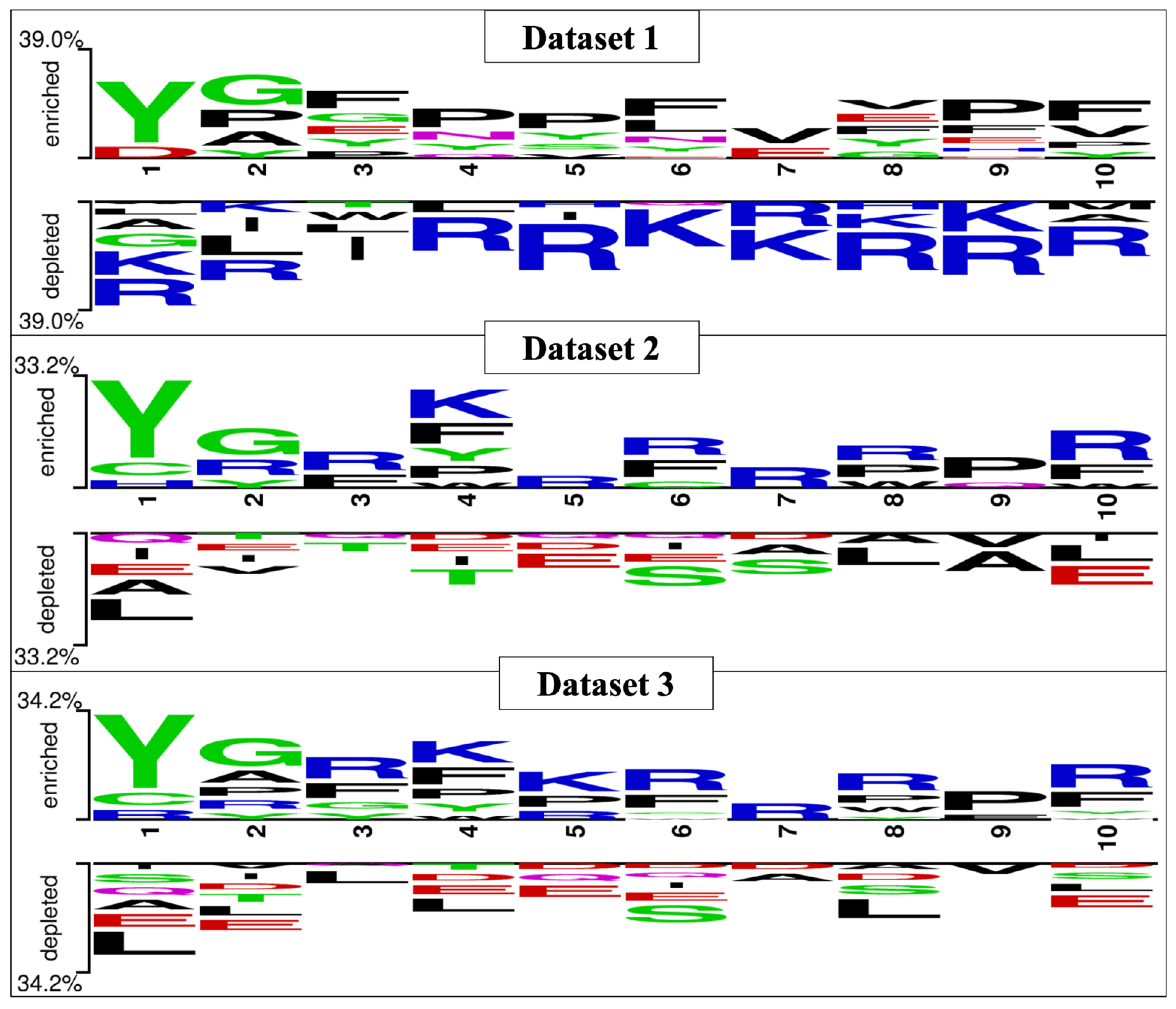
The preferential amino acid position is denoted in Figure 3, which was generated with the help of Two Sample Logo software. The preferred position of amino acids can be seen in the figure; tyrosine, glycine, and arginine are more prominent in the first three positions in B3PPs. The Two Sample Logos suggest that tyrosine, glycine, arginine, and lysine are more preferred throughout the B3PPs.
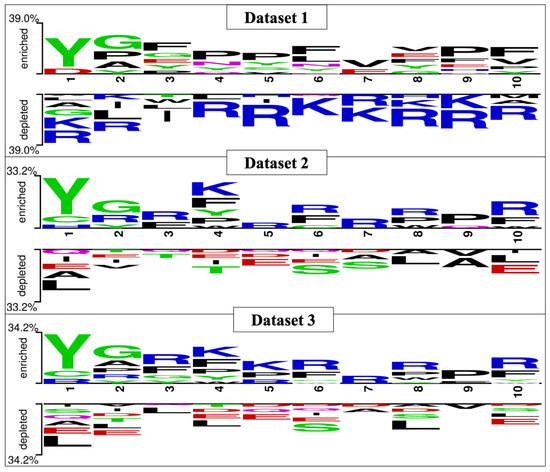
Figure 3.
Two Sample Logo (TSL) depiction of all the three datasets (i.e., Dataset_1, Dataset_2, and Dataset_3), preferred positions for amino acids can be seen in the TSLs.
