Synergistic Effect of Doxorubicin and siRNA-Mediated Silencing of Mcl-1 Using Cationic Niosomes against 3D MCF-7 Spheroids
Abstract
1. Introduction
2. Results
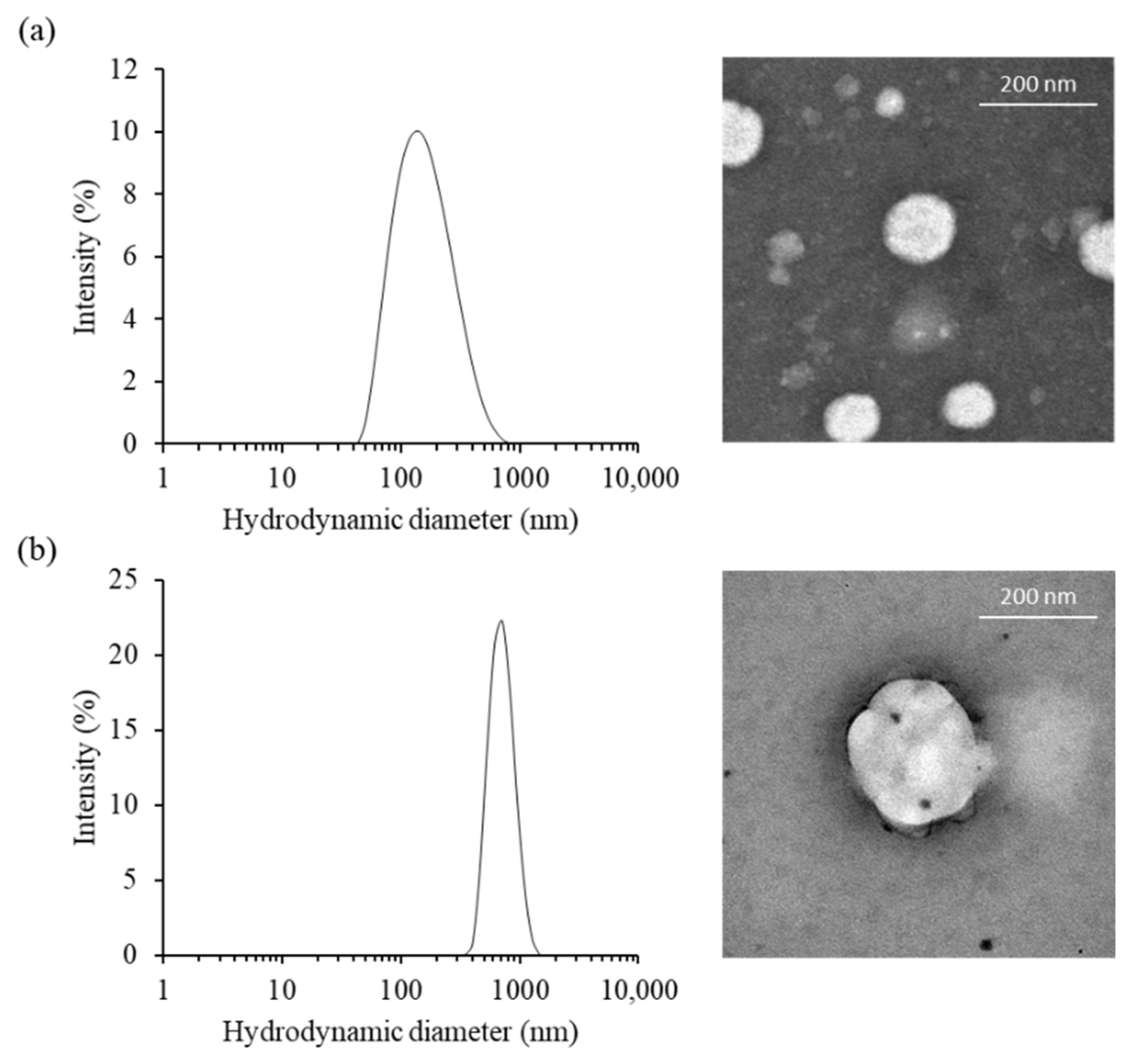
2.1. Characterization of Cationic Niosomes
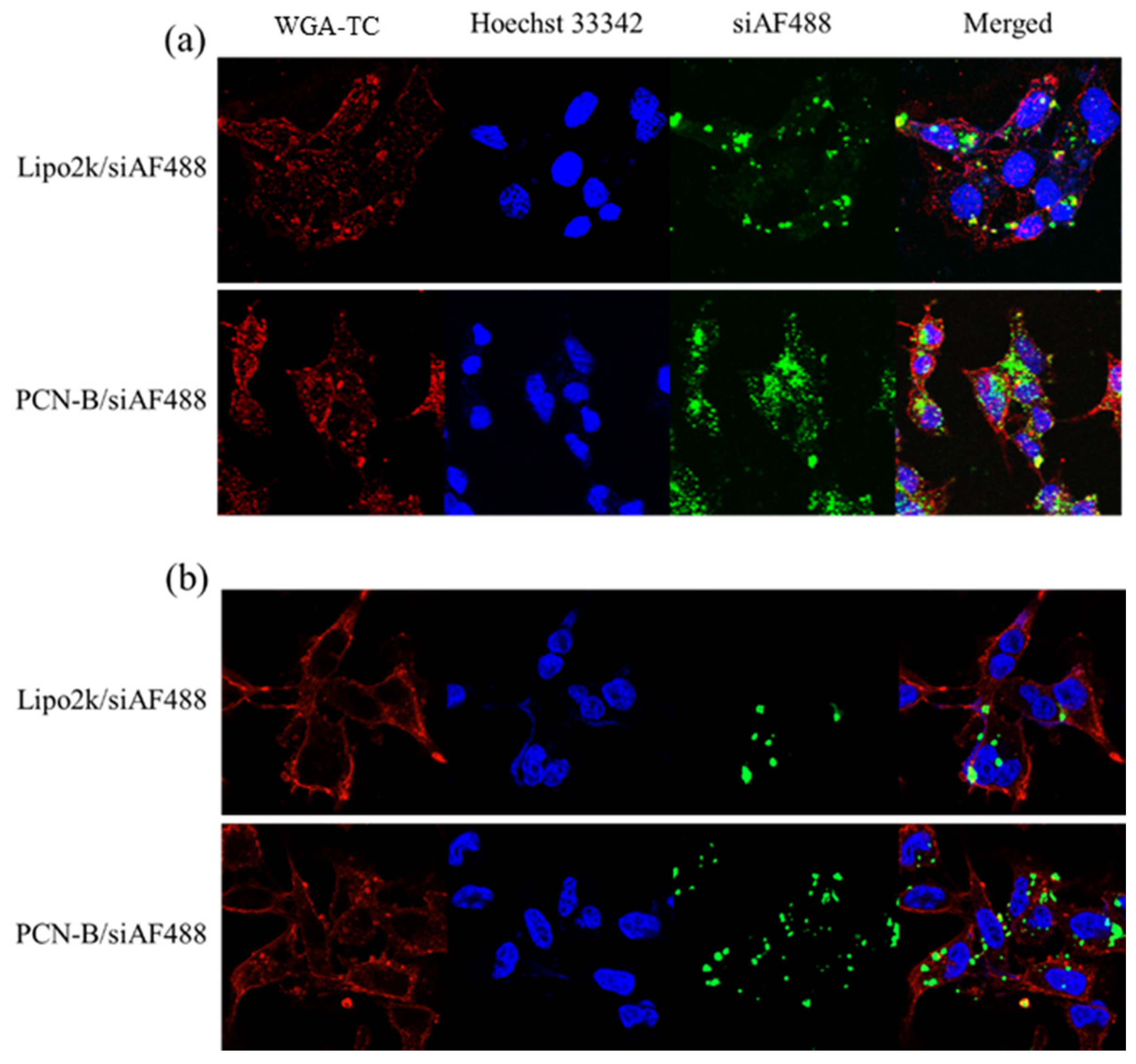
2.2. Cellular Uptakes and Intracellular Distribution of siRNA Complexes by Flow Cytometry and Confocal Laser Scanning Microscopy
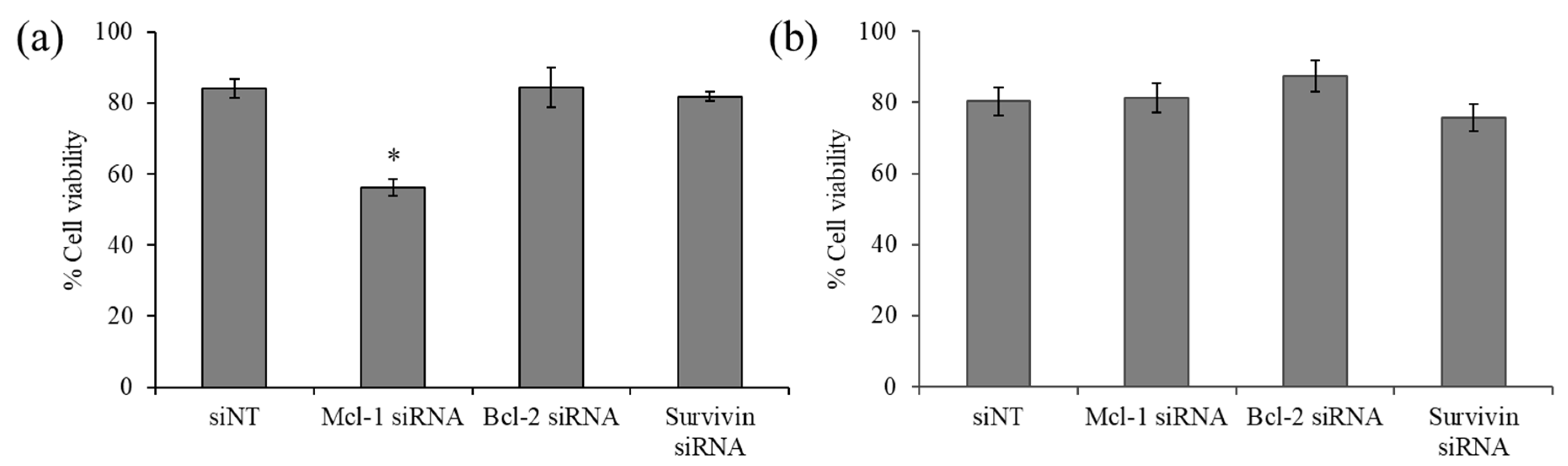
2.3. The Effect of Anti-Apoptosis siRNA Delivery on the Viability of Breast Cancer Cells
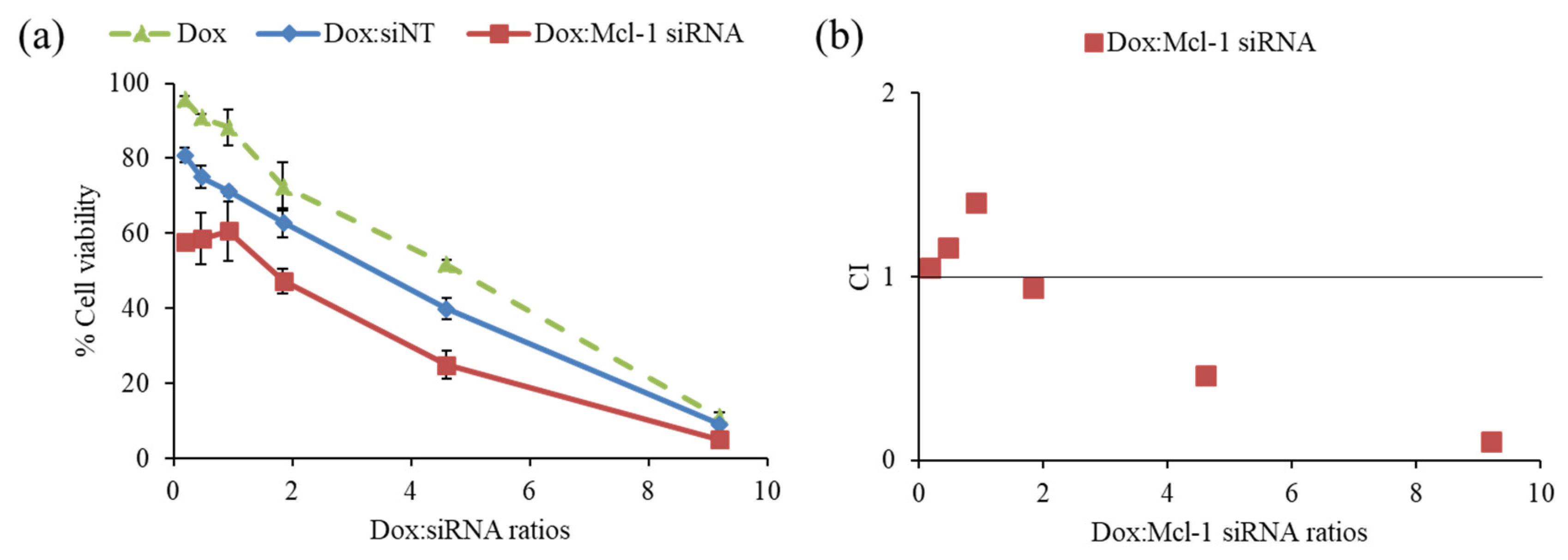
2.4. Combination of Dox and Mcl-1 siRNA with a Non-Constant Concentration Ratio
2.5. Combination of Dox and Mcl-1 siRNA with a Constant Concentration Ratio
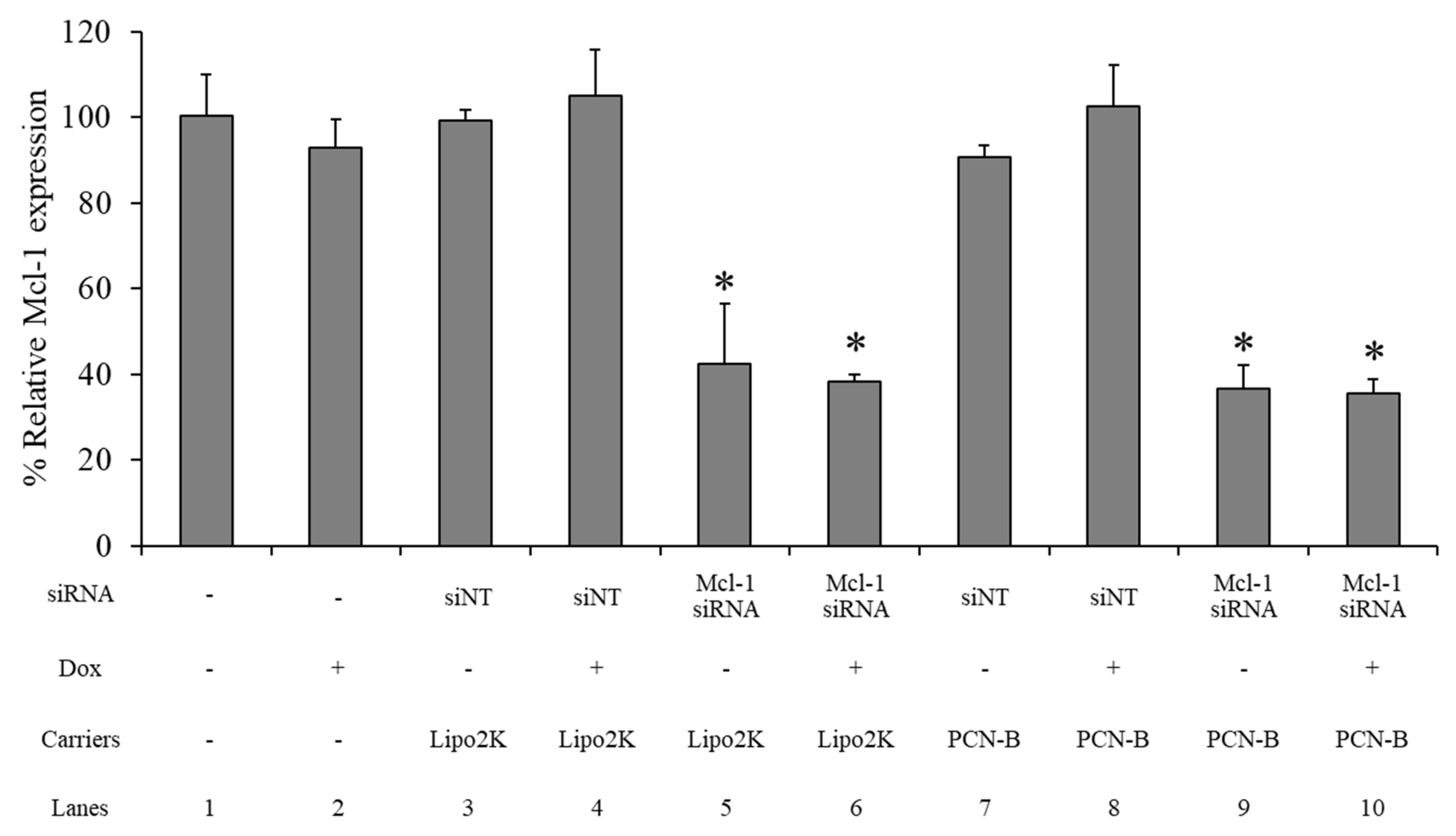
2.6. mRNA Expression
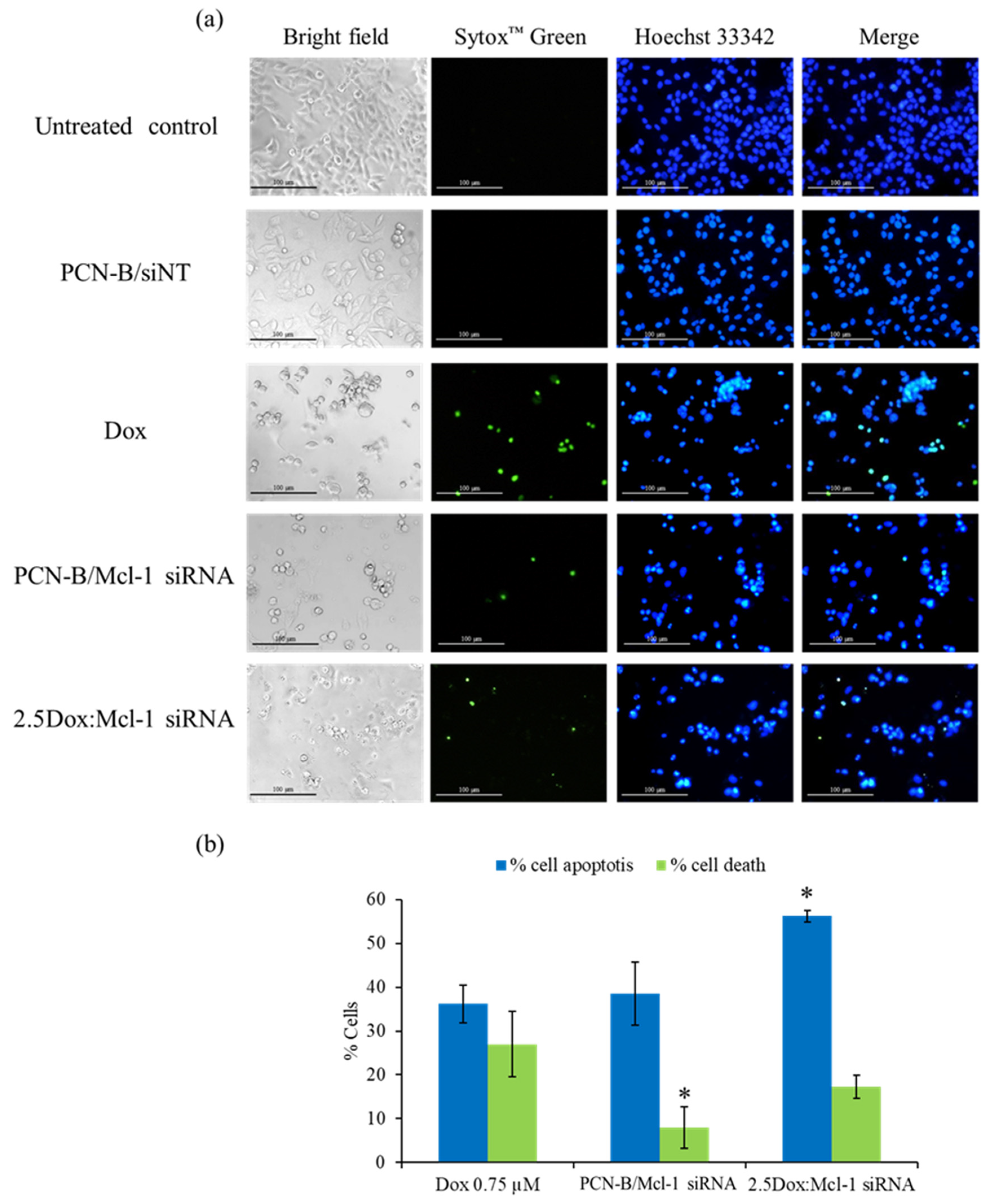
2.7. Detection of Apoptosis by Hoechst 33342 and SYTOX Green Double Staining Method
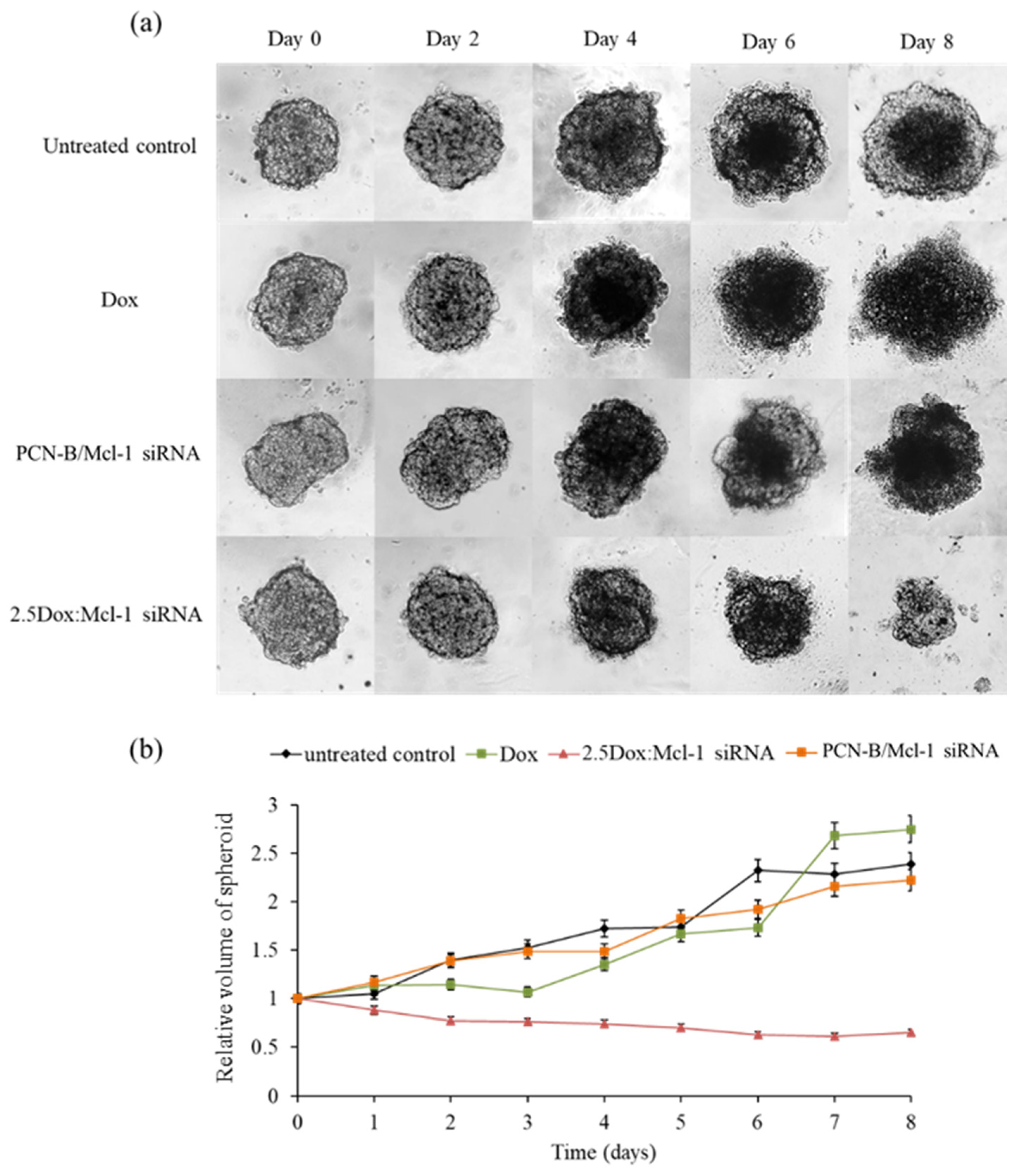
2.8. MCF-7 3D Spheroid Model
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. MCF-7 and MDA-MB-231 Cell Cultures
4.3. Cationic Niosome Preparation
4.4. siRNA Cellular Uptake Assessment
4.5. Cellular Imaging by Confocal Laser Scanning Microscope (CLSM)
4.6. siRNA Transfection
4.7. Evaluation of Cell Viability by the MTT Assay
4.8. Analysis of Dox/siRNA Synergistic Effect by CompuSyn Software
4.9. mRNA Expression Level Determination by Quantitative Real-Time PCR
4.10. Analysis of Cellular Apoptosis by Hoechst 33342 and SYTOX™ Green Double Staining
4.11. MCF-7 3D Spheroid Cell Culture
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pernaut, C.; Lopez, F.; Ciruelos, E. Standard Neoadjuvant Treatment in Early/Locally Advanced Breast Cancer. Breast Care 2018, 13, 244–249. [Google Scholar] [CrossRef] [PubMed]
- de Almeida, F.K.; Rosa, D.D. Adjuvant Dose-Dense Chemotherapy for Breast Cancer: Available Evidence and Recent Updates. Breast Care 2018, 13, 447–452. [Google Scholar] [CrossRef]
- Chopra, D.; Rehan, H.S.; Sharma, V.; Mishra, R. Chemotherapy-induced adverse drug reactions in oncology patients: A prospective observational survey. Indian J. Med. Paediatr. Oncol. 2016, 37, 42–46. [Google Scholar] [CrossRef] [PubMed]
