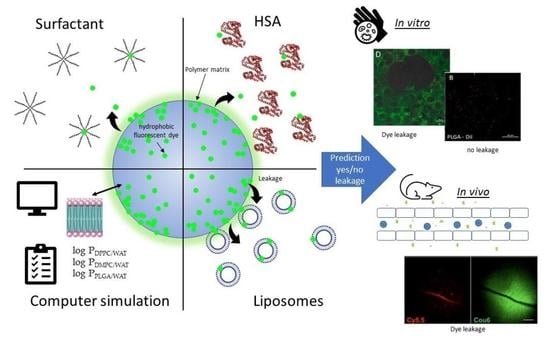
Fluorescently Labeled PLGA Nanoparticles for Visualization In Vitro and In Vivo: The Importance of Dye Properties
Abstract
1. Introduction
2. Experimental
2.1. Materials
2.2. Preparation of Fluorescently Labeled Nanoparticles
2.3. Physicochemical Characterization
2.4. Biological Studies
3. Results & Discussion
3.1. Preparation and Basic Properties of the Labeled Nanoparticles
3.2. Stability of the Nanoparticle Labeling
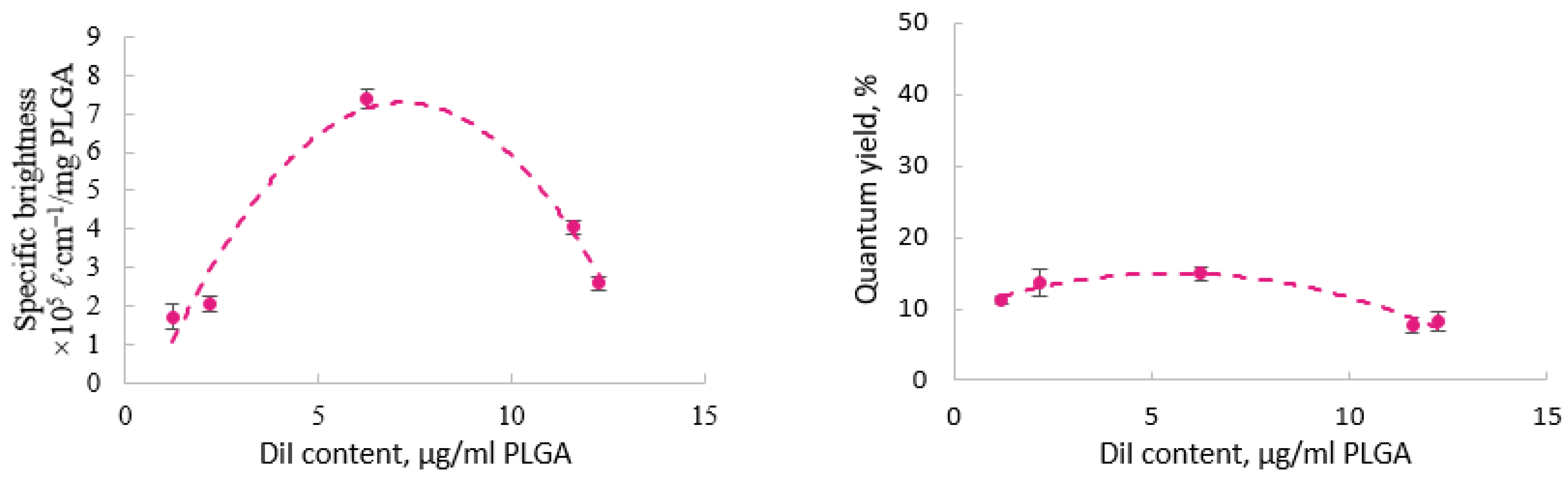
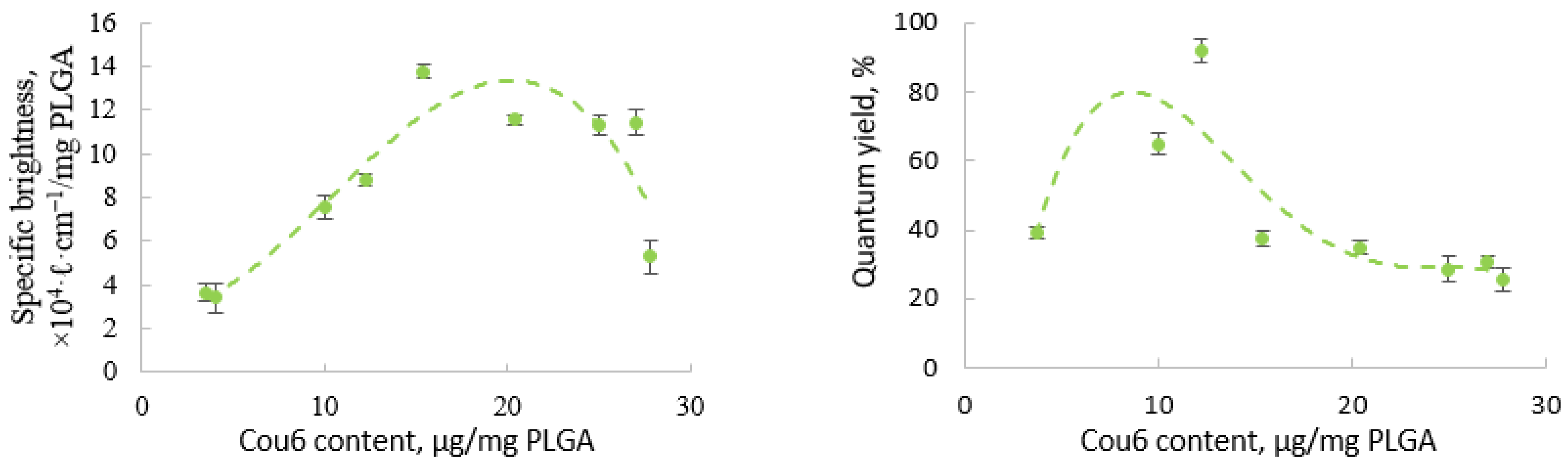
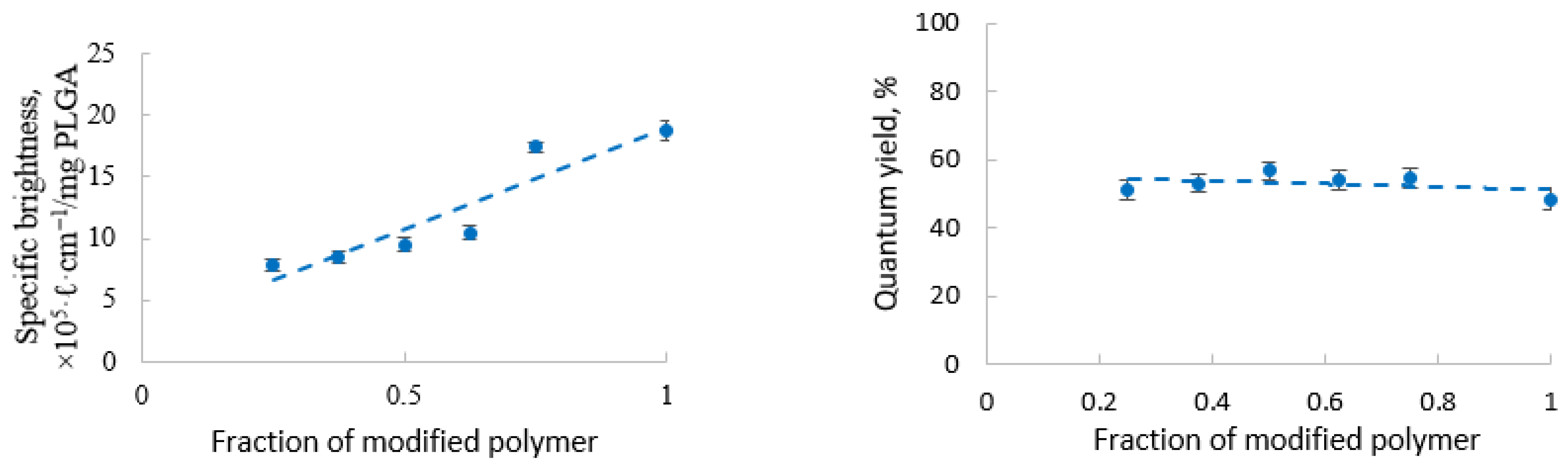
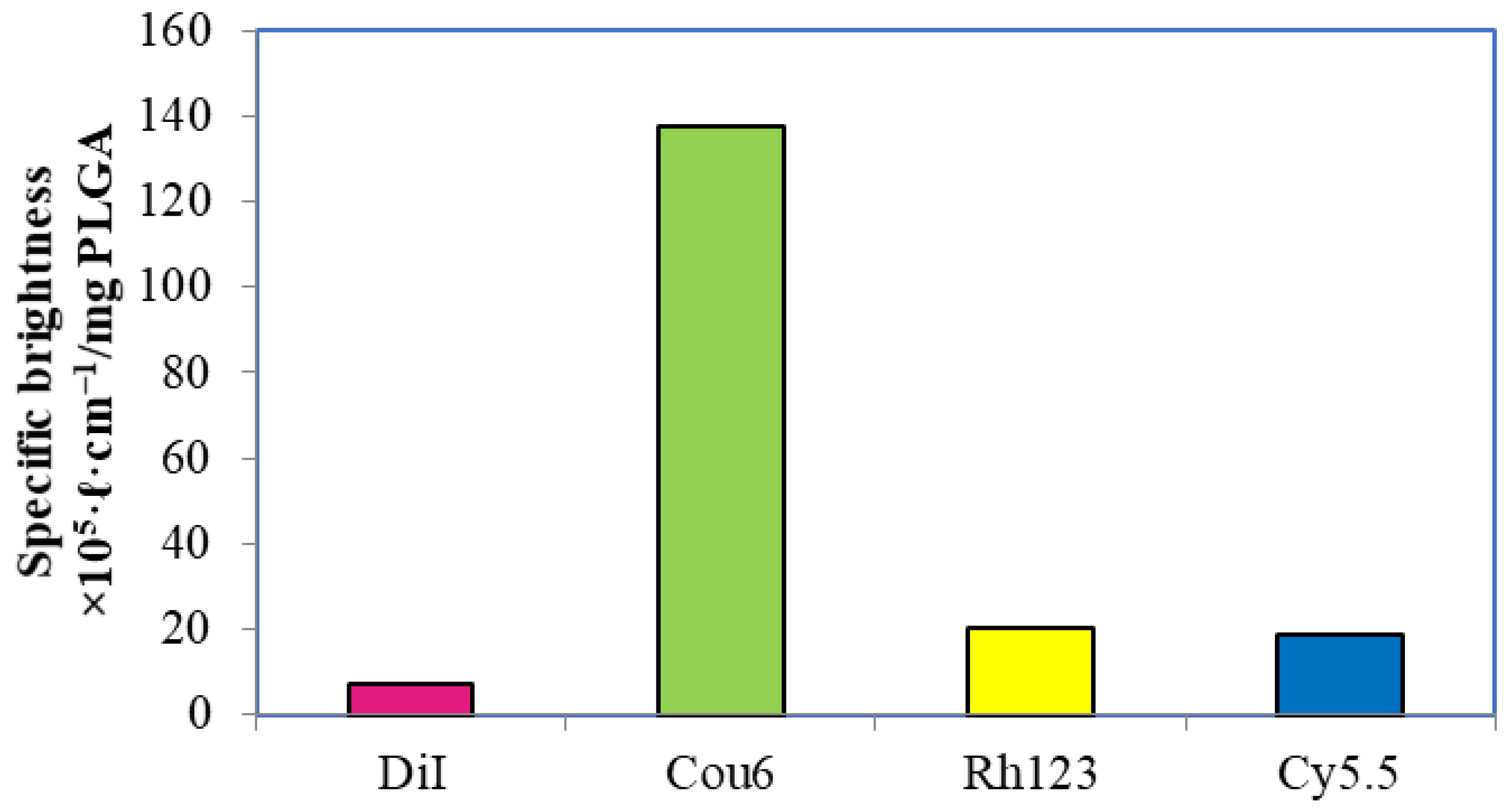
3.3. Fluorescent Properties of the Labeled Nanoparticles
3.3.1. PLGA Nanoparticles with a Single Label
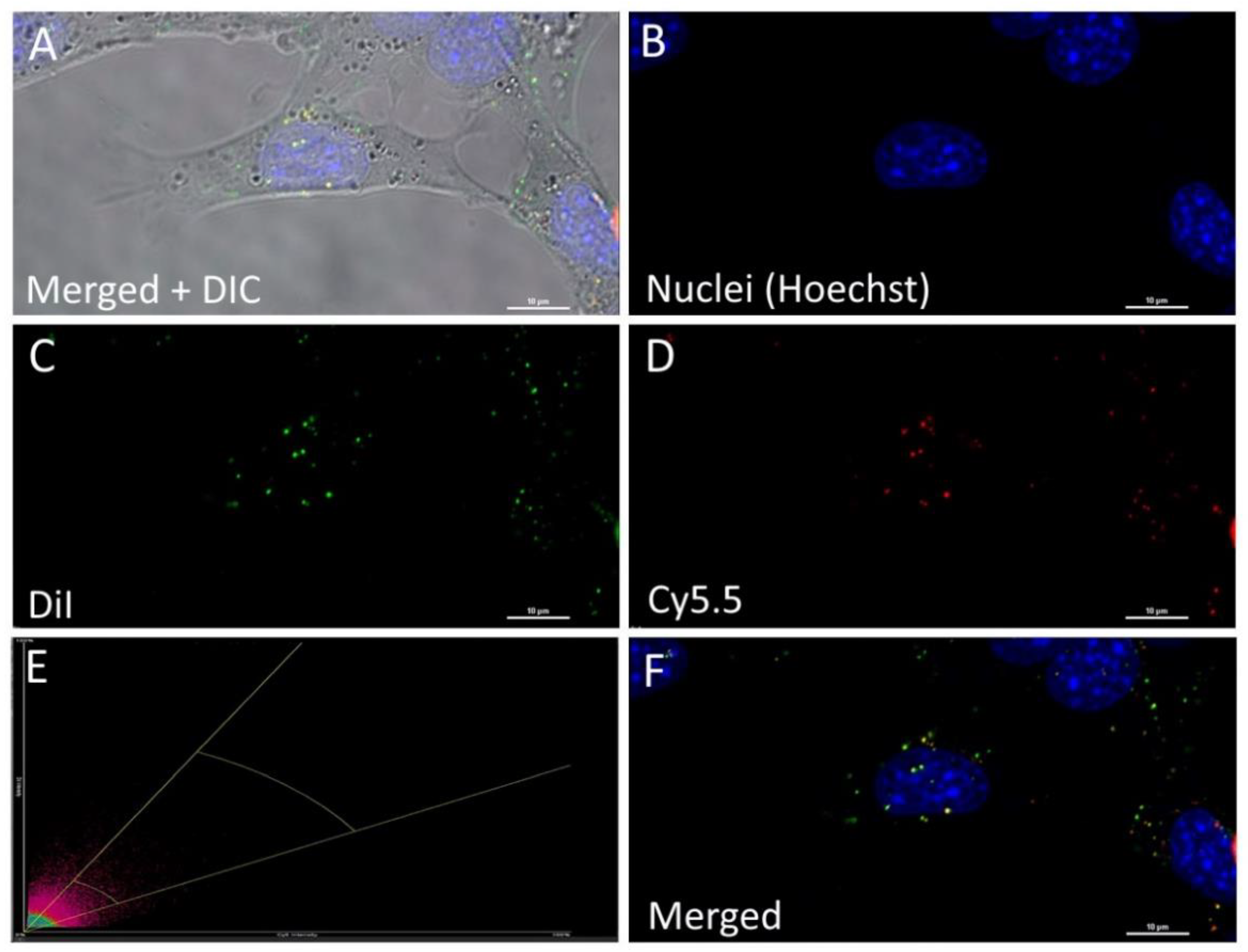
3.3.2. PLGA Nanoparticles with a Double Label
3.4. Evaluation of the Dyes’ Partitioning in Liposomes
3.5. Modeling of Dyes’ Interaction with Model Lipid Membranes
3.6. Differential Scanning Calorimetry Assay of Interaction between Dyes and PLGA
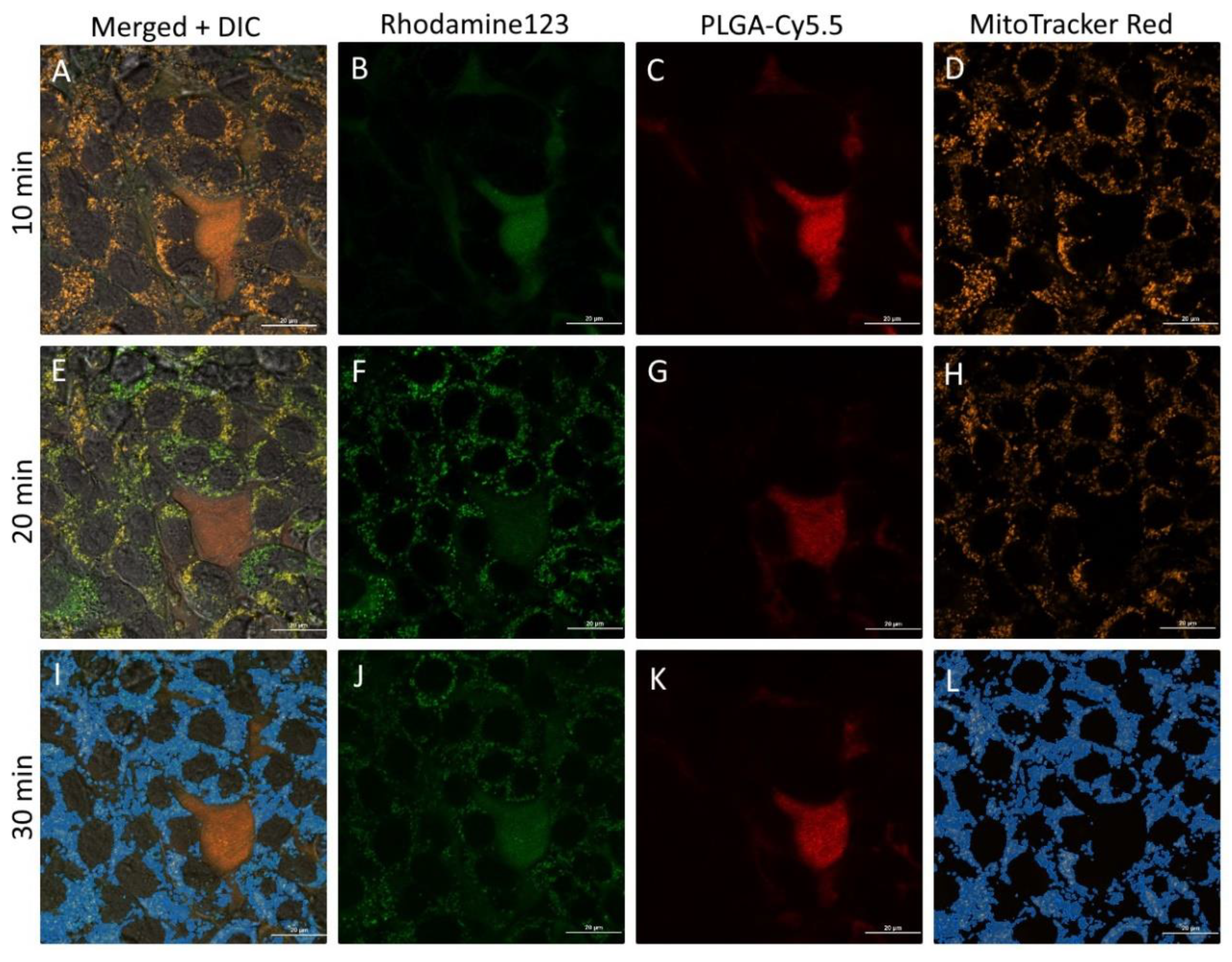
3.7. In Vitro Behavior of PLGA-Cou6 Nanoparticles
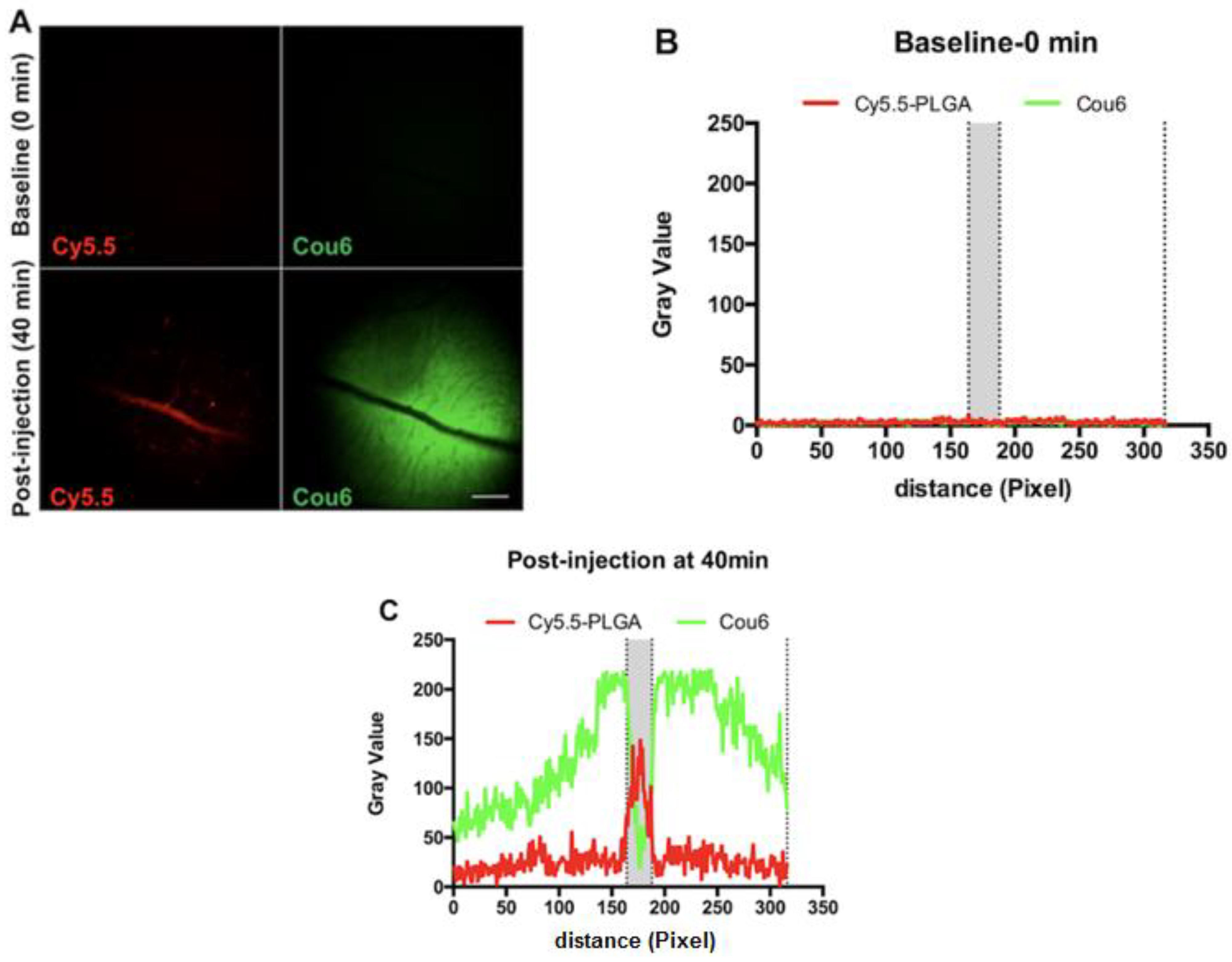
3.8. In Vivo Behavior of PLGA-Cou6 Nanoparticles
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Man, F.; Lammers, T.; de Rosales, R.T.M. Imaging Nanomedicine-Based Drug Delivery: A Review of Clinical Studies. Mol. Imaging Biol. 2018, 20, 683–695. [Google Scholar] [CrossRef]
- Moses, W.W. Fundamental Limits of Spatial Resolution in PET. Nucl. Instrum. Methods Phys. Res. Sect. A Accel. Spectrom. Detect. Assoc. Equip. 2011, 648, S236–S240. [Google Scholar] [CrossRef]
- Ostrowski, A.; Nordmeyer, D.; Boreham, A.; Holzhausen, C.; Mundhenk, L.; Graf, C.; Meinke, M.C.; Vogt, A.; Hadam, S.; Lademann, J.; et al. Overview about the Localization of Nanoparticles in Tissue and Cellular Context by Different Imaging Techniques. Beilstein J. Nanotechnol. 2015, 6, 263–280. [Google Scholar] [CrossRef]
- Li, K.; Liu, B. Polymer-Encapsulated Organic Nanoparticles for Fluorescence and Photoacoustic Imaging. Chem. Soc. Rev. 2014, 43, 6570–6597. [Google Scholar] [CrossRef]
- Thurner, G.C.; Debbage, P. Molecular Imaging with Nanoparticles: The Dwarf Actors Revisited 10 Years Later. Histochem. Cell Biol. 2018, 150, 733–794. [Google Scholar] [CrossRef]
- Shang, L.; Nienhaus, G.U. Small Fluorescent Nanoparticles at the Nano-Bio Interface. Mater. Today 2013, 16, 58–66. [Google Scholar] [CrossRef]
- Priem, B.; Tian, C.; Tang, J.; Zhao, Y.; Mulder, W.J.M. Fluorescent Nanoparticles for the Accurate Detection of Drug Delivery. Expert Opin. Drug Deliv. 2015, 12, 1881–1894. [Google Scholar] [CrossRef]
- Boschi, F.; de Sanctis, F. Overview of the Optical Properties of Fluorescent Nanoparticles for Optical Imaging. Eur. J. Histochem. 2017, 61, 2830. [Google Scholar] [CrossRef]
- Pratiwi, F.W.; Kuo, C.W.; Chen, B.C.; Chen, P. Recent Advances in the Use of Fluorescent Nanoparticles for Bioimaging. Nanomedicine 2019, 14, 1759–1769. [Google Scholar] [CrossRef]
