Anticancer Activity of Triazolo-Thiadiazole Derivatives and Inhibition of AKT1 and AKT2 Activation
Abstract
1. Introduction
2. Materials and Methods
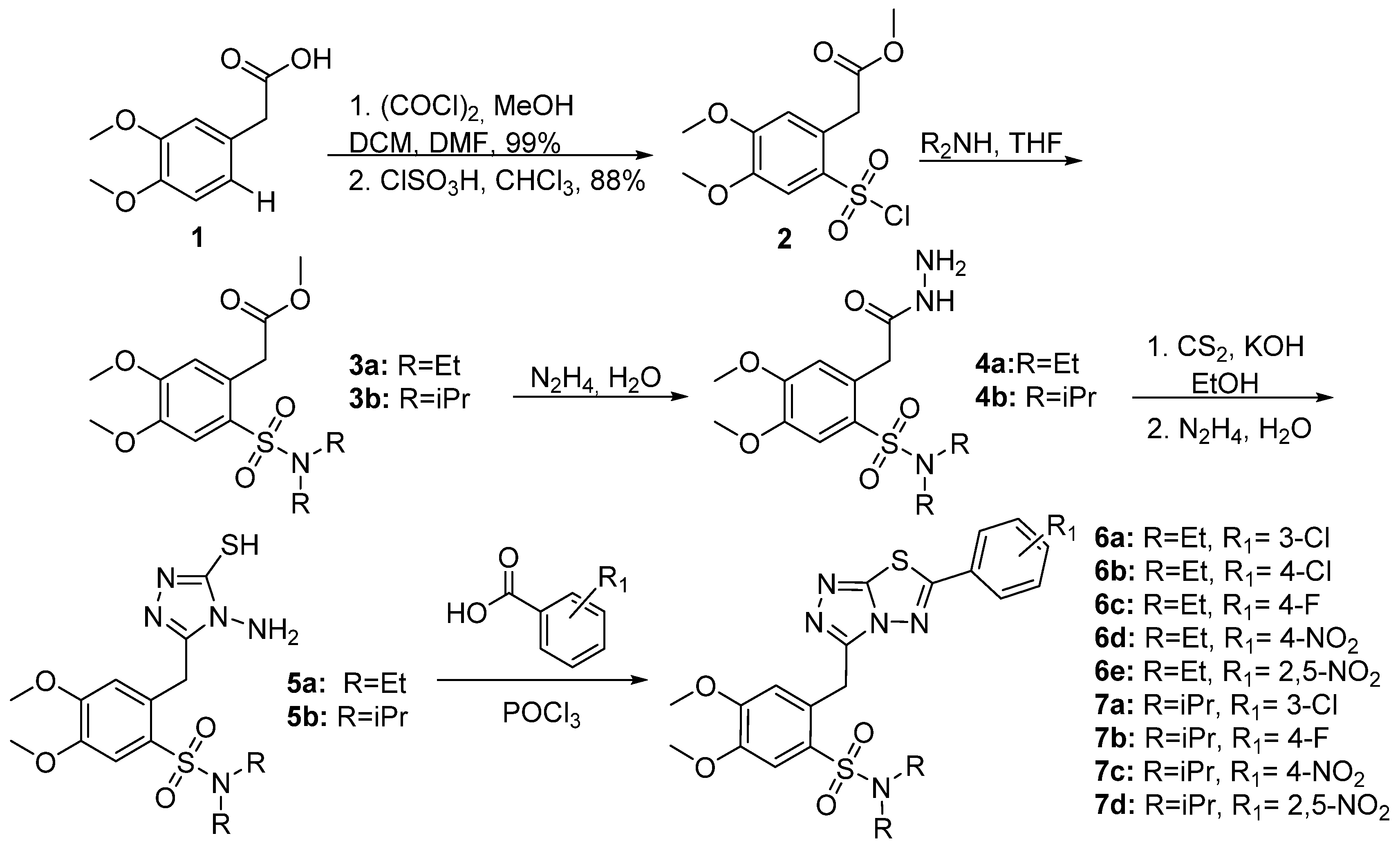
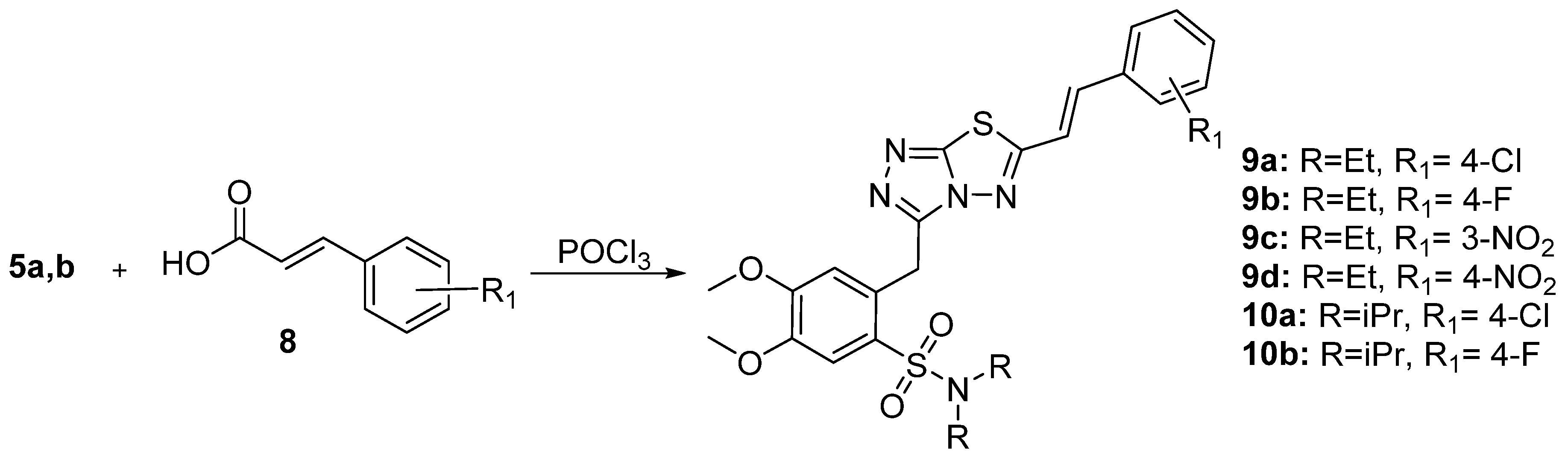
2.1. Synthesis of Triazolo-Thiadiazole Derivatives
2.2. Cell Lines and Culture Conditions
2.3. Drug Preparation for In Vitro Studies
2.4. In Vitro Antiproliferative Assay
2.5. Animals
2.6. In Vivo Acute Toxicity
2.7. HT-29 Colorectal Adenocarcinoma
2.8. Estimation of Tumor Growth
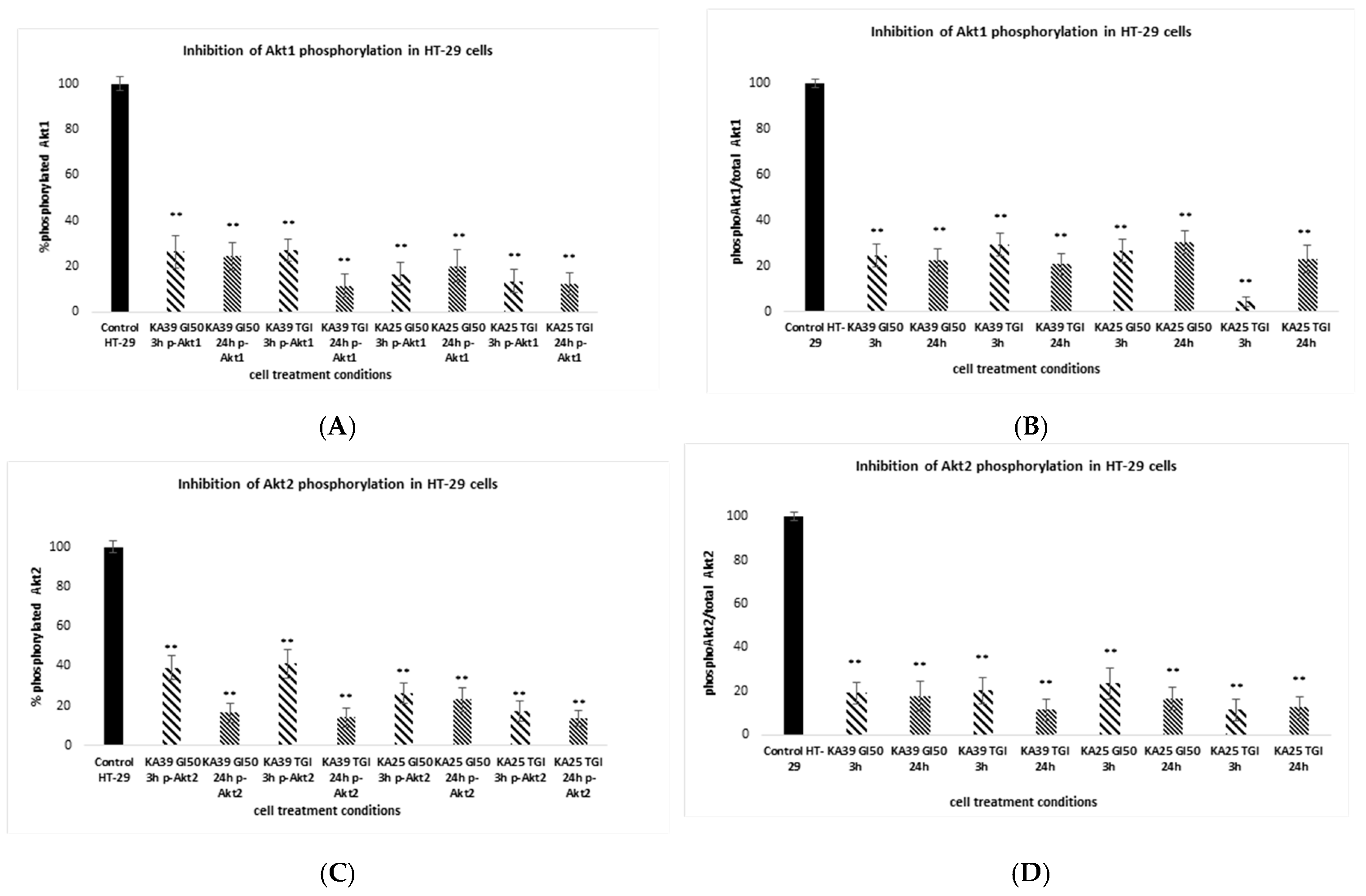
2.9. Detection of Akt1 and Akt2 Phosphorylation by Intracellular Flow Cytometry
2.10. In Silico Studies
2.11. Statistical Analysis
3. Results
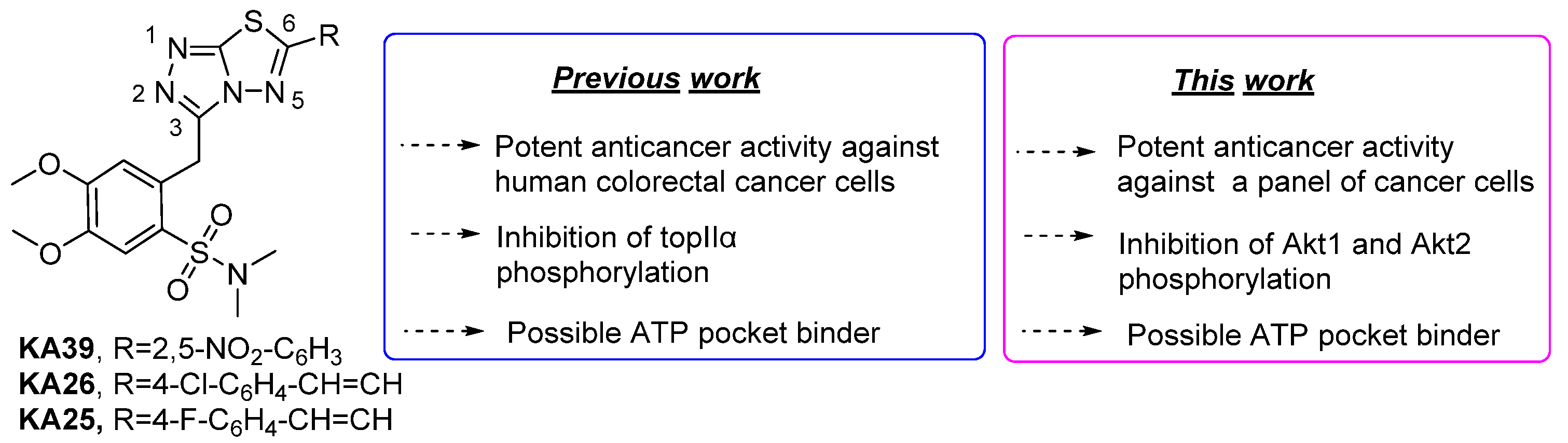
3.1. Synthesis of triazolo[3,4-b]thiadiazole Derivatives
3.1.1. Synthesis of 3,6-disubstituted 1,2,4-triazolo-[3,4-b]-[1,3,4]-thiadiazoles
3.1.2. In Vitro Anticancer Activity
3.1.3. In Vivo Acute Toxicity
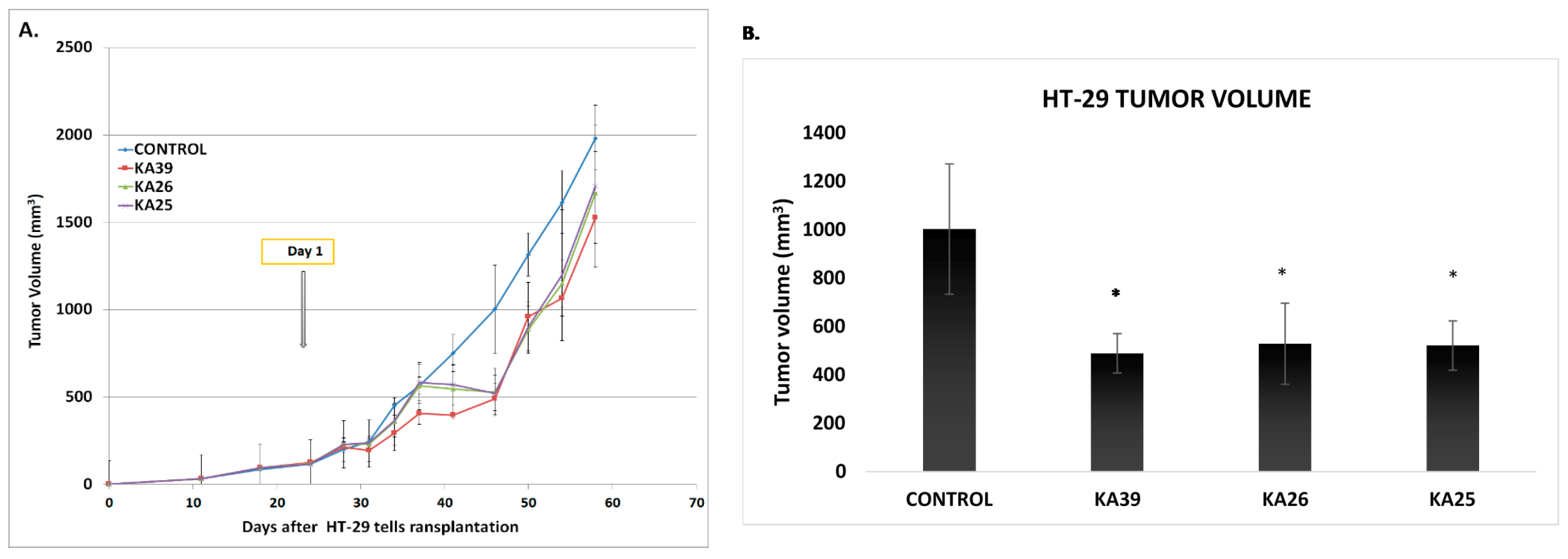
3.1.4. In Vivo Anticancer Activity
3.1.5. Phosphorylation of Akt1 and Akt2 Isoforms
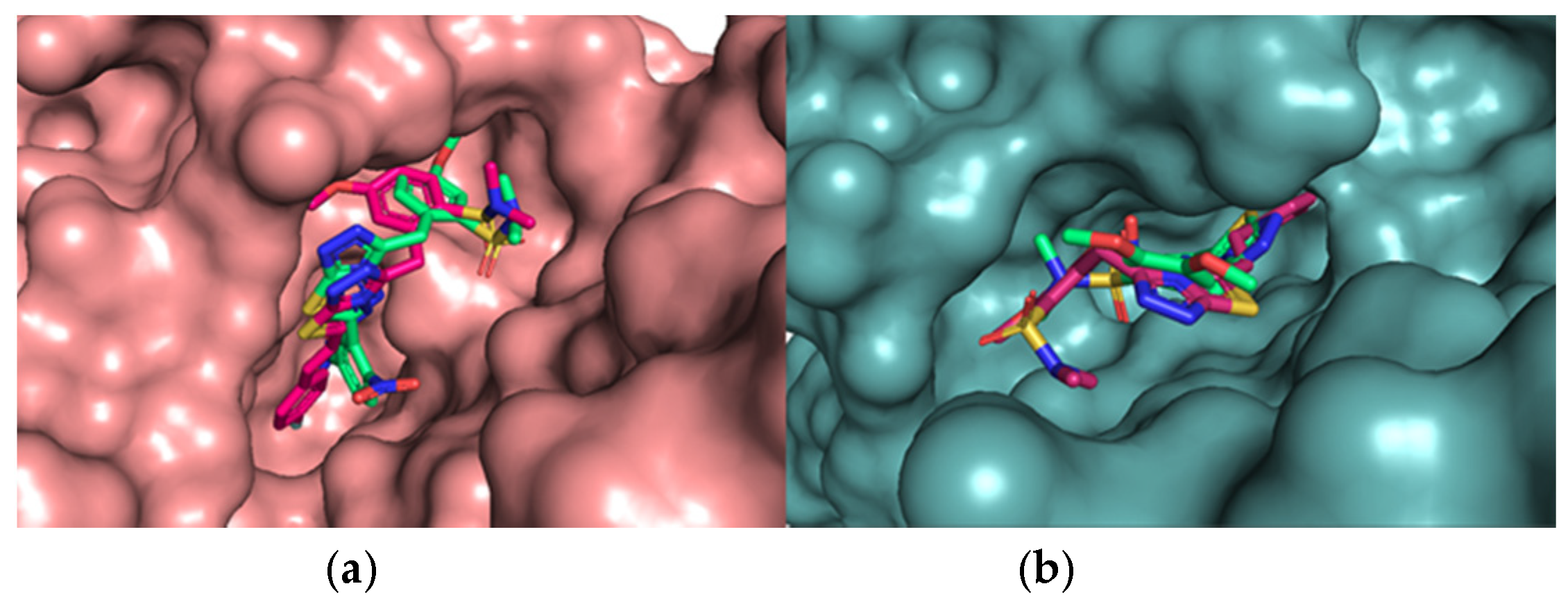
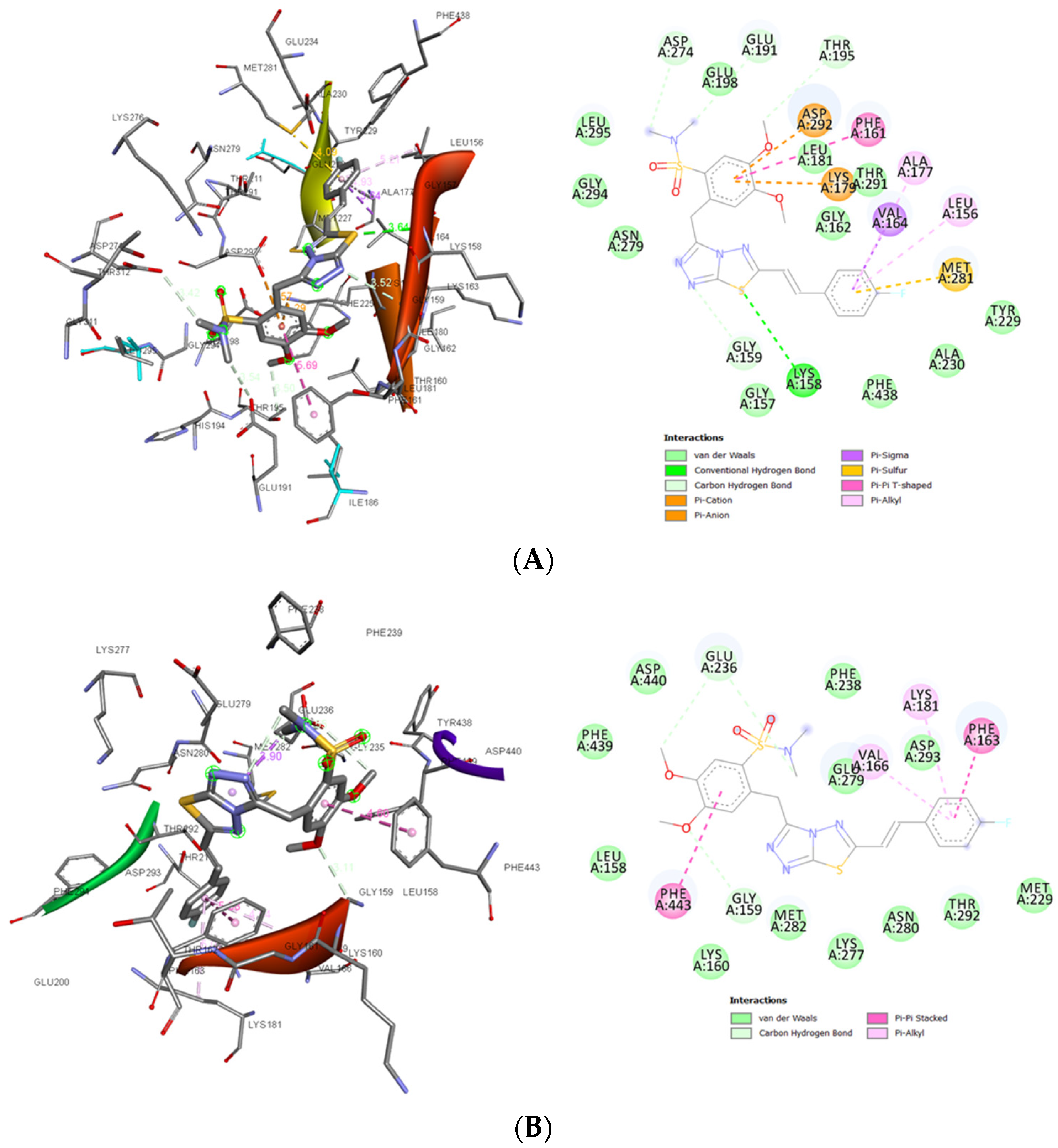
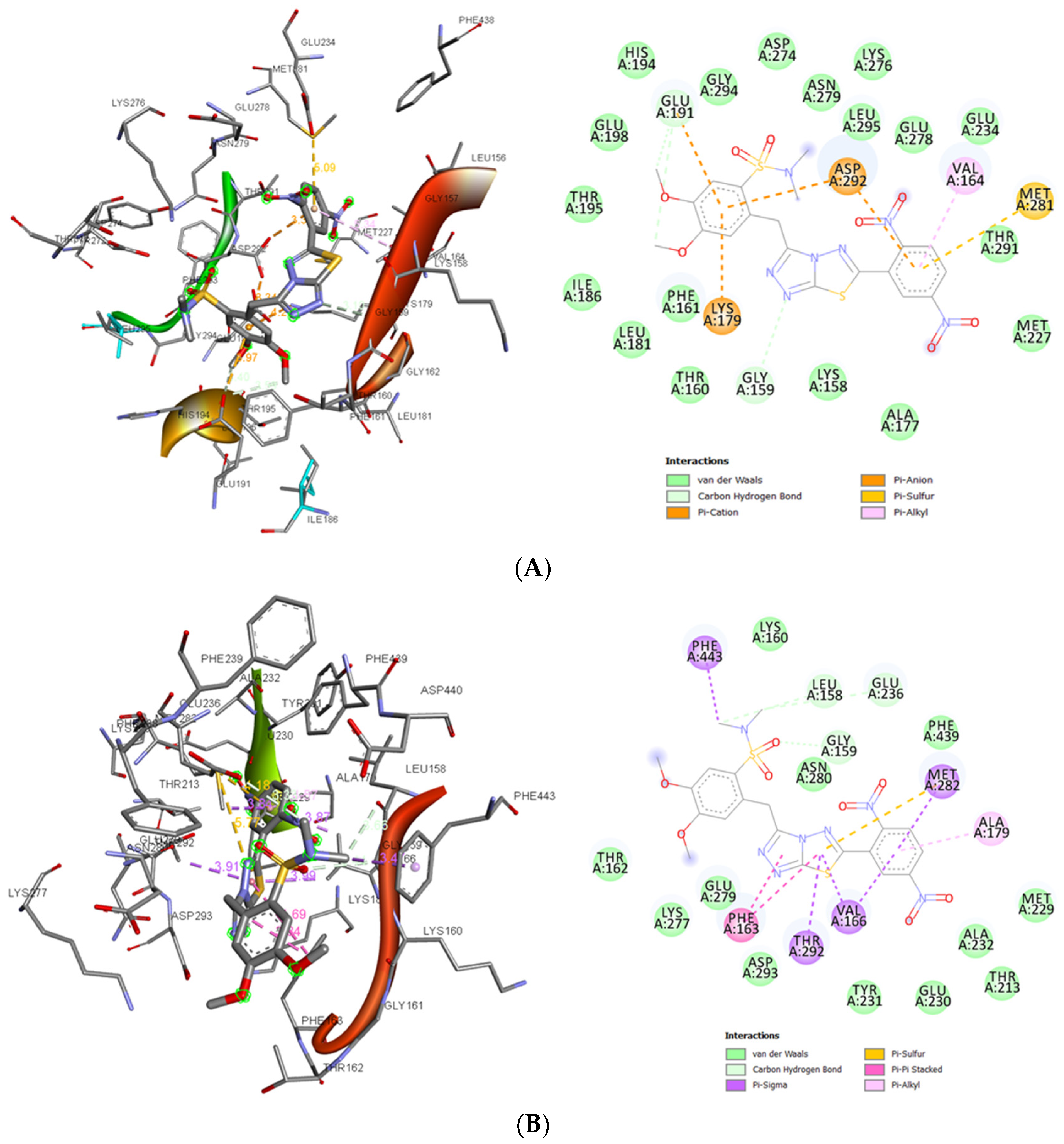
3.1.6. In Silico Studies
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Datta, S.R.; Brunet, A.; Greenberg, M.E. Cellular survival: A play in three Akts. Genes Dev. 1999, 13, 2905–2927. [Google Scholar] [CrossRef]
- Martelli, A.M.; Tabellini, G.; Bressanin, D.; Ognibene, A.; Goto, K.; Cocco, L.; Evangelisti, C. The emerging multiple roles of nuclear Akt. Biochim. Biophys. Acta 2012, 1823, 2168–2178. [Google Scholar] [CrossRef]
- Song, G.; Ouyang, G.; Bao, S. The activation of Akt/PKB signaling pathway and cell survival. J. Cell Mol. Med. 2005, 9, 59–71. [Google Scholar] [CrossRef] [PubMed]
- Andjelković, M.; Alessi, D.R.; Meier, R.; Fernandez, A.; Lamb, N.J.; Frech, M.; Cron, P.; Cohen, P.; Lucocq, J.M.; Hemmings, B.A. Role of translocation in the activation and function of protein kinase B. J. Biol. Chem. 1997, 272, 31515–31524. [Google Scholar] [CrossRef]
- Carnero, A.; Paramio, J.M. The PTEN/PI3K/AKT Pathway in vivo Cancer Mouse Models. Front Oncol. 2014, 4, 252. [Google Scholar] [CrossRef] [PubMed]