- Fisusi, F.A.; Akala, E.O. Drug Combinations in Breast Cancer Therapy. Pharm. Nanotechnol. 2019, 7, 3–23. [Google Scholar] [CrossRef]
- Treating Breast Cancer. Available online: https://www.cancer.org/cancer/breast-cancer/treatment/chemotherapy-for-breast-cancer.html (accessed on 3 April 2021).
- Chatterjee, K.; Zhang, J.; Honbo, N.; Karliner, J.S. Doxorubicin Cardiomyopathy. Cardiology 2010, 115, 155–162. [Google Scholar] [CrossRef]
- Volkova, M.; Russell, R., 3rd. Anthracycline cardiotoxicity: Prevalence, pathogenesis and treatment. Curr. Cardiol. Rev. 2011, 7, 214–220. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef]
- David, S.; Pitard, B.; Benoît, J.P.; Passirani, C. Non-viral nanosystems for systemic siRNA delivery. Pharmacol. Res. 2010, 62, 100–114. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pai, S.I.; Lin, Y.Y.; Macaes, B.; Meneshian, A.; Hung, C.F.; Wu, T.C. Prospects of RNA interference therapy for cancer. Gene Ther. 2006, 13, 464–477. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, Z.; Gemeinhart, R.A. Progress in microRNA delivery. J. Control. Release Off. J. Control. Release Soc. 2013, 172, 962–974. [Google Scholar] [CrossRef] [PubMed]
- Babu, A.; Munshi, A.; Ramesh, R. Combinatorial therapeutic approaches with RNAi and anticancer drugs using nanodrug delivery systems. Drug Dev. Ind. Pharm. 2017, 43, 1391–1401. [Google Scholar] [CrossRef]
- Favaloro, B.; Allocati, N.; Graziano, V.; Di Ilio, C.; De Laurenzi, V. Role of apoptosis in disease. Aging 2012, 4, 330–349. [Google Scholar] [CrossRef] [PubMed]
- Kerr, J.F.; Wyllie, A.H.; Currie, A.R. Apoptosis: A basic biological phenomenon with wide-ranging implications in tissue kinetics. Br. J. Cancer 1972, 26, 239–257. [Google Scholar] [CrossRef]
- Sharma, A.; Boise, L.H.; Shanmugam, M. Cancer Metabolism and the Evasion of Apoptotic Cell Death. Cancers 2019, 11, 1144. [Google Scholar] [CrossRef]
- Williams, M.M.; Cook, R.S. Bcl-2 family proteins in breast development and cancer: Could Mcl-1 targeting overcome therapeutic resistance? Oncotarget 2015, 6, 3519–3530. [Google Scholar] [CrossRef]
- Resnier, P.; Montier, T.; Mathieu, V.; Benoit, J.-P.; Passirani, C. A review of the current status of siRNA nanomedicines in the treatment of cancer. Biomaterials 2013, 34, 6429–6443. [Google Scholar] [CrossRef] [PubMed]