3.3. B3PPs Prediction Methods on Different Datasets
B3PPs prediction models were built using various machine learning techniques, such as random forest (RF), XG Boosting (XGB), Logistic Regression (LR), Support Vector Classifier (SVC), k-nearest neighbor (KNN), Gaussian Naive Bayes (GNB), and decision tree (DT) on various datasets. The best model was implemented in the webserver and standalone software. As we created three different datasets for the prediction of B3PPs, we generated 9189 peptide features by using Pfeature. These peptide features on each dataset were scrutinized and reduced by an SVC-L1 feature selection technique. The feature selection technique highlighted 73 features of Dataset_1, 73 features of Dataset_2, and 145 features of Dataset_3. After selecting features for the datasets, we developed prediction methods using different machine learning techniques. In order to classify B3PPs and CPPs, we developed models on Dataset_1, which contains 269 B3PPs and 269CPPs. Our random forest model achieved maximum performance using 73 selected features. Our RF-based method obtained an 85.12% accuracy with an AUROC of 0.92 on the training dataset, and an 84.25% accuracy with an AUROC of 0.89 on the validation dataset. KNN performed the worst and obtained a 65.58% accuracy with an AUROC of 0.74 on the training dataset, and a 50.92% accuracy with AUROC of 0.64 on the validation dataset (Table 1).

Table 1.
The performance of classification models developed on Dataset_1 for discriminating B3PPs and CPPs; models were developed using different machine learning techniques.
We developed classification models on Dataset_2 to classify B3PPs and non-B3PPs using different machine learning algorithms. Our RF-based model performed better than other models and achieved an 82.09% accuracy with an AUROC of 0.90 on the training dataset, and an 81.48% accuracy with an AUROC of 0.88 on the validation dataset (Table 2).

Table 2.
The performance of classification models developed on Dataset_2 for discriminating B3PPs and non-B3PPs; models were developed using different machine learning techniques.
Finally, classification models were developed on Dataset_3 for discriminating B3PPs and randomly generated non-B3PPs. Our RF-based model achieved the best performance with respect to the top 80 features (Supplementary Table S5). The performance of the RF model was an 85.25% accuracy with an AUROC of 0.93 on the training dataset, and an 82.93% accuracy with an AUROC of 0.90 on the validation dataset. It was the highest-performing among all the methods on all the datasets, so we incorporated this RF model into our webserver for the prediction of the B3PPs (Table 3).

Table 3.
The performance of classification models developed on Dataset_3 for discriminating B3PPs and CPPs; models were developed using different machine learning techniques.
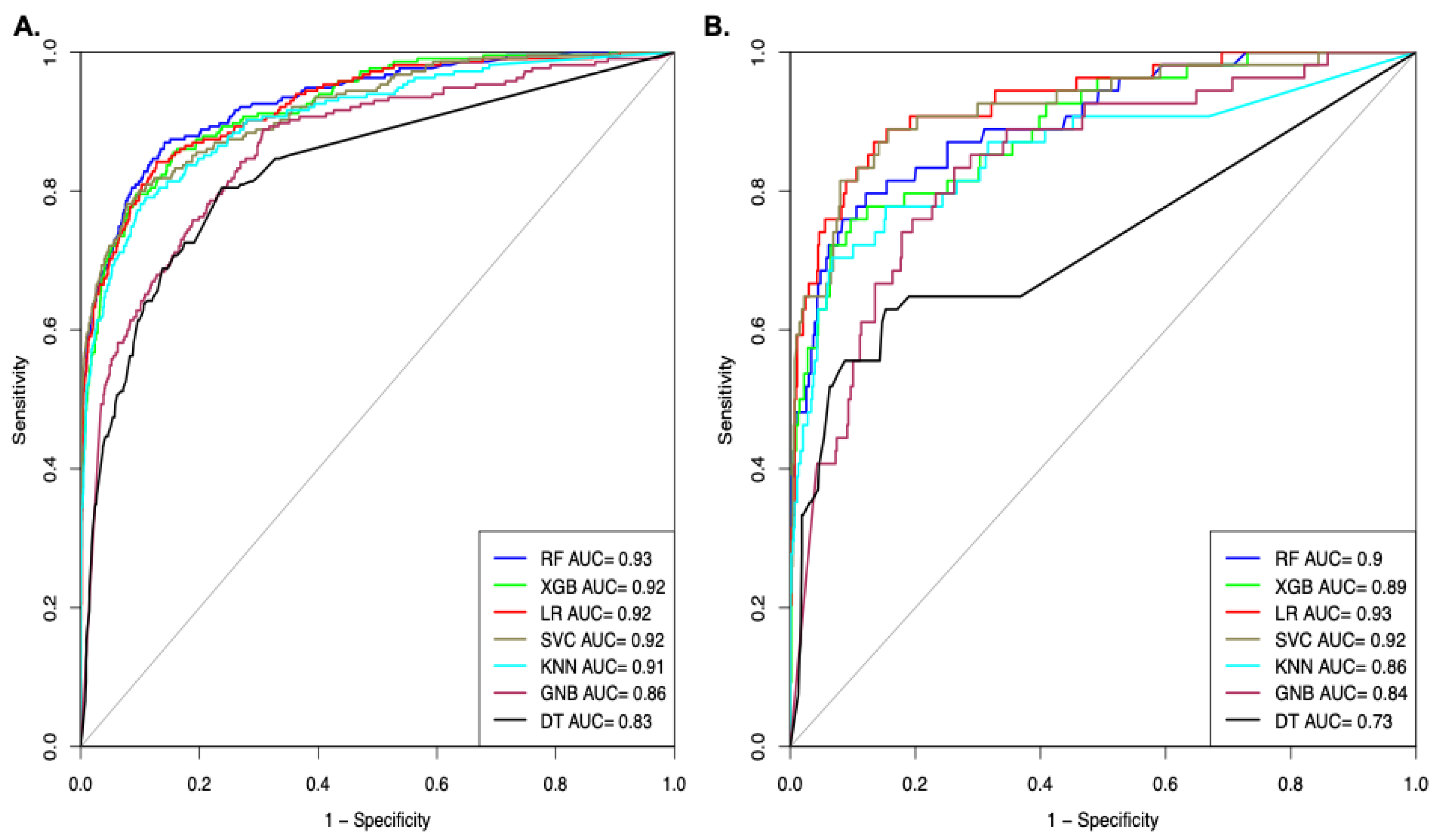
We also computed the performance of the models in terms of AUROC on Dataset_3; the models were developed using different machine learning techniques. As shown in Figure 4A, the RF-based model achieved the highest AUROC of 0.93 on the training dataset. As shown in Figure 4B, the SVC-based model achieved the maximal AUROC of 0.92 on the validation dataset (Figure 4).