- Conus, N.M.; Hannan, K.M.; Cristiano, B.E.; Hemmings, B.A.; Pearson, R.B. Direct identification of tyrosine 474 as a regulatory phosphorylation site for the Akt protein kinase. J. Biol. Chem. 2002, 277, 38021–38028. [Google Scholar] [CrossRef]
- Arcaro, A.; Guerreiro, A.S. The phosphoinositide 3-kinase pathway in human cancer: Genetic alterations and therapeutic implications. Curr. Genom. 2007, 8, 271–306. [Google Scholar] [CrossRef] [PubMed]
- Mitsiades, C.S.; Mitsiades, N.; Koutsilieris, M. The Akt pathway: Molecular targets for anti-cancer drug development. Curr. Cancer Drug Targets 2004, 4, 235–256. [Google Scholar] [CrossRef] [PubMed]
- Rychahou, P.G.; Kang, J.; Gulhati, P.; Doan, H.Q.; Chen, L.A.; Xiao, S.Y.; Chung, D.H.; Evers, B.M. Akt2 overexpression plays a critical role in the establishment of colorectal cancer metastasis. Proc. Natl. Acad. Sci. USA 2008, 105, 20315–20320. [Google Scholar] [CrossRef]
- Graff, J.R.; Konicek, B.W.; McNulty, A.M.; Wang, Z.; Houck, K.; Allen, S.; Paul, J.D.; Hbaiu, A.; Goode, R.G.; Sandusky, G.E.; et al. Increased AKT activity contributes to prostate cancer progression by dramatically accelerating prostate tumor growth and diminishing p27Kip1 expression. J. Biol. Chem. 2000, 275, 24500–24505. [Google Scholar] [CrossRef]
- Altomare, D.A.; Tanno, S.; De Rienzo, A.; Klein-Szanto, A.J.; Tanno, S.; Skele, K.L.; Hoffman, J.P.; Testa, J.R. Frequent activation of AKT2 kinase in human pancreatic carcinomas. J Cell Biochem. 2002, 87, 470–476. [Google Scholar] [CrossRef]
- Pal, S.K.; Reckamp, K.; Yu, H.; Figlin, R.A. AKT inhibitors in clinical development for the treatment of cancer. Expert Opi. Investig. Drugs 2010, 19, 1355–1366. [Google Scholar] [CrossRef]
- Brown, J.S.; Banerji, U. Maximising the potential of AKT inhibitors as anti-cancer treatments. Pharmacol. Ther. 2017, 172, 101–115. [Google Scholar]
- Yang, J.; Cron, P.; Thompson, V.; Good, V.M.; Hess, D.; Hemmings, B.A.; Barford, D. Molecular mechanism for the regulation of protein kinase B/Akt by hydrophobic motif 587 phosphorylation. Mol. Cell. 2002, 9, 1227–1240. [Google Scholar] [CrossRef]