- García-Aranda, M.; Pérez-Ruiz, E.; Redondo, M. Bcl-2 Inhibition to Overcome Resistance to Chemo- and Immunotherapy. Int. J. Mol. Sci. 2018, 19, 3950. [Google Scholar] [CrossRef]
- Akar, U.; Chaves-Reyez, A.; Barria, M.; Tari, A.; Sanguino, A.; Kondo, Y.; Kondo, S.; Arun, B.; Lopez-Berestein, G.; Ozpolat, B. Silencing of Bcl-2 expression by small interfering RNA induces autophagic cell death in MCF-7 breast cancer cells. Autophagy 2008, 4, 669–679. [Google Scholar] [CrossRef] [PubMed]
- Lindeman, G.J.; Visvader, J.E. Targeting BCL-2 in breast cancer: Exploiting a tumor lifeline to deliver a mortal blow? Breast Cancer Manag. 2012, 2, 1–4. [Google Scholar] [CrossRef]
- Oh, S.; Xiaofei, E.; Ni, D.; Pirooz, S.D.; Lee, J.Y.; Lee, D.; Zhao, Z.; Lee, S.; Lee, H.; Ku, B.; et al. Downregulation of autophagy by Bcl-2 promotes MCF7 breast cancer cell growth independent of its inhibition of apoptosis. Cell Death Differ. 2011, 18, 452–464. [Google Scholar] [CrossRef] [PubMed]
- Xiang, W.; Yang, C.Y.; Bai, L. MCL-1 inhibition in cancer treatment. OncoTargets Ther. 2018, 11, 7301–7314. [Google Scholar] [CrossRef] [PubMed]
- Akgul, C. Mcl-1 is a potential therapeutic target in multiple types of cancer. Cell. Mol. Life Sci. 2009, 66, 1326–1336. [Google Scholar] [CrossRef] [PubMed]
- Campbell, K.J.; Dhayade, S.; Ferrari, N.; Sims, A.H.; Johnson, E.; Mason, S.M.; Dickson, A.; Ryan, K.M.; Kalna, G.; Edwards, J.; et al. MCL-1 is a prognostic indicator and drug target in breast cancer. Cell Death Dis. 2018, 9, 19. [Google Scholar] [CrossRef]
- Aliabadi, H.M.; Maranchuk, R.; Kucharski, C.; Mahdipoor, P.; Hugh, J.; Uludağ, H. Effective response of doxorubicin-sensitive and -resistant breast cancer cells to combinational siRNA therapy. J. Control. Release 2013, 172, 219–228. [Google Scholar] [CrossRef] [PubMed]
- Long, J.; Ji, Z.; Jiang, K.; Wang, Z.; Meng, G. miR-193b Modulates Resistance to Doxorubicin in Human Breast Cancer Cells by Downregulating MCL-1. BioMed Res. Int. 2015, 2015, 373574. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, S.M.; O’Driscoll, L.; Purcell, R.; Fitz-simons, N.; McDermott, E.W.; Hill, A.D.; O’Higgins, N.J.; Parkinson, M.; Linehan, R.; Clynes, M. Prognostic importance of survivin in breast cancer. Br. J. Cancer 2003, 88, 1077–1083. [Google Scholar] [CrossRef]