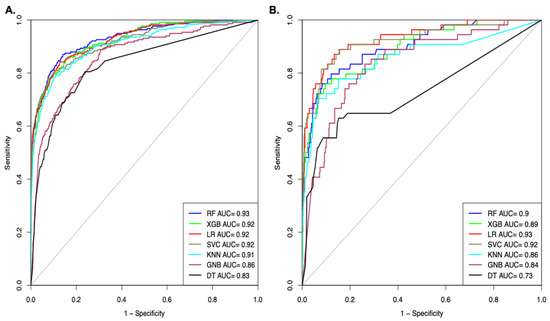
Figure 4.
AUROC plot shows the performance of models on Dataset_3 developed using top selected features (A). AUROC curve for the training dataset (B). AUROC curve for validation dataset.
3.4. Webserver and Standalone Software
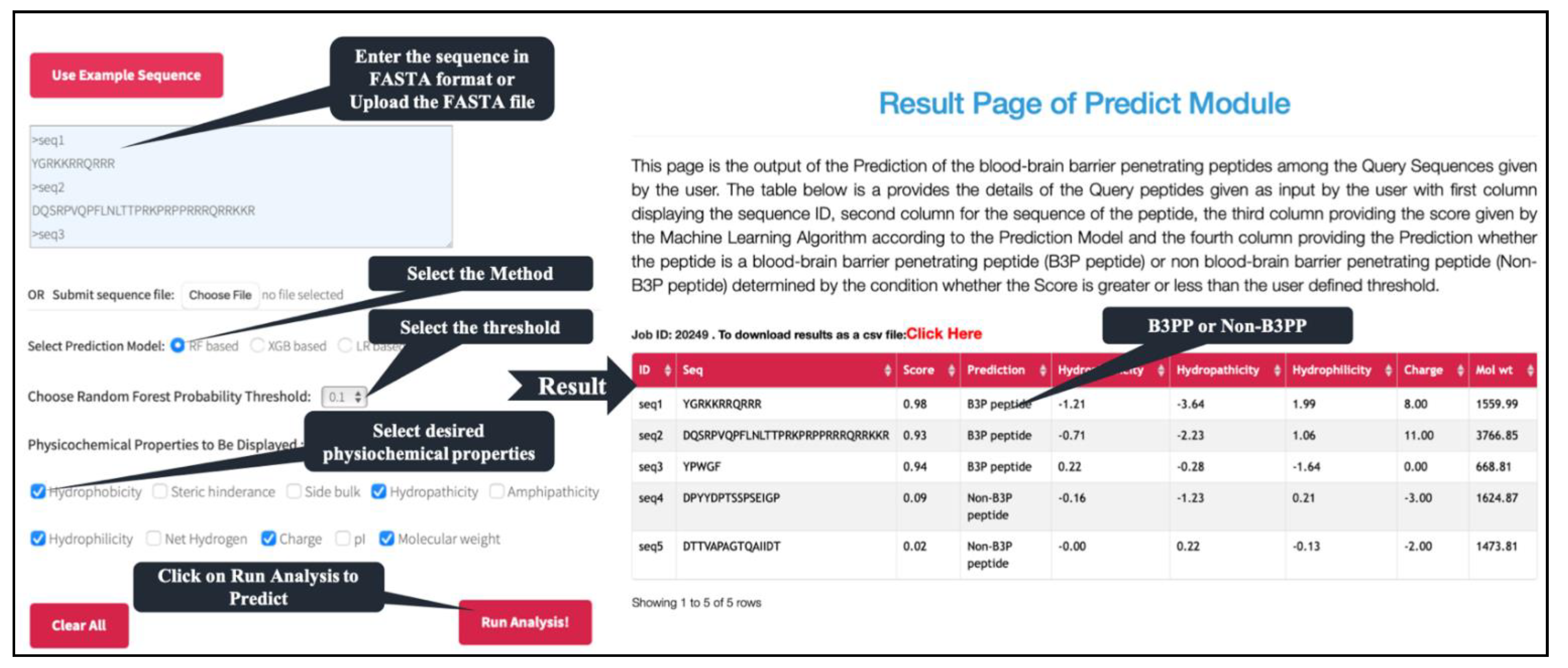
One of the major objectives of this study is to facilitate the scientific community in discovering B3PP-based drug delivery vehicles that can deliver cargo into brain tissues. Thus, we developed a standalone software as well as a web-based service to assist the researcher in finding new B3PPs or designing efficient B3PPs. Our webserver, B3Pred, has three major modules: predict, design, and scan. The predict module of B3pred allows users to predict B3PPs in a set of protein sequences submitted by the user. It allows users to select models developed on any dataset used in this study (Figure 5). The design module of B3pred was developed to discover the most promiscuous B3PPs for a given peptide. This module first generates all possible analogs of a peptide, then predicts the score for each analog. It also allows users to sort analogs, based on their score, and to select the best analog of a peptide. The scan module provides the facility to identify the B3PPs region in the user’s query protein. It allows the user to select the length of the peptide segment to be scanned in the protein sequence they submit. In addition to this web-based service, we also developed standalone software for searching B3PPs at a large scale, including searching B3PPs at the genome level.