- Hu, Y.; Qiao, L.; Wang, S.; Rong, S.B.; Meuillet, E.J.; Berggren, M.; Gallegos, A.; Powis, G.; Kozikowski, A.P. 3-(Hydroxymethyl)-bearing phosphatidylinositol ether lipid analogues and carbonate surrogates block PI3-K, Akt, and cancer cell growth. J. Med. Chem. 2000, 43, 3045–3051. [Google Scholar] [CrossRef]
- Lindsley, C.W.; Barnett, S.F.; Yaroschak, M.; Bilodeau, M.T.; Layton, M.E. Recent progress in the development of ATP-competitive and allosteric AKT kinase inhibitors. Curr. Top. Med. Chem. 2007, 7, 1349–1363. [Google Scholar]
- Manning, B.D.; Cantley, L.C. AKT/PKB signaling: Navigating downstream. Cell 2007, 129, 1261–1274. [Google Scholar] [CrossRef]
- Charitou, T.; Srihari, S.; Lynn, M.; Jarboui, M.-A.; Fasterius, E.; Moldovan, M.; Shirasawa, S.; Tsunoda, T.; Ueffing, M.; Xie, J.; et al. Transcriptional and metabolic rewiring of colorectal cancer cells expressing the oncogenic KRAS G13D mutation. Br. J. Cancer 2019, 121, 37–50. [Google Scholar] [CrossRef] [PubMed]
- Amir, M.; Kumar, H.; Javed, S.A. Condensed bridgehead nitrogen heterocyclic system: Synthesis and pharmacological activities of 1,2,4-triazolo-[3,4-b]-1,3,4-thiadiazole derivatives of ibuprofen and biphenyl-4-yloxy acetic acid. Eur. J. Med. Chem. 2008, 43, 2056–2066. [Google Scholar] [CrossRef] [PubMed]
- Karabasanagouda, T.; Adhikari, A.V.; Shetty, N.S. Synthesis and antimicrobial activities of some novel 1,2,4-triazolo[3,4-b]-1,3,4-thiadiazoles and 1,2,4-triazolo[3,4-b]-1,3,4-thiadiazines carrying thioalkyl and sulphonyl phenoxy moieties. Eur. J. Med. Chem. 2007, 42, 521–529. [Google Scholar] [CrossRef] [PubMed]
- Chawla, G.; Kumar, U.; Bawa, S.; Kumar, J. Syntheses and evaluation of anti-inflammatory, analgesic and ulcerogenic activities of 1,3,4-oxadiazole and 1,2,4-triazolo[3,4-b]-1,3,4-thiadiazole derivatives. J. Enz. Inhib. Med. Chem. 2012, 27, 658–665. [Google Scholar] [CrossRef] [PubMed]
- Kritsanida, M.; Mouroutsou, A.; Marakos, P.; Pouli, N.; Papakonstantinou-Garoufalias, S.; Pannecouque, C.; Witvrouw, M.; De Clercq, E. Synthesis and antiviral activity evaluation of some new 6-substituted 3-(1-adamantyl)-1,2,4-triazolo[3,4-b][1,3,4]thiadiazoles. Farmaco 2002, 57, 253–257. [Google Scholar] [CrossRef]