- Fili, N.; Toseland, C.P. Fluorescence and labelling: How to choose and what to do. In Fluorescent Methods for Molecular Motors; Springer: Berlin/Heidelberg, Germany, 2014; Volume 105, pp. 1–24. ISBN 9783034808569. [Google Scholar]
- Reisch, A.; Klymchenko, A.S. Fluorescent Polymer Nanoparticles Based on Dyes: Seeking Brighter Tools for Bioimaging. Small 2016, 12, 1968–1992. [Google Scholar] [CrossRef]
- Ma, X.; Sun, R.; Cheng, J.; Liu, J.; Gou, F.; Xiang, H.; Zhou, X. Fluorescence Aggregation-Caused Quenching versus Aggregation-Induced Emission: A Visual Teaching Technology for Undergraduate Chemistry Students. J. Chem. Educ. 2016, 93, 345–350. [Google Scholar] [CrossRef]
- Abdel-Mottaleb, M.M.A.; Beduneau, A.; Pellequer, Y.; Lamprecht, A. Stability of Fluorescent Labels in PLGA Polymeric Nanoparticles: Quantum Dots versus Organic Dyes. Int. J. Pharm. 2015, 494, 471–478. [Google Scholar] [CrossRef]
- Weiss, B.; Schaefer, U.F.; Zapp, J.; Lamprecht, A.; Stallmach, A.; Lehr, C.M. Nanoparticles Made of Fluorescence-Labelled Poly(L-Lactide-Co-Glycolide): Preparation, Stability, and Biocompatibility. J. Nanosci. Nanotechnol. 2006, 6, 3048–3056. [Google Scholar] [CrossRef]
- Malinovskaya, Y.; Melnikov, P.; Baklaushev, V.; Gabashvili, A.; Osipova, N.; Mantrov, S.; Ermolenko, Y.; Maksimenko, O.; Gorshkova, M.; Balabanyan, V.; et al. Delivery of Doxorubicin-Loaded PLGA Nanoparticles into U87 Human Glioblastoma Cells. Int. J. Pharm. 2017, 524, 77–90. [Google Scholar] [CrossRef]
- Bastiat, G.; Pritz, C.O.; Roider, C.; Fouchet, F.; Lignières, E.; Jesacher, A.; Glueckert, R.; Ritsch-Marte, M.; Schrott-Fischer, A.; Saulnier, P.; et al. A New Tool to Ensure the Fluorescent Dye Labeling Stability of Nanocarriers: A Real Challenge for Fluorescence Imaging. J. Control. Release 2013, 170, 334–342. [Google Scholar] [CrossRef] [PubMed]
- Snipstad, S.; Hak, S.; Baghirov, H.; Sulheim, E.; Mørch, Ý.; Lélu, S.; von Haartman, E.; Bäck, M.; Nilsson, K.P.R.; Klymchenko, A.S.; et al. Labeling Nanoparticles: Dye Leakage and Altered Cellular Uptake. Cytom. Part A 2017, 91, 760–766. [Google Scholar] [CrossRef]
- Tenuta, T.; Monopoli, M.P.; Kim, J.A.; Salvati, A.; Dawson, K.A.; Sandin, P.; Lynch, I. Elution of Labile Fluorescent Dye from Nanoparticles during Biological Use. PLoS ONE 2011, 6, e25556. [Google Scholar] [CrossRef]
- Andreozzi, P.; Martinelli, C.; Carney, R.P.; Carney, T.M.; Stellacci, F. Erythrocyte Incubation as a Method for Free-Dye Presence Determination in Fluorescently Labeled Nanoparticles. Mol. Pharm. 2013, 10, 875–882. [Google Scholar] [CrossRef]
- Chen, H.; Kim, S.; Li, L.; Wang, S.; Park, K.; Cheng, J.X. Release of Hydrophobic Molecules from Polymer Micelles into Cell Membranes Revealed by Förster Resonance Energy Transfer Imaging. Proc. Natl. Acad. Sci. USA 2008, 105, 6596–6601. [Google Scholar] [CrossRef] [PubMed]
- Tosi, G.; Rivasi, F.; Gandolfi, F.; Costantino, L.; Vandelli, M.A.; Forni, F. Conjugated Poly(D,L-Lactide-Co-Glycolide) for the Preparation of in Vivo Detectable Nanoparticles. Biomaterials 2005, 26, 4189–4195. [Google Scholar] [CrossRef]