Figure 5.
Depiction of predict module present in B3Pred webserver.
3.5. Comparison with the Existing Method
It is crucial to compare this newly developed method with existing methods to understand its benefits and drawbacks. BBPpred has been developed to predict B3PPs, which is trained on 100 B3PPs and 100 non-B3PPs, and the model is tested on only 19 B3PPs and 19 non-B3PPs. On the other hand, B3Pred is trained and tested on three different datasets: Dataset_1 contains 269 B3P peptides and 269 CPPs; Dataset_2 comprises 269 B3P peptides and 269 non-B3P peptides randomly generated using the Swiss-Prot database; and Dataset_3 accommodates 269 B3P peptides and 2690 non-B3P peptides randomly generated using the Swiss-Prot database. In terms of performance, BBPpred achieved a maximal AUROC of 0.87, whereas B3Pred achieved AUROCs of 0.92, 0.90, and 0.93 on Dataset_1, Dataset_2, and Dataset_3, respectively. BBPpred only provides the prediction facility; on the other hand, B3Pred provides a prediction, design, and scan facility. In addition, B3Pred is also available as standalone software, so that users can run it on their local machine at a large scale.
4. Discussion and Conclusions
The blood–brain barrier (BBB) is the natural guard of the brain, which inhibits unwanted molecules from crossing into brain tissue [69]. Unfortunately, neurological disease prevalence has increased tremendously in the last few decades. Thus, there is a need to discover new drugs that can be used to treat brain-associated diseases such as Alzheimer’s disease and Parkinson’s disease. Due to advancements in technology, researchers can discover drugs to treat these disorders in vitro. One of the major hurdles in treating brain-associated disease is delivering drugs into brain tissue, as the blood–brain barrier inhibits these drug molecules from reaching this tissue [70]. The transportation or delivery of the therapeutic molecules across the barriers of the brain is the major bottleneck in treating brain tumors and CNS diseases [71].
Several in silico methods have been developed to predict and improve the delivery of therapeutic molecules that circumvent the BBB. A study has shown that D-Ala-Peptide T-amide (DAPTA), or peptide T is an antiviral peptide that can cross the blood–brain barrier. Intranasal Peptide T can be obtained from the envelope protein of the human immunodeficiency virus (HIV). This peptide shows antiviral properties, usually inhibits chemokine (CCR5) receptors, and also acts as a B3PP [72,73]. Researchers have also found that AH-D, an amphipathic α-helical BBB-penetrating peptide, can act as a therapeutic agent for deadly viruses. It is used as a direct antiviral agent (DAA) to inhibit specific viral proteins. A recent study has suggested that potential antiviral AH-D is a target against deadly viruses, such as chikungunya virus, Zika, dengue, and yellow fever, with different inhibitory and cytotoxic concentrations [74,75,76,77]. These studies show that such peptides can be helpful in viral infections, along with any neurological complications that arise due to these viruses. These peptides can be used as therapeutic substitutes for antiviral drugs which are unable to cross the brain. This may help in controlling the neurological complications that arise due to COVID-19 [78].
In the present scenario, there is the utmost need to develop an efficient prediction tool that can accurately predict the peptides that have the property of penetrating through the blood–brain barrier. To facilitate the researchers working in this area, we proposed a method named B3pred for predicting B3PPs. We have also developed a free webserver, named B3pred, and have incorporated various modules to predict, design, scan for and analyze B3PPs. We believe that our method will help in the accurate prediction of B3PPs and aid the scientific community working in this area.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/pharmaceutics13081237/s1, Table S1: Description of all the 9189 features calculated using composition-based module of Pfeature; Table S2: Fold-wise performance of various machine learning algorithms on Dataset_1; Table S3: Fold-wise performance of various machine learning algorithms on Dataset_2; Table S4: Fold-wise performance of various machine learning algorithms on Dataset_3; Table S5: Top 80 features selected after implementation of SVC-L1 with their importance score calculated using feature-selector python library
Author Contributions
Collected and processed the datasets, V.K. and S.P.; created the prediction models, V.K. and S.P.; analyzed and interpreted the results, V.K., S.P., A.D., N.S. and G.P.S.R.; developed the webserver, V.K. and S.P.; prepared the manuscript, V.K., S.P., A.D. and G.P.S.R.; coordinated the project, G.P.S.R. All authors have read and agreed to the published version of the manuscript.