- Sagredou, S.; Dalezis, P.; Nikoleousakos, N.; Nikolaou, M.; Voura, M.; Almpanakis, K.; Panayiotidis, M.I.; Sarli, V.; Trafalis, D.T. 3,6-Disubstituted 1,2,4-Triazolo[3,4-b]Thiadiazoles with Anticancer Activity Targeting Topoisomerase II Alpha. Onco. Targets Ther. 2020, 13, 7369–7386. [Google Scholar] [CrossRef]
- Chang, D.J.; An, H.; Kim, K.-S.; Kim, H.H.; Jung, J.; Lee, J.M.; Kim, N.-J.; Han, Y.T.; Yun, H.; Lee, S.; et al. Design, synthesis, and biological evaluation of novel deguelin-based heat shock protein 90 (HSP90) inhibitors targeting proliferation and angiogenesis. J. Med. Chem. 2012, 55, 10863–10884. [Google Scholar] [CrossRef]
- Trafalis, D.T.; Camoutsis, C.; Papageorgiou, A. Research on the anti-tumour effect of steroid lactam alkylator (NSC-294859) in comparison with conventional chemotherapeutics in malignant melanoma. Melanoma Res. 2005, 15, 273–281. [Google Scholar] [CrossRef] [PubMed]
- Elkamhawy, A.; Park, J.; Cho, N.; Sim, T.; Pae, A.; Roh, E. Discovery of a broad spectrum antiproliferative agent with selectivity for DDR1 kinase: Cell line-based assay, kinase panel, molecular docking, and toxicity studies. J. Enzym. Inhib. Med. Chem. 2015, 31, 158–166. [Google Scholar] [CrossRef] [PubMed]
- Paull, K.D.; Shoemaker, R.H.; Hodes, L.; Monks, A.; Scudiero, D.A.; Rubinstein, L.; Plowman, J.; Boyd, M.R. Display and analysis of patterns of differential activity of drugs against human tumor cell lines: Development of mean graph and COMPARE algorithm. J. Natl. Cancer Inst. 1989, 81, 1088–1092. [Google Scholar] [CrossRef]
- Trafalis, D.T.P.; Geromichalos, G.D.; Koukoulitsa, C.; Papageorgiou, A.; Karamanakos, P.; Camoutsis, C. Lactandrate: A D-homo-aza-androsterone alkylator in the treatment of breast cancer. Breast Cancer Res. Treat. 2006, 97, 17–31. [Google Scholar] [CrossRef]
- Trafalis, D.; Geromichalou, E.; Dalezis, P.; Nikoleousakos, N.; Sarli, V. Synthesis and evaluation of new steroidal lactam conjugates with aniline mustards as potential antileukemic therapeutics. Steroids 2016, 115, 1–8. [Google Scholar] [CrossRef]