- Cong, H.; Xu, L.; Wu, Y.; Qu, Z.; Bian, T.; Zhang, W.; Xing, C.; Zhuang, C. Inhibitor of Apoptosis Protein (IAP) Antagonists in Anticancer Agent Discovery: Current Status and Perspectives. J. Med. Chem. 2019, 62, 5750–5772. [Google Scholar] [CrossRef]
- Wheatley, S.P.; Altieri, D.C. Survivin at a glance. J. Cell Sci. 2019, 132, jcs223826. [Google Scholar] [CrossRef]
- Chen, X.; Duan, N.; Zhang, C.; Zhang, W. Survivin and Tumorigenesis: Molecular Mechanisms and Therapeutic Strategies. J. Cancer 2016, 7, 314–323. [Google Scholar] [CrossRef]
- Jha, K.; Shukla, M.; Pandey, M. Survivin expression and targeting in breast cancer. Surg. Oncol. 2012, 21, 125–131. [Google Scholar] [CrossRef]
- Sarti, M.; Pinton, S.; Limoni, C.; Carbone, G.M.; Pagani, O.; Cavalli, F.; Catapano, C.V. Differential expression of testin and survivin in breast cancer subtypes. Oncol. Rep. 2013, 30, 824–832. [Google Scholar] [CrossRef]
- Salzano, G.; Riehle, R.; Navarro, G.; Perche, F.; De Rosa, G.; Torchilin, V.P. Polymeric micelles containing reversibly phospholipid-modified anti-survivin siRNA: A promising strategy to overcome drug resistance in cancer. Cancer Lett. 2014, 343, 224–231. [Google Scholar] [CrossRef]
- Hu, Y.; Xu, K.; Yagüe, E. miR-218 targets survivin and regulates resistance to chemotherapeutics in breast cancer. Breast Cancer Res. Treat. 2015, 151, 269–280. [Google Scholar] [CrossRef]
- Roma-Rodrigues, C.; Rivas-García, L.; Baptista, P.V.; Fernandes, A.R. Gene Therapy in Cancer Treatment: Why Go Nano? Pharmaceutics 2020, 12, 233. [Google Scholar] [CrossRef]
- Hu, B.; Zhong, L.; Weng, Y.; Peng, L.; Huang, Y.; Zhao, Y.; Liang, X.-J. Therapeutic siRNA: State of the art. Signal Transduct. Target. Ther. 2020, 5, 101. [Google Scholar] [CrossRef] [PubMed]
- Paecharoenchai, O.; Teng, L.; Yung, B.C.; Teng, L.; Opanasopit, P.; Lee, R.J. Nonionic surfactant vesicles for delivery of RNAi therapeutics. Nanomedicine 2013, 8, 1865–1873. [Google Scholar] [CrossRef]
- Grijalvo, S.; Puras, G.; Zárate, J.; Sainz-Ramos, M.; Qtaish, N.A.L.; López, T.; Mashal, M.; Attia, N.; Díaz, D.; Pons, R.; et al. Cationic Niosomes as Non-Viral Vehicles for Nucleic Acids: Challenges and Opportunities in Gene Delivery. Pharmaceutics 2019, 11, 50. [Google Scholar] [CrossRef] [PubMed]