Funding
Authors received the funding in terms of fellowships and financial support from the following funding agencies. Kumar V. received from University Grant Commission (UGC), Dhall A. and Sharma N. from Department of Science and Technology (DST), and Patiyal S. from Department of Biotechnology, Govt. of India.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The datasets are available at https://webs.iiitd.edu.in/raghava/b3pred/download.php (accessed on 7 July 2021).
Acknowledgments
Authors are thankful to funding agencies, University Grant Commission (UGC), Department of Science and Technology (DST) and Department of Biotechnology, Govt. of India for financial support and fellowships.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Abbott, N.J.; Rönnbäck, L.; Hansson, E. Astrocyte-endothelial interactions at the blood-brain barrier. Nat. Rev. Neurosci. 2006, 7, 41–53. [Google Scholar] [CrossRef]
- Kniesel, U.; Wolburg, H. Tight junctions of the blood-brain barrier. Cell. Mol. Neurobiol. 2000, 20, 57–76. [Google Scholar] [CrossRef]
- Fenstermacher, J.; Gross, P.; Sposito, N.; Acuff, V.; Pettersen, S.; Gruber, K. Structural and Functional Variations in Capillary Systems within the brain. Ann. N. Y. Acad. Sci. 1988, 529, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Rhea, E.M.; Banks, W.A. Role of the Blood-Brain Barrier in Central Nervous System Insulin Resistance. Front. Neurosci. 2019, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muoio, V.; Persson, P.B.; Sendeski, M.M. The neurovascular unit—Concept review. Acta Physiol. 2014, 210, 790–798. [Google Scholar] [CrossRef] [PubMed]
- Oldendorf, W.H. Brain uptake of radiolabeled amino acids, amines, and hexoses after arterial injection. Am. J. Physiol. 1971, 221, 1629–1639. [Google Scholar] [CrossRef] [Green Version]
- Tietz, S.; Engelhardt, B. Brain barriers: Crosstalk between complex tight junctions and adherens junctions. J. Cell Biol. 2015, 209, 493–506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pardridge, W.M. Blood-brain barrier delivery. Drug Discov. Today 2007, 12, 54–61. [Google Scholar] [CrossRef]
- Islam, Y.; Leach, A.G.; Smith, J.; Pluchino, S.; Coxon, C.; Sivakumaran, M.; Downing, J.E.; Fatokun, A.A.; Teixidò, M.; Ehtezazi, T. Peptide based drug delivery systems to the brain. Nano Express 2020, 1, 012002. [Google Scholar] [CrossRef]
- Banks, W.A. Peptides and the blood-brain barrier. Peptides 2015, 72, 16–19. [Google Scholar] [CrossRef] [Green Version]
- Funke, S.A.; Willbold, D. Peptides for Therapy and Diagnosis of Alzheimer’s Disease. Curr. Pharm. Des. 2012, 18, 755–767. [Google Scholar] [CrossRef] [Green Version]
- Baig, M.H.; Ahmad, K.; Saeed, M.; Alharbi, A.M.; Barreto, G.E.; Ashraf, G.M.; Choi, I. Peptide based therapeutics and their use for the treatment of neurodegenerative and other diseases. Biomed. Pharmacother. 2018, 103, 574–581. [Google Scholar] [CrossRef] [PubMed]
- Raucher, D. Tumor targeting peptides: Novel therapeutic strategies in glioblastoma. Curr. Opin. Pharmacol. 2019, 47, 14–19. [Google Scholar] [CrossRef] [PubMed]
- Oller-Salvia, B.; Sánchez-Navarro, M.; Giralt, E.; Teixidó, M. Blood-brain barrier shuttle peptides: An emerging paradigm for brain delivery. Chem. Soc. Rev. 2016, 45, 4690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, L.-P.; Ahmadvand, D.; Su, J.; Hall, A.; Tan, X.; Farhangrazi, Z.S.; Moghimi, S.M. Crossing the blood-brain-barrier with nanoligand drug carriers self-assembled from a phage display peptide. Nat. Commun. 2019, 10, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nosrati, H.; Tarantash, M.; Bochani, S.; Charmi, J.; Bagheri, Z.; Fridoni, M.; Abdollahifar, M.-A.; Davaran, S.; Danafar, H.; Manjili, H.K. Glutathione (GSH) Peptide Conjugated Magnetic Nanoparticles As Blood-Brain Barrier Shuttle for MRI-Monitored Brain Delivery of Paclitaxel. ACS Biomater. Sci. Eng. 2019, 5, 1677–1685. [Google Scholar] [CrossRef]
- Solbrig, M.V.; Koob, G.F. Epilepsy, CNS viral injury and dynorphin. Trends Pharmacol. Sci. 2004, 25, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Balasubramaniam, A. Clinical potentials of neuropeptide Y family of hormones. Am. J. Surg. 2002, 183, 430–434. [Google Scholar] [CrossRef]