- Chang, J.; Jallouli, Y.; Kroubi, M.; Yuan, X.; Feng, W.; Kang, C.; Pu, P.; Betbeder, D. Characterization of Endocytosis of Transferrin-Coated PLGA Nanoparticles by the Blood-Brain Barrier. Int. J. Pharm. 2009, 379, 285–292. [Google Scholar] [CrossRef] [PubMed]
- Sahin, A.; Esendagli, G.; Yerlikaya, F.; Caban-Toktas, S.; Yoyen-Ermis, D.; Horzum, U.; Aktas, Y.; Khan, M.; Couvreur, P.; Capan, Y. A Small Variation in Average Particle Size of PLGA Nanoparticles Prepared by Nanoprecipitation Leads to Considerable Change in Nanoparticles’ Characteristics and Efficacy of Intracellular Delivery. Artif. Cells Nanomed. Biotechnol. 2017, 45, 1657–1664. [Google Scholar] [CrossRef] [PubMed]
- Panyam, J.; Zhou, W.; Prabha, S.; Sahoo, S.K.; Labhasetwar, V. Rapid Endo-lysosomal Escape of Poly(DL-lactide-Co Glycolide) Nanoparticles: Implications for Drug and Gene Delivery. FASEB J. 2002, 16, 1217–1226. [Google Scholar] [CrossRef]
- Yan, F.; Zhang, C.; Zheng, Y.; Mei, L.; Tang, L.; Song, C.; Sun, H.; Huang, L. The Effect of Poloxamer 188 on Nanoparticle Morphology, Size, Cancer Cell Uptake, and Cytotoxicity. Nanomed. Nanotechnol. Biol. Med. 2010, 6, 170–178. [Google Scholar] [CrossRef]
- Lü, J.M.; Wang, X.; Marin-Muller, C.; Wang, H.; Lin, P.H.; Yao, Q.; Chen, C. Current Advances in Research and Clinical Applications of PLGA-Based Nanotechnology. Expert Rev. Mol. Diagn. 2009, 9, 325–341. [Google Scholar] [CrossRef]
- Zhang, J.; Shi, Y.; Zheng, Y.; Pan, C.; Yang, X.; Dou, T.; Wang, B.; Lu, W. Homing in on an Intracellular Target for Delivery of Loaded Nanoparticles Functionalized with a Histone Deacetylase Inhibitor. Oncotarget 2017, 8, 68242–68251. [Google Scholar] [CrossRef][Green Version]
- Kirthivasan, B.; Singh, D.; Bommana, M.M.; Raut, S.L.; Squillante, E.; Sadoqi, M. Active Brain Targeting of a Fluorescent P-Gp Substrate Using Polymeric Magnetic Nanocarrier System. Nanotechnology 2012, 23, 255102. [Google Scholar] [CrossRef]
- Weinheimer, M.; Fricker, G.; Burhenne, J.; Mylius, P.; Schubert, R. The Application of P-Gp Inhibiting Phospholipids as Novel Oral Bioavailability Enhancers—An in Vitro and in Vivo Comparison. Eur. J. Pharm. Sci. 2017, 108, 13–22. [Google Scholar] [CrossRef]
- Reimold, I.; Domke, D.; Bender, J.; Seyfried, C.A.; Radunz, H.E.; Fricker, G. Delivery of Nanoparticles to the Brain Detected by Fluorescence Microscopy. Eur. J. Pharm. Biopharm. 2008, 70, 627–632. [Google Scholar] [CrossRef] [PubMed]
- Saito, E.; Kuo, R.; Pearson, R.M.; Gohel, N.; Cheung, B.; King, N.J.C.; Miller, S.D.; Shea, L.D. Designing Drug-Free Biodegradable Nanoparticles to Modulate Inflammatory Monocytes and Neutrophils for Ameliorating Inflammation. J. Control. Release 2019, 300, 185–196. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Cano-Vega, M.A.; Yue, F.; Kuang, L.; Narayanan, N.; Uzunalli, G.; Merkel, M.P.; Kuang, S.; Deng, M. Dibenzazepine-Loaded Nanoparticles Induce Local Browning of White Adipose Tissue to Counteract Obesity. Mol. Ther. 2017, 25, 1718–1729. [Google Scholar] [CrossRef]