- Hemati, M.; Haghiralsadat, F.; Yazdian, F.; Jafari, F.; Moradi, A.; Malekpour-Dehkordi, Z. Development and characterization of a novel cationic PEGylated niosome-encapsulated forms of doxorubicin, quercetin and siRNA for the treatment of cancer by using combination therapy. Artif. Cells Nanomed. Biotechnol. 2019, 47, 1295–1311. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Gao, S.; Song, P.; Dagnæs-Hansen, F.; Jakobsen, M.; Kjems, J. Theranostic Niosomes for Efficient siRNA/MicroRNA Delivery and Activatable Near-Infrared Fluorescent Tracking of Stem Cells. ACS Appl. Mater. Interfaces 2018, 10, 19494–19503. [Google Scholar] [CrossRef]
- Pengnam, S.; Patrojanasophon, P.; Rojanarata, T.; Ngawhirunpat, T.; Yingyongnarongkul, B.-E.; Radchatawedchakoon, W.; Opanasopit, P. A novel plier-like gemini cationic niosome for nucleic acid delivery. J. Drug Deliv. Sci. Technol. 2019, 52, 325–333. [Google Scholar] [CrossRef]
- Pengnam, S.; Plainwong, S.; Patrojanasophon, P.; Rojanarata, T.; Ngawhirunpat, T.; Radchatawedchakoon, W.; Niyomtham, N.; Yingyongnarongkul, B.E.; Opanasopit, P. Effect of hydrophobic tails of plier-like cationic lipids on nucleic acid delivery and intracellular trafficking. Int. J. Pharm. 2020, 573, 118798. [Google Scholar] [CrossRef]
- Zhang, N.; Fu, J.N.; Chou, T.C. Synergistic combination of microtubule targeting anticancer fludelone with cytoprotective panaxytriol derived from panax ginseng against MX-1 cells in vitro: Experimental design and data analysis using the combination index method. Am. J. Cancer Res. 2016, 6, 97–104. [Google Scholar]
- Chonsut, P.; Mahalapbutr, P.; Pradubyat, N.; Chavasiri, W.; Wonganan, P.; Ketchart, W. Ethoxy mansonone G as an anticancer agent in estrogen receptor-positive and endocrine-resistant breast cancer. J. Pharm. Pharmacol. 2019, 71, 1839–1853. [Google Scholar] [CrossRef]
- Eid, S.Y.; El-Readi, M.Z.; Wink, M. Synergism of three-drug combinations of sanguinarine and other plant secondary metabolites with digitonin and doxorubicin in multi-drug resistant cancer cells. Phytomedicine Int. J. Phytother. Phytopharm. 2012, 19, 1288–1297. [Google Scholar] [CrossRef] [PubMed]