- Tomayko, M.M.; Reynolds, C.P. Determination of subcutaneous tumor size in athymic (nude) mice. Cancer Chemother. Pharmcol. 1989, 24, 148–154. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Reid, J.R.; Heindel, N.D. Improved syntheses of 5-substituted-4-amino-3-mercapto-(4H)-1,2,4-triazoles. J. Heterocycl. Chem. 1976, 13, 925–926. [Google Scholar] [CrossRef]
- You, I.; Erickson, E.C.; Donovan, K.A.; Eleuteri, N.A.; Fischer, E.S.; Gray, N.S.; Toker, A. Discovery of an AKT Degrader with Prolonged Inhibition of Downstream Signaling. Cell Chem Biol. 2020, 27, 66–73. [Google Scholar] [CrossRef]
- Hennessy, B.T.; Smith, D.L.; Ram, P.T.; Lu, Y.; Mills, G.B. Exploiting the PI3K/AKT pathway for cancer drug discovery. Nat. Rev. Drug Discov. 2005, 4, 988–1004. [Google Scholar] [CrossRef] [PubMed]
- Rhodes, N.; Heerding, D.A.; Duckett, D.R.; Eberwein, D.J.; Knick, V.B.; Lansing, T.J.; McConnell, R.T.; Gilmer, T.M.; Zhang, S.-J.; Robell, K.; et al. Characterization of an Akt kinase inhibitor with potent pharmacodynamic and antitumor activity. Cancer Res. 2008, 68, 2366–2374. [Google Scholar] [CrossRef] [PubMed]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, l1. [Google Scholar] [CrossRef]
- Shoji, K.; Oda, K.; Nakagawa, S.; Hosokawa, S.; Nagae, G.; Uehara, Y.; Sone, K.; Miyamoto, Y.; Hiraike, H.; Hiraike-Wada, O.; et al. The oncogenic mutation in the pleckstrin homology domain of AKT1 in endometrial carcinomas. Br. J. Cancer 2009, 101, 145–148. [Google Scholar] [CrossRef]
- Carpten, J.D.; Faber, A.L.; Horn, C.; Donoho, G.P.; Briggs, S.L.; Robbins, C.M.; Hostetter, G.; Boguslawski, S.; Moses, T.Y.; Savage, S.; et al. A transforming mutation in the pleckstrin homology domain of AKT1 in cancer. Nature 2007, 448, 439–444. [Google Scholar] [CrossRef]
- Landgraf, K.E.; Pilling, C.; Falke, J.J. Molecular mechanism of an oncogenic mutation that alters membrane targeting: Glu17Lys modifies the PIP lipid specificity of the AKT1 PH domain. Biochemistry 2008, 47, 12260–12269. [Google Scholar] [CrossRef]