- Yao, H.; Ashiba, K. Highly Fluorescent Organic Nanoparticles of Thiacyanine Dye: A Synergetic Effect of Intermolecular H-Aggregation and Restricted Intramolecular Rotation. RSC Adv. 2011, 1, 834–838. [Google Scholar] [CrossRef]
- Rhys Williams, A.T.; Winfield, S.A.; Miller, J.N. Relative Fluorescence Quantum Yields Using a Computer-Controlled Luminescence Spectrometer. Analyst 1983, 108, 1067–1071. [Google Scholar] [CrossRef]
- Brouwer, A.M. Standards for Photoluminescence Quantum Yield Measurements in Solution; IUPAC Technical Report. Pure Appl. Chem. 2011, 83, 2213–2228. [Google Scholar] [CrossRef]
- Magde, D.; Wong, R.; Seybold, P.G. Fluorescence Quantum Yields and Their Relation to Lifetimes of Rhodamine 6G and Fluorescein in Nine Solvents: Improved Absolute Standards for Quantum Yields. Photochem. Photobiol. 2002, 75, 327–334. [Google Scholar] [CrossRef]
- Grabolle, M.; Spieles, M.; Lesnyak, V.; Gaponik, N.; Eychmüller, A.; Resch-Genger, U. Determination of the Fluorescence Quantum Yield of Quantum Dots: Suitable Procedures and Achievable Uncertainties. Anal. Chem. 2009, 81, 6285–6294. [Google Scholar] [CrossRef]
- Majoul, I.; Jia, Y.; Duden, R. Practical Fluorescence Resonance Energy Transfer or Molecular Nanobioscopy of Living Cells. In Handbook of Biological Confocal Microscopy, 3rd ed.; Springer: Boston, MA, USA, 2006; pp. 788–808. [Google Scholar] [CrossRef]
- Harder, E.; Damm, W.; Maple, J.; Wu, C.; Reboul, M.; Xiang, J.Y.; Wang, L.; Lupyan, D.; Dahlgren, M.K.; Knight, J.L.; et al. OPLS3: A Force Field Providing Broad Coverage of Drug-like Small Molecules and Proteins. J. Chem. Theory Comput. 2016, 12, 281–296. [Google Scholar] [CrossRef] [PubMed]
- Watts, K.S.; Dalal, P.; Murphy, R.B.; Sherman, W.; Friesner, R.A.; Shelley, J.C. ConfGen: A Conformational Search Method for Efficient Generation of Bioactive Conformers. J. Chem. Inf. Model. 2010, 50, 534–546. [Google Scholar] [CrossRef]
- Řezáč, J.; Hobza, P. Advanced Corrections of Hydrogen Bonding and Dispersion for Semiempirical Quantum Mechanical Methods. J. Chem. Theory Comput. 2012, 8, 141–151. [Google Scholar] [CrossRef]
- Becke, A.D. Density-Functional Exchange-Energy Approximation with Correct Asymptotic Behavior. Phys. Rev. A 1988, 38, 3098–3100. [Google Scholar] [CrossRef]
- Vosko, S.H.; Wilk, L.; Nusair, M. Accurate Spin-Dependent Electron Liquid Correlation Energies for Local Spin Density Calculations: A Critical Analysis. Can. J. Phys. 1980, 58, 1200–1211. [Google Scholar] [CrossRef]
- Perdew, J.P. Density-Functional Approximation for the Correlation Energy of the Inhomogeneous Electron Gas. Phys. Rev. B 1986, 33, 8822–8824. [Google Scholar] [CrossRef]
- Schäfer, A.; Huber, C.; Ahlrichs, R. Fully Optimized Contracted Gaussian Basis Sets of Triple Zeta Valence Quality for Atoms Li to Kr. J. Chem. Phys. 1994, 100, 5829–5835. [Google Scholar] [CrossRef]
- Klamt, A. The COSMO and COSMO-RS Solvation Models. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2018, 8, 699–709. [Google Scholar] [CrossRef]