- Chou, T.C. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 2010, 70, 440–446. [Google Scholar] [CrossRef]
- Chou, T.-C.; Talalay, P. Quantitative analysis of dose-effect relationships: The combined effects of multiple drugs or enzyme inhibitors. Adv. Enzym. Regul. 1984, 22, 27–55. [Google Scholar] [CrossRef]
- Wei, L.; Surma, M.; Gough, G.; Shi, S.; Lambert-Cheatham, N.; Chang, J.; Shi, J. Dissecting the Mechanisms of Doxorubicin and Oxidative Stress-Induced Cytotoxicity: The Involvement of Actin Cytoskeleton and ROCK1. PLoS ONE 2015, 10, e0131763. [Google Scholar] [CrossRef]
- Pilco-Ferreto, N.; Calaf, G.M. Influence of doxorubicin on apoptosis and oxidative stress in breast cancer cell lines. Int. J. Oncol. 2016, 49, 753–762. [Google Scholar] [CrossRef]
- Beh, C.W.; Seow, W.Y.; Wang, Y.; Zhang, Y.; Ong, Z.Y.; Ee, P.L.; Yang, Y.Y. Efficient delivery of Bcl-2-targeted siRNA using cationic polymer nanoparticles: Downregulating mRNA expression level and sensitizing cancer cells to anticancer drug. Biomacromolecules 2009, 10, 41–48. [Google Scholar] [CrossRef]
- Pattingre, S.; Tassa, A.; Qu, X.; Garuti, R.; Liang, X.H.; Mizushima, N.; Packer, M.; Schneider, M.D.; Levine, B. Bcl-2 Antiapoptotic Proteins Inhibit Beclin 1-Dependent Autophagy. Cell 2005, 122, 927–939. [Google Scholar] [CrossRef]
- Decuypere, J.-P.; Parys, J.B.; Bultynck, G. Regulation of the Autophagic Bcl-2/Beclin 1 Interaction. Cells 2012, 1, 284–312. [Google Scholar] [CrossRef]
- Talaiezadeh, A.; Jalali, F.; Galehdari, H.; Khodadadi, A. Time depended Bcl-2 inhibition might be useful for a targeted drug therapy. Cancer Cell Int. 2015, 15, 105. [Google Scholar] [CrossRef]
- Ciechomska, I.A.; Goemans, G.C.; Skepper, J.N.; Tolkovsky, A.M. Bcl-2 complexed with Beclin-1 maintains full anti-apoptotic function. Oncogene 2009, 28, 2128–2141. [Google Scholar] [CrossRef] [PubMed]
- Trabulo, S.; Cardoso, A.M.; Santos-Ferreira, T.; Cardoso, A.L.; Simões, S.; Pedroso de Lima, M.C. Survivin silencing as a promising strategy to enhance the sensitivity of cancer cells to chemotherapeutic agents. Mol. Pharm. 2011, 8, 1120–1131. [Google Scholar] [CrossRef]
- Adams, K.W.; Cooper, G.M. Rapid turnover of mcl-1 couples translation to cell survival and apoptosis. J. Biol. Chem. 2007, 282, 6192–6200. [Google Scholar] [CrossRef] [PubMed]