- Kumar, A.; Purohit, R. Cancer associated E17K mutation causes rapid conformational drift in AKT1 pleckstrin homology (PH) domain. PLoS ONE 2013, 8, e64364. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.Q.; Ruggeri, B.; Klein, W.M.; Sonoda, G.; Altomare, D.A.; Watson, D.K.; Testa, J.R. Amplification of AKT2 in human pancreatic cells and inhibition of AKT2 expression and tumorigenicity by antisense RNA. Proc. Natl. Acad. Sci. USA 1996, 93, 3636–3641. [Google Scholar] [CrossRef] [PubMed]
- Nitulescu, G.M.; Margina, D.; Juzenas, P.; Peng, Q.; Olaru, O.T.; Saloustros, E.; Fenga, C.; Spandidos, D.Α.; Libra, M.; Tsatsakis, A.M. Akt inhibitors in cancer treatment: The long journey from drug discovery to clinical use (Review). Int. J. Oncol. 2016, 48, 869–885. [Google Scholar] [CrossRef] [PubMed]
- Ikenoue, T.; Kanai, F.; Hikiba, Y.; Obata, T.; Tanaka, Y.; Imamura, J.; Ohta, M.; Jazag, A.; Guleng, B.; Tateishi, K.; et al. Functional Analysis of PIK3CA Gene Mutations in Human Colorectal Cancer. Cancer Res. 2005, 65, 4562–4567. [Google Scholar] [CrossRef]
- Tsubaki, M.; Takeda, T.; Noguchi, M.; Jinushi, M.; Seki, S.; Morii, Y.; Shimomura, K.; Imano, M.; Satou, T.; Nishida, S. Overactivation of Akt Contributes to MEK Inhibitor Primary and Acquired Resistance in Colorectal Cancer Cells. Cancers 2019, 11, 1866. [Google Scholar] [CrossRef]
- Gymnopoulos, M.; Elsliger, M.; Vogt, P. Rare cancer-specific mutations in PIK3CA shows gain of function. Proc. Natl. Acad. Sci. USA 2007, 104, 5569–5574. [Google Scholar] [CrossRef]
- Ahmed, D.; Eide, P.; Eilertsen, I.; Danielsen, S.A.; Eknæs, M.; Hektoen, M.; Lind, G.E.; Lothe, R.A. Epigenetic and genetic features of 24 colon cancer cell lines. Oncogenesis 2013, 2, e71. [Google Scholar] [CrossRef]
- Berg, K.; Eide, P.; Eilertsen, I.; Johannessen, B.; Bruun, J.; Danielsen, S.A.; Bjørnslett, M.; Meza-Zepeda, L.A.; Eknæs, M.; Lind, G.E.; et al. Multi-omics of 34 colorectal cancer cell lines—A resource for biomedical studies. Mol. Cancer 2017, 16, 116. [Google Scholar] [CrossRef]
- Portals.broadinstitute.org. Broad Institute Cancer Cell Line Encyclopedia (CCLE). Available online: https://portals.broadinstitute.org/ccle/page?cell_line=SW403_LARGE_INTESTINE (accessed on 16 September 2020).