- Claes, S.J. Corticotropin-releasing hormone (CRH) in psychiatry: From stress to psychopathology. Ann. Med. 2004, 36, 50–61. [Google Scholar] [CrossRef]
- Ströhle, A.; Holsboer, F. Stress Responsive Neurohormones in Depression and Anxiety. Pharmacopsychiatry 2003, 36, 207–214. [Google Scholar]
- Li, C.; Wu, X.; Liu, S.; Zhao, Y.; Zhu, J.; Liu, K. Roles of Neuropeptide Y in Neurodegenerative and Neuroimmune Diseases. Front. Neurosci. 2019, 13, 869. [Google Scholar] [CrossRef]
- Dwibhashyam, V.S.N.M.; Nagappa, A. Strategies for enhanced drug delivery to the central nervous system. Indian J. Pharm. Sci. 2008, 70, 145–153. [Google Scholar]
- Reese, T.S.; Karnovsky, M.J. Fine structural localization of a blood-brain barrier to exogenous peroxidase. J. Cell Biol. 1967, 34, 207–217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kapoor, P.; Singh, H.; Gautam, A.; Chaudhary, K.; Kumar, R.; Raghava, G.P.S. TumorHoPe: A database of tumor homing peptides. PLoS ONE 2012, 7, e35187. [Google Scholar] [CrossRef] [Green Version]
- Gautam, A.; Singh, H.; Tyagi, A.; Chaudhary, K.; Kumar, R.; Kapoor, P.; Raghava, G.P.S. CPPsite: A curated database of cell penetrating peptides. Database 2012, 2012, bas015. [Google Scholar] [CrossRef] [Green Version]
- Sharma, A.; Kapoor, P.; Gautam, A.; Chaudhary, K.; Kumar, R.; Chauhan, J.S.; Tyagi, A.; Raghava, G.P.S. Computational approach for designing tumor homing peptides. Sci. Rep. 2013, 3, 1607. [Google Scholar] [CrossRef] [PubMed]
- Gautam, A.; Sharma, M.; Vir, P.; Chaudhary, K.; Kapoor, P.; Kumar, R.; Nath, S.K.; Raghava, G.P.S. Identification and characterization of novel protein-derived arginine-rich cell-penetrating peptides. Eur. J. Pharm. Biopharm. 2015, 89, 93–106. [Google Scholar] [CrossRef] [PubMed]
- Shergalis, A.; Bankhead, A.; Luesakul, U.; Muangsin, N.; Neamati, N. Current challenges and opportunities in treating glioblastomas. Pharmacol. Rev. 2018, 70, 412–445. [Google Scholar] [CrossRef] [Green Version]
- Stalmans, S.; Wynendaele, E.; Bracke, N.; Gevaert, B.; D’Hondt, M.; Peremans, K.; Burvenich, C.; De Spiegeleer, B. Chemical-Functional Diversity in Cell-Penetrating Peptides. PLoS ONE 2013, 8, e71752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stalmans, S.; Bracke, N.; Wynendaele, E.; Gevaert, B.; Peremans, K.; Burvenich, C.; Polis, I.; De Spiegeleer, B. Cell-penetrating peptides selectively cross the blood-brain barrier in vivo. PLoS ONE 2015, 10, e0139652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamano, S.; Dai, J.; Hanatani, S.; Haku, K.; Yamanaka, T.; Ishioka, M.; Takayama, T.; Yuvienco, C.; Khapli, S.; Moursi, A.M.; et al. Long-term efficient gene delivery using polyethylenimine with modified Tat peptide. Biomaterials 2014, 35, 1705–1715. [Google Scholar] [CrossRef]
- Huwyler, J.; Wu, D.; Pardridge, W.M. Brain drug delivery of small molecules using immunoliposomes. Proc. Natl. Acad. Sci. USA 1996, 93, 14164–14169. [Google Scholar] [CrossRef] [Green Version]
- Knight, A.; Carvajal, J.; Schneider, H.; Coutelle, C.; Chamberlain, S.; Fairweather, N. Non-viral neuronal gene delivery mediated by the H(C) fragment of tetanus toxin. Eur. J. Biochem. 1999, 259, 762–769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El-Andaloussi, S.; Holm, T.; Langel, U. Cell-Penetrating Peptides: Mechanisms and Applications. Curr. Pharm. Des. 2005, 11, 3597–3611. [Google Scholar] [CrossRef] [PubMed]
- Milletti, F. Cell-penetrating peptides: Classes, origin, and current landscape. Drug Discov. Today 2012, 17, 850–860. [Google Scholar] [CrossRef] [PubMed]
- Stewart, K.M.; Horton, K.L.; Kelley, S.O. Cell-penetrating peptides as delivery vehicles for biology and medicine. Org. Biomol. Chem. 2008, 6, 2242–2255. [Google Scholar] [CrossRef]
- Mueller, J.; Kretzschmar, I.; Volkmer, R.; Boisguerin, P. Comparison of cellular uptake using 22 CPPs in 4 different cell lines. Bioconjug. Chem. 2008, 19, 2363–2374. [Google Scholar] [CrossRef] [PubMed]
- Meade, A.J.; Meloni, B.P.; Mastaglia, F.L.; Knuckey, N. The application of cell penetrating peptides for the delivery of neuroprotective peptides/proteins in experimental cerebral ischaemia studies. J. Exp. Stroke Transl. Med. 2009, 2, 22–40. [Google Scholar] [CrossRef]
- Mathur, D.; Prakash, S.; Anand, P.; Kaur, H.; Agrawal, P.; Mehta, A.; Kumar, R.; Singh, S.; Raghava, G.P.S. PEPlife: A Repository of the Half-life of Peptides. Sci. Rep. 2016, 6, 36617. [Google Scholar] [CrossRef] [Green Version]