- Eckert, F.; Klamt, A. Fast Solvent Screening via Quantum Chemistry: COSMO-RS Approach. AIChE J. 2002, 48, 369–385. [Google Scholar] [CrossRef]
- Marenich, A.V.; Cramer, C.J.; Truhlar, D.G. Universal Solvation Model Based on Solute Electron Density and on a Continuum Model of the Solvent Defined by the Bulk Dielectric Constant and Atomic Surface Tensions. J. Phys. Chem. B 2009, 113, 6378–6396. [Google Scholar] [CrossRef]
- Loschen, C.; Klamt, A. Prediction of Solubilities and Partition Coefficients in Polymers Using COSMO-RS. Ind. Eng. Chem. Res. 2014, 53, 11478–11487. [Google Scholar] [CrossRef]
- Prilloff, S.; Fan, J.; Henrich-Noack, P.; Sabel, B.A. In Vivo Confocal Neuroimaging (ICON): Non-Invasive, Functional Imaging of the Mammalian CNS with Cellular Resolution. Eur. J. Neurosci. 2010, 31, 521–528. [Google Scholar] [CrossRef] [PubMed]
- Sabel, B.A.; Engelmann, R.; Humphrey, M.F. In Vivo Confocal Neuroimaging (ICON) of CNS Neurons. Nat. Med. 1997, 3, 244–247. [Google Scholar] [CrossRef]
- Finke, J.H.; Richter, C.; Gothsch, T.; Kwade, A.; Büttgenbach, S.; Müller-Goymann, C.C. Coumarin 6 as a Fluorescent Model Drug: How to Identify Properties of Lipid Colloidal Drug Delivery Systems via Fluorescence Spectroscopy? Eur. J. Lipid Sci. Technol. 2014, 116, 1234–1246. [Google Scholar] [CrossRef]
- Rivolta, I.; Panariti, A.; Lettiero, B.; Sesana, S.; Gasco, P.; Gasco, M.R.; Masserini, M.; Miserocchi, G. Cellular Uptake of Coumarin-6 as a Model Drug Loaded in Solid Lipid Nanoparticles. J. Physiol. Pharmacol. 2011, 62, 45–53. [Google Scholar] [PubMed]
- Guerrini, L.; Alvarez-Puebla, R.A.; Pazos-Perez, N. Surface Modifications of Nanoparticles for Stability in Biological Fluids. Materials 2018, 11, 1154. [Google Scholar] [CrossRef] [PubMed]
- Abouelmagd, S.A.; Sun, B.; Chang, A.C.; Ku, Y.J.; Yeo, Y. Release Kinetics Study of Poorly Water-Soluble Drugs from Nanoparticles: Are We Doing It Right? Mol. Pharm. 2015, 12, 997–1003. [Google Scholar] [CrossRef] [PubMed]
- Simonsson, C.; Bastiat, G.; Pitorre, M.; Klymchenko, A.S.; Béjaud, J.; Mély, Y.; Benoit, J.P. Inter-Nanocarrier and Nanocarrier-to-Cell Transfer Assays Demonstrate the Risk of an Immediate Unloading of Dye from Labeled Lipid Nanocapsules. Eur. J. Pharm. Biopharm. 2016, 98, 47–56. [Google Scholar] [CrossRef]
- Shabbits, J.A.; Chiu, G.N.C.; Mayer, L.D. Development of an in Vitro Drug Release Assay That Accurately Predicts in Vivo Drug Retention for Liposome-Based Delivery Systems. J. Control. Release 2002, 84, 161–170. [Google Scholar] [CrossRef]
- Pretor, S.; Bartels, J.; Lorenz, T.; Dahl, K.; Finke, J.H.; Peterat, G.; Krull, R.; Al-Halhouli, A.T.; Dietzel, A.; Büttgenbach, S.; et al. Cellular Uptake of Coumarin-6 under Microfluidic Conditions into HCE-T Cells from Nanoscale Formulations. Mol. Pharm. 2015, 12, 34–45. [Google Scholar] [CrossRef]
- Tian, Y.; Halle, J.; Wojdyr, M.; Sahoo, D.; Scheblykin, I.G. Quantitative Measurement of Fluorescence Brightness of Single Molecules. Methods Appl. Fluoresc. 2014, 2, 035003. [Google Scholar] [CrossRef]