- Tun, J.O.; Salvador-Reyes, L.A.; Velarde, M.C.; Saito, N.; Suwanborirux, K.; Concepcion, G.P. Synergistic Cytotoxicity of Renieramycin M and Doxorubicin in MCF-7 Breast Cancer Cells. Mar Drugs 2019, 17, 536. [Google Scholar] [CrossRef]
- Joshi, P.; Lee, M.-Y. High Content Imaging (HCI) on Miniaturized Three-Dimensional (3D) Cell Cultures. Biosensors 2015, 5, 768–790. [Google Scholar] [CrossRef]
- Baek, N.; Seo, O.W.; Kim, M.; Hulme, J.; An, S.S. Monitoring the effects of doxorubicin on 3D-spheroid tumor cells in real-time. OncoTargets Ther. 2016, 9, 7207–7218. [Google Scholar] [CrossRef]
- Quinn, B.A.; Dash, R.; Azab, B.; Sarkar, S.; Das, S.K.; Kumar, S.; Oyesanya, R.A.; Dasgupta, S.; Dent, P.; Grant, S.; et al. Targeting Mcl-1 for the therapy of cancer. Expert Opin. Investig. Drugs 2011, 20, 1397–1411. [Google Scholar] [CrossRef] [PubMed]
- Belmar, J.; Fesik, S.W. Small molecule Mcl-1 inhibitors for the treatment of cancer. Pharmacol. Ther. 2015, 145, 76–84. [Google Scholar] [CrossRef]
- Han, J.; Goldstein, L.A.; Gastman, B.R.; Froelich, C.J.; Yin, X.M.; Rabinowich, H. Degradation of Mcl-1 by granzyme B: Implications for Bim-mediated mitochondrial apoptotic events. J. Biol. Chem. 2004, 279, 22020–22029. [Google Scholar] [CrossRef]
- Opferman, J.T.; Letai, A.; Beard, C.; Sorcinelli, M.D.; Ong, C.C.; Korsmeyer, S.J. Development and maintenance of B and T lymphocytes requires antiapoptotic MCL-1. Nature 2003, 426, 671–676. [Google Scholar] [CrossRef]
- Podar, K.; Gouill, S.L.; Zhang, J.; Opferman, J.T.; Zorn, E.; Tai, Y.T.; Hideshima, T.; Amiot, M.; Chauhan, D.; Harousseau, J.L.; et al. A pivotal role for Mcl-1 in Bortezomib-induced apoptosis. Oncogene 2008, 27, 721–731. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Willis, S.N.; Chen, L.; Dewson, G.; Wei, A.; Naik, E.; Fletcher, J.I.; Adams, J.M.; Huang, D.C. Proapoptotic Bak is sequestered by Mcl-1 and Bcl-xL, but not Bcl-2, until displaced by BH3-only proteins. Genes Dev. 2005, 19, 1294–1305. [Google Scholar] [CrossRef] [PubMed]
- Pengnam, S.; Plianwong, S.; Singpanna, K.; Ni-yomtham, N.; Radchatawedchakoon, W.; Yingyongnarongkul, B.E.; Opanasopit, P. PEGylated Plier-Like Cationic Niosomes on Gene Delivery in HeLa Cells. Key Eng. Mater. 2019, 819, 151–156. [Google Scholar] [CrossRef]