- Gautam, A.; Chaudhary, K.; Kumar, R.; Sharma, A.; Kapoor, P.; Tyagi, A.; Raghava, G.P.S. In silico approaches for designing highly effective cell penetrating peptides. J. Transl. Med. 2013, 11, 74. [Google Scholar] [CrossRef] [Green Version]
- Wei, L.; Tang, J.; Zou, Q. SkipCPP-Pred: An improved and promising sequence-based predictor for predicting cell-penetrating peptides. BMC Genom. 2017, 18, 742. [Google Scholar] [CrossRef] [Green Version]
- Wei, L.; Xing, P.; Su, R.; Shi, G.; Ma, Z.S.; Zou, Q. CPPred-RF: A Sequence-based Predictor for Identifying Cell-Penetrating Peptides and Their Uptake Efficiency. J. Proteome Res. 2017, 16, 2044–2053. [Google Scholar] [CrossRef]
- Pandey, P.; Patel, V.; George, N.V.; Mallajosyula, S.S. KELM-CPPpred: Kernel Extreme Learning Machine Based Prediction Model for Cell-Penetrating Peptides. J. Proteome Res. 2018, 17, 3214–3222. [Google Scholar] [CrossRef]
- Kumar, V.; Agrawal, P.; Kumar, R.; Bhalla, S.; Usmani, S.S.; Varshney, G.C.; Raghava, G.P.S. Prediction of cell-penetrating potential of modified peptides containing natural and chemically modified residues. Front. Microbiol. 2018, 9, 725. [Google Scholar] [CrossRef]
- Qiang, X.; Zhou, C.; Ye, X.; Du, P.-F.; Su, R.; Wei, L. CPPred-FL: A sequence-based predictor for large-scale identification of cell-penetrating peptides by feature representation learning. Brief. Bioinform. 2018, 21, 11–23. [Google Scholar] [CrossRef] [PubMed]
- Shaker, B.; Yu, M.-S.; Song, J.S.; Ahn, S.; Ryu, J.Y.; Oh, K.-S.; Na, D. LightBBB: Computational prediction model of blood–brain-barrier penetration based on LightGBM. Bioinformatics 2021, 37, 1135–1139. [Google Scholar] [CrossRef] [PubMed]
- Carpenter, T.; Kirshner, D.A.; Lau, E.Y.; Wong, S.E.; Nilmeier, J.P.; Lightstone, F.C. A Method to Predict Blood-Brain Barrier Permeability of Drug-Like Compounds Using Molecular Dynamics Simulations. Biophys. J. 2014, 107, 630–641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mensch, J.; Oyarzabal, J.; Mackie, C.; Augustijns, P. In vivo, in vitro and in silico methods for small molecule transfer across the BBB. J. Pharm. Sci. 2009, 98, 4429–4468. [Google Scholar] [CrossRef] [PubMed]
- Dai, R.; Zhang, W.; Tang, W.; Wynendaele, E.; Zhu, Q.; Bin, Y.; De Spiegeleer, B.; Xia, J. BBPpred: Sequence-Based Prediction of Blood-Brain Barrier Peptides with Feature Representation Learning and Logistic Regression. J. Chem. Inf. Model. 2021, 61, 525–534. [Google Scholar] [CrossRef]
- Kumar, V.; Patiyal, S.; Kumar, R.; Sahai, S.; Kaur, D.; Lathwal, A.; Raghava, G.P.S. B3Pdb: An archive of blood–brain barrier-penetrating peptides. Brain Struct. Funct. 2021. [Google Scholar] [CrossRef]
- Agrawal, P.; Bhalla, S.; Usmani, S.S.; Singh, S.; Chaudhary, K.; Raghava, G.P.S.; Gautam, A. CPPsite 2.0: A repository of experimentally validated cell-penetrating peptides. Nucleic Acids Res. 2016, 44, D1098–D1103. [Google Scholar] [CrossRef] [PubMed]
- Boutet, E.; Lieberherr, D.; Tognolli, M.; Schneider, M.; Bansal, P.; Bridge, A.; Poux, S.; Bougueleret, L.; Xenarios, I.; Boutet, E. Uniprotkb/swiss-prot, the manually annotated section of the uniprot knowledgebase: How to use the entry view. Methods Mol. Biol. 2016, 1374, 23–54. [Google Scholar] [PubMed]
- Agrawal, P.; Raghava, G.P.S. Prediction of Antimicrobial Potential of a Chemically Modified Peptide From Its Tertiary Structure. Front. Microbiol. 2018, 9, 2551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vacic, V.; Iakoucheva, L.M.; Radivojac, P. Two Sample Logo: A graphical representation of the differences between two sets of sequence alignments. Bioinformatics 2006, 22, 1536–1537. [Google Scholar] [CrossRef] [Green Version]
- Pande, A.; Patiyal, S.; Lathwal, A.; Arora, C.; Kaur, D.; Dhall, A.; Mishra, G.; Kaur, H.; Sharma, N.; Jain, S.; et al. Computing wide range of protein/peptide features from their sequence and structure. bioRxiv 2019. [Google Scholar] [CrossRef]
- Tang, J.; Alelyani, S.; Liu, H. Feature selection for classification: A review. Data Classif Algorithms Appl. 2014, 37–64. Available online: https://www.cvs.edu.in/upload/feature_selection_for_classification.pdf (accessed on 22 February 2021).