- Zou, P.; Chen, H.; Paholak, H.J.; Sun, D. Noninvasive Fluorescence Resonance Energy Transfer Imaging of in Vivo Premature Drug Release from Polymeric Nanoparticles. Mol. Pharm. 2013, 10, 4185–4194. [Google Scholar] [CrossRef]
- Wang, H.; Chen, J.; Xu, C.; Shi, L.; Tayier, M.; Zhou, J.; Zhang, J.; Wu, J.; Ye, Z.; Fang, T.; et al. Cancer Nanomedicines Stabilized by π-π Stacking between Heterodimeric Prodrugs Enable Exceptionally High Drug Loading Capacity and Safer Delivery of Drug Combinations. Theranostics 2017, 7, 3638. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, T.; Fukushima, S.; Okamoto, K.; Suzuki, M.; Matsumura, Y.; Yokoyama, M.; Okano, T.; Sakurai, Y.; Kataoka, K. Development of the Polymer Micelle Carrier System for Doxorubicin. J. Control. Release 2001, 74, 295–302. [Google Scholar] [CrossRef]
- Behnke, T.; Würth, C.; Laux, E.M.; Hoffmann, K.; Resch-Genger, U. Simple Strategies towards Bright Polymer Particles via One-Step Staining Procedures. Dye. Pigment. 2012, 94, 247–257. [Google Scholar] [CrossRef]
- Sharafy, S.; Muszkat, K.A. Viscosity Dependence of Fluorescence Quantum Yields. J. Am. Chem. Soc. 1971, 93, 4119–4125. [Google Scholar] [CrossRef]
- Hall, L.M.; Gerowska, M.; Brown, T. A Highly Fluorescent DNA Toolkit: Synthesis and Properties of Oligonucleotides Containing New Cy3, Cy5 and Cy3B Monomers. Nucleic Acids Res. 2012, 40, e108. [Google Scholar] [CrossRef] [PubMed]
- Edetsberger, M.; Knapp, M.; Gaubitzer, E.; Miksch, C.; Gvichiya, K.E.; Köhler, G. Effective Staining of Tumor Cells by Coumarin-6 Depends on the Stoichiometry of Cyclodextrin Complex Formation. J. Incl. Phenom. Macrocycl. Chem. 2011, 70, 327–331. [Google Scholar] [CrossRef]
- Li, Y.; Song, Y.; Zhao, L.; Gaidosh, G.; Laties, A.M.; Wen, R. Direct Labeling and Visualization of Blood Vessels with Lipophilic Carbocyanine Dye DiI. Nat. Protoc. 2008, 3, 1703–1708. [Google Scholar] [CrossRef] [PubMed]
- Tian, B.-C.; Zhang, W.-J.; Xu, H.-M.; Hao, M.-X.; Liu, Y.-B.; Yang, X.-G.; Pan, W.-S.; Liu, X.-H. Further Investigation of Nanostructured Lipid Carriers as an Ocular Delivery System: In Vivo Transcorneal Mechanism and in Vitro Release Study. Colloids Surf. B Biointerfaces 2013, 102, 251–256. [Google Scholar] [CrossRef]
- Jensen, K.H.R.; Berg, R.W. CLARITY-Compatible Lipophilic Dyes for Electrode Marking and Neuronal Tracing. Sci. Rep. 2016, 6, 32674. [Google Scholar] [CrossRef]
- Poulain, F.E.; Gaynes, J.A.; Hörndli, C.S.; Law, M.Y.; Chien, C.B. Analyzing retinal axon guidance in zebrafish. Methods Cell Biol. 2010, 100, 2–26. [Google Scholar]
- Goss, K.U. Predicting Equilibrium Sorption of Neutral Organic Chemicals into Various Polymeric Sorbents with COSMO-RS. Anal. Chem. 2011, 83, 5304–5308. [Google Scholar] [CrossRef] [PubMed]
- Klamt, A.; Eckert, F.; Reinisch, J.; Wichmann, K. Prediction of Cyclohexane-Water Distribution Coefficients with COSMO-RS on the SAMPL5 Data Set. J. Comput. Aided Mol. Des. 2016, 30, 959–967. [Google Scholar] [CrossRef]