- Cancer.sanger.ac.uk. COSMIC—Catalogue of Somatic Mutations in Cancer. Available online: https://cancer.sanger.ac.uk/cosmic (accessed on 16 September 2020).
- Cancer.sanger.ac.uk. Mutation Overview Page PIK3CA—P.Q546K (Substitution—Missense). Available online: https://cancer.sanger.ac.uk/cell_lines/mutation/overview?id=94232292 (accessed on 4 September 2020).
- Johnson, C.W.; Lin, Y.-J.; Reid, D.; Parker, J.; Pavolopoulos, S.; Dischinger, P.; Graveel, C.; Aguirre, A.J.; Steensma, M.; Haigis, K.M.; et al. Isoform-Specific Destabilization of the Active Site Reveals a Molecular Mechanism of Intrinsic Activation of KRas G13D. Cell Rep. 2019, 28, 1538–1550.e7. [Google Scholar] [CrossRef]
- Stolze, B.; Reinhart, S.; Bullinger, L.; Fröhling, S.; Scholl, C. Comparative analysis of KRAS codon 12, 13, 18, 61, and 117 mutations using humanMCF10A isogenic cell lines. Sci. Rep. 2015, 5, 8535. [Google Scholar] [CrossRef]
- Ihle, N.T.; Byers, L.A.; Kim, E.S.; Saintigny, P.; Lee, J.J.; Blumenschein, G.R.; Tsao, A.; Liu, S.; Larsen, J.E.; Wang, J.; et al. Effect of KRAS oncogene substitutions on protein behavior: Implications for signaling and clinical outcome. J. Natl. Cancer Inst. 2012, 104, 228–239. [Google Scholar] [CrossRef] [PubMed]
- Dumble, M.; Crouthamel, M.-C.; Zhang, S.-Y.; Schaber, M.; Levy, D.; Robell, K.; Liu, Q.; Figueroa, D.J.; Minthorn, E.A.; Seefeld, M.A.; et al. Discovery of Novel AKT Inhibitors with Enhanced Anti-Tumor Effects in Combination with the MEK Inhibitor. PLoS ONE 2014, 9, e100880. [Google Scholar] [CrossRef] [PubMed]
- She, Q.; Chandarlapaty, S.; Ye, Q.; Lobo, J.; Haskell, K.M.; Leander, K.R.; DeFeo-Jones, D.; Huber, H.E.; Rosen, N. Breast Tumor Cells with PI3K Mutation or HER2 Amplification Are Selectively Addicted to Akt Signaling. PLoS ONE 2008, 3, e3065. [Google Scholar] [CrossRef] [PubMed]
- Vasudevan, K.M.; Barbie, D.A.; Davies, M.A.; Rabinovsky, R.; McNear, C.J.; Kim, J.J.; Hennessy, B.T.; Tseng, H.; Pochanard, P.; Kim, S.Y.; et al. AKT-Independent Signaling Downstream of Oncogenic PIK3CA Mutations in Human Cancer. Cancer Cell 2009, 16, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Tolcher, A.W.; Khan, K.; Ong, M.; Banerji, U.; Papadimitrakopoulou, V.; Gandara, D.R.; Patnaik, A.; Baird, R.D.; Olmos, D.; Garrett, C.R.; et al. Antitumor activity in RAS-driven tumors by blocking AKT and MEK. Clin. Cancer Res. 2015, 21, 739–748. [Google Scholar] [CrossRef] [PubMed]
- Davies, B.; Greenwood, H.; Dudley, P.; Crafter, C.; Yu, D.-H.; Zhang, J.; Li, J.; Gao, B.; Ji, Q.; Maynard, J.; et al. Preclinical Pharmacology of AZD5363, an Inhibitor of AKT: Pharmacodynamics, Antitumor Activity, and Correlation of Monotherapy Activity with Genetic Background. Mol. Cancer Ther. 2012, 11, 873–887. [Google Scholar] [CrossRef]
- She, Q.; Halilovic, E.; Ye, Q.; Zhen, W.; Shirasawa, S.; Sasazuki, T.; Solit, D.; Rosen, N. 4E-BP1 Is a Key Effector of the Oncogenic Activation of the AKT and ERK Signaling Pathways that Integrates Their Function in Tumors. Cancer Cell 2010, 18, 39–51. [Google Scholar] [CrossRef] [PubMed]
- Gamba, S.; Camaj, P.; Heinemann, V.; Laubender, R.P.; Wang, Y.; Zhao, Y.; Stintzing, S.; Giessen, C.; Boeck, S.; Haertl, C.; et al. Effect of KRAS exon 2 mutations on antitumor activity of afatinib and gefitinib. Anticancer Drugs 2015, 26, 371–378. [Google Scholar] [CrossRef] [PubMed]
- Song, Q.; Sun, X.; Guo, H.; Yu, Q. Concomitant inhibition of receptor tyrosine kinases and downstream AKT synergistically inhibited growth of KRAS/BRAF mutant colorectal cancer cells. Oncotarget 2017, 8, 5003–5015. [Google Scholar] [CrossRef][Green Version]
| Cell Line | KA25 | KA26 | KA39 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| GI50 (μΜ) | TGI (μΜ) | IC50 (μΜ) | GI50 (μΜ) | TGI (μΜ) | IC50 (μΜ) | GI50 (μΜ) | TGI (μΜ) | IC50 (μΜ) | |
| SKOV-3 | 15 ± 0.5 | 50 ± 1.5 | 85 ± 2.1 | 20 ± 0.5 | >100 | >100 | 7 ± 0.2 | 12 ± 0.8 | 25 ± 1.4 |
| UWB 1.289 | 2 ± 0.5 | 5 ± 0.7 | 62 ± 1.2 | 29 ± 0.7 | 53 ± 1 | 70 ± 1.4 | 6 ± 0.2 | 10 ± 0.8 | 22 ± 1.2 |
| UWB1.289+BRCA1 | 4.0 ± 0.5 | 13 ± 0.8 | 38 ± 1.1 | 42 ± 0.5 | 65 ± 0.7 | >100 | 6 ± 0.5 | 11 ± 0.7 | 94 ± 2.3 |