- Parmar, M.B.; Arteaga Ballesteros, B.E.; Fu, T.; KC, R.B.; Montazeri Aliabadi, H.; Hugh, J.C.; Löbenberg, R.; Uludağ, H. Multiple siRNA delivery against cell cycle and anti-apoptosis proteins using lipid-substituted polyethylenimine in triple-negative breast cancer and nonmalignant cells. J. Biomed. Mater. Res. Part A 2016, 104, 3031–3044. [Google Scholar] [CrossRef] [PubMed]
- Chou, T.-C. Theoretical Basis, Experimental Design, and Computerized Simulation of Synergism and Antagonism in Drug Combination Studies. Pharmacol. Rev. 2006, 58, 621–681. [Google Scholar] [CrossRef] [PubMed]
- Dong, D.; Gao, W.; Liu, Y.; Qi, X.R. Therapeutic potential of targeted multifunctional nanocomplex co-delivery of siRNA and low-dose doxorubicin in breast cancer. Cancer Lett. 2015, 359, 178–186. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pengnam, S.; Plianwong, S.; Patrojanasophon, P.; Radchatawedchakoon, W.; Yingyongnarongkul, B.-e.; Opanasopit, P.; Charoensuksai, P. Synergistic Effect of Doxorubicin and siRNA-Mediated Silencing of Mcl-1 Using Cationic Niosomes against 3D MCF-7 Spheroids. Pharmaceutics 2021, 13, 550. https://doi.org/10.3390/pharmaceutics13040550
Pengnam S, Plianwong S, Patrojanasophon P, Radchatawedchakoon W, Yingyongnarongkul B-e, Opanasopit P, Charoensuksai P. Synergistic Effect of Doxorubicin and siRNA-Mediated Silencing of Mcl-1 Using Cationic Niosomes against 3D MCF-7 Spheroids. Pharmaceutics. 2021; 13(4):550. https://doi.org/10.3390/pharmaceutics13040550
Chicago/Turabian StylePengnam, Supusson, Samarwadee Plianwong, Prasopchai Patrojanasophon, Widchaya Radchatawedchakoon, Boon-ek Yingyongnarongkul, Praneet Opanasopit, and Purin Charoensuksai. 2021. "Synergistic Effect of Doxorubicin and siRNA-Mediated Silencing of Mcl-1 Using Cationic Niosomes against 3D MCF-7 Spheroids" Pharmaceutics 13, no. 4: 550. https://doi.org/10.3390/pharmaceutics13040550
APA StylePengnam, S., Plianwong, S., Patrojanasophon, P., Radchatawedchakoon, W., Yingyongnarongkul, B.-e., Opanasopit, P., & Charoensuksai, P. (2021). Synergistic Effect of Doxorubicin and siRNA-Mediated Silencing of Mcl-1 Using Cationic Niosomes against 3D MCF-7 Spheroids. Pharmaceutics, 13(4), 550. https://doi.org/10.3390/pharmaceutics13040550