- Loschen, C.; Reinisch, J.; Klamt, A. COSMO-RS Based Predictions for the SAMPL6 LogP Challenge. J. Comput. Aided Mol. Des. 2020, 34, 385–392. [Google Scholar] [CrossRef] [PubMed]
- Biltonen, R.L.; Lichtenberg, D. The Use of Differential Scanning Calorimetry as a Tool to Characterize Liposome Preparations. Chem. Phys. Lipids 1993, 64, 129–142. [Google Scholar] [CrossRef]
- Steuer, H.; Jaworski, A.; Elger, B.; Kaussmann, M.; Keldenich, J.; Schneider, H.; Stoll, D.; Schlosshauer, B. Functional Characterization and Comparison of the Outer Blood-Retina Barrier and the Blood-Brain Barrier. Investig. Ophthalmol. Vis. Sci. 2005, 46, 1047–1053. [Google Scholar] [CrossRef] [PubMed]
- Pietzonka, P.; Rothen-Rutishauser, B.; Langguth, P.; Wunderli-Allenspach, H.; Walter, E.; Merkle, H.P. Transfer of Lipophilic Markers from PLGA and Polystyrene Nanoparticles to Caco-2 Monolayers Mimics Particle Uptake. Pharm. Res. 2002, 19, 595–601. [Google Scholar] [CrossRef] [PubMed]
- Zhang, E.; Zhukova, V.; Semyonkin, A.; Osipova, N.; Malinovskaya, Y.; Maksimenko, O.; Chernikov, V.; Sokolov, M.; Grigartzik, L.; Sabel, B.A.; et al. Release Kinetics of Fluorescent Dyes from PLGA Nanoparticles in Retinal Blood Vessels: In Vivo Monitoring and Ex Vivo Localization. Eur. J. Pharm. Biopharm. 2020, 150, 131–142. [Google Scholar] [CrossRef]
- Demuth, P.C.; Garcia-Beltran, W.F.; Ai-Ling, M.L.; Hammond, P.T.; Irvine, D.J. Composite Dissolving Microneedles for Coordinated Control of Antigen and Adjuvant Delivery Kinetics in Transcutaneous Vaccination. Adv. Funct. Mater. 2013, 23, 161–172. [Google Scholar] [CrossRef]
- Tahara, K.; Karasawa, K.; Onodera, R.; Takeuchi, H. Feasibility of Drug Delivery to the Eye’s Posterior Segment by Topical Instillation of PLGA Nanoparticles. Asian J. Pharm. Sci. 2017, 12, 394–399. [Google Scholar] [CrossRef]
- Zhang, E.; Osipova, N.; Sokolov, M.; Maksimenko, O.; Semyonkin, A.; Wang, M.; Grigartzik, L.; Gelperina, S.; Sabel, B.A.; Henrich-Noack, P. Exploring the Systemic Delivery of a Poorly Water-Soluble Model Drug to the Retina Using PLGA Nanoparticles. Eur. J. Pharm. Sci. 2021, 105905. [Google Scholar] [CrossRef]
- Hofmann, D.; Messerschmidt, C.; Bannwarth, M.B.; Landfester, K.; Mailänder, V. Drug Delivery without Nanoparticle Uptake: Delivery by a Kiss-and-Run Mechanism on the Cell Membrane. Chem. Commun. 2014, 50, 1369–1371. [Google Scholar] [CrossRef]





| Type of Nanoparticles | Fraction of Released Dye, % | ||
|---|---|---|---|
| 1% Solution of Poloxamer 188 | 1% Solution of HSA | ||
| 2 h | 2 h | 24 h | |
| PLGA-DiI | 9.8 ± 1.5 | 1.2 ± 0.1 | 42.3 ± 1.7 |
| PLGA-Cy5.5/DiI | 4.1 ± 0.3 | <0.1 | 15.2 ± 0.7 |
| PEG-PLGA-Cou6 | 17.0 ± 0.4 | 42.4 ± 2.1 | 65.0 ± 2.9 |
| PLGA-Rh123 | 57.0 ± 3.5 | n/d | n/d |
| Type of Nanoparticles | Fraction of a Dye Transitioned to Liposomes, % | Amount of Dye Found in Liposomes, μg/mg |
|---|---|---|
| PLGA-Cou6 | 62.4 ± 7.1 | 0.93 |
| PLGA-DiI | 38.7 ± 0.6 | 1.45 |
| PLGA-DiI * | 0 | 0 |
| PLGA-Rh123 | 8.1 ± 1.4 | 0.20 |