- Chang, K.-W.; Hsieh, C.-J.; Lin, C.-J. LIBLINEAR: A Library for Large Linear Classification Rong-En Fan Xiang-Rui Wang. J. Mach. Learn. Res. 2008, 9, 1871–1874. [Google Scholar]
- Ke, G.; Meng, Q.; Finley, T.; Wang, T.; Chen, W.; Ma, W.; Ye, Q.; Liu, T.-Y. LightGBM: A Highly Efficient Gradient Boosting Decision Tree. Adv. Neural Inf. Process. Syst. 2017, 30, 3146–3154. [Google Scholar]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Webb, G.I. Decision Tree Grafting From the All-Tests-But-One Partition. In Proceedings of the Sixteenth International Joint Conference on Artificial Intelligence, IJCAI 99, Stockholm, Sweden, 31 July–6 August 1999. [Google Scholar]
- Zhang, H. Exploring conditions for the optimality of naïve bayes. Int. J. Pattern Recognit. Artif. Intell. 2005, 19, 183–198. [Google Scholar] [CrossRef]
- Geurts, P.; Ernst, D.; Wehenkel, L. Extremely randomized trees. Mach. Learn. 2006, 63, 3–42. [Google Scholar] [CrossRef] [Green Version]
- Tolles, J.; Meurer, W.J. Logistic regression: Relating patient characteristics to outcomes. J. Am. Med. Assoc. 2016, 316, 533–534. [Google Scholar] [CrossRef] [PubMed]
- Mucherino, A.; Papajorgji, P.J.; Pardalos, P.M. Data Mining in Agriculture. Dyn. Disasters 2009, 34. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. XGBoost: A Scalable Tree Boosting System. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016. [Google Scholar]
- Chang, C.-C.; Lin, C.-J. LIBSVM: A Library for Support Vector Machines. ACM Trans. Intell. Syst. Technol. 2011, 2, 1–27. [Google Scholar] [CrossRef]
- Kumar, V.; Kumar, R.; Agrawal, P.; Patiyal, S.; Raghava, G.P.S. A Method for Predicting Hemolytic Potency of Chemically Modified Peptides From Its Structure. Front. Pharmacol. 2020, 11, 54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dhall, A.; Patiyal, S.; Sharma, N.; Usmani, S.S.; Raghava, G.P.S. Computer-aided prediction and design of IL-6 inducing peptides: IL-6 plays a crucial role in COVID-19. Brief. Bioinform. 2021, 22, 936–945. [Google Scholar] [CrossRef] [PubMed]
- Thangudu, S.; Cheng, F.-Y.; Su, C.-H. Advancements in the Blood-Brain Barrier Penetrating Nanoplatforms for Brain Related Disease Diagnostics and Therapeutic Applications. Polymers 2020, 12, 55. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Liu, J.; Liang, J.; Liu, X.; Li, W.; Liu, Z.; Ding, Z.; Tuo, D. Towards Improvements for Penetrating the Blood-Brain Barrier-Recent Progress from a Material and Pharmaceutical Perspective. Cells 2018, 7, 24. [Google Scholar] [CrossRef] [Green Version]
- Banks, W.A. From blood-brain barrier to blood-brain interface: New opportunities for CNS drug delivery. Nat. Rev. Drug Discov. 2016, 15, 275–292. [Google Scholar] [CrossRef]
- Polianova, M.T.; Ruscetti, F.W.; Pert, C.B.; Tractenberg, R.E.; Leoung, G.; Strang, S.; Ruff, M.R. Antiviral and immunological benefits in HIV patients receiving intranasal peptide T (DAPTA). Peptides 2003, 24, 1093–1098. [Google Scholar] [CrossRef]
- Barrera, C.M.; Kastin, A.J.; Banks, W.A. D-[Ala1]-peptide T-Amide is transported from blood to brain by a saturable system. Brain Res. Bull. 1987, 19, 629–633. [Google Scholar] [CrossRef]
- Jackman, J.A.; Costa, V.V.; Park, S.; Real, A.L.C.V.; Park, J.H.; Cardozo, P.L.; Ferhan, A.R.; Olmo, I.G.; Moreira, T.P.; Bambirra, J.L.; et al. Therapeutic treatment of Zika virus infection using a brain-penetrating antiviral peptide. Nat. Mater. 2018, 17, 971–977. [Google Scholar] [CrossRef] [PubMed]
- Cho, N.-J.; Dvory-Sobol, H.; Xiong, A.; Cho, S.-J.; Frank, C.W.; Glenn, J.S. Mechanism of an amphipathic α-helical peptide’s antiviral activity involves size-dependent virus particle lysis. ACS Chem. Biol. 2009, 4, 1061–1067. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Shi, P.Y. Targeting vesicle size. Nat. Mater. 2018, 17, 955–956. [Google Scholar] [CrossRef]
- Boldescu, V.; Behnam, M.; Vasilakis, N.; Klein, C.D. Broad-spectrum agents for flaviviral infections: Dengue, Zika and beyond. Nat. Rev. Drug Discov. 2017, 16, 565–586. [Google Scholar] [CrossRef] [Green Version]
- Mao, X.Y.; Jin, W.L. The COVID-19 Pandemic: Consideration for Brain Infection. Neuroscience 2020, 437, 130–131. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).