| HT-29 | 1 ± 0.3 | >100 | >100 | 2 ± 0.76 | >100 | >100 | 11.5 ± 0.8 | 15.9 ± 0.55 | 19.5 ± 0.9 |
| LS174T | 9.3 ± 2.0 | 100 ± 0.1 | >100 | 8 ± 1.5 | >100 | >100 | 12 ± 1.5 | 17 ± 1.3 | 21.5 ± 1.5 |
| SW403 | 4.8 ± 0.8 | >100 | >100 | 11 ± 0.8 | >100 | >100 | 5.2 ± 0.2 | 7.9 ± 0.7 | 10 ± 0.76 |
| LoVo | 8 ± 0.76 | 13.8 ± 0.52 | >100 | 15 ± 0.8 | >100 | >100 | 2.2 ± 0.2 | 5.5 ± 0.1 | 10.5 ± 0.15 |
| PC-3 | 14 ± 1.0 | 22 ± 0.8 | >100 | 42.5 ± 1.5 | >100 | >100 | 5 ± 0.15 | 8 ± 0.1 | 12 ± 0.1 |
| DU-145 | 12.8 ± 1.3 | 13.2 ± 0.5 | 27 ± 0.3 | 7 ± 1.0 | 70 ± 1.3 | >100 | 5.8 ± 0.2 | 8 ± 0.4 | 10.3 ± 1.8 |
| Compound | PC-3 | SKOV-3 | ||||
|---|---|---|---|---|---|---|
| GI50 (μΜ) | TGI (μΜ) | IC50 (μΜ) | GI50 (μΜ) | TGI (μΜ) | IC50 (μΜ) | |
| 9b | 42 ± 1.0 | >100 | >100 | >100 | >100 | >100 |
| 10b | 31 ± 1.1 | >100 | >100 | 50 ± 2.0 | >100 | >100 |
| 9a | 11 ± 1.0 | >100 | >100 | 84 ± 2.0 | >100 | >100 |
| 10a | 2 ± 0.5 | 84 ± 2.0 | >100 | 76 ± 6.0 | >100 | >100 |
| 6b | 32 ± 0.4 | 56 ± 0.6 | 104 ± 0.8 | 36 ± 0.1 | 60 ± 0.8 | >100 |
| 6a | 25 ± 1.4 | 112 ± 2.5 | >100 | 6 ± 0.3 | 14 ± 2.5 | >100 |
| 7a | 20 ± 0.8 | >100 | >100 | 16 ± 0.8 | 46 ± 1.5 | >100 |
| 6c | 90 ± 2.0 | >100 | >100 | >100 | >100 | >100 |
| 7b | 25 ± 0.5 | 80 ± 1 | >100 | 80 ± 0.8 | >100 | >100 |
| 6d | 50 ± 0.5 | >100 | >100 | 80 ± 0.5 | >100 | >100 |
| 7c | 23 ± 0.8 | 102 ± 1.2 | >100 | 15 ± 1.4 | 28 ± 2.2 | 66 ± 2.5 |
| 9c | 38 ± 2.0 | >100 | >100 | >100 | >100 | >100 |
| 9d | 30 ± 0.5 | >100 | >100 | >100 | >100 | >100 |
| Cell Lines | 6e | 7d | ||||
|---|---|---|---|---|---|---|
| GI50 (μΜ) | TGI (μΜ) | IC50 (μΜ) | GI50 (μΜ) | TGI (μΜ) | IC50 (μΜ) | |
| SKOV-3 | 18 ± 0.7 | 24 ± 1.2 | 38 ± 1.8 | 13 ± 0.5 | 17 ± 1.0 | 23 ± 1.4 |
| PC-3 | 11 ± 1.0 | 17 ± 1.0 | 26.5 ± 0.9 | 8 ± 1.0 | 11 ± 1.04 | 22 ± 0.3 |
| DU-145 | 13 ± 0.5 | 18.3 ± 0.2 | 28 ± 0.2 | 6.8 ± 0.8 | 14 ± 0.5 | 21.3 ± 0.9 |
| DLD-1 | 25 ± 2.0 | 37 ± 2.0 | 53.6 ± 1.05 | 7.3 ± 0.75 | 12 ± 0.5 | 22 ± 1.0 |
| HT-29 | 18 ± 1.0 | 29 ± 0.5 | 44 ± 1.0 | 17.9 ± 2.51 | 26 ± 1.52 | 40 ± 1.0 |
| Compound | LD10 (mg/kg) | LD50 (mg/kg) |
|---|---|---|
| KA39 | 70 | 100 |
| KA25 | 200 | 360 |
| KA26 | 100 | 160 |
| Compounds | Akt1 | Akt2 |
|---|---|---|
| Docking Score (Kcal/mol) | Docking Score (Kcal/mol) | |
| KA25 | −9.5 | −8.9 |
| KA39 | −8.7 | −8.6 |
| Cancer Type | Human Cell Line Designation | KRAS Status | PIK3CA Status | References |
|---|---|---|---|---|
| Colorectal adenocarcinoma | HT-29 | wild-type | p.P449T | [47,48] |
| Colorectal adenocarcinoma, Duke’s type C, grade IV | LoVo | p.G13D | wild-type | [47,48] |
| Colorectal adenocarcinoma Dukes’ type C, grade III | SW403 | p.G12V | p.Q546K | [49,50,51] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Trafalis, D.T.; Sagredou, S.; Dalezis, P.; Voura, M.; Fountoulaki, S.; Nikoleousakos, N.; Almpanakis, K.; Deligiorgi, M.V.; Sarli, V. Anticancer Activity of Triazolo-Thiadiazole Derivatives and Inhibition of AKT1 and AKT2 Activation. Pharmaceutics 2021, 13, 493. https://doi.org/10.3390/pharmaceutics13040493
Trafalis DT, Sagredou S, Dalezis P, Voura M, Fountoulaki S, Nikoleousakos N, Almpanakis K, Deligiorgi MV, Sarli V. Anticancer Activity of Triazolo-Thiadiazole Derivatives and Inhibition of AKT1 and AKT2 Activation. Pharmaceutics. 2021; 13(4):493. https://doi.org/10.3390/pharmaceutics13040493
Chicago/Turabian StyleTrafalis, Dimitrios T., Sofia Sagredou, Panayiotis Dalezis, Maria Voura, Stella Fountoulaki, Nikolaos Nikoleousakos, Konstantinos Almpanakis, Maria V. Deligiorgi, and Vasiliki Sarli. 2021. "Anticancer Activity of Triazolo-Thiadiazole Derivatives and Inhibition of AKT1 and AKT2 Activation" Pharmaceutics 13, no. 4: 493. https://doi.org/10.3390/pharmaceutics13040493
APA StyleTrafalis, D. T., Sagredou, S., Dalezis, P., Voura, M., Fountoulaki, S., Nikoleousakos, N., Almpanakis, K., Deligiorgi, M. V., & Sarli, V. (2021). Anticancer Activity of Triazolo-Thiadiazole Derivatives and Inhibition of AKT1 and AKT2 Activation. Pharmaceutics, 13(4), 493. https://doi.org/10.3390/pharmaceutics13040493