| PLGA-Cy5.5 | 0 | 0 |
| Parameter | DiI | Coumarin 6 | Rhodamine 123 |
|---|---|---|---|
| Topological polar surface area, Å2 (“polarity”) | 6.25 * | 42.43 * | 91.48 * |
| log POCT/WAT ** (hydrophobicity) | 10.3 | 5.0 | 0.04 |
| log PDPPC/WAT ** (affinity to a lipid bilayer) | 8.6 | 5.2 | −0.6 |
| log PDMPC/WAT ** (affinity to lipid bilayer) | 8.7 | 5.2 | −0.6 |
| log PPLGA/WAT ** (affinity to NPs’ polymeric matrix) | 5.3 | 1.2 | −3.6 |
| log PDCM/PLGA ** | 0 | 0 | 0 |
| log PDPPC/PLGA ** | 2.3 | 0.7 | 2.6 |
| log PDMPC/PLGA ** | 1.6 | 0.7 | 2.1 |
| Sample | Tg, °C |
|---|---|
| PLGA (bulk) | 43.27 |
| Non-loaded PLGA nanoparticles | 44.90 |
| Cou6-PLGA nanoparticles | 43.91 |
| DiI-PLGA nanoparticles | 42.35 |
| Type of NPs | Dye Content, µg/mg PLGA | EE, % | Mean Diameter Z-ave, nm | PDI | Zeta Potential, mV | |
|---|---|---|---|---|---|---|
| PLGA-DiI | 6.4 | 76.6 | 156 ± 5 | 0.156 ± 0.016 | −27 ± 2 | |
| Non-polymeric DiI formulation | 23.6 µg/mL * | - | 333 ± 93 | 0.355 ± 0.077 | −21 ± 6 | |
| PEG-PLGA/Cou6 | 27.2 | 99.7 | 139 ± 2 | 0.036 ± 0.028 | −15 ± 3 | |
| Non-polymeric Cou6 formulation | 32.6 µg/mL * | - | 1086 ± 166 | 0.830 ± 0.111 | −6.6 ± 0.1 | |
| PLGA-Cy5.5/DiI | DiI | 6.2 | 77 | 137 ± 1 | 0.136 ± 0.022 | −18 ± 1 |
| Cy5.5 | 0.8 | |||||
| PEG-PLGA-Cy5.5/Cou6 NPs | Cou6 | 5.7 | 91 | 128 ± 1 | 0.137 ± 0.022 | −22 ± 1 |
| Cy5.5 | 0.8 | |||||
| PLGA-Cy5.5/Rh123 | Rh123 | 6.8 | 96.6 | 98 ± 1 | 0.196 ± 0.031 | −24 ± 4 |
| Cy5.5 | 0.8 | |||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhukova, V.; Osipova, N.; Semyonkin, A.; Malinovskaya, J.; Melnikov, P.; Valikhov, M.; Porozov, Y.; Solovev, Y.; Kuliaev, P.; Zhang, E.; et al. Fluorescently Labeled PLGA Nanoparticles for Visualization In Vitro and In Vivo: The Importance of Dye Properties. Pharmaceutics 2021, 13, 1145. https://doi.org/10.3390/pharmaceutics13081145
Zhukova V, Osipova N, Semyonkin A, Malinovskaya J, Melnikov P, Valikhov M, Porozov Y, Solovev Y, Kuliaev P, Zhang E, et al. Fluorescently Labeled PLGA Nanoparticles for Visualization In Vitro and In Vivo: The Importance of Dye Properties. Pharmaceutics. 2021; 13(8):1145. https://doi.org/10.3390/pharmaceutics13081145
Chicago/Turabian StyleZhukova, Vasilisa, Nadezhda Osipova, Aleksey Semyonkin, Julia Malinovskaya, Pavel Melnikov, Marat Valikhov, Yuri Porozov, Yaroslav Solovev, Pavel Kuliaev, Enqi Zhang, and et al. 2021. "Fluorescently Labeled PLGA Nanoparticles for Visualization In Vitro and In Vivo: The Importance of Dye Properties" Pharmaceutics 13, no. 8: 1145. https://doi.org/10.3390/pharmaceutics13081145
APA StyleZhukova, V., Osipova, N., Semyonkin, A., Malinovskaya, J., Melnikov, P., Valikhov, M., Porozov, Y., Solovev, Y., Kuliaev, P., Zhang, E., Sabel, B. A., Chekhonin, V., Abakumov, M., Majouga, A., Kreuter, J., Henrich-Noack, P., Gelperina, S., & Maksimenko, O. (2021). Fluorescently Labeled PLGA Nanoparticles for Visualization In Vitro and In Vivo: The Importance of Dye Properties. Pharmaceutics, 13(8), 1145. https://doi.org/10.3390/pharmaceutics13081145